Introduction
Gliomas are intracranial tumors that are thought to
develop from astrocytes or oligodendrocytes (1,2). A
genome-wide mutation analysis of human brain tumors revealed
predominant somatic mutations in the gene encoding isocitrate
dehydrogenase 1 (IDH1) in glioblastoma (GBM) (3,4). Further
analysis of the IDH1 gene structure confirmed these findings,
identifying IDH1 mutations in over 70% of secondary GBM or
low-grade gliomas, but infrequently in primary GBM (5,6). Almost
all IDH1 mutations were identified at the 132th arginine residue
(R132). In addition, R132 mutations of IDH1 have been identified in
acute myeloid leukemia (AML) and chondrosarcoma (CS) (7–9). R132
mutants of IDH1 induced the reduction of α-ketoglutarate (α-KG) and
increased the 2-hydroxyglutarate (2-HG) production, whereas
converting nicotinamide adenine dinucleotide phosphate (NADPH) to
NADP+ (10–12). The 2-HG increase in the brain
propagates reactive oxygen species, leading to a variety of
after-effects (13). A decrease in
α-KG with an increase in 2-HG causes the reduction of
α-KG-dependent prolyl hydroxylases, such as those that regulate
hypoxia-inducible factor-1α subunit (HIF-1α) levels. Alterations in
HIF-1α have been reported to result from mutant IDH1 protein
expression (14), causing oncogenic
transformation. In addition, the loss or gain of IDH1 function
without R132 mutation is also related to cancer progression and
resistance to chemotherapy as mediated by NADPH biosynthesis
(15–17). Therefore, it is important to focus on
mutations other than the R132H mutation, in order to further
investigate IDH1 molecular function and cancer development.
Although intracranial tumors in dogs, such as
meningiomas and gliomas, are relatively common brain diseases
(18), partial sequencing analysis
targeting R132 in IDH1 has been performed, but no mutations have
been identified in canine gliomas (19). We recently cloned full-length cDNA of
cIDH1 and performed artificial R132 mutation analysis of cIDH1
(20). The antibodies used to detect
the specific R132 mutation in humans could also be used to detect
R132 mutations in cIDH1 (21,22), and
the production ability of NADPH was attenuated by the R132 mutation
of cIDH1. Furthermore, the R132H mutant of cIDH1 intensified HIF-1α
expression, showing that the R132 mutation in cIDH1 plays a
potential role in tumor predisposition. The most recent study to
show that the R132C mutation was found in canine glioma cases
(23) did not found any other
mutations in cIDH1.
In this study, we found the novel mutation Y208C in
cIDH1 by sequencing formalin-fixed paraffin-embedded (FFPE) canine
chondrosarcoma tissues. We compared the production ability of NADPH
and induction of HIF-1α between the wild-type (WT) and cIDH1
mutants. Furthermore, the dimerization ability necessary to exert
the enzyme activity of IDH1 was estimated in silico and
measured by cell biology analysis.
Materials and methods
Sample preparation and sequencing
The genomic DNA of the FFPE tissue from paraffin
scrolls (Table SI) was extracted
from canine tumor samples using the QIAamp DNA FFPE Tissue Kit
(Qiagen) according to the manufacturer's instructions. PCR
amplification was performed using PrimeSTAR (Takara). Primer pairs
used for amplifying cIDH1 exons are listed in Table I. Sequence data were directly
determined using an ABI 3100-Avant Genetic Analyzer (Applied
Biosystems). For the sequence analysis, human IDH1 (GenBank
accession no. NP_005887.2) and cIDH1 (BBC43078.1) were compared
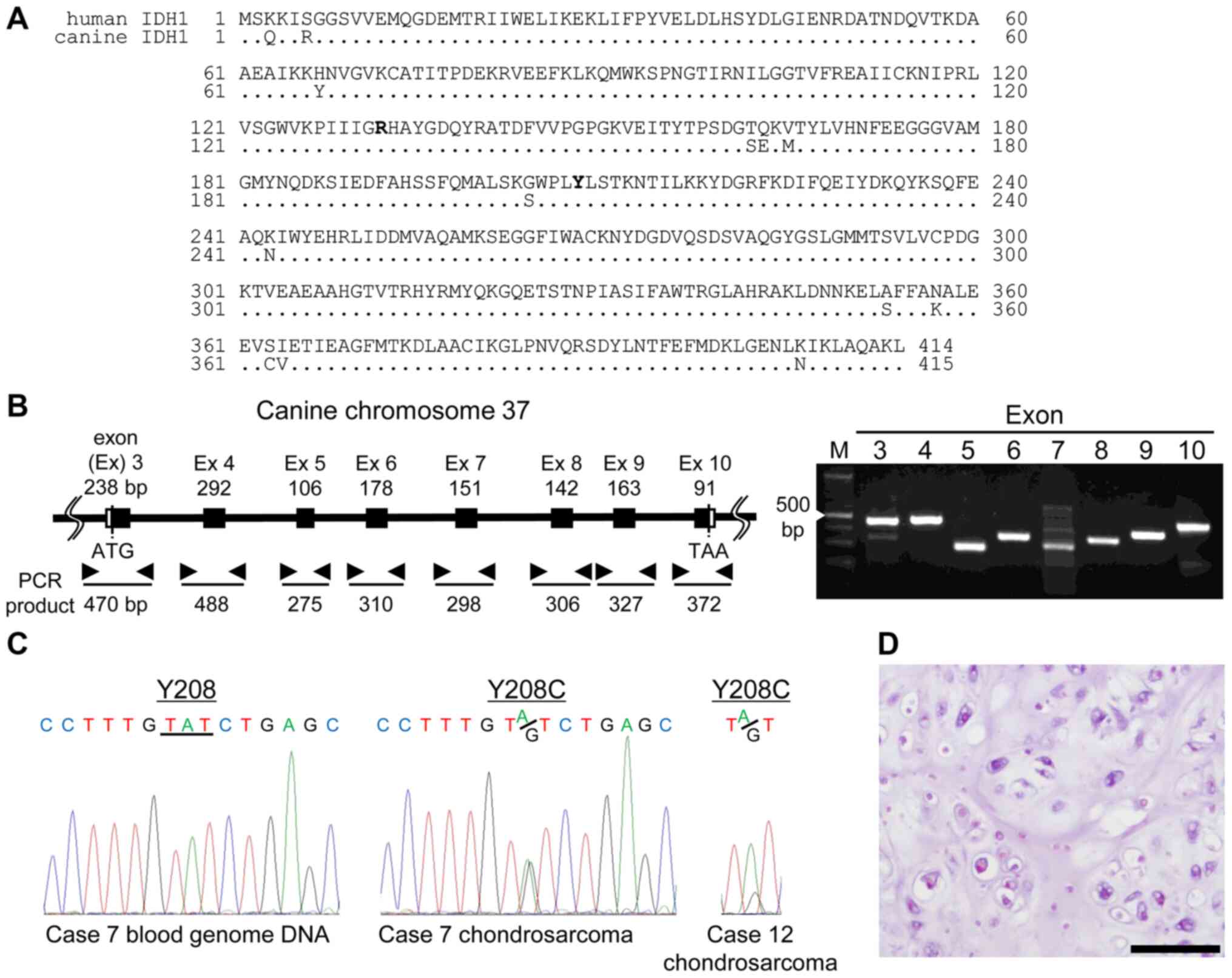
accordingly (Fig. 1A).
