Introduction
Pediatric brain tumor is one of the leading causes
of cancer-associated mortality in children (1). The incidence rate of childhood and
adolescent brain tumors in the United States is approximately 5.67
per 100,000 person-years (2).
Medulloblastoma (MB) is the most common malignant pediatric brain
tumor, but can also rarely occur in adults. MB is classified into 4
subgroups based on molecular characteristics, as follows: WNT, SHH,
Group 3 (G3) and Group 4 (G4) (3).
WNT-MBs frequently contain a CTNNB1 mutation, while the SHH subtype
contains mutations that activate the SHH pathway (3). The Group 3 subtype is characterized by
overexpression of MYC, while Group 4 is the only subgroup lacking
upregulated MYC expression (4).
Currently, the standard of care is maximal surgical resection
followed by risk adapted cranio-spinal irradiation (CSI) and
adjuvant chemotherapy, and the 5-year overall survival rates for
patients with average- and high-risk MBs range from 65–85%
(5). However, the adverse effects of
CSI, including permanent neurocognitive disability, growth
disturbances, infertility and hearing loss, continue to affect the
improving prognosis of MB (6). Thus,
it remains critical to identify alternative therapies to delay or
omit CSI in children.
Immunotherapy is effective for patients with
melanoma, leukemia and other solid tumors (7–10). The
importance of lymphocytes and tumor-associated macrophages (TAMs)
in the tumor microenvironment has been assessed and is considered a
key factor of immunotherapy response (11,12).
However, little is known about the comprehensive immune
infiltration in brain tumor, particularly in MB. Due to limited
patient sample sizes and preclinical models, the status and
function of immune microenvironment remains controversial.
Vermeulen et al (13)
reported no programmed cell death protein ligand 1 (PD-L1)
expression in 26 MB cohorts, suggesting limited or no added value
for immunotherapy with PD1/PD-L1 blockers in MB. Conversely, Martin
et al (14) demonstrated that
PD-L1 is expressed at low levels in MB to facilitate immune
escape.
A comprehensive heterogeneity within four subgroups
is one of the most actively studied fields in MB (15). However, the characterization of the
immune microenvironment in the four subgroups remains unclear.
Bockmayr et al (16) assessed
the subgroup-specific immune microenvironment in MB using an open
gene expression database. However, data only based on gene
expression provides limited reliability and may fail to provide
information on the functional status. For example, cytokines and
chemokines play an important role in modulating the MB immune
microenvironment and tumor progression (17,18).
Thus, the present study compared 69 cytokines/chemokines within the
four MB subgroups and glioblastoma multiform (GBM). Taken together,
the results of the present study provide a novel comprehensive
description of the immune microenvironment of MB, which may help
guide clinical translation of immune-related therapies.
Materials and methods
Human tissue samples
The present study was approved by the Ethics
Committee of Sanbo Brain Hospital, Capital Medical University
(Beijing, China; approval no. SBNK-YJ-2018-013-01). and all
procedures were performed in accordance with the 1964 Declaration
of Helsinki (19). All patients or
their guardians provided written informed consent prior to the
study start. A total of 19 MB tissues, three GBM tissues and two
peritumoral normal brain tissue samples were collected from
patients who underwent surgical resection at Sanbo Brain Hospital,
Capital Medical University between January 2017 and December 2018.
Snap-frozen tissues were labeled and stored at −80°C until
subsequent experimentation. Patients characteristics, including
sex, age and collection date are listed in Table I.
 | Table I.Clinicopathological
characteristics. |
Table I.
Clinicopathological
characteristics.
|
| Sex |
|
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Male, n | Female, n | Age range,
(years) | Collection
date |
|---|
| MB subtype |
|
WNT | 0 | 1 | 8 | March 2017 |
|
SHH | 4 | 1 | 3-11 | January
2017-December 2018 |
| G3 | 3 | 2 | 8-24 | January 2017-March
2018 |
| G4 | 6 | 2 | 7-18 | February
2017-December 2018 |
| GBM | 1 | 2 | 37-58 | October
2018-December 2018 |
| Normal | 1 | 1 | 17-31 | May 2018-August
2018 |
RNA sequencing
Total RNA was extracted from snap-frozen tissues
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). RNA quality was assessed using the Bioanalyzer
2200 (Agilent Technologies, Inc.) and RNA integrity number >6.0
was qualified for cDNA library construction. The cDNA libraries
were constructed for each pooled RNA sample using the NEBNext
UltraTM Directional RNA Library Prep kit (cat. no. E7420S; New
England BioLabs, Inc.) for Illumina, according to the manufacturers
instructions. The following steps were performed: mRNA was
fragmented into 150–200 bp using divalent cations at 94°C for 8
min, the cleaved RNA fragments were reverse transcribed into first-
and second-strand cDNA, respectively, fragments were end repaired,
and A-tailed and ligated with indexed adapters. Target bands were
harvested using AMPure XP Beads (Beckman Coulter, Inc.). The
products were purified and enriched via PCR to create the final
cDNA libraries, and quantified using Agilent2200 software (version
A.02.01; Agilent Technologies, Inc.). The tagged cDNA libraries
were pooled in an equal ratio and used for 150 bp paired-end
sequencing in a single lane of the Illumina Nova Seq (https://www.illumina.com/systems/sequencing-platforms/novaseq.html).
K-means clustering
To identify the MB subgroups in the 19 patients with
MB, previously reported subgroup-specific signature genes were used
for K-means clustering (4). The
cluster number was set to four because MB was reported to comprise
four distinct molecular variants (4). Each cluster was then analyzed by
hierarchical clustering algorithms with squared Pearson correlation
as similarity measurement (20).
Gene Set Enrichment Analysis
(GSEA)
Due to the limited sample size of WNT MB (n=1), GSEA
analysis was only preformed in SHH, Group 3 and Group 4. GSEA
(version 4.0.3; Broad Institute; www.broadinstitute.org) was used according to the
manufacturers instructions. Briefly, gene expression data matrix
was classified into three subgroups (SHH, Group 3 and Group 4). H:
hallmark gene sets were downloaded from the MSigDB database
(http://software.broadinstitute.org/gsea/msigdb), and
the number of permutations was set to 1,000. Gene set enrichment
was considered significant at false discovery rate <0.25.
