Introduction
Breast cancer is the most frequently diagnosed type
of tumor and the second most common cause of mortality among women
worldwide (1). As breast cancer is
considered a systemic disease, comprehensive treatment with surgery
as the main component, in combination with chemotherapy,
radiotherapy, endocrine therapy, molecular targeted therapy and
other auxiliary interventions, has become the standard for breast
cancer treatment. Clinically, chemotherapy serves crucial roles in
the control and reduction of lesions before surgery and the
prevention of recurrence and metastasis after surgery. For advanced
and triple-negative breast cancer, chemotherapy remains the main
means of reducing recurrence and metastasis following surgery
(2,3). However, as highly heterogeneous tumors,
breast cancers with identical pathological and molecular types may
differ in their sensitivity to the same chemotherapy regimen. Thus,
not all patients will benefit from the same chemotherapy regimen.
This variation may be due to the differential expression of certain
genes associated with chemotherapy. Consequently, detecting the
expression of these genes to guide the selection of
chemotherapeutic drugs is of great significance for improving the
efficacy of chemotherapy and reducing the associated toxicity.
Numerous studies have suggested that the
differential expression of several genes, including excision repair
cross complementing 1 (ERCC1), ribonucleotide reductase M1
(RRM1), thymidylate synthetase (TYMS), β-tubulin III
(TUBB3) and topoisomerase IIα (TOP2A), in tumor
tissues is closely associated with chemoresistance and prognosis in
patients with cancer. For example, the expression level of
ERCC1, which is crucial for the repair of platinum-DNA
adducts, has been reported to negatively affect the effectiveness
of platinum drugs and suggested to be a major predictor of the
response of cancer to platinum-based chemotherapy (4,5).
Furthermore, a randomized prospective clinical study confirmed that
customized cisplatin chemotherapy based on quantitative
ERCC1 mRNA expression improved the survival of patients with
non-small-cell lung cancer (6).
These studies indicate that the assessment of ERCC1 mRNA
expression is feasible in a clinical setting and is able to predict
the response to cisplatin-based treatment. The expression level of
RRM1, which is the main target of gemcitabine, has been
reported to be negatively correlated with the efficacy of
gemcitabine (6,7). TUBB3 is thought to be a marker
of taxane resistance, and high expression levels of TUBB3
are reported to correlate with low response rates in patients
treated with taxane-containing regimens (8,9). The
expression level of TYMS, which is a central enzyme in the
folate metabolic pathway and a major target for cytotoxic
antifolate chemotherapeutic agents, such as 5-fluorouracil and
capecitabine, is negatively associated with the efficacy of
antimetabolic drugs (10,11). TOP2A is an essential nuclear
enzyme that changes DNA topology and is the primary molecular
target of various cytotoxic agents, including anthracyclines. The
expression level of TOP2A has been demonstrated to be
positively correlated with the efficacy of anthracycline drugs
(12,13). Therefore, the assessment of the
expression levels of these drug-associated genes in the tumor
tissues of patients prior to chemotherapy is useful for therapeutic
decision-making.
Although mounting evidence indicates their important
roles in the evaluation of chemoresistance, to the best of our
knowledge, no study on the combined detection of ERCC1, RRM1,
TUBB3, TYMS and TOP2A gene expression for the guidance
of chemotherapy in breast cancer patients has yet been reported.
Therefore, the present prospective study was carried out to with
the aim of providing new suggestions and clinical evidence for the
individualized treatment of breast cancer.
Materials and methods
Data collection
All 140 breast cancer patients, who were treated by
the same medical team from January 1, 2012 to December 31, 2013 at
the Department of Thyroid and Breast Surgery, the General Hospital
of Western Theater Command (Chengdu, China) were enrolled in the
study. The patients included an individualized chemotherapy group
(n=70) and a classic chemotherapy group (n=70). The mechanism, cost
and expected efficacy of the two chemotherapy methods were
explained in detail to the patients, and each patient decided which
method of treatment to receive. All patients had complete medical
records and none of them had received neoadjuvant therapy prior to
surgery. All patients had primary operable breast cancer with no
distant metastasis. Details of multiple clinicopathological
parameters were collected, including age, body mass index (BMI),
menstrual status, histological grade, tumor size, axillary lymph
node status, TNM stage, estrogen receptor status, progesterone
receptor status, human epidermal growth factor receptor 2 status,
Ki67 index, molecular classification, type of surgery, and hormonal
and radioactive therapy status. All patients provided written
informed consent for tissue sample retention and analysis for
research purposes and publication in the present article. This
retrospective study was approved by the ethics committee of the
General Hospital of Western Theater Command (registration no.
