Introduction
Although the management and therapy of cancers has
improved in the past decades, cancer remains one of the leading
causes of death worldwide, especially breast cancer among women
(1,2). Globally, over 1,000,000 people are
diagnosed with breast cancer annually and 400,000 females died from
breast cancer every year (3). In
China, the incidence of breast cancer among women was ~20-30% in
2016 and annually grows by 3–5% according to Chinese urban cancer
registries (4). The number of
patients newly diagnosed with breast cancer is still increasing
annually, while the therapeutic effect of breast cancer is
unsatisfactory (5,6). Distant metastasis and recurrence are
the main factors that result in death and poor prognosis of
patients with breast cancer (7).
Therefore, an improved understanding of tumor progression in breast
cancer and the identification of novel biomarkers, which are
associated with the development of breast cancer are essential for
improved disease stratification and clinical management
choices.
Studies have demonstrated that microRNA (miRNA)
expression profiling studies have provided a lot of evidence for
the regulatory role of miRNAs in tumor progression (8–10).
miRNAs are a group of small non-coding RNAs, which are involved in
tumorgenesis and regulate a variety of cellular pathways including
proliferation, differentiation, migration, and invasion (11,12).
miRNA expression profiling can screen dysregulated miRNAs in
various cancers, such as prostate cancer, hepatocellular carcinoma,
and gastric cancer, which provides potential functional miRNAs for
the diagnosis and prognosis of cancers (13). Differentially expressed miRNAs being
expressed in distinguishable patterns allows them to be used as
potentially novel clinical and prognostic biomarkers (14). Downregulation of miR-143 can promote
cell apoptosis and regulate the progression of pancreatic cancer
(15). miR-558 serves as a biomarker
for gastric cancer as its upregulation promotes tumorigenesis and
aggressiveness of gastric cancer by targeting heparinase (16). miR-425, miR-132, miR-145 have been
reported to serve roles in the progression of breast cancer with
different degrees of dysregulation reported (17–19).
miR-623 is a demonstrated downregulated miRNA in
breast cancer and has also been reported to serve roles in a number
of other cancers, such as gastric cancer, pancreatic cancer, and
lung adenocarcinoma (20–22). Reduced or increased expression of
miR-623 regulates the progression of various cancers; hence, it was
hypothesized that the dysregulation of miR-623 may act as a
regulator during the development of breast cancer. The present
study aimed to estimate the clinical significance and functional
role of miR-623 in breast cancer.
Materials and methods
Patients and samples
A total of 121 paired samples were used the present
study, which included breast cancer tissues and adjacent normal
tissues (>5 cm from tumor tissues) with histopathological
diagnosis. The inclusion criteria for the patients were: i) Female
confirmed diagnosed of breast cancer; ii) underwent mastectomy or
breast-conserving surgery; and iii) complete clinical data and
follow-up status. The exclusion criteria were as follows: i)
Patients underwent chemotherapy, radiotherapy or other types of
anticancer therapy; ii) diagnosis with other malignant tumor; and
iii) family history of breast cancer. Patients had an average age
range of 34–66 years with an average age of 50.18±6.82 years. All
samples were obtained from patients with breast cancer who
underwent surgery at The Second Hospital of Liaocheng affiliated to
Shandong First Medical University (Linqing, China) from January
2011 to December 2013. The characteristics of patients are
summarized in Table I. The TNM stage
was recorded according to the 2010 tumor-node metastasis
classification recommended by the American Joint Committee on
Cancer (AJCC 7th edition) (23).
Samples were immediately frozen in liquid nitrogen and stored at
−80°C for subsequent experimentation and analysis. Written informed
consent was obtained from every patient, and the present study was
approved by the Ethics Committee of The Second Hospital of
Liaocheng affiliated to Shandong First Medical University (approval
no. 201033). In addition, all patients participated in a 5-year
follow-up survey for the collection of the survival information.
Patients were followed-up at 6, 9, 12, 15, 18, 21, 24, 30, 36, 42,
48, and 60 months after surgery over the telephone.
 | Table I.Association between miR-623
expression and the characteristics of patients with breast
cancer. |
Table I.
Association between miR-623
expression and the characteristics of patients with breast
cancer.