 | Table I.Primer pairs to amplify canine
Isocitrate dehydrogenase 1 exons. |
Table I.
Primer pairs to amplify canine
Isocitrate dehydrogenase 1 exons.
| Exon | Primer
sequences |
|---|
| 3 | F:
5′-GCAGCCTCAAAAGCCACACACGC-3′ |
|
| R:
5′-TGTACTTATCTTTAAGCATCCC-3′ |
| 4 | F:
5′-CGTTGTGCGCCATCACACAG-3′ |
|
| R:
5′-CACTTAAAGGGAGTAGTCAC-3′ |
| 5 | F:
5′-TGATCTTGAGTCTATACCAG-3′ |
|
| R:
5′-TGGCTAGTTCCCTTTGTGTC-3′ |
| 6 | F:
5′-GACTTTCTTCCAATCACGTG-3 |
|
| R:
5′-TATGCCCTTAACTTTATGGG-3′ |
| 7 | F:
5′-GCCTGATGCAAGACTCGATC-3′ |
|
| R:
5′-TTCATTGATGACTACACATGC-3′ |
| 8 | F:
5′-GGACCCTGCTTCCTGAGAGG-3′ |
|
| R:
5′-GGACCCTGCTTCCTGAGAGG-3′ |
| 9 | F:
5′-TCTGCTCAACAGCAAGACAG-3′ |
|
| R:
5′-TGACTGTGCTCCTTCCACAG-3′ |
| 10 | F:
5′-GTGGCCGAGCTGCCAGTGCAGGC-3′ |
|
| R:
5′-CCTGCCACGTTCACGAGGGTG-3′ |
Histological analysis
With permission from the University Ethics
Committee, we obtained tissue samples from the Department of
Veterinary Pathology, School of Veterinary Science, Nippon
Veterinary and Life Science University (approval no. 11-50, 27 May
2018). All samples were classified by veterinary pathologists
according to the World Health Organization classification (24) (Table
SI). FFPE cancer tissues were sliced at a thickness of 4 µm,
the sections were placed on slides, and hematoxylin and eosin
(H&E) staining was performed.
Cell cultures
HeLa and MDCK cells were purchased from the American
Type Culture Collection (ATCC). The cell lines were maintained in
Dulbecco's modified Eagles medium (Wako) supplemented with 10%
fetal bovine serum, penicillin, and streptomycin (Applied
Biosystems) and incubated at 37°C in a 5% CO2
atmosphere.
Measurement of the isocitrate
dehydrogenase activity
To measure the production of NADH and NADPH,
cIDH1-transfected cells (5×104) were processed using the
Isocitrate Dehydrogenase Activity Colorimetric Assay Kit
(BioVision) according to the manufacturer's instructions. The
reaction mix was treated for 10 min, and the optical density at 450
nm was measured using an iMark microplate reader (Bio-Rad
Laboratories).
Measurement of the HIF-1α promoter
activity
HeLa cell transfection was performed in a 96-well
plate at 80% confluency. The vector containing the HIF-1α response
element pGL4.42[luc2P/HRE/Hygro] (Promega) was
co-transfected with the control vector phRL-TK (Promega) as a
transfection efficiency control. Forty-eight hours later,
luciferase activity was measured using the Dual-Glo Luciferase
Assay System (Promega). Luciferase activity was normalized to that
of Renilla luciferase activity.
α-KG assays
HeLa cells were harvested in a 24-well plate at a
density of 1×105 cells/well and transfected with 250 ng
of HA-tagged, full-length WT, R132H, or Y208C mutant of cIDH1 in
pMACS Kk.HA-C (Miltenyi Biotec). The assay was performed using the
coupled enzymatic assay method according to the manufacturer's
instructions (Sigma-Aldrich; Merck KGaA, catalog no. MAK054). In
this method, α-KG concentration is determined by a coupled enzyme
assay, which results in a colorimetric (570 nm) product that, in
turn, is proportional to the amount of α-KG present in the
sample.
Induction of HIF-1α expression by
CoCl2
For the CoCl2 (Wako) experiments to
induce HIF-1α expression, 2×105 HeLa cells were seeded
in 6-well plates for 24 h before being treated with 100 µM
CoCl2 for an additional 24 h.
Immunoblotting
Immunoblotting was performed using the following
primary antibodies: Rabbit polyclonal anti-HA (561, 1:1,000; MBL),
anti-β-actin (PM053, 1:2,000; MBL), rabbit polyclonal anti-HIF-1α
(#3716, 1:1,000; Cell Signaling Technology), and anti-Halo antibody
(G9281, 1:1000; Promega). Horseradish peroxidase-conjugated
secondary antibodies and EzWestLumi plus (ATTO) were used for
detecting antibody-bound proteins.
