Introduction
The detection of oncogenic driver mutations, such as
epidermal growth factor receptor (EGFR) mutations, is
essential for determining treatment strategies for advanced
non-small cell lung cancer (NSCLC). The EGFR-tyrosine kinase
inhibitor (TKI) is among the most successful treatments for NSCLC
with EGFR mutations, as it extends the median overall
survival by over 30 months (1,2). A
third-generation EGFR-TKI, osimertinib, has been clinically
approved for lung cancers harboring the acquired resistance
mutation EGFR T790M (3).
However, the invasive tissue biopsy required for EGFR
testing is often challenging. Indeed, data show that re-biopsy was
not performed in 20–50% of patients treated with EGFR-TKIs, raising
questions on the viability of this method (4–6).
Liquid biopsies that assess circulating tumor DNA
from blood samples have been developed, providing an alternative
for biomarker identification (7,8). Other
samples, such as exhaled breath condensate (EBC) and urine, have
also been tested as alternatives to liquid biopsy (9–12).
We previously reported the detection of EGFR
exon 19 deletion (Ex19del) mutations in EBC by conventional
polymerase chain reaction (PCR) (13). Recently, a highly sensitive droplet
digital PCR (ddPCR) was developed for liquid biopsy (14–16). In
this study, we investigated whether EBC testing using ddPCR
(EBC-ddPCR) for EGFR mutations is viable in patients with
NSCLC.
Patients and methods
Clinical samples and lung cancer cell
lines
Patients with lung cancer harboring the EGFR
mutations L858R, Ex19del, or T790M were enrolled in this study
between June 1, 2014 and December 31, 2017 after obtaining written
informed consent. All patients were diagnosed with NSCLC using
surgical tissue samples, biopsy specimens, or cytology samples. The
diagnosis was based on the General Rules for the Clinical and
Pathological Classification of Lung Cancer of the Japan Lung Cancer
Society (8th edition) and TNM staging system of the International
Association for the Study of Lung Cancer (8th edition) (17). Written informed consent was also
obtained from three healthy volunteers. The baseline EGFR
mutations of lung cancers were confirmed by clinically approved
methods in Japan by practically examining surgical tissues, biopsy
specimens, or cytology samples. Approximately 1–2 ml of EBC was
collected using RTube™ (Respiratory Research), according to the
manufacturer's protocol. After collecting the EBC, 1-ml aliquots
were dispensed and stored at −80°C. This study was approved by the
Ethics Committee of Okayama University (Authorization number:
2221).
The lung cancer cell line H3255 harboring
EGFR L858R was kindly provided by Dr William Pao (Vanderbilt
University, Nashville, TN, USA) (18). The gefitinib-resistant lung cancer
cell lines RPC-9 harboring EGFR Ex19del and T790M were
previously established in our laboratory (19).
Droplet digital PCR assay for EGFR
mutation detection
DNA was extracted using the QIAamp DNA Mini kit
(Qiagen GmbH) according to the manufacturer's protocol. DNA
qualification was performed with a NanoDrop spectrometer (Thermo
Fisher Scientific, Inc.). The following primer and probe kits were
purchased from Bio-Rad): ddPCR Mutation assay: EGFR p.L858R
c.2573T>G (#10049550); ddPCR EGFR Exon 19 Deletions Screening
kit (#12002392) and ddPCR Mutation assay: EGFR p.T790M, Human
(#10049550).
ddPCR was performed at Biobank (Okayama University
Hospital, Okayama, Japan) using the QX200 Droplet Digital PCR
system (Bio-Rad) according to the manufacturer's protocol. The
following conditions were used for ddPCR: i) An initial
denaturation step at 95°C for 10 min followed by: ii) 45 cycles at
94°C for 30 sec; and iii) 45 cycles at °C for 1 min, with a 4)
final enzyme deactivation step at 98°C for 10 min. The ramp rate
for all steps was 2°C/sec. PCR products were then subjected to
analysis with a QX-200 Droplet reader and QuantaSoft analysis
software (Bio-Rad), with a EGFR Specific Multiplex DNA Reference
Standard (#HD802, Horizon Discovery) used as a positive control.
The accuracy of the ddPCR was confirmed using serially diluted DNA
of lung cancer cell lines (Fig.
S1A-F). The patient samples and control samples including EBC
samples from three healthy-volunteers or elution buffer AE from the
QIAamp DNA Mini kit (Qiagen) as negative controls were tested to
determine the threshold (Fig.
S2A-D). No droplets were detected for EGFR L858R or
EGFR T790M, whereas positive reactions were observed for
EGFR Ex19del in the negative control samples. Therefore, the
threshold for positive results for EGFR Ex19del were
analyzed with the receiver operating characteristic (ROC) using
patient samples (Fig. S3). As a
result, the threshold for EGFR mutation-positive was defined
as follows: EGFR L858R (≥0.01 copies/µl), EGFR
Ex19del (≥0.5 copies/µl), and EGFR T790M (≥0.01
copies/µl).