2011ky020).
Detection of mRNA expression
levels
The mRNA expression levels of ERCC1, RRM1, TUBB3,
TYMS and TOP2A in the breast cancer tissues were
measured simultaneously using multiplex branched DNA liquidchip
(MBL) technology (Guangzhou SurExam Bio-Tech Co., Ltd.) as
previously reported (14–16). The main steps in this analysis were
as follows: i) Samples were lysed in buffer at 56°C for 2 h; ii)
the lysed product was added to each well of a 96-well plate
containing blocking reagent, target gene-specific probe sets and
capture beads; iii) the plate was sealed, and then incubated for 18
h at 54°C on a shaker, followed by the addition of hybridization
mixture; iv) the unbound mRNA and other debris in each well were
removed by washing three times with buffer; v) signals for bound
target mRNA were amplified with streptavidin-phycoerythrin at 50°C
for 30 min; vi) the fluorescence value of each sample was measured
and analyzed using the Luminex® 200 system™ (Luminex
Corporation) to determine the mRNA expression level of each gene.
Compared with the cut-off value of each gene, the mRNA expression
level was categorized as low (<25%), low-to-medium (25–49%),
medium (50%), medium-to-high (51–75%) and high expression (>75%)
(17).
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from cryopreserved tissue
using TRIzol reagent (Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. Total RNA was reverse transcribed into
cDNA using the RevertAid™ First Strand cDNA Synthesis kit (cat. no.
k1622; Fermentas, Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The following thermocycling conditions
were used for qPCR: 50°C for 2 min, 95°C for 10 min, 40 cycles at
95°C for 20 sec, and 60°C for 1 min. A total of 40 cycles of
nucleic acid amplification were applied using Fast SYBR™ Green
Master Mix 4385612 (Applied Biosystems; Thermo Fisher Scientific,
Inc.) in an ABI PRISM® 7900HT Sequence Detection System
(Applied Biosystems; Thermo Fisher Scientific, Inc.), and the cycle
threshold (CT) value of the target gene was identified. Target
genes were normalized to the internal reference gene GAPDH,
and quantified using the comparative 2−ΔΔCq method
(18). Gene expression levels were
measured in triplicate, with a good reproducibility, and the
average was calculated. The following primer sequences were used
for qPCR: ERCC1 forward, 5′-GGGAATTTGGCGACGTAATTC-3′, and
reverse, 5′-GCGGAGGCTGAGGAACAG-3′; RRM1 forward,
5′-TGGCCTTGTACCGATGCTG-3′ and reverse,
5′-GCTGCTCTTCCTTTCCTGTGTT-3′; TUBB3 forward,
5′-AGTCGCCCACGTAGTTGC-3′ and reverse, 5′-CGCCCAGTATGAGGGAGAT-3′;
TYMS forward, 5′-GCCTCGGTGTGCCTTTCA-3′ and reverse,
5′-CGTGATGTGCGCAATCATG-3′; TOP2A forward,
5′-CATTGAAGACGCTTCGTTATGG-3′ and reverse,
5′-CCAGTTGTGATGGATAAAATTAATCAG-3′; and GAPDH forward,
5′-GCCACATCGCTCAGACACC-3′, and reverse,
5′-GATGGCAACAATATCCACTTTACC-3′.
Selection and implementation of
chemotherapy schemes
The regimen of each patient in the individualized
chemotherapy group was based on their genetic report. The
principles of selection were as follows: i) Platinum drugs, such as
cisplatin and oxaliplatin, are recommended for patients with low
ERCC1 expression; this regimen can be used in patients with
low-to-medium expression but should be avoided in patients with
medium-to-high and high expression (6). ii) Gemcitabine is recommended for
patients with low RRM1 expression; this regimen can be used
in patients with low-to-medium expression but should be avoided in
patients with medium-to-high and high expression (6). iii) Anti-microtubule drugs, such as
docetaxel and paclitaxel, are recommended for patients with low
TUBB3 expression; this regimen can be used in patients with
low-to-medium expression but should be avoided in patients with
medium-to-high and high expression (9). iv) Capecitabine is recommended for
patients with low TYMS expression; this regimen can be used
in patients with low-to-medium expression but should be avoided in
patients with medium-to-high and high expression (11). v) Anthracycline drugs, such as
epirubicin and doxorubicin, are recommended for patients with high
TOP2A expression (13); this
regimen can be used in patients with medium-to-high expression but
should be avoided in patients with low-to-medium expression and low
expression. Although multiple treatments may be recommended based
on these principles, only treatments that meet the guideline for
diagnosis and treatment of breast cancer (version 2011) will be
used for individualized chemotherapy (19). For the classic chemotherapy group,
the docetaxel + epirubicin + cyclophosphamide (TEC) regimen was
used. Details of the implementation of the chemotherapy regimens
are shown in Table I.
 | Table I.Implementation of chemotherapy
regimens. |
Table I.