|
|
| miR-623
expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Patients
(n=121) | Low (n=68) | High (n=53) | P-value |
|---|
| Age, years |
|
|
| 0.162 |
|
≤50 | 54 | 30 | 24 |
|
|
>50 | 67 | 38 | 29 |
|
| Tumor size, cm |
|
|
| 0.247 |
| ≤5 | 62 | 32 | 30 |
|
|
>5 | 59 | 36 | 23 |
|
| Lymph node
metastasis |
|
|
| 0.162 |
|
Negative | 69 | 40 | 29 |
|
|
Positive | 52 | 28 | 24 |
|
| TNM stage |
|
|
| 0.028 |
|
I–II | 71 | 36 | 35 |
|
|
III–IV | 50 | 32 | 18 |
|
| HER 2 status |
|
|
| 0.255 |
|
Negative | 55 | 29 | 26 |
|
|
Positive | 66 | 39 | 27 |
|
| ER status |
|
|
| 0.341 |
|
Negative | 58 | 35 | 23 |
|
|
Positive | 63 | 33 | 30 |
|
| PR status |
|
|
| 0.276 |
|
Negative | 57 | 37 | 23 |
|
|
Positive | 64 | 31 | 33 |
|
Cell culture and transfection
Four human breast cancer cell lines, MCF-7,
MDA-MB-231, HCC1954, and HCC1937, and the normal human breast
epithelial cell line MCF-10A were used in the present study. All
cell lines were purchased from Shanghai Cell Bank of the Chinese
Academy of Medical Sciences and cultured in Dulbecco's modified
Eagle's medium (DMEM) supplemented with 10% fetal bovine serum
(FBS) (both Gibco; Thermo Fisher Scientific Inc.). All cell
cultures were maintained at 37°C in a humidified incubator with 5%
CO2 for 24 h.
To regulate the expression of miR-623 and explore
its effects on the cellular processes of breast cancer, breast
cancer cells were transfected with 20 nM miR-623 mimic, miR-623
inhibitor, mimic negative control (mimic NC), or inhibitor negative
control (inhibitor NC) purchased from Guangzhou RiboBio Co., Ltd.
Lipofectamine 2000® reagent (Invitrogen; Thermo Fisher
Scientific Inc.) was used for transfection at 37°C for 24 h.
Untransfected cells were defined as control group. The sequences of
transfections are as follows: miR-623 mimic,
5′-AUCCCUUGCAGGGGCUGUUGGGU-3′; miR-623 inhibitor,
5′-ACCCAACAGCCCCUGCAAGGGAU-3′; mimic NC,
5′-UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT-3′ and inhibitor NC,
5′-CAGUACUUUUGUGUAGUACAA-3′. After 48 h of transfection, subsequent
experimentation was performed.
RNA isolation and
reverse-transcription quantitative (RT-q)PCR assay
Total RNA from collected tissues and cultured cells
was extracted by using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific Inc.) and reverse transcribed into cDNA by using miRNA
First-Strand cDNA Synthesis kit (Invitrogen; Thermo Fisher
Scientific Inc.). The reverse transcription protocol was: 37°C for
1 h and 85°C for 5 min. SYBR Green I Master Mix kit (Invitrogen;
Thermo Fisher Scientific Inc.) was used to perform RT-qPCR on the
7300 Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific Inc.). The thermocycling conditions were as follows:
95°C for 10 min followed by 40 cycles of 95°C for 15 sec and 60°C
for 15 sec. The expression of miR-623 was normalized to that of U6
with relative quantification by the 2−ΔΔCq calculation
method (24). The primer sequences
of miR-623 were: Forward. 5′-ATCCCTTGCAGGGGCTGTTGGGT-3′ and
reverse, 5′-GCCAGCACAGAATTAATACGAC-3′. The primer sequences of U6
were: Forward 5′-CTCGCTTCGGCAGCACA-3′; reverse
5′-AACGCTTCACGAATTTGCGT-3′.
CCK8 assay
To measure the proliferation ability of breast
cancer cells, a CCK8 assay was performed according to the
manufacturer's instructions. Cells were plated in 96-well plates at
a density of 5×103 cells/well and cultured at 37°C with
5% CO2 for 0, 24, 48 and 72 h. Cells were then incubated
with 10 µl cell counting kit-8 (CCK-8) reagent (Dojindo Molecular
Technologies Inc.) per well for 4 h at 37°C with 5% CO2.
Absorbance at 450 nm was measured with a microplate reader (Thermo
Fisher Scientific Inc.).
Transwell assay
Matrigel-uncoated and coated transwell (for
invasion) inserts (8-mm pore size; Thermo Fisher Scientific Inc.)
were used for the detection of cell migration and invasion.
Matrigel precoating was performed at 37°C for 1 h for the invasion
assay. A total of 2×105 transfected cells were seeded
into the upper chamber with serum-free medium and culture medium
with 10% FBS was placed in the lower chamber as a chemoattractant.
The Transwell was incubated at 37°C for 24 h, after that,
transwells were removed and stained with 0.1% crystal violet
(Sigma; Merck KGaA) at 37°C for 5 min. The number of migrated and
invaded cells in the lower chamber was detected by a fluorescence
microscope (magnification, ×100).
Statistical analysis
All data were represented as the mean value ± SD
obtained from 3 repeats and were analyzed by SPSS 20.0 software
(IBM Corp.) and GraphPad Prism 5.0 software (GraphPad Software,
Inc.) The differences in the expression of miR-623 in breast cancer
tissues and adjacent normal tissues were analyzed using a paired
Student's t-test. The differences between multiple groups was
assessed by one-way ANOVA followed by the post hoc Tukey's test.
The association between miR-623 expression and clinical
characteristics of patients was evaluated by the χ2
test. The survival curves of patients were generated using the
Kaplan-Meier method and were compared using the log-rank test.