Statistical analysis
Statistical analysis was performed using STATA
software version 15.1 (StataCorp). ROC analysis was performed to
determine the optimal threshold for epidermal growth factor
receptor exon 19 deletion. The 95% confidence interval (CI) was
calculated by using the Clopper-Pearson exact method for binomial
proportions.
Results
Patient characteristics and EBC
samples
Patient and healthy control characteristics are
detailed in Tables I and II, respectively. The median patient age
was 66 years (range, 54–81), comprising 3 males and 9 females:
10/12 were never-smokers and 9/12 were in stage IV of the disease.
Four lung tumors harbored EGFR L858R, whereas 8 tumors
showed the EGFR Ex19del mutation (Table I). In total, 21 samples were
collected from the 12 patients. Of these 21 samples, 11 were from
patients with lung cancers harboring EGFR L858R and 10 were
from patients with lung cancers harboring EGFR Ex19del. The
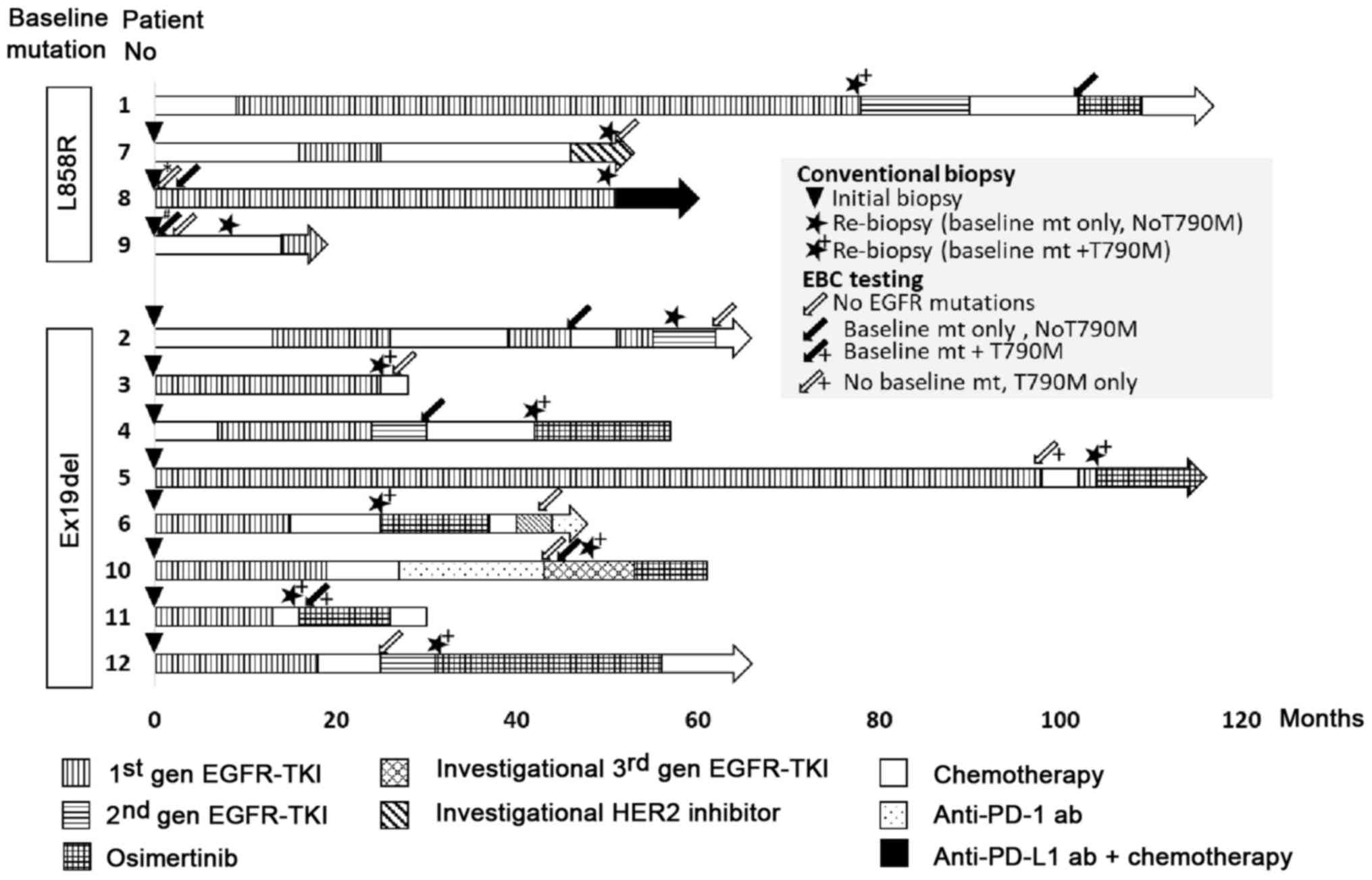
timing of EBC collection and treatment history are shown in
Fig. 1.
 | Table I.Patient characteristics. |
Table I.