Immunohistochemistry (IHC)
Lymphocyte infiltration and PD-L1 expression were
assessed via IHC analysis on 4 µm formalin-fixed and
paraffin-embedded (FFPE) tissues. Human glioblastoma tissue was
used as the positive control for PD-L1 staining. Tissue slides were
deparaffinized in xylene for 20 min at room temperature and
rehydrated in a descending ethanol series (90-50%). For antigen
retrieving, the tissue slides were incubated in EDTA buffer (pH
9.0) (cat. no. E673003; Sangon Biotech, Co., Ltd.) for 8 min at
room temperature. Tissue sections were washed three times with
phosphate-buffered saline, prior to incubation with 3%
H2O2 for 10 min at room temperature to
inhibit endogenous peroxidase activity. The sections were
subsequently blocked with 5% normal goat serum (cat. no. E510009;
Sangon Biotech, Co., Ltd.) for 30 min at room temperature and
incubated with primary antibodies against CD3 (cat. no. ab16669,
1:100; Abcam) and PD-L1 (cat. no. ab213524, 1:100; Abcam) overnight
at 4°C. The Dako REAL EnVision Detection System (Dako; Agilent
Technologies, Inc.) was used as a secondary antibody, according to
the manufacturer's protocol. Tissue sections were subsequently
dehydrated in an ascending ethanol series (50-90%), cleared with
xylene and covered with a coverslip. The sections were observed
under a Nikon Eclipse light microscope (magnifications, ×20 and
×40). The IHC score for CD3 staining was calculated based on the
CD3 positive cells/field.
Immunofluorescence (IF)
Microglia/macrophage recruitment was assessed via IF
analysis on FFPE tissues. Tissue slides were deparaffinized,
rehydrated following antigen retrieving and blocked as described
for IHC. The sections were incubated with rabbit anti-IBA1
(1:1,000; Wako Pure Chemical Industries, Ltd.) overnight at 4°C.
Subsequently, tissue sections were incubated with AlexaFluor 488
conjugated secondary antibodies (cat. no. R37116, 1:500,
Invitrogen; Thermo Fisher Scientific, Inc.) at room temperature for
4 h to detect the primary labeling. Tissue sections were mounted
using ProLong Diamond Antifade Mountant with DAPI (cat. no. P36962;
Thermo Fisher Scientific, Inc.), and the mounted samples were
observed under a confocal microscope (LSM 700; Carl Zeiss AG;
magnifications, ×40 and ×60). Z-stack images were acquired and
stacked via maximum intensity project using ImageJ software
(version 1.0; National Institutes of Health). IBA-1 positive cells
were manually counted and quantified using SPSS 17.0 software
(SPSS, Inc.).
Estimating immune signatures
Immune signatures of mRNA markers from 10 cell
populations were used to estimate the microenvironment cell
populations (MCP), including T cells, CD8+ T cells,
cytotoxic T lymphocytes (CTL), B cells, NK cells, monocytic lineage
cells, myeloid dendritic cells, neutrophils, fibroblasts and
endothelial cells in MB tissues. Microenvironment Cell Population
Counter (MCP-Counter) (15) scores
were defined as the log2 average expression of the transcriptomic
markers for each population.
Luminex assay
MB samples were collected during surgery,
snap-frozen in liquid nitrogen and stored at −80°C. The Bio-plex