Additionally, the prognostic value of miR-623 was further assessed
by the Cox regression analyses. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression level of miR-623 in breast
cancer tissue and cells
In the collected breast cancer and adjacent normal
tissues, the expression of miR-623 was detected by RT-qPCR. miR-623
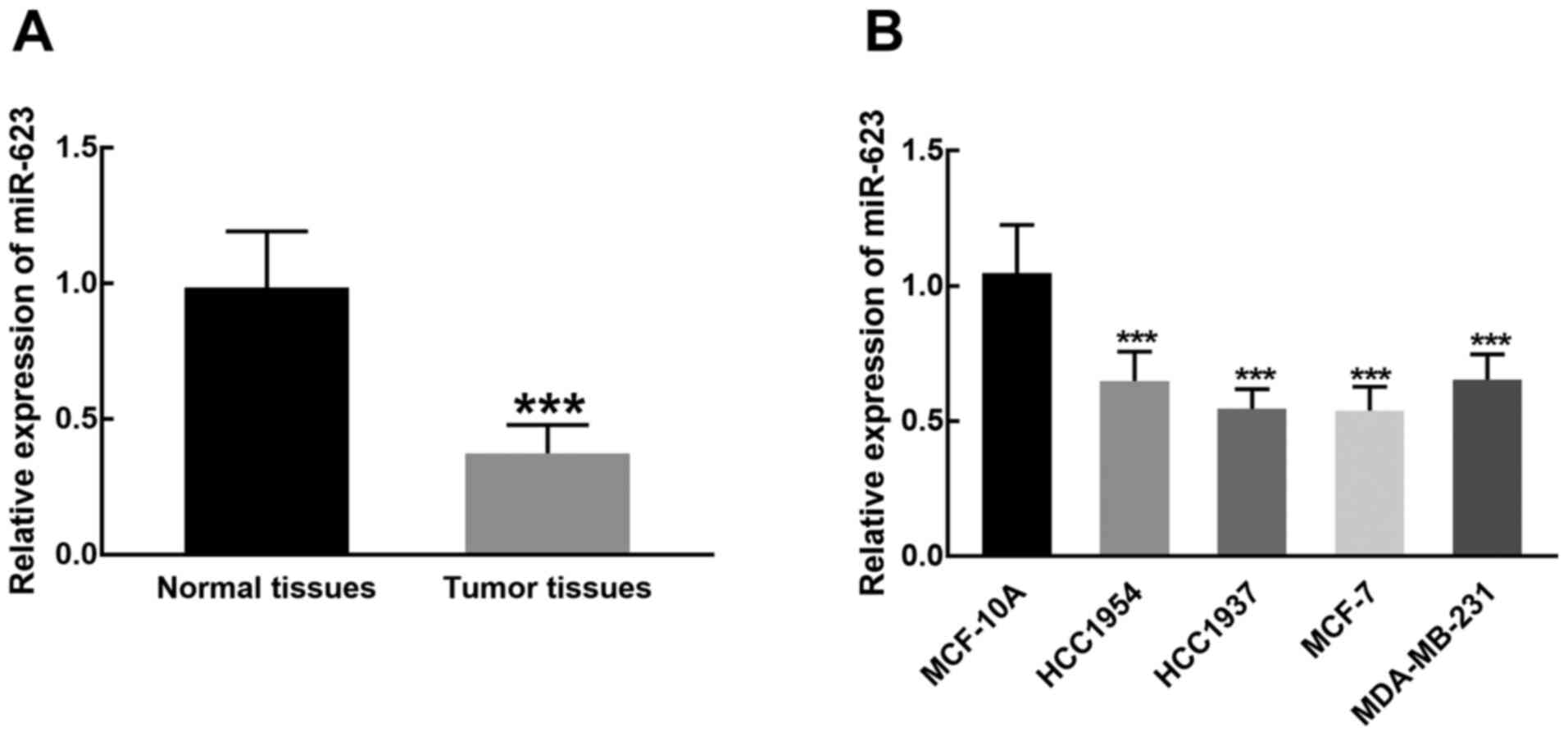
demonstrated significantly decreased expression in breast cancer
tissues compared with adjacent normal tissues (P<0.001; Fig. 1A). Similarly, miR-623 was
significantly downregulated in breast cancer cell lines (HCC1954,
HCC1937, MCF-7 and MDA-MB-231) compared with MCF-10A cells
(P<0.001; Fig. 1B). The
relatively lower expression of miR-623 in HCC1937 and MCF-7 cells
compared to the other cancer cell lines indicated that they are
more sensitive to the dysregulation of miR-623, hence they were
chosen for subsequent cell experiments.
Association between miR-623 expression
and clinical characteristics of patients with breast cancer
A total of121 breast cancer patients were divided
into miR-623 high or low expression groups based on the mean value
of the miR-623 level (0.375) in breast cancer tissues. Results of
the χ2 test demonstrated a significant association
between miR-623 expression and the TNM stage of patients (P=0.028),
while age, tumor size, lymph node metastasis, and the status of
human epidermal growth factor receptor 2, estrogen receptor (ER)
and progesterone receptor of patients demonstrated no significant
association with the expression of miR-623 (P>0.05, Table I).