Patient characteristics.
| Patient number | Sex | Age (years) | Smoking | Stageb | Baseline
EGFR mts | Biopsy site |
|---|
| 1 | F | 60 | Never | rec | L858R | -a |
| 2 | M | 68 | Former | IVB | Ex19del | RL lobe |
| 3 | F | 81 | Never | IVB | Ex19del | Pleural fluid |
| 4 | F | 74 | Never | IVB | Ex19del | RU lobe |
| 5 | F | 66 | Never | rec | Ex19del | RU lobe |
| 6 | F | 58 | Never | IVB | Ex19del | Pleural fluid |
| 7 | M | 76 | Never | IVA | L858R | Pleura |
| 8 | F | 67 | Never | rec | L858R | Middle lobe |
| 9 | F | 70 | Former | IVB | L858R | Pleural fluid |
| 10 | M | 56 | Never | IVB | Ex19del | RU lobe |
| 11 | F | 56 | Never | IVB | Ex19del | LL lobe |
| 12 | F | 54 | Never | IVB | Ex19del | RL lobe |
 | Table II.Healthy volunteer
characteristics. |
Table II.
Healthy volunteer
characteristics.
| Healthy
volunteer | Sex | Age | Smoking |
|---|
| 1 | M | 41 | Never |
| 2 | M | 33 | Never |
| 3 | M | 37 | Never |
Nine EBC samples were collected from two patients at
the time of initiation of EGFR-TKI (Table III). In contrast, 12 EBC samples
were collected from ten patients at the time of the second biopsy
(Table IV). There were no adverse
events due to EBC sampling.
 | Table III.EBC-ddPCR. |
Table III.
EBC-ddPCR.
| Patient no. | Baseline
EGFR mts | Biopsy site | T-factor | Localization of
lung tumors | Sample no. | DNA (ng) | 260/280 ratio | EBC-ddPCR |
|---|
| 8 | L858R | RL lobe | 2b | Peri | 8-1 | 279.5 | 1.46 | (−) |
|
|
|
|
|
| 8-2 | 133.5 | 1.37 | (−) |
|
|
|
|
|
| 8-3 | 503.0 | 1.73 | (−) |
|
|
|
|
|
| 8-4 | 250.0 | 1.86 | L858R |
| 9 | L858R | Pleural fluid | 4 | Center | 9-1 | 138.0 | 1.66 | (−) |
|
|
|
|
|
| 9-2 | 257.0 | 1.53 | (−) |
|
|
|
|
|
| 9-3 | 253.5 | 2.06 | (−) |
|
|
|
|
|
| 9-4 | 267 | 1.99 | L858R |
|
|
|
|
|
| 9-5 | 181.5 | 1.37 | (−) |
 | Table IV.EBC-ddPCR. |
Table IV.
EBC-ddPCR.
| Patient no. | 2nd-biopsy