Pro Human Cytokine kit (cat. no. 12007283; Bio-Rad Laboratories,
Inc.) and the Bio-Plex Pro Human Chemokine Panel 40-plex kit (cat.
no. 171AK99MR2; Bio-Rad Laboratories, Inc.) were used according to
the manufacturers recommendations. Briefly, beads were added to a
96-well plate and washed with Assay Buffer for 30 sec. Samples were
added to the plate containing the mixed antibody-linked beads and
incubated at room temperature for 1 h, at 850 rpm on a plate
shaker. Following the primary incubation, the plates were washed
three times with Assay Buffer and subsequently incubated with
Detection Antibody for 75 min at room temperature, on a plate
shaker. The plates were re-washed, followed by addition of
streptavidin-PE. Following incubation for 30 min at room
temperature, the plates were washed prior to addition of reading
buffer. The plates were analyzed on the Luminex 200 platform (Wayen
Biotechnologies, Shanghai, Inc.; www.wayenbio.com). Each sample was measured in
duplicate.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 7.0 software (GraphPad Software, Inc.) and SPSS 17.0 software
(SPSS, Inc.). All experiments were performed in triplicate and data
are presented as the mean ± standard error of the mean. One-way
analysis of variance followed by Bonferronis post hoc test was
performed to compare differences between multiple groups. Pearsons
correlation analysis was performed to determine the association
between cytokine/chemokine protein and RNA expression. P<0.05
was considered to indicate a statistically significant
difference.
Results
Identification of molecular subgroups
of MBs using transcriptional profiling data
To identify WNT, SHH, Group 3 and Group 4 MB, RNA
sequencing (RNA-Seq) was performed to obtain gene expression of all
MB samples (n=19). According to the previously published 84
subgroup-specific signature genes (4), the present study identified 1 WNT, 5
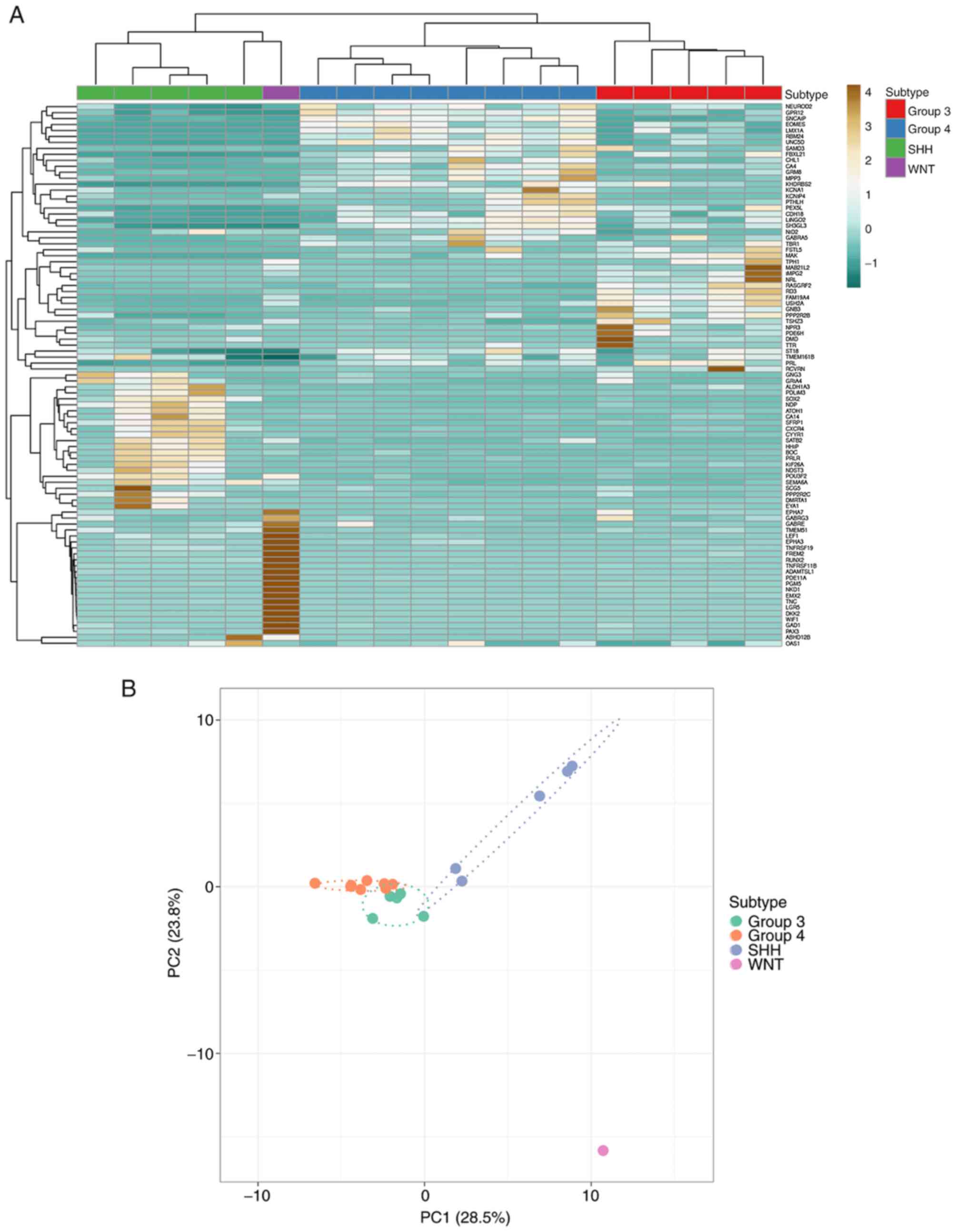
SHH, 5 Group 3 and 8 Group 4 MBs using K-means clustering. The
heatmap demonstrated the distinct patterns of expression of
subgroup-specific signature genes among the four MB subgroups
(Fig. 1A). The degree of separation
of the four subgroups was assessed via principal component
analysis, with the WNT subtype demonstrating a distanced separation
away from the other three groups. While Group 3 and Group 4 were
closer to each other, the SHH subtype demonstrated a clear
demarcation from the Group 3 and Group 4 subtypes (Fig. 1B).
Immune response is predominantly
elevated in the SHH subgroup
To assess and compare gene expression of the
different subgroups of MB, GSEA was performed with RNA-Seq data.
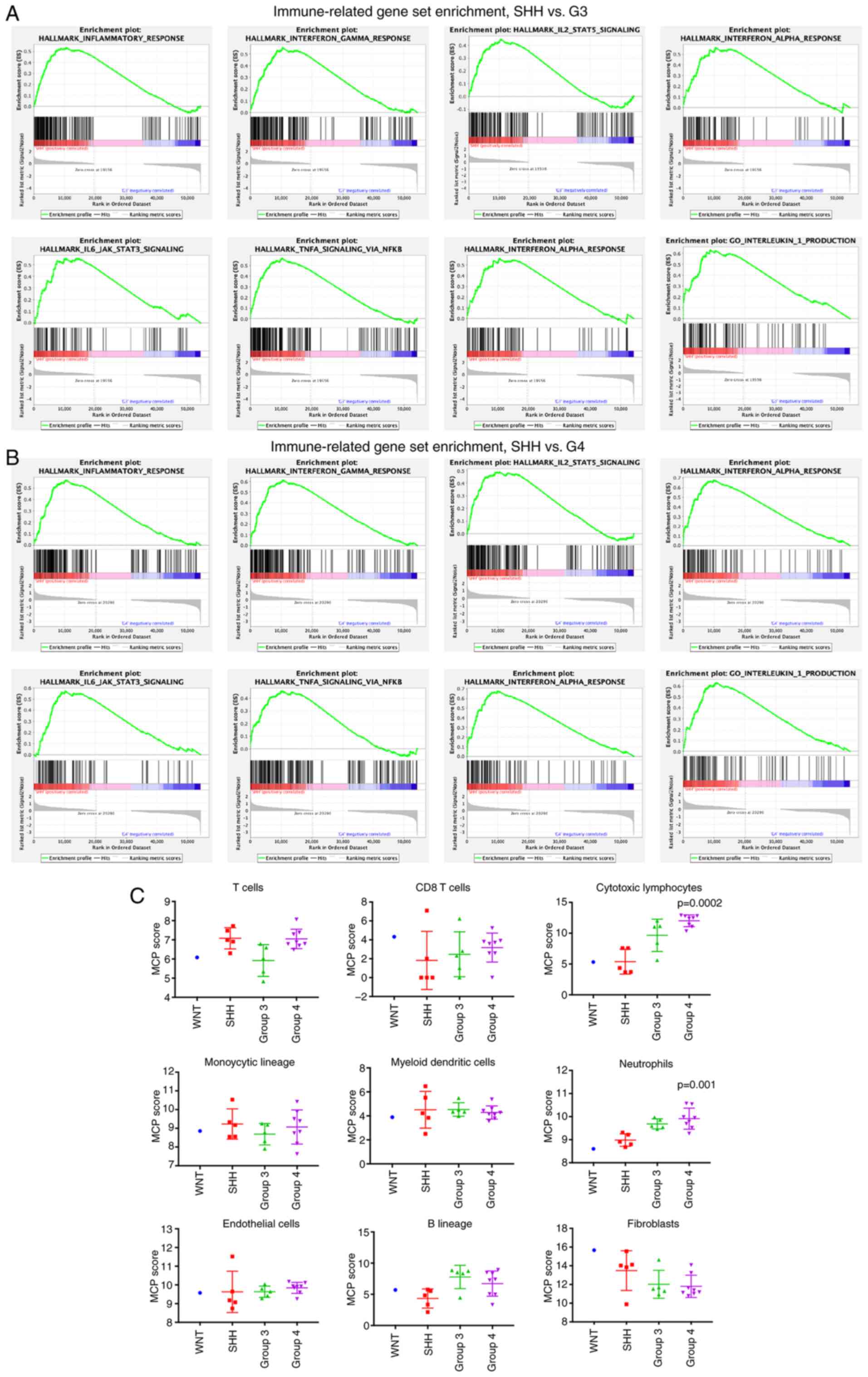
Due to the limited sample size of WNT MB, analysis was only
performed in the SHH, Group 3 and Group 4 subtypes. A total of 50
hallmark gene sets were downloaded from the MSigDB database, with 6
immune-related gene sets significantly enriched in the SHH subgroup
compared with Group 3 in the top 20 enrichment score (ES) gene
sets, including inflammatory response, interferon γ response,
IL6-JAK-STAT3 signaling, TNF-α signaling via NFKB, interferon α
response and IL2-STAT5 signaling (Fig.