Prognostic value of miR-623 in breast
cancer
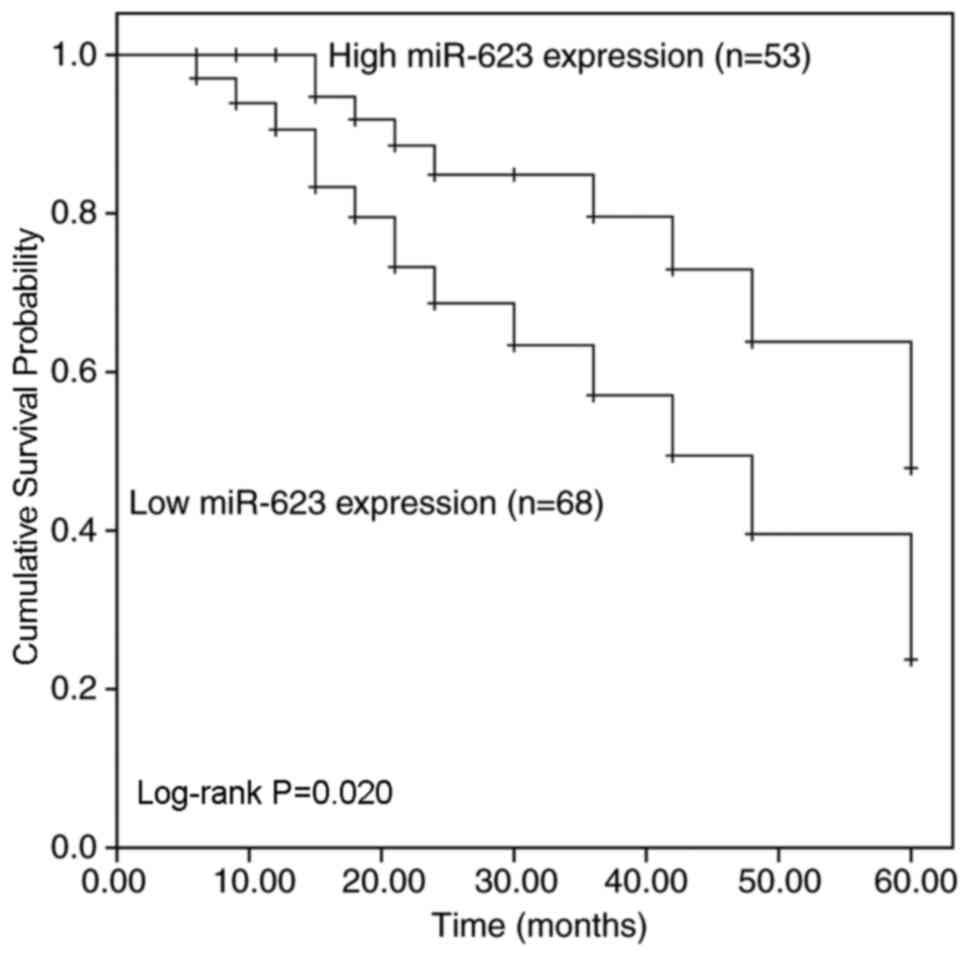
Fig. 2 demonstrates
the survival rate of patients with breast cancer with low or high
miR-623 expression levels 5 years after surgery. Patients with low
miR-623 expression demonstrated a shorter overall survival time
compared with patients with high miR-623 expression (Log-rank
P=0.020; Fig. 2). Additionally, the
results of Cox regression analysis indicated that miR-623
expression [hazard ratio (HR) factor=2.743; 95% confidence interval
(CI)=1.260–5.971; P=0.011] and TNM stage (HR factor=2.191; 95%
CI=1.082–4.434; P=0.029) are two independent prognostic factors for
breast cancer due to their close association with the survival rate
of patients (Table II).
 | Table II.Cox regression analysis of the
association between characteristics of patients with breast cancer
and survival rate. |
Table II.
Cox regression analysis of the
association between characteristics of patients with breast cancer
and survival rate.