Crystal structure modeling
We retrieved the crystal structure of the human IDH1
dimer from the Research Collaboratory for Structural Bioinformatics
Protein Data Bank at http://www.rcsb.org/ (PDB ID: 5YFM) and analyzed it
using the University of California, San Francisco Chimera software
(http://www.cgl.ucsf.edu/chimera/)
(25).
Mammalian cell two-hybrid assay
For the mammalian cell two-hybrid assay (MTH), WT
and Y208C cIDH1 cDNA were cloned into the EcoRI and
MluI sites of the pM GAL4 DNA-binding domain of the GAL4-DBD
plasmid (pM) (Clontech Laboratories) and the pVP16 transactivation
domain of the VP16-AD plasmid (pVP16) (Clontech Laboratories),
respectively. Approximately 2×105 HeLa cells were placed
in a 24-well plate and were co-transfected with 100 ng of pM, 100
ng of pVP16, 100 ng of pFR-Luc firefly luciferase reporter plasmid
(Promega), and 2 ng of phRL-TK Renilla luciferase reporter
plasmid (Promega). The cells were harvested 48 h after
transfection, and luciferase activity was measured using a Dual-Glo
Luciferase Assay System (Promega). Luciferase activity was
normalized to that of Renilla luciferase activity.
Pull-down assay
We cloned the Halo- or HA-tagged, full-length WT,
R132H or Y208C mutant into the pFN21A (Promega) or pMACS Kk.HA-C
vector, respectively. The expression of the Halo- and HA-tagged
constructs in the HeLa cells was induced using FuGENE HD
Transfection Reagent (Promega), and the transfected cells were
grown for 48 h. The cells were then lysed and pulled down using
Mammalian Lysis Buffer (Promega) containing Protease Inhibitor
Cocktail (Promega) for 15 min, and cellular debris was cleared by
centrifugation at 12,000 × g for 10 min. In total, 50 µl of Magne
Halo-Tag Beads (Promega) equilibrated with TBS containing 0.05%
IGEPAL CA-630 (TBS+) was added to the supernatant. The samples were
incubated for 20 min at 22°C with rotation. The supernatant was
discarded, and the protein-captured beads were washed thrice with
TBS+ and suspended in SDS-PAGE loading buffer. The samples were
analyzed by immunoblotting using anti-Halo or anti-HA antibody and
horseradish peroxidase-conjugated anti-rabbit IgG antibody (GE
Healthcare). The blots were developed using EzWestLumi plus
reagents.
Sample preparation, cross-linking
procedure and detection of cIDH1 dimer
HeLa cells in 6-well plates were transfected with
either WT, R132H or Y208C of cIDH1 expression plasmids (1 µg/well).
After 48 h of transfection, the cells were lysed with mammalian
lysis buffer (Promega) supplemented with a protease inhibitor
cocktail (Promega). Post lysis, the samples were centrifuged
(15,000 × g for 15 min at 4°C) to obtain the supernatant
accordingly. Total protein levels were measured using BCA (Nacalai
Tesque). Equal amounts of proteins (100 µg/condition) were then
incubated with glutaraldehyde at different concentrations (0, 0.04,
0.1, 0.25 or 0.5%) and incubated on ice for 30 min accordingly. To
make a working solution of glutaraldehyde, commercially available
25% glutaraldehyde solution was diluted in PBS and discarded after
use. To quench the reaction, sample buffer was added to obtain the
following final concentrations: 250 mM Tris-HCl, pH 8.5; 2% lithium
dodecyl sulfate; 100 mM DTT; 0.4 mM EDTA; 10% glycerol; and 0.2 mM
bromophenol blue. Samples were then separated by SDS-PAGE and the
monomer and dimer of cIDH1 was detected using anti-HA antibody
accordingly.
NADPH oxidase (NOX) activity
assay
NOX activity was evaluated by assessing the
superoxide production by lucigenin-enhanced chemiluminescence
(26). WT or mutant
cIDH1-transfected HeLa cells were disrupted in 0.2 ml of extraction
buffer (20 mM sodium phosphate buffer (pH 7.0), 1 mM EDTA, 0.5 mM
PMSF, 1 µg/ml Aprotinin, 0.5 µg/ml leupeptin) using a homogenizer
on ice. After the homogenates were centrifuged at 1,000 × g for 10
min at 4°C, the supernatant (20 µl) was added to 0.2 ml of assay
buffer (50 mM sodium phosphate buffer (pH 7.0), 1 mM EDTA, 150 mM
sucrose, and 50 mM lucigenin). After the addition of 0.1 mM NADPH,
luminescence was measured as relative light units (RLUs) at 15-sec
intervals for 1,000 msec in a luminometer (GLOMAX; Promega). NOX
activity was indicated as RLUs per minute per mg of protein.
Statistical analysis
Data are expressed as mean ± standard deviation
(SD). Analysis of variance (ANOVA) with a Tukey's post-hoc test was
used when multiple comparisons were required. P<0.01 was
considered statistically significant.
Results
Isolation of the Y208C mutation from
canine CS
Genomic DNA was isolated from tumor tissues and
blood. PCR amplification of the coding exons of cIDH1 (Fig. 1B) and sequence analysis of IDH1 in 45
tumor samples (Table SI) revealed
two genetic alternations in CS, from tyrosine to cysteine at
residue 208 (Y208C). No other somatic mutations in cIDH1 were found
in these samples. These two case's genome DNA, isolated from blood,
were not found to be mutated (Fig.
1C). Fig. 1D shows
representative CS samples with Y208C (case no. 7 in Table SI) H&E-stained. The
proliferation of neoplastic chondrocytes with abundant chondroid
matrix was observed.