2A). Similar results were observed following comparison of the
SHH and Group 4 subgroups, suggesting that the SHH subgroup MBs are
more involved in the immune microenvironment compared with Group 3
and Group 4 MBs (Fig. 2B).
Tissue-infiltrating immune cell
population in the MB subgroups
The MCP-Counter method was used to estimate the
population abundance of tissue-infiltrating immune cells in the MB
subgroups, using the RNA-Seq data. The expression scores were
significantly different between the subgroups in cytotoxic
lymphocytes (P=0.0002) and neutrophils (P=0.001). In addition,
Group 4 tumors had more cytotoxic lymphocytes and neutrophils
compared with tumors from the other subgroups (Fig. 2C).
Lymphocyte infiltration in the MB
subgroups
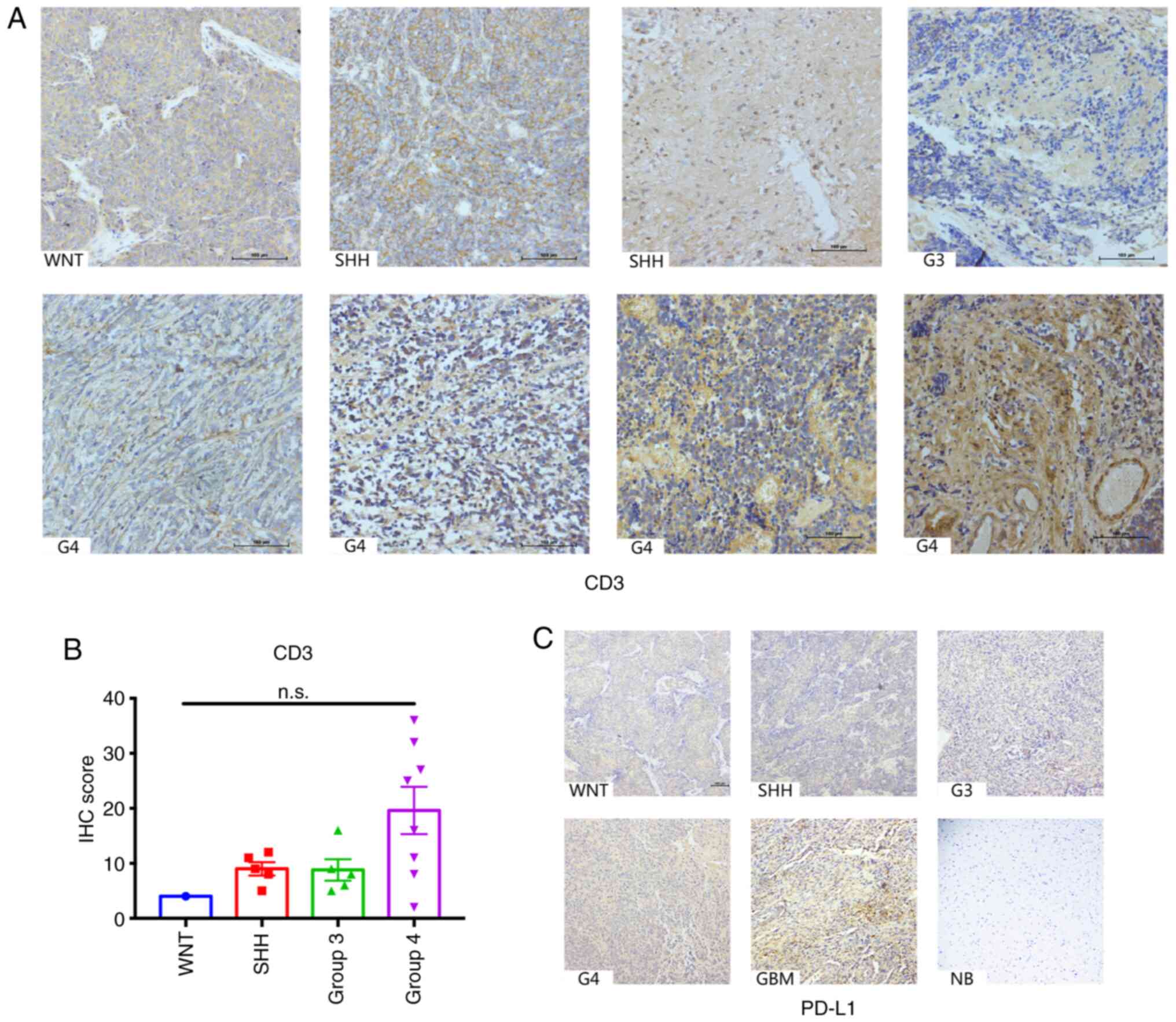
To validate the diverse pattern of lymphocyte
infiltration in the four MB subgroups, IHC analysis was performed
on the tissue samples for CD3. The results demonstrated that
overall CD3 staining in the MB tissues from the four subgroups were
minimal, which was consistent with previous studies (13,16).
Although the number of CD3 positive cells between the subgroups was
not significantly different (P=0.0886), tumors from the Group 4
subtype exhibited more CD3 positive lymphocytes infiltration
compared with the other subgroups, which was concordant with the
MCP-Counter data of the cohort in the present study (Fig. 3A and B). Notably, the majority of CD3
positive cells were localized in the perivascular, suggesting the
lack of infiltration of T cells in the tumor mass (Fig. 3A).
The PD1/PD-L1 axis has been investigated and is
considered a promising target for immunotherapy in MBs (13,14,21). The
present study investigated PD-L1 expression in the patient cohort;
however, no positive staining was observed (Fig. 3C). Collectively, these results
suggest that lymphocyte infiltration is relatively low in MBs, and
the PD1/PD-L1 axis may not be a promising therapeutic target in
clinical practice.