|
Characteristics | HR factor | 95% CI | P-value |
|---|
| miR-623
expression | 2.743 | 1.260–5.971 | 0.011 |
| Age | 1.250 | 0.602–2.598 | 0.549 |
| Tumor size | 1.633 | 0.789–3.377 | 0.186 |
| Lymph node
metastasis | 1.694 | 0.831–3.453 | 0.147 |
| TNM stage | 2.191 | 1.082–4.434 | 0.029 |
| HER2 status | 1.594 | 0.765–3.320 | 0.213 |
| ER status | 1.447 | 0.698–3.003 | 0.321 |
| PR status | 1.504 | 0.741–3.049 | 0.258 |
Effect of miR-623 expression on cell
proliferation of breast cancer cells
miR-623 mimic, miR-623 inhibitor and corresponding
negative controls were transfected into HCC1937 and MCF-7 cells for
the overexpression or knockdown of miR-623. In addition, the
proliferation ability of transfected cells was analyzed by the CCK8
assay to investigate the effect of miR-623 expression on cell
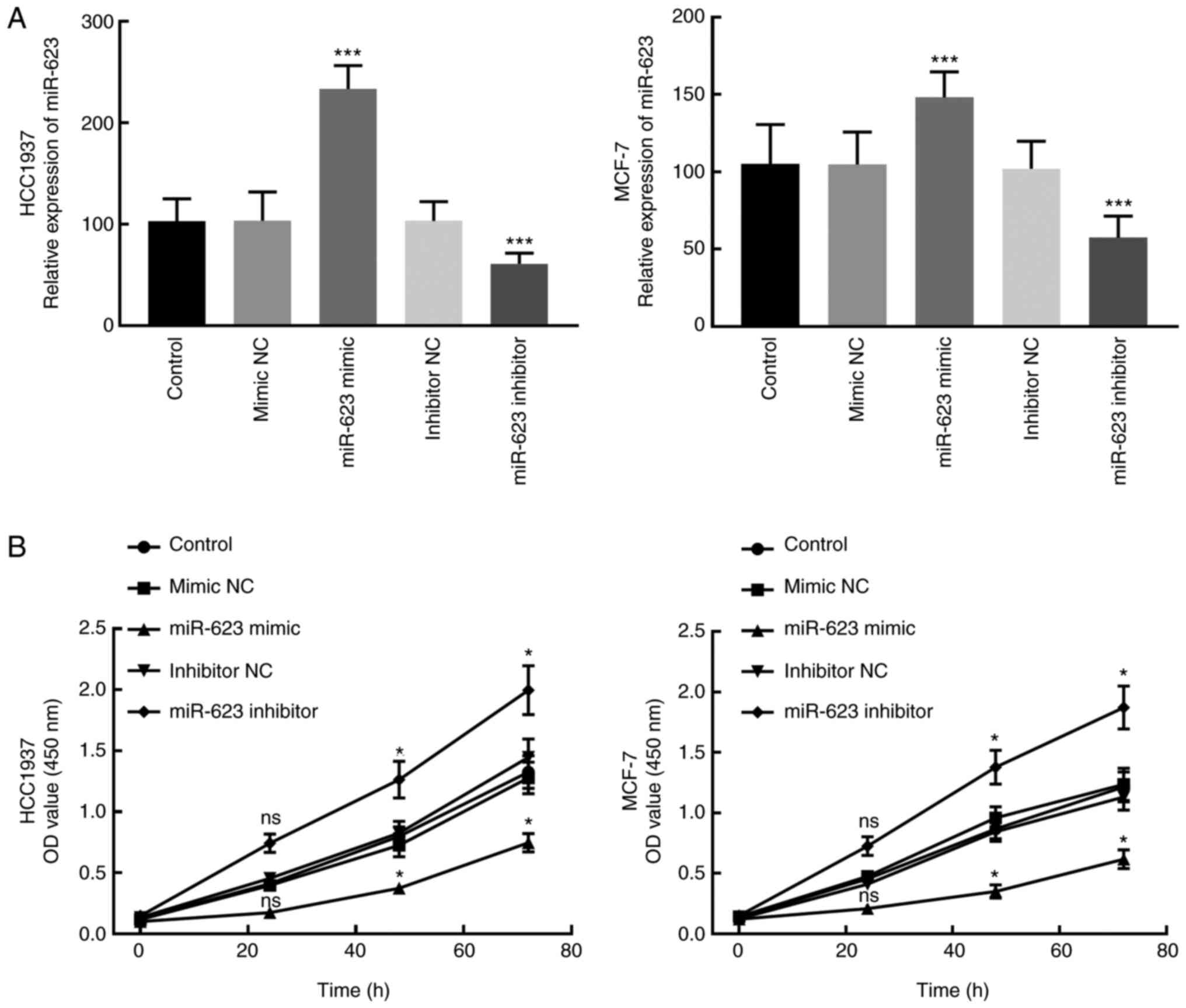
proliferation of breast cancer cells. Following the transfection of
miR-623 mimic, miR-623 was significantly upregulated, while the
transfection of miR-623 inhibitor significantly downregulated
miR-623 in HCC1937 and MCF-7 cells compared with cells in control
group (P<0.001; Fig. 3A).
The results of CCK8 assay revealed that the
proliferation of HCC1937 and MCF-7 cells was significantly
inhibited by the miR-623 mimic and promoted by the miR-623
inhibitor (P<0.05 relative to control group; Fig. 3B). This suggests that miR-623 may be
involved in the proliferation of breast cancer cells.