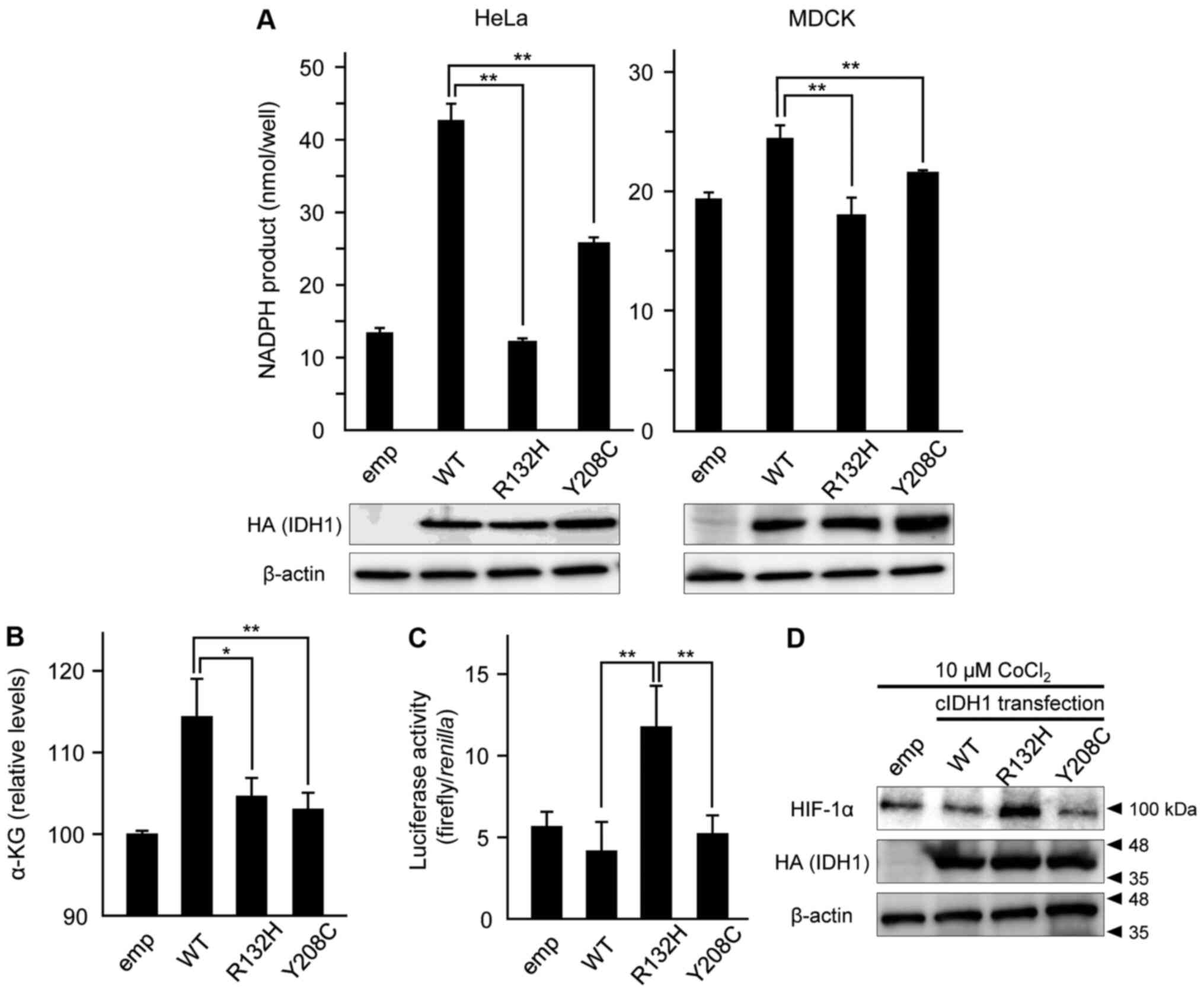
Attenuation of NADPH and α-KG
production in Y208C-mutant IDH1 cells
The formation of NADPH in multiple types of
cIDH1-overexpressing HeLa and MDCK cells was measured using
colorimetric analysis. The productivity of NADPH in the R132H
mutant of cIDH1 was significantly lower than that in WT, and Y208C
showed moderate productivity in both HeLa and MDCK cells (Fig. 2A). Production of α-KG in HeLa cells
was weakened in both R132H and Y208C mutant transfection compared
to in WT transfection (Fig. 2B).
Overexpression of the Y208C mutant did
not lead to HIF-1α induction
To evaluate HIF-1α induction by overexpression of
the cIDH1 mutant, a reporter assay was performed to measure under
the control of a promoter containing hypoxic response element sites
(27). Hypoxia caused by 10 µM
CoCl2 stimulation and overexpression of the R132H mutant
led to the induction of HIF-1α reporter activities, but the Y208C
mutant did not (Fig. 2C). HIF-1α
protein expression was also induced by CoCl2 stimulation
and R132H expression, but not Y208C (Fig. 2D).
Y208C mutation results in a
conformational change in the dimerization form of IDH1
To predict the functional alteration based on the
IDH1 mutation, the protein structure editing tool in the UCSF
Chimera software package was used to analyze the possible
structural outcomes of Y208C substitutions. Y208 is located
adjacent to the binding surface of the IDH1 dimerization form. Y208
showed hydrogen bonds with amino acids belonging to the
intra-strand W245, E247, R249, M254, Q257, and W267 (Fig. 3A). The Rotamers tool allows amino
acid side chain rotamers to be viewed and evaluated (25). The best rotamers for C208 were
selected based on their side-chain torsion as well as on the
probability values in the rotamer library and in the context of the
structural environment. These calculations revealed that the Y208C
substitution disrupted or reduced the inter-strand hydrogen bond
with the side chains of W245, E247, and Q257 (Fig. 3B). MTH and PD assays, which examined
the binding activities between WT and mutant cIDH1, showed no
significant change in WT-R132H heterodimerization and attenuated
binding activity in the WT-Y208C mutant (Fig. 3C, D). The dimerization ability of
cIDH1 transfected into HeLa cells was assessed using a
glutaraldehyde cross-linking assay. Cell lysates from HeLa cells
expressing HA-tagged WT, R132H or Y208 mutant were treated first
with glutaraldehyde crosslinker and then analyzed by western
blotting method. Both WT and R132H mutants formed dimers following
the glutaraldehyde treatment, however, the Y208C mutant formed a
weak dimer (Fig. 3E).