Activated macrophage/microglia
recruitment in the MB subgroups
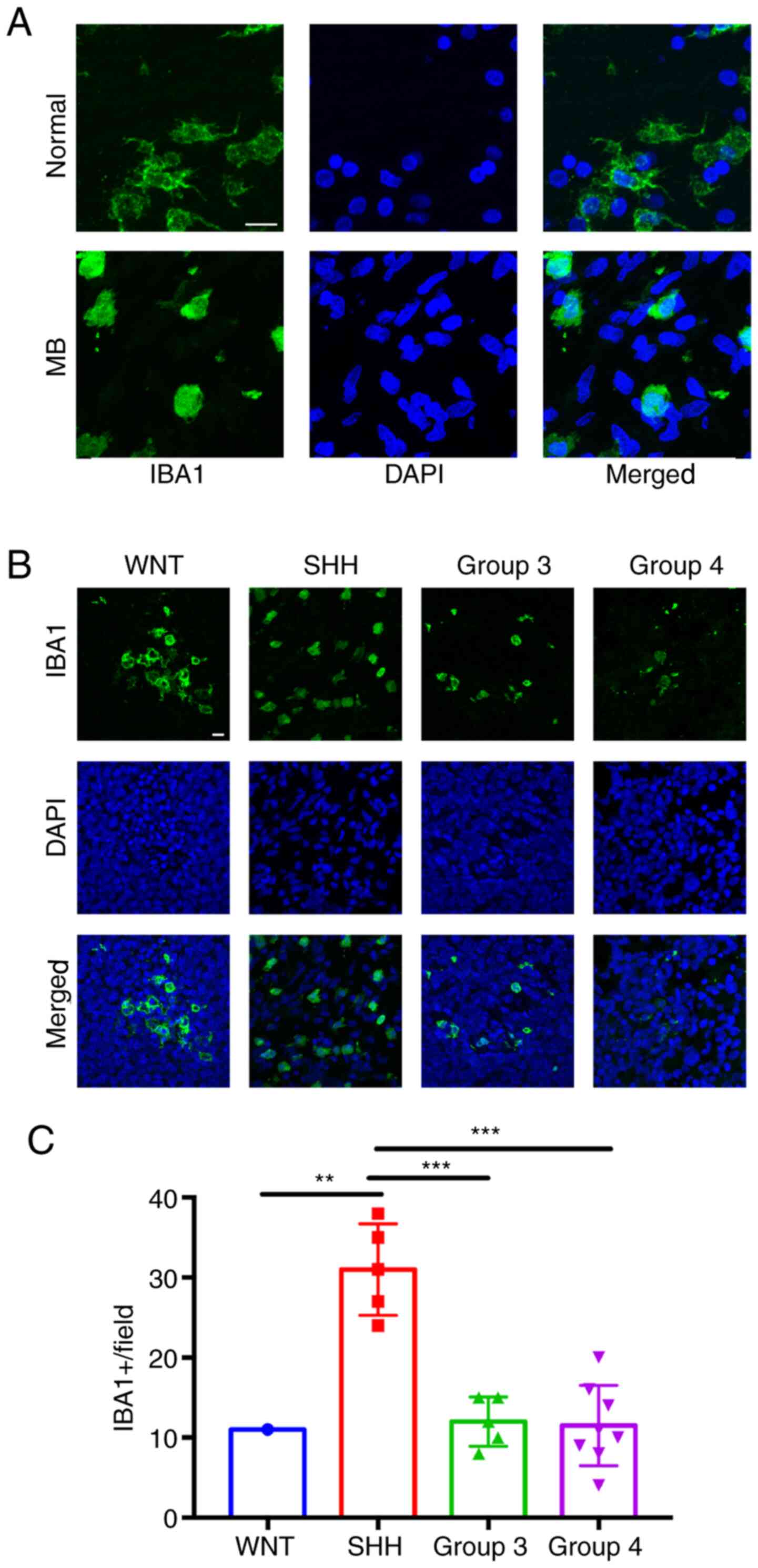
Macrophages and microglia have been reported to play
vital roles in central nervous system (CNS) inflammation (22). The present study initially identified
macrophage/microglia via IF IBA1 staining in both MB tissues and
normal cerebellum tissues. While the microglia in normal cerebellum
exhibited a resting phenotype with long processes, the MB TAM
exhibited an activated phenotype with short or absent processes
(Fig. 4A). The morphology of
activated TAMs in the four MB subtypes were similar (data not
shown), so only the SHH subtype is presented in Fig. 4A. The density of activated TAMs in
the MB subgroups was subsequently assessed. The results
demonstrated that SHH tumors had more TAMs recruitment compared
with tumors in the other subgroups (Fig.
4B and C).
Cytokine profiling of MB tumors across
the four subgroups
Cytokines play a key role in modulating the tumor
microenvironment and immune cells infiltration (17). Given that the SHH tumors exhibited
more inflammatory compared with the other subgroups, it was
speculated that the inflammatory-related cytokines of SHH tumors
may differ from the other subgroups. A total of three tumors were
collected from the SHH, Group 3 and Group 4 subtypes, respectively,
a WNT tumor, three GBM tumors and two normal cerebellum tissues,
and multiple cytokine/chemokine was analyzed using Luminex array.
K-means hierarchical clustering analysis demonstrated that MB
tumors were similar to normal cerebellum, and secreted less
cytokines and chemokines compared with GBM tumors (Fig. 5A). These results were consistent with
the low expression of cytokine/chemokine-encoding genes. Given the
importance of inferring the microenvironment to guide immunotherapy
for clinical practice (23), the
correlation between the concentration of cytokine/chemokine protein
and RNA expression was assessed. Pearsons correlation analysis
demonstrated a positive correlation between cytokine/chemokine
protein and RNA expression (Fig.
5B). Collectively, these results may explain why MB recruits
fewer immune cells to the tumor microenvironment compared with GBM.
In addition, the overall expression of cytokine-encoding genes in
the RNA-Seq data were low. Most of the Fragments Per Kilobase per
Million values were <5, which was considered negative (Fig. 5C).