Role of miR-623 in cell migration and
invasion of breast cancer cells
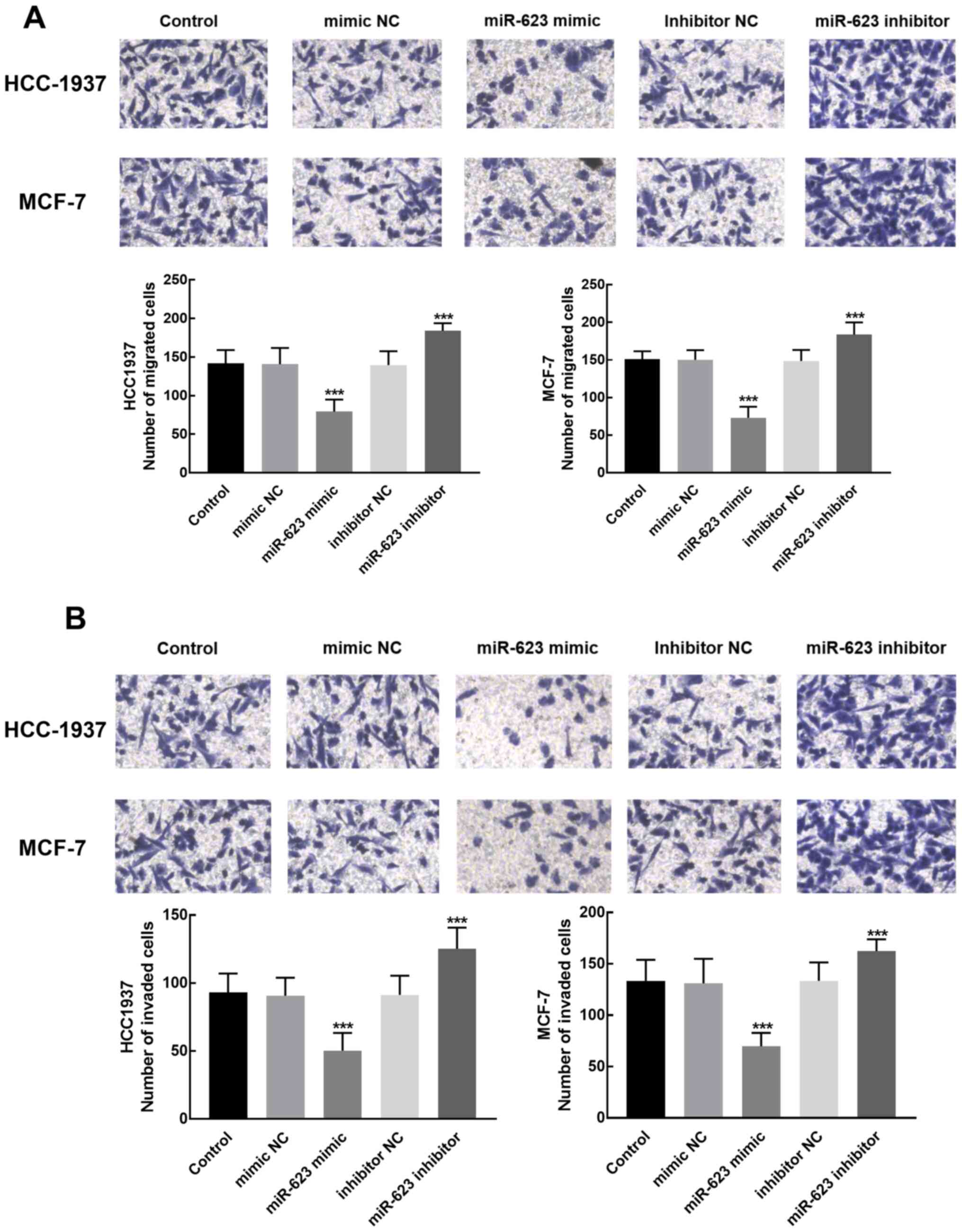
Transwell assays were performed to evaluate the
migration and invasion of HCC1937 and MCF-7 cells with different
expression levels of miR-623. The results demonstrated that the
migration and invasion of cells transfected with the miR-623 mimic
was significantly lower compared with cells transfected with
miR-623 inhibitor, which revealed the inhibitory role of miR-623 in
the migration and invasion of breast cancer cells (P<0.001;
Fig. 4A and B).
Discussion
Due to the increasing number of new cases and the
death rate resulting from recurrence and metastasis, breast cancer
is still considered as the most malignant tumor in women (25). The exploration of novel biomarkers in
breast cancer has received increasing attention in recent decades
(26). miRNAs have been identified
as biomarkers for the diagnosis or prognosis of various cancers,
such as gastric cancer, lung cancer, colorectal cancer, and
numerous other cancers and diseases, such as acute myeloid leukemia
and Alzheimer's disease (16,27–29).
For example, overexpressed miR-675 in non-small cell lung cancer
promotes the progression of the disease by activating the NF-κB
signaling pathway (30). A number of
miRNAs, such as miR-145-5p, miR-940, and miR-205-3p (19,31,32),
which serve roles in the progression of breast cancer have been
considered as potential biomarkers for breast cancer.