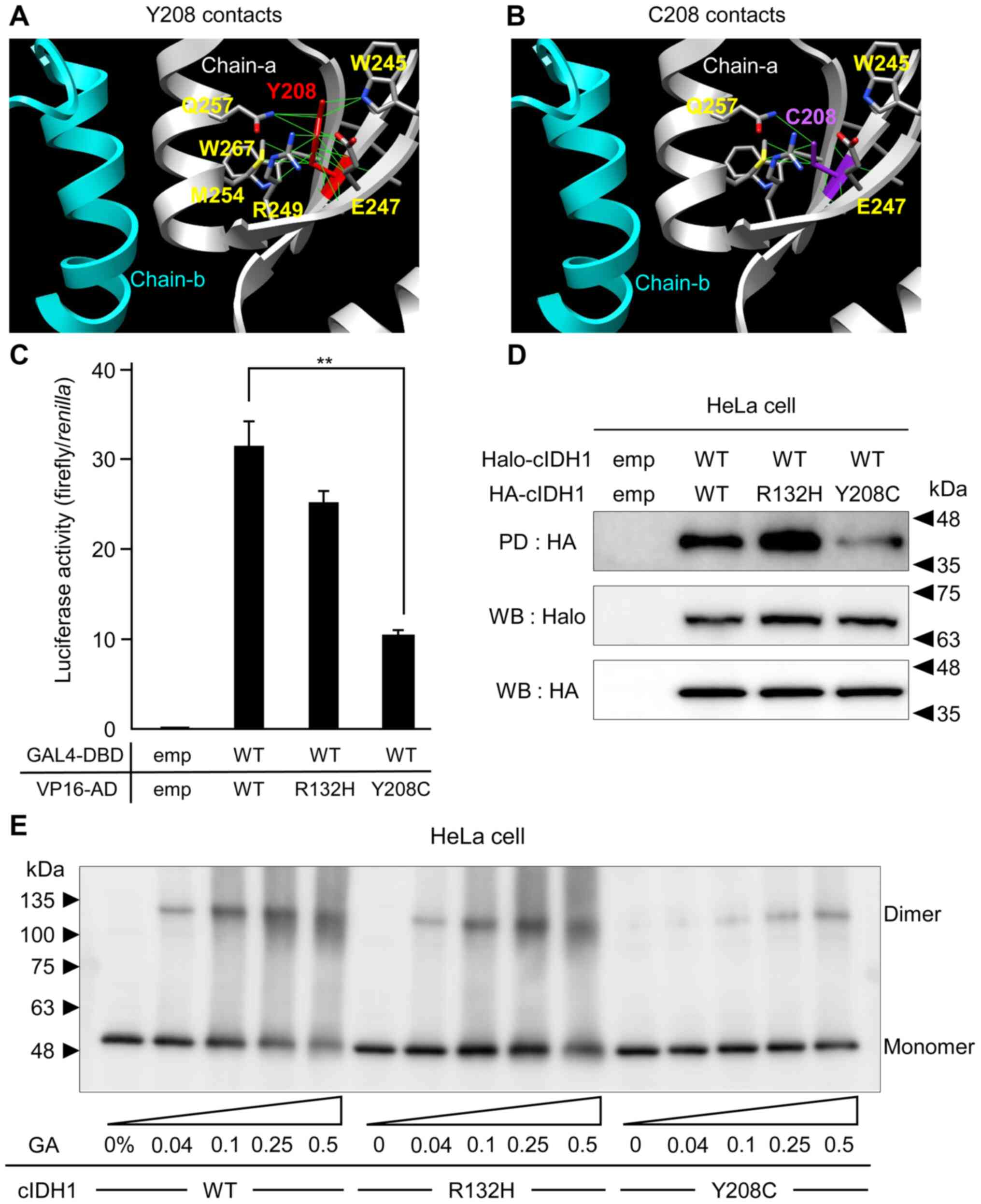 | Figure 3.In silico and cell biology
analysis of the effect of Y208C mutation dimerization ability. Two
cIDH1 proteins are depicted as magenta (chain-a) and gray (chain-b)
ribbons. The Y208 residue (red) was mutated to Cys (purple). The
contacts between residues at the (A) Y208 or (B) C208 positions in
the chain were calculated. The amino acid residues linking Y208 and
C208 are colored gray. The solid green lines denote stable
contacts, as determined using the Chimera program. (C) Binding
intensities between GAL4-DBD and VP16-AD fused with the WT and
cIDH1 mutants. Data are presented as the mean ± SD (n=4).
**P<0.01 as indicated. (D) A pull-down assay was performed for
WT and mutated cIDH1 cloned into pFN21A and pMACS Kk.HA-C plasmid
vectors. Halo-tagged cIDH1 and HA-tagged cIDH1 were subsequently
analyzed using the indicated antibodies. (E) Glutaraldehyde
cross-linking analysis of WT, R132H and Y208C cIDH1 mutants.