EGFR mutations | EGFR test | Biopsy site | T-factor | Localization of
lung tumors | Sample no. | DNA (ng) | 260/280 ratio | EBC-ddPCR |
|---|
| 1 | L858R + T790M | Clamp | LL lobe | Txa | Peri | 1-1 | 214.5 | 1.37 | L858R |
| 2 | Ex19del | Clamp | RL lobe | 4 | Center | 2-1 | 268.5 | 2.33 | Ex19del |
|
|
|
|
|
|
| 2-2 | 234.0 | 1.88 | (−) |
| 3 | Ex19del +
T790M | Clamp | Pleural fluid | 2a | Peri | 3-1 | 272.0 | 192.0 | (−) |
| 4 | Ex19del +
T790M | TaqMan | Blood | 2a | Peri | 4-1 | 244.5 | 2.17 | Ex19del |
| 5 | Ex19del +
T790M | Clamp | CSF | Txb | Peri | 5-1 | 277.0 | 1.64 | T790M |
| 6 | Ex19del +
T790M | Clamp | Pleural fluid | 1b | Peri | 6-1 | 280.5 | 2.07 | (−) |
| 7 | L858R | Clamp | Pleural fluid | 2a | Peri | 7-1 | 420.5 | 1.55 | (−) |
| 10 | Ex19del +
T790M | Clamp | Pleural fluid | 1b | Peri | 10-1 | 123.5 | 10.48 | (−) |
|
|
|
|
|
|
| 10-2 | 163.5 | 4.41 | (−) |
| 11 | Ex19del +
T790M | Clamp | LL lobe | 3 | Peri | 11-1 | 152.0 | 4.01 | Ex19del +
T790M |
| 12 | Ex19del +
T790M | Clamp | RU lobe | 2a | Peri | 12-1 | 323.0 | 1.66 | (−) |
EBC-ddPCR for EGFR mutations in
patients
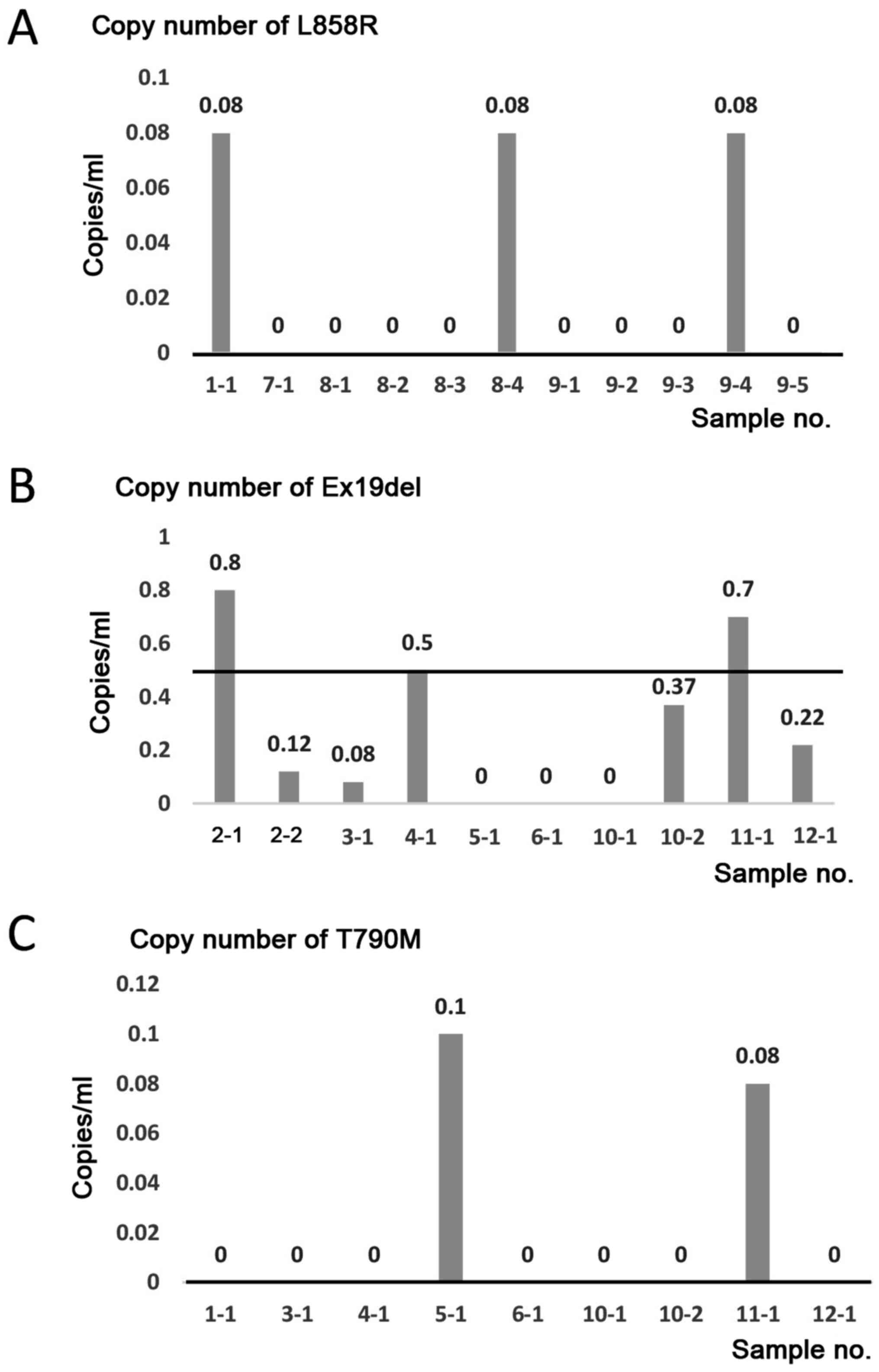
The 21 EBC samples from 12 patients were analyzed by
ddPCR using EGFR L858R, Ex19del. or T790M primer sets. In
the four patients with lung cancer harboring EGFR L858R, 3
of 11 EBC samples (sample nos. 1–1, 8–4, and 9–4) were positive for
EGFR L858R (Fig. 2A and
Tables II and IV). In the 8 patients with lung cancer
harboring EGFR Ex19del, 3/10 EBC samples (sample nos. 2-1,
4-1, and 11-1) were positive for EGFR Ex19del (Fig. 2B and Table IV). In the 8 patients with lung
cancer harboring EGFR T790M, 2/9 EBC samples (sample nos.
5-1 and 11-1) were positive for EGFR T790M (Fig. 2C and Table IV). No association was detected
between the detection of mutations and T-factor/tumor localization
in the lung or quality/quantity of DNA in the EBC samples (Tables III and IV).