Previously, miR-623 was reported to be dysregulated
in breast cancer (33) and to serve
roles in various cancers (20–22). For
example, miR-623 was demonstrated to inhibit cell migration,
invasion, and metastasis of pancreatic cancer in vitro and
in vivo by directly targeting MMP1 (22). In gastric cancer, miR-623 can
function as a tumor suppressor and enhance chemosensitivity
(20). miR-623 has also been
demonstrated to suppress the progression of lung adenocarcinoma by
targeting MMP-2 and MMP-9 (21). The
present study was performed to identify the role of miR-623 in the
progression and prognosis of breast cancer. miR-623 was found to be
downregulated in breast cancer tissues and cell lines compared with
normal breast tissues and cell lines. In the present study,
overexpression of miR-623 by miR-623 mimic transfection was
demonstrated to exert inhibitory effects on cell proliferation,
migration and invasion of breast cancer, indicating the tumor
suppressor role of miR-623 in the progression of breast cancer.
Recently, the biological function of miR-623 has
been revealed by Li et al (34) in MCF-7 and MDA-MB-453 cells. The
exogenous overexpression of miR-623 inhibited cell proliferation
and promoted cell apoptosis of breast cancer by targeting X-ray
repair cross-complementing protein 5 (34), which is consistent with the
biological function of miR-623 in MCF-7 and HCC1937 cells in the
present study. In the present study, in addition, to miR-623
function in cell progression of breast cancer, the clinical
significance of miR-623 was also assessed. miR-623 expression had a
close relationship with the TNM stage of patients. Additionally, in
the present study patients who had low miR-623 mRNA expression
demonstrated poorer prognosis compared with patients with high
miR-623 expression, indicating that the downregulation of miR-623
was associated with poor prognosis of breast cancer. Based on the
findings of the present study, miR-623 expression and TNM stage
were considered as two independent prognostic factors for breast
cancer. miRNAs have been reported to predict the prognosis of a
variety of cancers and serve as independent prognostic factors for
cancers. For instance, overexpression of miR-153 in prostate cancer
can predicate the poor prognosis of patients (35). Downregulation of miR-1247-5p in
breast cancer is associated with poor prognosis of patients
(36). All the aforementioned
findings of the present study, indicate that miR-623 participated
in and suppressed the progression of breast cancer.
However, in addition to the investigation of the
function of miR-623 in the progression of breast cancer, it is also
necessary to determine the mechanism underlying the functional role
of miR-623. Previously, MMP1 and Cyclin D1 were demonstrated to be
direct targets of miR-623, regulating how miR-623 participates in
the progression of pancreatic cancer and gastric cancer (20,22).
Hence, it was speculated that miR-623 may regulate the progression
of breast cancer by targeting these genes or other potential
targets, which is a limitation of this study and needs further
experiments to validate this hypothesis.
In conclusion, the results of the present study
demonstrated that miR-623 was downregulated in breast cancer
tissues and cells, which was associated with the TNM stage of
patients and predicted poor prognosis of patients with breast
cancer. miR-623 participated in the progression of breast cancer
and was an independent prognostic factor together with TNM stage.
In addition, knocking down of miR-623 significantly promoted cell
proliferation, migration and invasion of breast cancer, while its
overexpression significantly suppressed these cellular processes,
which indicated the tumor inhibitor role of miR-623 in breast
cancer. Malignant tumor is a multifaceted disease with important
differences among diverse types of cancers (37). This research is an expanding study
that applied regular methods to investigate the role of miR-623 in
breast cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The data used during the current study are available
from the corresponding author on reasonable request.
Authors' contributions
CW, JW, JZ, YL, QS, FG and XA made substantial
contributions to conception and design. CW, JW, JZ, YL, QS and FG
contributed to acquisition of data. CW and XA analyzed and
interpreted the data. All the authors were involved in writing the
manuscript. All authors have read and approved the manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from every
patient and this study was approved by the Ethics Committees of The
Second Hospital of Liaocheng affiliated to Shandong First Medical
University (Linqing, China) (approval number, 201033).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Espie M: The management of breast cancer.
Diagn Interv Imaging. 95:753–757. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shimizu C: Breast cancer in young women:
Its biological and clinical uniqueness and needs of comprehensive
care. Breast Cancer. 21:641–642. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li T, Mello-Thoms C and Brennan PC:
Descriptive epidemiology of breast cancer in China: Incidence,
mortality, survival and prevalence. Breast Cancer Res Treat.
159:395–406. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sorscher S: Prognosis of patients with
multifocal breast cancer. Hum Pathol. 84:335–336. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Plichta JK: Breast cancer prognostic
staging and internal mammary lymph node metastases: A brief
overview. Chin Clin Oncol. 8:S112019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Van Langenberg A: Prognosis after breast
recurrence following conservative surgery and radiotherapy in
patients with node-negative breast cancer. Br J Surg. 87:681–683.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hwang J, Min BH, Jang J, Kang SY, Bae H,
Jang SS, Kim JI and Kim KM: MicroRNA expression profiles in gastric
carcinogenesis. Sci Rep. 8:143932018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mohr AM and Mott JL: Overview of microRNA
biology. Semin Liver Dis. 35:3–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kallioniemi A: Molecular signatures of
breast cancer-predicting the future. N Engl J Med. 347:2067–2068.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu B, Liu J, Xiang X, Liu S, Zhong P, Xie
F, Mou T and Lai L: Expression of miRNA-143 in pancreatic cancer
and its clinical significance. Cancer Biother Radiopharm.