Samples were separated by polyacrylamide gel electrophoresis and
immunoblotted with an anti-HA antibody. cIDH1, canine isocitrate
dehydrogenase 1; GAL4-DBD, GAL4-DNA binding domain; VP16-AD, VP16
activation domain; HA, hemagglutinin; WT, wild-type; emp, empty;
WB, western blotting; PD, pull-down; GA, glutaraldehyde. |
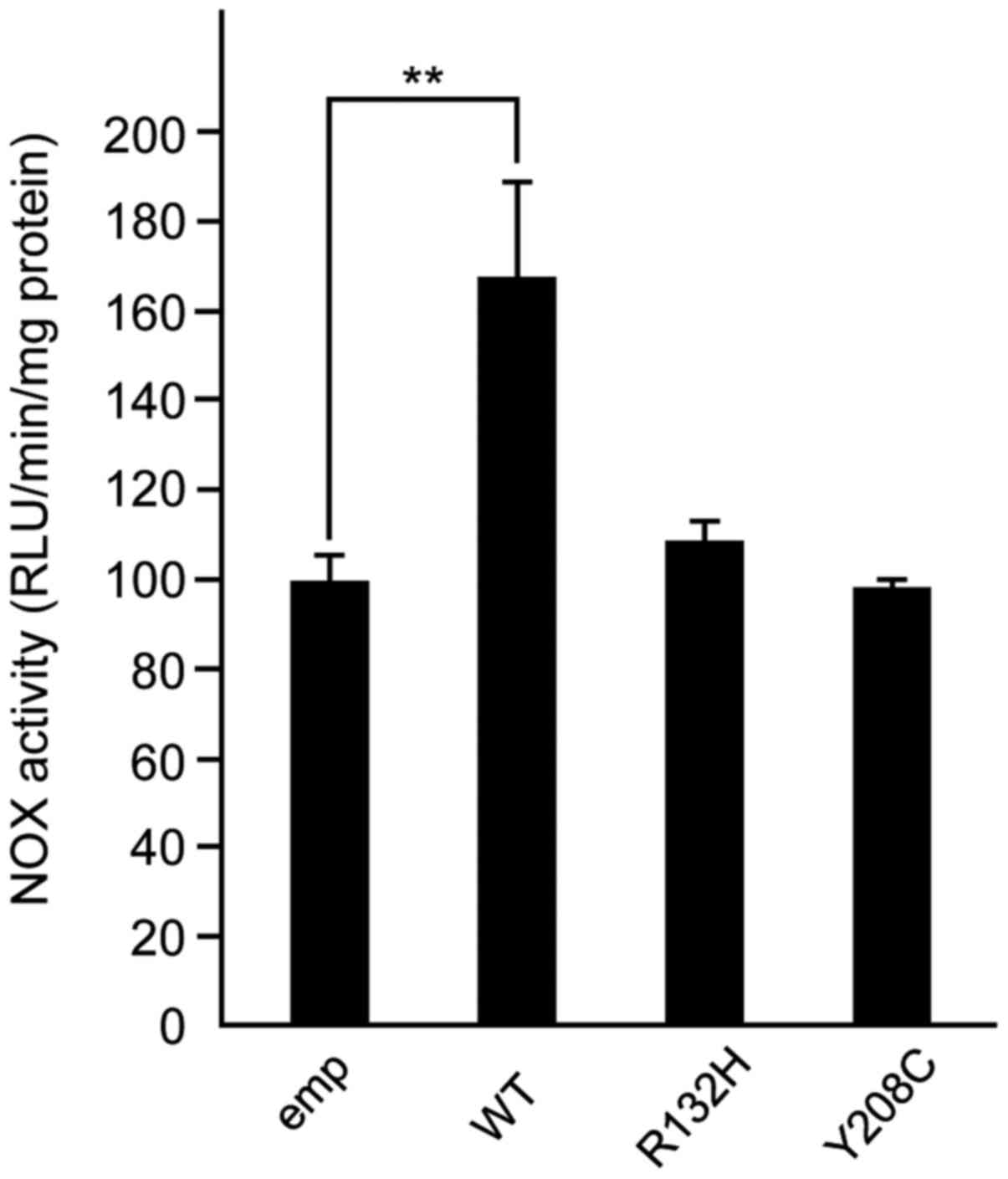
R132H and Y208C mutations did not
affect the alteration of NOX activities
NOX activity in WT or mutant cIDH1-transfected HeLa
cells was assessed. The WT of cIDH1-transfected cells showed a
significant increase compared with the empty vector-transfected
control, but the R132H and Y208C mutant transfectants did not show
(Fig. 4).
Discussion
Two cases of novel Y208C mutations in CS tissues
were not detected in the genomic DNA isolated from blood;
therefore, the Y208C mutation appeared to be spontaneous somatic
mutations. Although the sensitivity for detection by Sanger
sequencing of R132 mutation in human cases is sometimes low
(28), the Sanger sequencing peak of
mutated alleles was definitive in the two cases of Y208C mutation
in this study. Since there are more sensitive detection methods,
such as allele-specific oligonucleotide–PCR and pyrosequencing
(29,30), using these methods may further
increase the positive mutation rate. Deep sequencing analysis has
been performed in canine glioma cases (23,31), but
the Y208C mutation was not detected; therefore, the Y208C mutation
may be a unique mutation in CS. Two cases of c.623A>C and
p.Y208C mutations in human liver cancer were archived in the
Catalog of Somatic Mutations In Cancer (COSMIC; http://cancer.sanger.ac.uk/cosmic) database
(32), but the gene function
alterations were not described. Y208H (rs587778402) and Y208C
(rs186787509) mutations in human IDH1 were also studied in the NCBI
SNP database (https://www.ncbi.nlm.nih.gov/snp). Histological
analysis showed that there was no remarkable difference compared
with other CS tissues. Because the number of cases is still small,
it is possible that further histological analysis of CS with Y208C
mutation in IDH1 would lead to the identification of the peculiar
characteristics of the Y208C mutation. The production amount of
α-KG was also measured in the cIDH1 transfectant HeLa cell
accordingly. The R132H and Y208C mutant transfectant showed
significant attenuation of α-KG production when compared with WT
transfection. This data shows that the Y208C mutation of cIDH1
leads to the loss-of catalytic function from isocitrate to α-KG,
and decreases the NADPH biosynthesis accordingly.
The R132H mutated IDH1 proteins lose normal
catalytic activity for α-KG and produce less NADPH. Instead, the
abnormal enzymatic activity produces 2-HG and consumes NADPH
(33). Therefore, we investigated
the formation of NADPH in multiple types of cIDH1-overexpressing
HeLa and MDCK cells using colorimetric analysis. Previous studies
have used HeLa and MDCK cell lines for the analysis of the
biological reaction against IDH1 overexpression (20,14,34).
Compared with the R132H mutation transfectant, which showed a
significant decrease in NADPH production, the Y208C transfectant
showed moderate reduction of products in both cell types. These
data indicate that the Y208C mutation diminished the isocitrate
dehydrogenation activity, but not at lower levels than the R132H
mutation.
The R132H mutation of IDH1 produces 2-HG from α-KG,
which reduces α-KG-dependent prolyl hydroxylases, which regulate
HIF-1α levels (35). HeLa cells
transfected with the R132H mutant of cIDH1 showed an increase in
HIF-1α promoter activity and retention of HIF1-α protein, but Y208C
mutant transfection did not change compared with parental cells.
These data suggest that the Y208C mutation has a different
mechanism for tumorigenesis than the R132 mutation.