Consequently, the sensitivity of the EBC test for
EGFR mutations was as follows: 27.3% (95% CI, 6.0–61.0%) for
EGFR L858R, 30.0% (95% CI, 6.7–65.2%) for EGFR
Ex19del, and 22.2% (95% CI, 2.8–60.0%) for EGFR T790M. The
specificity was as follows: 80.0% (95% CI, 44.4–97.7%) for
EGFR L858R, 90.9% (95% CI, 58.7–99.8%) for EGFR
Ex19del, and 100% (95% CI, 73.5–100%) for EGFR T790M
(Table V).
 | Table V.Sensitivity, specificity and
predictive values of exhaled breath concentrate-droplet digital
PCR. |
Table V.
Sensitivity, specificity and
predictive values of exhaled breath concentrate-droplet digital
PCR.
| Parameter | L858R | Ex19del | T790M |
|---|
| Sensitivity (95%
CI) | 27.3
(6.0–61.0) | 30.0
(6.7–65.2) | 22.2
(2.8–60.0) |
| Specificity (95%
CI) | 80.0
(44.4–97.5) | 90.9
(58.7–99.8) | 100 (73.5–100) |
| Positive predictive
value (95% CI) | 60.0
(14.7–94.7) | 75.0
(19.4–99.4) | 100 (15.8–100) |
| Negative predictive
value (95% CI) | 50.0
(24.7–75.3) | 58.8
(32.9–81.6) | 63.2
(38.4–83.7) |
Clinical course of highlighted cases
with lung adenocarcinoma, whose EGFR mutations were detected by
EBC-ddPCR
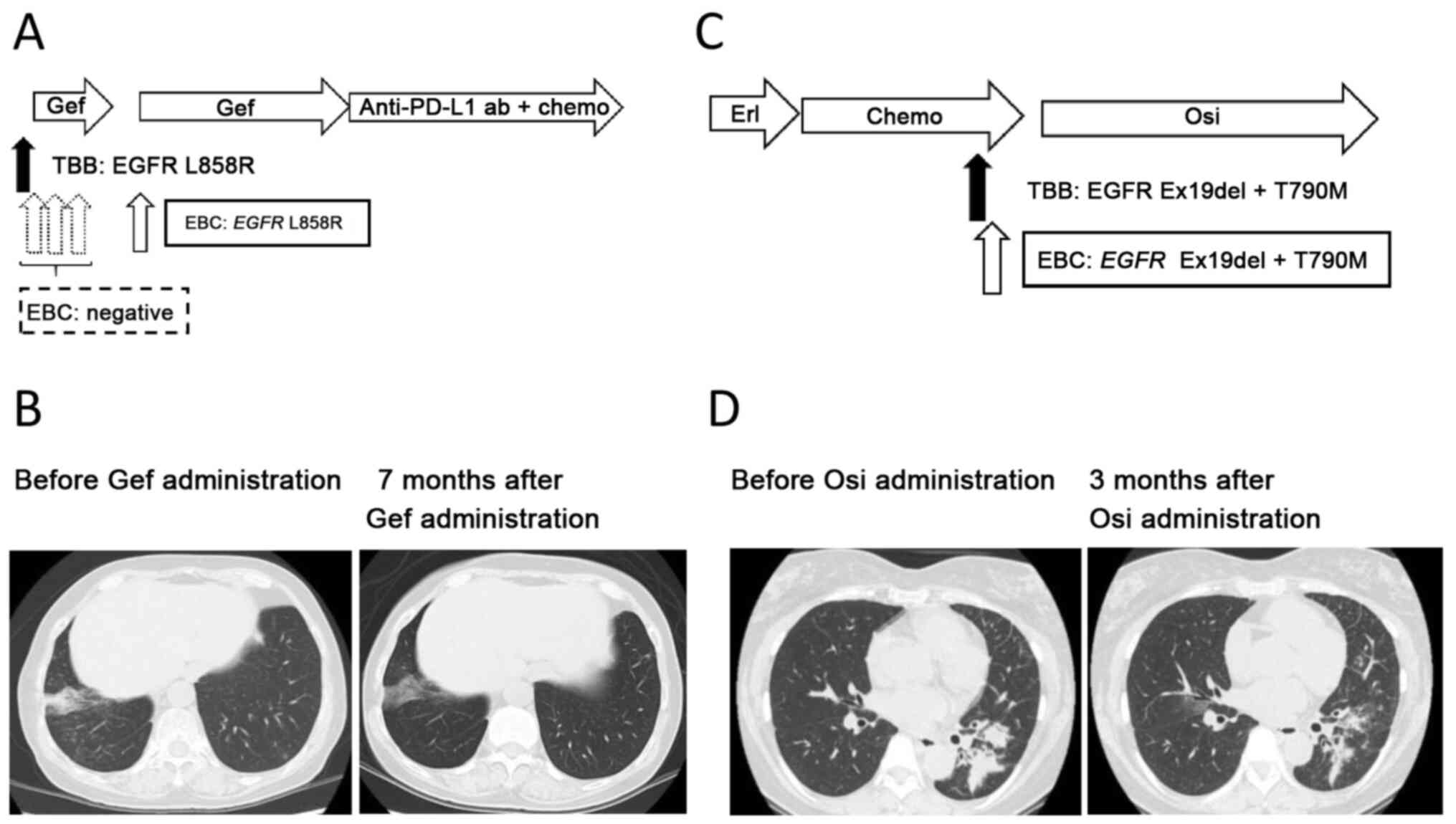
Patient number 8. A non-smoking 67-year-old woman
was diagnosed with synchronous double primary lung cancer composed
of adenocarcinoma harboring EGFR L858R in the right lower
lobe of the lung, and adenocarcinoma harboring EGFR Ex19del
in the right upper lobe of the lung. A right lower lobectomy
(pathological stage T2bN0M0 cStage IIA) and right segment 3a
partial resection (pathological stage T1aN0M0 cStage IA1) were
performed, with four subsequent courses of adjuvant chemotherapy
with a combination of cisplatin and vinorelbine. However, at 12
months post-surgery, a new lesion appeared in the middle lobe.