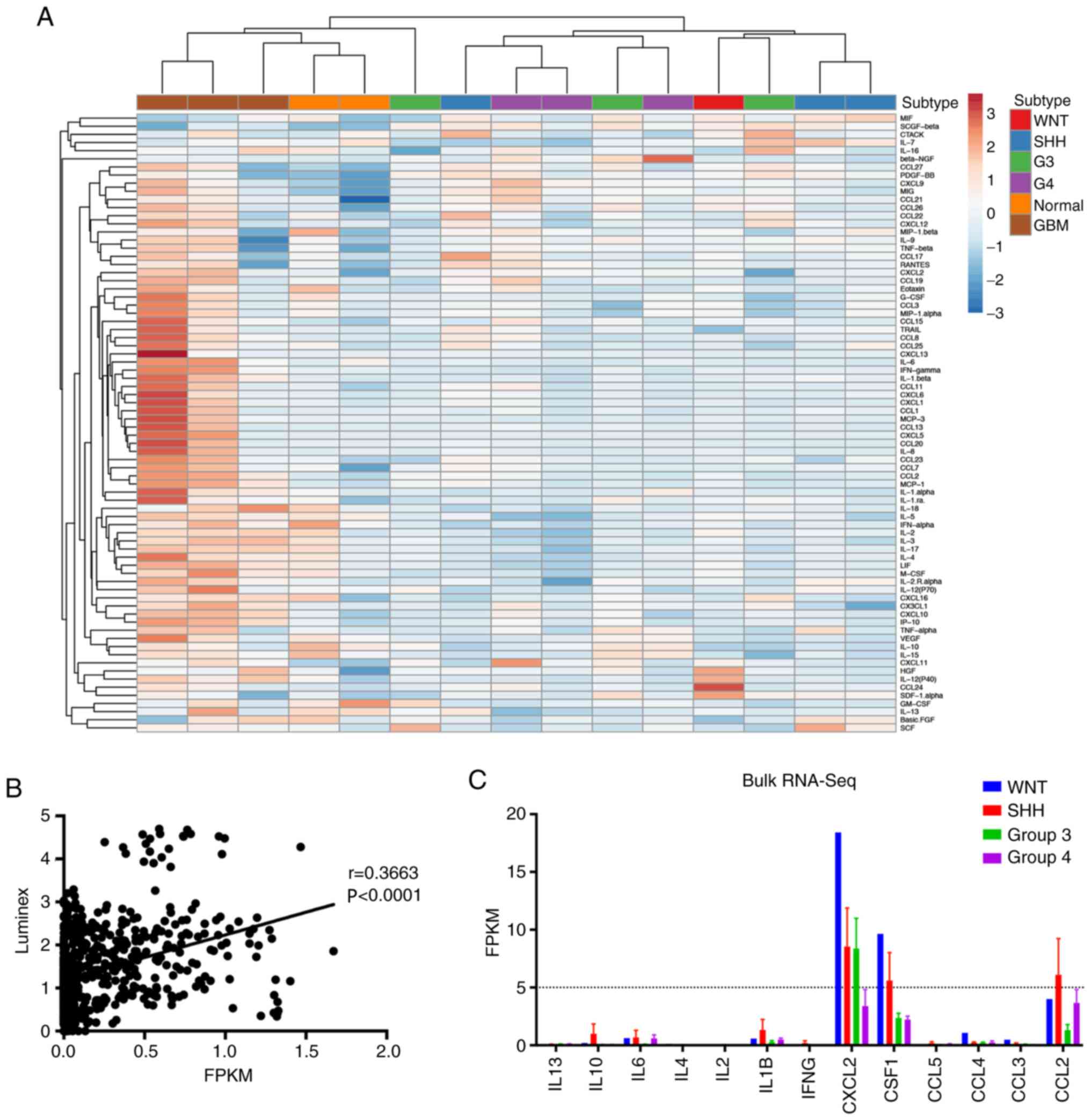 | Figure 5.Cytokines and chemokines profile in
the MB subgroups. (A) Hierarchical clustering of mean values of
cytokines and chemokines in the MB subgroups, normal brain tissues
and glioma samples. Each column represents the average score
assessed in triplicate, and each row represents a separately
measured cytokine or chemokine. (B) Pearsons correlation analysis
of cytokine concentration and gene expression. y-axis represents
the cytokine concentration measured by Luminex, while the x-axis
represents the cytokine-encoding gene expression by RNA-Seq. (C)
Bar chart of FPKM of cytokine-encoding genes. Dashed line
represents FPKM=5. MB, medulloblastoma; RNA-Seq, RNA sequencing;
G3, Group 3; G4, Group 4; GBM, glioblastoma multiform; IL,
interleukin; IFNγ, interferon-gamma; CXCL2, C-X-C motif ligand 2;
CSF1, colony stimulating factor 1; CCL, C-C motif ligand 2; FPKM,
fragments per kilobase per million. |
Discussion
MB is the most common pediatric brain tumor
originating from the posterior fossa, and is a leading cause of
cancer-associated mortality in children in the United States
(1,24). MB is currently classified into four
groups based on the molecular pathway characteristics, as follows:
WNT, SHH, Group 3 and Group 4 subtypes. In the WNT subtype, the
canonical Wnt signaling pathway is upregulated (24). The SHH subtype is defined by
activation of the Sonic hedgehog signaling pathway (24). The Group 3 subtype, which has the
worst outcome among all MB subtypes, is characterized by
amplification of multiple proto-oncogenes, including MYC, PVT1 and
SMARCA4 (4). The Group 4 MB subtype
is characterized by molecular abnormalities associated with a
frequent mutation in the KDM6A gene (25). As suggested in the NCCN guideline
(26), patients who have a high risk
of relapse should be administrated radiotherapy, which may cause
severe cognitive dysfunction (27).
Thus, alternative therapeutic strategies are required, among which
immunotherapy proves promising (28). Compared with adult brain tumors, less
is known about the immune microenvironment of MB due to the limited
sample size, which in turn impedes clinical translation attempt for
precision immunotherapy. Thus, the present study aimed to
investigate the immune microenvironment of the four subtypes of MB
in 19 freshly collected MB samples. Bioinformatics and IHC
analyses, along with multiple cytokine/chemokine Luminex assay were
performed in the hope to provide a comprehensive overview from
different dimensions.
In order to compare the distinct immune
microenvironment within the four MB subtypes, RNA sequence of the
19 freshly collected MB samples was initially acquired. Based on
previously published subtype-related gene signature (4), hierarchical clustering analysis
effectively separated the samples into the four subgroups. However,
only one WNT MB was identified, which is compatible with a low
incidence (~10%) of all MB diagnosis (24). Thus, most of the statistical analyses
excluded the WNT subtype due to the limited sample size.
GSEA was performed to compare hallmark pathways in
the different subgroups. The results demonstrated that the
immune-related gene sets were significantly enriched in the SHH
subgroup compared with the G3 and G4 subtypes, suggesting an
activated immune phenotype in the SHH subgroup. The immune cell
populations were subsequently estimated using the MCP-counter,
which uses specific gene signatures to identify eight immune cell
populations and two stromal cell populations (29). Among the 10 populations, only
cytotoxic lymphocytes and neutrophils exhibited statistical
differences. Notably, the G4 subgroup had more immune cell
infiltration compared with the SHH subgroup, which exhibited an
activated immune phenotype in GSEA. Both analyses were based on
transcriptome data; however, the biological implications varied.
GESA hallmark gene sets represent specific well-defined biological
states, while MCP-Counter estimates the infiltration of immune cell
populations (29,30). These bioinformatics data indicated
that tumor cells, rather than infiltrated immune cells, activate
immune-related pathways in the MB tumor microenvironment. Notably,
a similar phenotype was reported in another study on the
microenvironment of diffuse intrinsic pontine glioma, which is
another non-inflammatory pediatric brain tumor (31).
Given that the G4 subtype exhibited the highest
levels of lymphocytes and neutrophil infiltration (Fig. 2C), the present study stained all
samples for CD3, PD-L1 and MPO. The results demonstrated that the
G4 subtype contained more CD3 positive T cells; however, there were
no statistical significant differences compared with the other
subgroups. More samples should be involved in the future study to
validate if G4 MB have more CD3 positive T cells infiltration than
other subgroups.