In Fig. 1A, as seen
clearly, the amino acids sequence of human and canine IDH1 are
highly conserved; hence, we thought that the protein structure data
of human IDH1 (PDB ID: 5YFM) could be extrapolated to analyze the
Y208C mutation in cIDH1. Our in silico simulation of amino
acid substitution from tyrosine to cysteine showed the loss of
contact against intra-strand amino acids located in an α-helix
structure (amino acid residues 251 to 261 of GenBank accession no.
BBC43078.1), which forms the binding surface of the homodimer
(36), because the side chain of
cysteine was shorter than that of the tyrosine side chain. As
predicted by in silico analysis, Y208C mutation attenuated
the binding ability against WT of IDH1 by MTH, PD assay and protein
cross-linked electrophoresis, however not for the WT-R132H
interaction. Both R132H and Y208C mutants attenuate isocitrate
dehydrogenase activity. The R132H mutant produces carcinogenic 2-HG
from α-KG, which causes a reduction in the NADPH production. On the
other hand, the Y208C mutant cannot form dimers, which causes
attenuation of enzymatic activity. These phenomena suggest that the
carcinogenic mechanisms are different between R132H and Y208C
mutations in IDH1.
IDH1 mutation causes a change in NADPH production,
so we predicted that IDH1 mutation may affect NOX activity, which
reflects the amount of reactive oxygen species (ROS). To elucidate
the cause of tumorigenesis due to the Y208C mutation, we analyzed
NOX activity in cIDH1-transfected cells. NOX activity was
significantly higher in WT IDH1-transfected cells, and there was no
significant difference between the negative control and both R132H
and Y208C mutant transfected cells. This result suggests that WT
IDH1 could produce more NADPH, a source of ROS, but IDH1 mutants
lose the ability to produce more NADPH; therefore, mutant
transfected cells could not produce ROS. Since there was no
difference in the results of ROS productivity between R132H and
Y208C, the mechanism of carcinogenesis of the Y208C mutation
remains unclear. This study is the first experimental report to
describe the relationship between canine IDH1 mutation and NOX
activity. Future studies will need to elucidate the mechanism of
tumorigenesis of the Y208C mutation. Furthermore, future studies
will have to look for Y208C mutations in various tumors.
In conclusion, we identified for the first time
Y208C spontaneous somatic mutations of canine IDH1 in
chondrosarcomas and assessed the impact of these mutations on IDH1
functions. Y208C mutation attenuated the NADPH production ability
but did not enhance HIF-1α retention in CoCl2-treated
cells. This phenomenon was caused by the attenuation of the
dimerization ability of the Y208C mutation. We hope that the
precise analysis of IDH1 functional changes can help elucidate the
tumorigenesis involvement of the Y208C IDH1 mutation.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by KAKENHI scientific
research grants from the Ministry of Education, Culture, Sports,
Science and Technology of Japan (grant nos. 18H02334 and
19K06390).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SK, MS, MMi, MMo, KI, MW and KO designed the study
and performed the bioinformatics analysis. SK, MU, NK, MMa, YM, DA,
ASE, EO and TO performed the laboratory experiments. KO and YT
performed statistical analysis and wrote the manuscript. MW, YT, TO
and KO supervised the study. All authors have read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
The current study was approved by the University
Ethics Committee of the Department of Veterinary Pathology, School
of Veterinary Science, Nippon Veterinary and Life Science
University (approval no. 11-50, 27 May 2018).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sahm F, Reuss D, Koelsche C, Capper D,
Schittenhelm J, Heim S, Jones DT, Pfister SM, Herold-Mende C, Wick
W, et al: Farewell to oligoastrocytoma: In situ molecular genetics
favor classification as either oligodendroglioma or astrocytoma.
Acta Neuropathol. 128:551–559. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lapointe S, Perry A and Butowski NA:
Primary brain tumours in adults. Lancet. 392:432–446. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Parsons DW, Jones S, Zhang X, Lin JC,
Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et
al: An integrated genomic analysis of human glioblastoma
multiforme. Science. 321:1807–1812. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Reitman ZJ and Yan H: Isocitrate
dehydrogenase 1 and 2 mutations in cancer: Alterations at a
crossroads of cellular metabolism. J Natl Cancer Inst. 102:932–941.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bleeker FE, Lamba S, Rodolfo M, Scarpa A,
Leenstra S, Vandertop WP and Bardelli A: Mutational profiling of
cancer candidate genes in glioblastoma, melanoma and pancreatic
carcinoma reveals a snapshot of their genomic landscapes. Hum
Mutat. 30:E451–E459. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yan H, Parsons DW, Jin G, McLendon R,
Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ,
et al: IDH1 and IDH2 mutations in gliomas. N Engl J Med.
360:765–773. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mardis ER, Ding L, Dooling DJ, Larson DE,
McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath
SD, et al: Recurring mutations found by sequencing an acute myeloid
leukemia genome. N Engl J Med. 361:1058–1066. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lugowska I, Teterycz P, Mikula M, Kulecka
M, Kluska A, Balabas A, Piatkowska M, Wagrodzki M, Pienkowski A,
Rutkowski P and Ostrowski J: IDH1/2 mutations predict shorter
survival in chondrosarcoma. J Cancer. 9:998–1005. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Waitkus MS, Diplas BH and Yan H:
Biological role and therapeutic potential of IDH mutations in
cancer. Cancer Cell. 34:186–195. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dang L, White DW, Gross S, Bennett BD,
Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et
al: Cancer-associated IDH1 mutations produce 2-hydroxyglutarate.