33:373–379. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zheng L, Jiao W, Song H, Qu H, Li D, Mei
H, Chen Y, Yang F, Li H, Huang K and Tong Q: miRNA-558 promotes
gastric cancer progression through attenuating Smad4-mediated
repression of heparanase expression. Cell Death Dis. 7:e23822016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xiao S, Zhu H, Luo J, Wu Z and Xie M:
miR425-5p is associated with poor prognosis in patients with breast
cancer and promotes cancer cell progression by targeting PTEN.
Oncol Rep. 42:2550–2560. 2019.PubMed/NCBI
|
|
18
|
Li S, Xu JJ and Zhang QY: MicroRNA-132-3p
inhibits tumor malignant progression by regulating
lysosomal-associated protein transmembrane 4 beta in breast cancer.
Cancer Sci. 110:3098–3109. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tang W, Zhang X, Tan W, Gao J, Pan L, Ye
X, Chen L and Zheng W: miR-145-5p suppresses breast cancer
progression by inhibiting SOX2. J Surg Res. 236:278–287. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang L, Yang W, Bian W, Yang H, Wu X, Li
Y, Feng W and Liu X: MicroRNA-623 targets cyclin D1 to inhibit cell
proliferation and enhance the chemosensitivity of cells to
5-fluorouracil in gastric cancer. Oncol Res. 27:19–27. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wei S, Zhang ZY, Fu SL, Xie JG, Liu XS, Xu
YJ, Zhao JP and Xiong WN: Hsa-miR-623 suppresses tumor progression
in human lung adenocarcinoma. Cell Death Dis. 7:e23882016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen Y, Peng S, Cen H, Lin Y, Huang C,
Chen Y, Shan H, Su Y and Zeng L: MicroRNA hsa-miR-623 directly
suppresses MMP1 and attenuates IL-8-induced metastasis in
pancreatic cancer. Int J Oncol. 55:142–156. 2019.PubMed/NCBI
|
|
23
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li G, Hu J and Hu G: Biomarker studies in
early detection and prognosis of breast cancer. Adv Exp Med Biol.
1026:27–39. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Q, Wang Y, Hu R and Yang G:
Dysregulation of SPRR3/miR-876-3p axis contributes to tumorigenesis
in non-small-cell lung cancer. Onco Targets Ther. 13:2411–2419.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sahami-Fard MH, Kheirandish S and Sheikhha
MH: Expression levels of miR-143-3p and −424-5p in colorectal
cancer and their clinical significance. Cancer Biomark. 24:291–297.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang Z, Zhang J, Li J, Geng H, Zhou B,
Zhang B and Chen H: miR-320/ELF3 axis inhibits the progression of
breast cancer via the PI3K/AKT pathway. Oncol Lett. 19:3239–3248.
2020.PubMed/NCBI
|
|
30
|
Feng Y, Yang C, Hu D, Wang X and Liu X:
miR-675 promotes disease progression of non-small cell lung cancer
via activating NF-κB signaling pathway. Cell Mol Biol
(Noisy-le-grand). 63:7–10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu W, Xu Y, Guan H and Meng H: Clinical
potential of miR-940 as a diagnostic and prognostic biomarker in
breast cancer patients. Cancer Biomark. 22:487–493. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qiu C, Huang F, Zhang Q, Chen W and Zhang
H: miR-205-3p promotes proliferation and reduces apoptosis of
breast cancer MCF-7 cells and is associated with poor prognosis of
breast cancer patients. J Clin Lab Anal. 33:e229662019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hamam R, Ali AM, Alsaleh KA, Kassem M,
Alfayez M, Aldahmash A and Alajez NM: microRNA expression profiling
on individual breast cancer patients identifies novel panel of
circulating microRNA for early detection. Sci Rep. 6:259972016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Q, Liu J, Jia Y, Li T and Zhang M:
miR-623 suppresses cell proliferation, migration and invasion
through direct inhibition of XRCC5 in breast cancer. Aging (Albany
NY). 12:10246–10258. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bi CW, Zhang GY, Bai Y, Zhao B and Yang H:
Increased expression of miR-153 predicts poor prognosis for
patients with prostate cancer. Medicine (Baltimore). 98:e167052019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zeng B, Li Y, Feng Y, Lu M, Yuan H, Yi Z,
Wu Y, Xiang T, Li H and Ren G: Downregulated miR-1247-5p associates
with poor prognosis and facilitates tumor cell growth via
DVL1/Wnt/β-catenin signaling in breast cancer. Biochem Biophys Res
Commun. 505:302–308. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Das J, Gayvert KM and Yu H: Predicting
cancer prognosis using functional genomics data sets. Cancer
Inform. 13 (Suppl 5):S85–S88. 2014.
|