Previous studies have demonstrated that PD-L1
expression in MB is associated with prognosis and response to
immune checkpoint inhibitor (21,32).
However, in the present cohort, none of 19 MB samples exhibited
positive PD-L1 staining. Consistent with previous findings
(13,33), the results of the present study
suggest that PD-L1 expression is absent in MB samples. Thus, PD-L1
blockade may not be a feasible strategy for MB therapy. Neutrophils
have been reported to promote adult malignant glioma progression,
and have been observed in high grade gliomas (34). The present study failed to identify
any MPO positive neutrophils in the assessed MB samples however,
the MCP-counter score demonstrated positive infiltration. Taken
together, these results suggest that neutrophil infiltration is
more common in malignant glioma. Prospective studies are required
to validate the role of neutrophils in MB.
Microglia are the resident immune cells in the CNS
(35). TAMs play an important role
in the MB microenvironment, whereby TAM-secreted cytokines,
chemokines and growth factors regulate immune function and modulate
the interaction of multiple cellular populations within the tumor
microenvironment, thus contribute to tumor progression (35). A recent study by Yao et al
(18) demonstrated that
tumor-cell-derived astrocytes produce IL-4 that stimulates
microglia to produce IGF1, which in turn induces progression of the
SHH subtype in a preclinical mouse model (18). Consistent with previous findings
(36,37), the results of the present study
suggest that the SHH subtype has a distinct pattern of TAMs
infiltration. Thus, it was hypothesized that the SHH subtype
secretes distinct cytokines/chemokines. However, the results of the
present study demonstrated that the cytokines/chemokines were not
associated with this molecular subgroup. The most plausible
explanation is that MB secrets none to very low levels of
cytokines/chemokines, thus, the difference between subtypes is not
significant. To the best of our knowledge, the present study was
the first to compare cytokine secretion within MB subtypes and GBM.
Although the overall expression of cytokines is very low, exogenous
administration of cytokines or inhibitors may be a useful target
for immunotherapy. For example, TGF-β neutralization facilitated
NK-cell induces MB suppression (38).
Collectively, the results of the present study
suggest a ‘cold’ immune microenvironment of MB, which guides the
strategy of MB immunotherapy towards inducing more immune cells
into the tumor microenvironment. The capacity of CTL infiltration
in MB is another issue due to the blood brain barrier (28). Thus, intratumorally delivering T
cells may prove useful. For example, intratumorally delivered
anti-human epidermal growth factor receptor 2 CAR-T cells have
demonstrated effective preclinical efficacy in vitro and in
mouse medulloblastoma treatment (39).
Taken together, the results of the present study
suggest that the immune microenvironment of pediatric MB is
non-inflammatory and does not recruit as much immune cells compared
with glioblastoma. Targeting the PD1/PD-L1 axis for MB treatment
may not be a plausible strategy due to the absence of PD-L1
expression. Taken together, the results of the present study
provide novel insight into the design of immunotherapeutic
approaches for MB, such as CAR-T therapy which may induce more
cytotoxic lymphocytes into the tumor microenvironment.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Beijing
Postdoctoral Research Foundation (grant no. ZZ2019-02).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors contributions
SD and CG performed the experiments, analyzed the
data and drafted the initial manuscript. HZ acquired the patient
samples and performed the histology scoring. CY conceived and
designed the present study, and supervised the project. All authors
have read and approved the final manuscript.
Ethical approval and consent to
participate
The present study was approved by the Ethics
Committee of Sanbo Brain Hospital Capital Medical University
(Beijing, China; approval no. SBNK-YJ-2018-013-01), and all
procedures were performed in accordance with the 1964 Declaration
of Helsinki (19). All patients or
their guardians provided written informed consent prior to the
study start.
Patients consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Burstein R, Henry NJ, Collison ML, Marczak
LB, Sligar A, Watson S, Marquez N, Abbasalizad-Farhangi M, Abbasi
M, Abd-Allah F, et al: Mapping 123 million neonatal, infant and
child deaths between 2000 and 2017. Nature. 574:353–358. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ostrom QT, Gittleman H, Xu J, Kromer C,
Wolinsky Y, Kruchko C and Barnholtz-Sloan JS: CBTRUS Statistical
Report: Primary Brain and Other Central Nervous System Tumors
Diagnosed in the United States in 2009–2013. Neuro Oncol. 18
(Suppl_5):v1–v75. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Millard NE and De Braganca KC:
Medulloblastoma. J Child Neurol. 31:1341–1353. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Northcott PA, Korshunov A, Witt H,
Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins
CE, French P, et al: Medulloblastoma comprises four distinct
molecular variants. J Clin Oncol. 29:1408–1414. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sirachainan N, Nuchprayoon I,
Thanarattanakorn P, Pakakasama S, Lusawat A, Visudibhan A,
Dhanachai M, Larbcharoensub N, Amornfa J, Shotelersuk K, et al:
Outcome of medulloblastoma in children treated with reduced-dose
radiation therapy plus adjuvant chemotherapy. J Clin Neurosci.
18:515–519. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Doger de Speville E, Robert C,
Perez-Guevara M, Grigis A, Bolle S, Pinaud C, Dufour C, Beaudré A,
Kieffer V, Longaud A, et al: Relationships between regional
radiation doses and cognitive decline in children treated with
cranio-Spinal irradiation for posterior fossa tumors. Front Oncol.
7:1662017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cuevas LM and Daud AI: Immunotherapy for
melanoma. Semin Cutan Med Surg. 37:127–131. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Barsan V, Ramakrishna S and Davis KL:
Immunotherapy for the treatment of acute lymphoblastic leukemia.
Curr Oncol Rep. 22:112020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Naylor EC, Desani JK and Chung PK:
Targeted therapy and immunotherapy for lung cancer. Surg Oncol Clin
N Am. 25:601–609. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Morrison AH, Byrne KT and Vonderheide RH:
Immunotherapy and prevention of pancreatic cancer. Trends Cancer.