Transbronchial biopsy of the lesion revealed an adenocarcinoma
harboring EGFR L858R. The patient's Eastern Cooperative
Group Performance Status was grade 1, and thus gefitinib was
administered at 250 mg daily. EBC was collected 1 day prior
starting gefitinib and at 2, 16 and 62 days after gefitinib
initiation. Gefitinib was discontinued from days 41–62 because of
liver damage, but later re-administered at a reduced dosage of 250
mg every 2 days. EBC-ddPCR detected EGFR L858R only in the
fourth sampling (21 days after gefitinib cessation). The maximum
therapeutic effect of gefitinib was a partial response (Response
Evaluation Criteria in Solid Tumors version 1.1), with a
progression-free survival of 48 months (Fig. 3A and B).
Patient number 11
A non-smoking 56-year-old woman was diagnosed with
adenocarcinoma in the left lower lobe of the lung (clinical stage
T3N1M1b cStage IVB, multiple brain metastases, bone metastases).
The patient's Eastern Cooperative Group Performance Status was
grade 1. EGFR Ex19del was detected in the biopsied tissue,
after which erlotinib was administered. However, at 12 months after
initiating erlotinib, the primary tumor increased. Two subsequent
cycles of cytotoxic chemotherapy with cisplatin and pemetrexed were
administered, but the tumor regrew following treatment. Re-biopsy
was performed on primary lung tumor and EBC was collected at the
same time. In addition to the baseline EGFR Ex19del
mutation, EGFR T790M was also detected in the biopsy sample.
Similarly, EGFR Ex19del and T790M were detected by the
EBC-ddPCR method. Subsequently osimertinib was administered and
continued over 6 months, which caused a partial response (Response
Evaluation Criteria in Solid Tumors version 1.1) (Fig. 3C and D).
Discussion
This study demonstrated that using EBC-ddPCR to
detect EGFR mutations is feasible and shows a modest
sensitivity (20-30%) and acceptable sensitivity (80-100%) in
patients with lung cancers harboring EGFR mutations.
Up to 60% of lung adenocarcinomas treated with
first- or second-generation EGFR-TKIs develop resistant EGFR
T790M mutations (20,21). However, third-generation EGFR-TKIs
were only administered in 23.7% of patients (22). A negative result for EGFR
T790M in re-biopsied samples should not prevent physicians from
performing a repeat biopsy; however, it is not always possible to
carry out repeat biopsies (21,23). The
sensitivity of EBC testing for EGFR mutations was modest
compared to that of blood tests (8);
however, EBC testing is much more easily repeated because of its
minimal invasiveness. In this study, we performed three or four
repeated EBC samplings without any adverse effects in both case 8
and 9. Although we did not assess the concordance between tissue
biopsy and EBC testing on a larger scale, gefitinib and osimertinib
inhibited the lung tumor in cases 8 and 11, respectively, with both
the tissue biopsy and EBC testing detecting EGFR L858R or
EGFR Ex19del and T790M. These cases suggest the potential of
EBC testing as a complementary option for patients in whom tissue
biopsy is difficult or for those who refuse repeated blood
sampling.
Although ddPCR is thought to detect 0.005–0.1% of
target DNA (24), this study
revealed a modest sensitivity for EGFR mutations in EBC
samples (20-30%). Smyth et al reported that EGFR
T790M was detected in 9/10 EBC samples by UltraSEEK™ technology
(12). Possible explanations for the
discrepancy between the previous report and our study are
differences in sample quality and sensitivity of the detection
methods.
This study had several limitations. The sample size
was small (n=21 in 12 patients) and the number of EBC samples from
treatment naïve patients and samples prior to initiating EGFR-TKI
were from only 1 case; therefore, patient bias should be
considered. Furthermore, concordance among EBC testing, blood
testing, and tissue biopsy was not assessed in detail. Therefore,
these data should be considered as exploratory.