Nature. 462:739–744. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gross S, Cairns RA, Minden MD, Driggers
EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, et
al: Cancer-associated metabolite 2-hydroxyglutarate accumulates in
acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2
mutations. J Exp Med. 207:339–344. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schnittger S, Haferlach C, Ulke M,
Alpermann T, Kern W and Haferlach T: IDH1 mutations are detected in
6.6% of 1414 AML patients and are associated with intermediate risk
karyotype and unfavorable prognosis in adults younger than 60 years
and unmutated NPM1 status. Blood. 116:5486–5496. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wajner M, Latini A, Wyse AT and
Dutra-Filho CS: The role of oxidative damage in the neuropathology
of organic acidurias: insights from animal studies. J Inherit Metab
Dis. 27:427–448. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang
P, Yu W, Li Z, Gong L, Peng Y, et al: Glioma-derived mutations in
IDH1 dominantly inhibit IDH1 catalytic activity and induce
HIF-1alpha. Science. 324:261–265. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Calvert AE, Chalastanis A, Wu Y, Hurley
LA, Kouri FM, Bi Y, Kachman M, May JL, Bartom E, Hua Y, et al:
Cancer-associated IDH1 promotes growth and resistance to targeted
therapies in the absence of mutation. Cell Rep. 19:1858–1873. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wahl DR, Dresser J, Wilder-Romans K,
Parsels JD, Zhao SG, Davis M, Zhao L, Kachman M, Wernisch S, Burant
CF, et al: Glioblastoma therapy can be augmented by targeting
IDH1-mediated NADPH biosynthesis. Cancer Res. 77:960–970. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zarei M, Lal S, Parker SJ, Nevler A,
Vaziri-Gohar A, Dukleska K, Mambelli-Lisboa NC, Moffat C, Blanco
FF, Chand SN, et al: Posttranscriptional upregulation of IDH1 by
HuR establishes a powerful survival phenotype in pancreatic cancer
cells. Cancer Res. 77:4460–4471. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dobson JM, Samuel S, Milstein H, Rogers K
and Wood JL: Canine neoplasia in the UK: Estimates of incidence
rates from a population of insured dogs. J Small Anim Pract.
43:240–246. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Reitman ZJ, Olby NJ, Mariani CL, Thomas R,
Breen M, Bigner DD, McLendon RE and Yan H: IDH1 and IDH2 hotspot
mutations are not found in canine glioma. Int J Cancer.
127:245–246. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kawakami S, Ochiai K, Azakami D, Kato Y,
Michishita M, Morimatsu M, Ishiguro-Oonuma T, Onozawa E, Watanabe M
and Omi T: R132 mutations in canine isocitrate dehydrogenase 1
(IDH1) lead to functional changes. Vet Res Commun. 42:49–56. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kaneko MK, Tian W, Takano S, Suzuki H,
Sawa Y, Hozumi Y, Goto K, Yamazaki K, Kitanaka C and Kato Y:
Establishment of a novel monoclonal antibody SMab-1 specific for
IDH1-R132S mutation. Biochem Biophys Res Commun. 406:608–613. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kato Y: Specific monoclonal antibodies
against IDH1/2 mutations as diagnostic tools for gliomas. Brain
Tumor Pathol. 32:3–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Amin SB, Anderson KJ, Boudreau CE,
Martinez-Ledesma E, Kocakavuk E, Johnson KC, Barthel FP, Varn FS,
Kassab C, Ling X, et al: Comparative molecular life history of
spontaneous canine and human gliomas. Cancer Cell. 37:243–257.e7.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Misdorp W, Else RW, Hellmen E and Lipscomb
TP: Histological classification of mammary tumors of the dog and
the cat. World Health Organization International Histological
Classification of Tumors of Domestic Animals. Schulman FY: 7. 2nd
Series. Armed Forces Institute of Pathology; Washington, DC: pp.
581999
|
|
25
|
Pettersen EF, Goddard TD, Huang CC, Couch
GS, Greenblatt DM, Meng EC and Ferrin TE: UCSF Chimera-a
visualization system for exploratory research and analysis. J
Comput Chem. 25:1605–1612. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jalil JE, Perez A, Ocaranza MP, Bargetto
J, Galaz A and Lavandero S: Increased aortic NADPH oxidase activity
in rats with genetically high angiotensin-converting enzyme levels.
Hypertension. 46:1362–1367. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Emerling BM, Platanias LC, Black E,
Nebreda AR, Davis RJ and Chandel NS: Mitochondrial reactive oxygen
species activation of p38 mitogen-activated protein kinase is
required for hypoxia signaling. Mol Cell Biol. 25:4853–4862. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Preusser M, Wohrer A, Stary S, Hoftberger
R, Streubel B and Hainfellner JA: Value and limitations of
immunohistochemistry and gene sequencing for detection of the
IDH1-R132H mutation in diffuse glioma biopsy specimens. J
Neuropathol Exp Neurol. 70:715–723. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ashraf S, Noguera NI, Di Giandomenico J,
Zaza S, Hasan SK and Lo-Coco F: Rapid detection of IDH2 (R140Q and
R172K) mutations in acute myeloid leukemia. Ann Hematol.
92:1319–1323. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Catteau A, Girardi H, Monville F,
Poggionovo C, Carpentier S, Frayssinet V, Voss J, Jenkins R,
Boisselier B, Mokhtari K, et al: A new sensitive PCR assay for
one-step detection of 12 IDH1/2 mutations in glioma. Acta
Neuropathol Commun. 2:582014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Truve K, Dickinson P, Xiong A, York D,
Jayashankar K, Pielberg G, Koltookian M, Muren E, Fuxelius HH,
Weishaupt H, et al: Utilizing the dog genome in the search for
novel candidate genes involved in glioma development-genome wide
association mapping followed by targeted massive parallel
sequencing identifies a strongly associated locus. PLoS Genet.
12:e10060002016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tate JG, Bamford S, Jubb HC, Sondka Z,
Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E,
et al: COSMIC: The catalogue of somatic mutations in cancer.
Nucleic Acids Res. 47:D941–D947. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang LE: Friend or foe-IDH1 mutations in
glioma 10 years on. Carcinogenesis. 40:1299–1307. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Koyasu S, Shimizu Y, Morinibu A, Saga T,
Nakamoto Y, Togashi K and Harada H: Increased
14C-acetate accumulation in IDH-mutated human
glioblastoma: Implications for detecting IDH-mutated glioblastoma
with 11C-acetate PET imaging. J Neuro Oncol.
145:441–447. 2019. View Article : Google Scholar
|
|
35
|
Zhao S and Guan KL: IDH1 mutant structures
reveal a mechanism of dominant inhibition. Cell Res. 20:1279–1281.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vinekar R, Verma C and Ghosh I: Functional
relevance of dynamic properties of Dimeric NADP-dependent
isocitrate dehydrogenases. BMC Bioinformatics. 13 (Suppl
17):S22012. View Article : Google Scholar : PubMed/NCBI
|