4:418–428. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lorenzo-Sanz L and Muñoz P:
Tumor-infiltrating immunosuppressive cells in cancer-cell
plasticity, tumor progression and therapy response. Cancer
Microenviron. 12:119–132. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kennedy BC, Showers CR, Anderson DE,
Anderson L, Canoll P, Bruce JN and Anderson RC: Tumor-associated
macrophages in glioma: Friend or foe? J Oncol. 2013:4869122013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vermeulen JF, Van Hecke W, Adriaansen EJM,
Jansen MK, Bouma RG, Villacorta Hidalgo J, Fisch P, Broekhuizen R,
Spliet WGM, Kool M, et al: Prognostic relevance of
tumor-infiltrating lymphocytes and immune checkpoints in pediatric
medulloblastoma. OncoImmunology. 7:e13988772017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Martin AM, Nirschl CJ, Polanczyk MJ, Bell
WR, Nirschl TR, Harris-Bookman S, Phallen J, Hicks J, Martinez D,
Ogurtsova A, et al: PD-L1 expression in medulloblastoma: An
evaluation by subgroup. Oncotarget. 9:19177–19191. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cavalli FMG, Remke M, Rampasek L, Peacock
J, Shih DJH, Luu B, Garzia L, Torchia J, Nor C, Morrissy AS, et al:
Intertumoral heterogeneity within medulloblastoma subgroups. Cancer
Cell. 31:737–754.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bockmayr M, Mohme M, Klauschen F, Winkler
B, Budczies J, Rutkowski S and Schüller U: Subgroup-specific immune
and stromal microenvironment in medulloblastoma. Oncoimmunology.
7:e14624302018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gieryng A, Pszczolkowska D, Walentynowicz
KA, Rajan WD and Kaminska B: Immune microenvironment of gliomas.
Lab Invest. 97:498–518. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yao M, Ventura PB, Jiang Y, Rodriguez FJ,
Wang L, Perry JSA, Yang Y, Wahl K, Crittenden RB, Bennett ML, et
al: Astrocytic trans-differentiation completes a multicellular
paracrine feedback loop required for medulloblastoma tumor growth.
Cell. 180:502–520.e19. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gandevia B and Tovell A: Declaration of
Helsinki. Med J Aust. 2:320–321. 1964. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Eisen MB, Spellman PT, Brown PO and
Botstein D: Cluster analysis and display of genome-wide expression
patterns. Proc Natl Acad Sci USA. 95:14863–14868. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pham CD, Flores C, Yang C, Pinheiro EM,
Yearley JH, Sayour EJ, Pei Y, Moore C, McLendon RE, Huang J, et al:
Differential immune microenvironments and response to immune
checkpoint blockade among molecular subtypes of murine
medulloblastoma. Clin Cancer Res. 22:582–595. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Graeber MB, Scheithauer BW and Kreutzberg
GW: Microglia in brain tumors. Glia. 40:252–259. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
McGranahan T, Therkelsen KE, Ahmad S and
Nagpal S: Current state of immunotherapy for treatment of
glioblastoma. Curr Treat Options Oncol. 20:242019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Northcott PA, Robinson GW, Kratz CP,
Mabbott DJ, Pomeroy SL, Clifford SC, Rutkowski S, Ellison DW,
Malkin D, Taylor MD, et al: Medulloblastoma. Nat Rev Dis Primers.
5:112019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pugh TJ, Weeraratne SD, Archer TC,
Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter
SL, Cibulskis K, Erlich RL, et al: Medulloblastoma exome sequencing
uncovers subtype-specific somatic mutations. Nature. 488:106–110.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nabors LB, Portnow J, Ammirati M, Baehring
J, Brem H, Brown P, Butowski N, Chamberlain MC, Fenstermaker RA,
Friedman A, et al: Central Nervous System Cancers, Version 1.2015.
J Natl Compr Canc Netw. 13:1191–1202. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Palmer SL, Reddick WE and Gajjar A:
Understanding the cognitive impact on children who are treated for
medulloblastoma. J Pediatr Psychol. 32:1040–1049. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lim M, Xia Y, Bettegowda C and Weller M:
Current state of immunotherapy for glioblastoma. Nat Rev Clin
Oncol. 15:422–442. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Becht E, Giraldo NA, Lacroix L, Buttard B,
Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautès-Fridman
C, Fridman WH, et al: Estimating the population abundance of
tissue-infiltrating immune and stromal cell populations using gene
expression. Genome Biol. 17:2182016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liberzon A, Birger C, Thorvaldsdóttir H,
Ghandi M, Mesirov JP and Tamayo P: The Molecular Signatures
Database (MSigDB) hallmark gene set collection. Cell Syst.
1:417–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lin GL, Nagaraja S, Filbin MG, Suvà ML,
Vogel H and Monje M: Non-inflammatory tumor microenvironment of
diffuse intrinsic pontine glioma. Acta Neuropathol Commun.
6:512018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Murata D, Mineharu Y, Arakawa Y, Liu B,
Tanji M, Yamaguchi M, Fujimoto KI, Fukui N, Terada Y, Yokogawa R,
et al: High programmed cell death 1 ligand-1 expression:
Association with CD8+ T-cell infiltration and poor
prognosis in human medulloblastoma. J Neurosurg. 128:710–716. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Majzner RG, Simon JS, Grosso JF, Martinez
D, Pawel BR, Santi M, Merchant MS, Geoerger B, Hezam I, Marty V, et
al: Assessment of programmed death-ligand 1 expression and
tumor-associated immune cells in pediatric cancer tissues. Cancer.
123:3807–3815. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liang J, Piao Y, Holmes L, Fuller GN,
Henry V, Tiao N and de Groot JF: Neutrophils promote the malignant
glioma phenotype through S100A4. Clin Cancer Res. 20:187–198. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Quail DF and Joyce JA: The
microenvironmental landscape of brain tumors. Cancer Cell.
31:326–341. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Margol AS, Robison NJ, Gnanachandran J,
Hung LT, Kennedy RJ, Vali M, Dhall G, Finlay JL, Erdreich-Epstein
A, Krieger MD, et al: Tumor-associated macrophages in SHH subgroup
of medulloblastomas. Clin Cancer Res. 21:1457–1465. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Maximov V, Chen Z, Wei Y, Robinson MH,
Herting CJ, Shanmugam NS, Rudneva VA, Goldsmith KC, MacDonald TJ,
Northcott PA, et al: Tumour-associated macrophages exhibit
anti-tumoural properties in Sonic Hedgehog medulloblastoma. Nat
Commun. 10:24102019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Powell AB, Yadavilli S, Saunders D, Van
Pelt S, Chorvinsky E, Burga RA, Albihani S, Hanley PJ, Xu Z, Pei Y,
et al: Medulloblastoma rendered susceptible to NK-cell attack by
TGFβ neutralization. J Transl Med. 17:3212019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nellan A, Rota C, Majzner R,
Lester-McCully CM, Griesinger AM, Mulcahy Levy JM, Foreman NK,
Warren KE and Lee DW: Durable regression of medulloblastoma after
regional and intravenous delivery of anti-HER2 chimeric antigen
receptor T cells. J Immunother Cancer. 6:302018. View Article : Google Scholar : PubMed/NCBI
|