EBC-ddPCR for EGFR mutations by ddPCR was
feasible and showed moderate sensitivity and acceptable
specificity. EBC sampling is minimally invasive and replicable;
therefore, EBC tests could be a complementary option for patients
in whom tissue biopsy is difficult or for those who refuse repeated
blood sampling. Further studies are needed to explore the potential
of the EBC test.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Mrs. Hiromi
Nakashima (Okayama University Graduate School of Medicine,
Dentistry and Pharmaceutical Sciences) and Mrs. Kyoko Maeda
(Okayama University Graduate School of Medicine, Dentistry and
Pharmaceutical Sciences) for providing technical support.
Funding
The present study was supported by JSPS for
Scientific Research Grant-in-Aid for Young Scientists (B): KAKEN
(grant no. 16K19454). The current research was also supported by
Chugoku Occupational Health Association (grant no. CRE 17-2) and
Okayama health foundation (grant no. 33K-2017-03).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
Data interpretation and presentation were the sole
responsibility of the authors. KaN and KO had full access to all
data and assume responsibility for data integrity and the accuracy
of data analysis. KaN and KO contributed to the study design, data
collection, analyses, and manuscript writing. TT, KiN, TM, SS, HK,
HW, NO, GM, HH, YK, TN, TK, HY, STom, KH, MT, SToy, YM, KK
collected the data and prepared the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institute
Research Ethics Committee of the Okayama University Hospital and
written informed consent was obtained from all patients.
Patient consent for publication
Written informed consent regarding publication was
obtained from all patients.
Competing interests
Dr Kadoaki Ohashi reports research funding from
Boehringer Ingelheim, Novartis, AstraZeneca, Eli Lilly, MSD, and
Daiichi-Sankyo outside the submitted work. Dr Kadoaki Ohashi
reports personal fees from AstraZeneca, MSD, and Chugai
Pharmaceutical outside the submitted work.
Glossary
Abbreviations
Abbreviations:
|
EGFR
|
epidermal growth factor receptor
|
|
PCR
|
polymerase chain reaction
|
|
ddPCR
|
droplet digital polymerase chain
reaction
|
|
NSCLC
|
non-small cell lung cancer
|
|
EBC
|
exhale breath condensate
|
|
DNA
|
deoxyribonucleic acid
|
|
PNA
|
peptide nucleic acid
|
|
LNA
|
locked nucleic acid
|
|
TKI
|
tyrosine kinase inhibitor
|
|
HER2
|
human epidermal growth factor receptor
type 2
|
|
EBC-ddPCR
|
ddPCR using EBC
|
References
|
1
|
Kris MG, Johnson BE, Berry LD, Kwiatkowski
DJ, Iafrate AJ, Wistuba II, Varella-Garcia M, Franklin WA, Aronson
SL, Su PF, et al: Using multiplexed assays of oncogenic drivers in
lung cancers to select targeted drugs. JAMA. 311:1998–2006. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Okamoto I, Morita S, Tashiro N, Imamura F,
Inoue A, Seto T, Yamamoto N, Ohe Y, Nakagawa K and Fukuoka M:
Real-world treatment and outcomes in EGFR mutation-positive
non-small cell lung cancer: Long-term follow-up of a large patient
cohort. Lung Cancer. 117:14–19. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mok TS, Wu Y-L, Ahn M-J, Garassino MC, Kim
HR, Ramalingam SS, Shepherd FA, He Y, Akamatsu H, Theelen WS, et
al: Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung
cancer. N Engl J Med. 376:629–640. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nosaki K, Satouchi M, Kurata T, Yoshida T,
Okamoto I, Katakami N, Imamura F, Tanaka K, Yamane Y, Yamamoto N,
et al: Re-biopsy status among non-small cell lung cancer patients
in Japan: A retrospective study. Lung Cancer. 101:1–8. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chouaid C, Dujon C, Do P, Monnet I,
Madroszyk A, Le Caer H, Auliac JB, Berard H, Thomas P, Lena H, et
al: Feasibility and clinical impact of re-biopsy in advanced
non-small-cell lung cancer: A prospective multicenter study in a
real-world setting (GFPC study 12-01). Lung Cancer. 86:170–173.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim TO, Oh IJ, Kho BG, Park HY, Chang JS,
Park CK, Shin HJ, Lim JH, Kwon YS, Kim YI, et al: Feasibility of
re-biopsy and EGFR mutation analysis in patients with non-small
cell lung cancer. Thorac Cancer. 9:856–864. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Crowley E, Di Nicolantonio F, Loupakis F
and Bardelli A: Liquid biopsy: Monitoring cancer-genetics in the
blood. Nat Rev Clin Oncol. 10:472–484. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Oxnard GR, Thress KS, Alden RS, Lawrance
R, Paweletz CP, Cantarini M, Yang JC, Barrett JC and Jänne PA:
Association between plasma genotyping and outcomes of treatment
with osimertinib (AZD9291) in advanced non-small-cell lung cancer.
J Clin Oncol. 34:3375–3382. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Husain H, Melnikova VO, Kosco K, Woodward
B, More S, Pingle SC, Weihe E, Park BH, Tewari M, Erlander MG, et
al: Monitoring daily dynamics of early tumor response to targeted
therapy by detecting circulating tumor DNA in urine. Clin Cancer
Res. 23:4716–4723. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Carpagnano GE, Foschino-Barbaro MP, Mulé
G, Resta O, Tommasi S, Mangia A, Carpagnano F, Stea G, Susca A, Di
Gioia G, et al: 3P microsatellite alterations in exhaled breath
condensate from patients with non-small cell lung cancer. Am J
Respir Crit Care Med. 172:738–744. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Youssef O, Knuuttila A, Piirilä P, Böhling
T, Sarhadi V and Knuutila S: Hotspot mutations detectable by
next-generation sequencing in exhaled breath condensates from
patients with lung cancer. Anticancer Res. 38:5627–5634. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Smyth RJ, Toomey SM, Sartori A, O'Hanrahan
E, Cuffe SD, Breathnach OS, Morgan RK and Hennessy BT: Brief report
on the detection of the EGFR T790M mutation in exhaled breath
condensate from lung cancer patients. J Thorac Oncol. 13:1213–1216.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang D, Takigawa N, Ochi N, Tanimoto Y,
Noujima D, Chen YY, Tanimoto M and Kiura K: Detection of the EGFR
mutation in exhaled breath condensate from a heavy smoker with
squamous cell carcinoma of the lung. Lung Cancer. 73:379–380. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sacher AG, Paweletz C, Dahlberg SE, Alden
RS, O'Connell A, Feeney N, Mach SL, Jänne PA and Oxnard GR:
Prospective validation of rapid plasma genotyping for the detection
of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol.
2:1014–1022. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Suzawa K, Yamamoto H, Ohashi K, Hashida S,
Tomida S, Kubo T, Maki Y, Soh J, Tsukuda K, Kiura K, et al: Optimal
method for quantitative detection of plasma EGFR T790M mutation
using droplet digital PCR system. Oncol Rep. 37:3100–3106. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Senoo S, Ohashi K, Nishii K, Hara N, Kano
H, Ninomiya K, Maeda Y and Kiura K: Osimertinib depletes EGFR T790M
in the spinal fluid of patients with carcinomatous meningitis of
lung adenocarcinoma harboring de novo EGFR T790M. J Thorac Oncol.
13:e140–e142. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Goldstraw P, Chansky K, Crowley J,
Rami-Porta R, Asamura H, Eberhardt WE, Nicholson AG, Groome P,
Mitchell A, Bolejack V, et al: The IASLC lung cancer staging
project: Proposals for revision of the TNM stage groupings in the
forthcoming (eighth) edition of the TNM Classification for lung
cancer. J Thorac Oncol. 11:39–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ohashi K, Sequist LV, Arcila ME, Moran T,
Chmielecki J, Lin YL, Pan Y, Wang L, de Stanchina E, Shien K, et
al: Lung cancers with acquired resistance to EGFR inhibitors
occasionally harbor BRAF gene mutations but lack mutations in KRAS,
NRAS, or MEK1. Proc Natl Acad Sci USA. 109:E2127–E2133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ogino A, Kitao H, Hirano S, Uchida A,
Ishiai M, Kozuki T, Takigawa N, Takata M, Kiura K and Tanimoto M:
Emergence of epidermal growth factor receptor T790M mutation during
chronic exposure to gefitinib in a non-small cell lung cancer cell
line. Cancer Res. 67:7807–7814. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ohashi K, Maruvka YE, Michor F and Pao W:
Epidermal growth factor receptor tyrosine kinase
inhibitor-resistant disease. J Clin Oncol. 31:1070–1080. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee K, Kim Y, Jung HA, Lee SH, Ahn JS, Ahn
MJ, Park K, Choi YL and Sun JM: Repeat biopsy procedures and T790M
rates after afatinib, gefitinib, or erlotinib therapy in patients
with lung cancer. Lung Cancer. 130:87–92. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Seto T, Nogami N, Yamamoto N, Atagi S,
Tashiro N, Yoshimura Y, Yabuki Y and Saka H: Real-world EGFR T790M
testing in advanced non-small-cell lung Cancer: A prospective
observational study in Japan. Oncol Ther. 6:203–215. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ichihara E, Hotta K, Kubo T, Higashionna
T, Ninomiya K, Ohashi K, Tabata M, Maeda Y and Kiura K: Clinical
significance of repeat rebiopsy in detecting the EGFR T790M
secondary mutation in patients with non-small cell lung cancer.
Oncotarget. 9:29525–29531. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vogelstein B and Kinzler KW: Digital PCR.
Proc Natl Acad Sci USA. 96:9236–9241. 1999. View Article : Google Scholar : PubMed/NCBI
|