Implementation of chemotherapy
regimens.
|
| No. of cycles |
|
|---|
|
|
|
|
|---|
| Chemotherapy
regimens | Four | Five | Six | Seven | Eight | n |
|---|
| Individualized
chemotherapy |
|
|
|
|
|
|
| E (90
mg/m2) + P (80 mg/m2) | 1 | 1 | 2 |
|
| 4 |
| E (90
mg/m2) + G (1,000 mg/m2) |
|
| 1 |
|
| 1 |
| E (90
mg/m2) + X (950 mg/m2) |
|
| 1 |
|
| 1 |
| T (75
mg/m2) + P (80 mg/m2) |
| 1 | 4 |
|
| 5 |
| T (75
mg/m2) + C (500 mg/m2) |
|
| 1 |
|
| 1 |
| T (75
mg/m2) + G (1,000 mg/m2) | 1 | 2 | 14 | 3 | 4 | 24 |
| T (75
mg/m2) + X (950 mg/m2) | 1 |
| 8 | 1 |
| 10 |
| T (75
mg/m2) + E (90 mg/m2) + C (500 mg/m2) | 1 | 2 | 17 | 1 | 3 | 24 |
| Classic
chemotherapy |
|
|
|
|
|
|
| T (75
mg/m2) + E (90 mg/m2) + C (500
mg/m2) | 4 | 6 | 60 |
|
| 70 |
Prognosis and safety evaluation
The endpoints of the study were disease-free
survival (DFS) and overall survival (OS). DFS time was calculated
as the length of time between the first confirmed diagnosis to
tumor recurrence or metastasis. OS time was calculated as the
length of time between the first confirmed diagnosis and mortality
from any cause. Censoring was defined as being lost to follow-up or
alive without relapse (local or distant) or mortality at the end of
follow-up. Breast ultrasound, liver-focused abdominal ultrasound,
axillary and neck lymph node ultrasound, chest computed tomography
(CT), skull enhanced magnetic resonance imaging/CT, bone emission
computed tomography, serum tumor markers and pathological
examinations were performed as appropriate to detect whether local
tumor recurrence or distant metastasis occurred. Survival data were
obtained in follow-ups with all patients conducted via telephone
contact or outpatient visits; the deadline was January 1, 2019.
Adverse events associated with chemotherapy were evaluated and
graded according to the National Cancer Institute Common Toxicity
Criteria 4 (NCI-CTC version 4.0).
Statistical analysis
Categorical variables are presented as numbers and
corresponding percentages, while continuous variables are presented
as mean ± standard deviation. Student's t-test was applied to
compare differences in age and BMI between the individualized and
classic groups. The differences in other baseline characteristics
and adverse events between the groups were evaluated using
Pearson's χ2 test. The Kaplan-Meier method was employed
for survival analysis, and the curves were compared using the
log-rank test. DFS time and OS time were analyzed using
Kruskal-Wallis and Dunn's post hoc test. Cox's proportional hazards
regression model was used to identify the independent predictors of
DFS and OS. Univariate predictors with P≤0.10 were entered into a
stepwise multivariate model to identify factors that independently
predicted DFS and OS. For all analyses, a two-tailed P≤0.05 was
considered to indicate a statistically significant result. All
statistical analyses were performed using SPSS 17.0 software (SPSS,
Inc.).
Results
Comparison of baseline
characteristics
A total of 140 well-matched female patients with
breast cancer were analyzed. All patients were histologically
confirmed as having invasive ductal carcinoma and none of them had
received targeted therapy or traditional Chinese medicine prior to
surgery. There were no significant differences in baseline
characteristics between the individualized chemotherapy and classic
chemotherapy groups. Details of the baseline characteristics of the
two groups of patients are summarized in Table II.
 | Table II.Baseline characteristics of the
patients. |
Table II.
Baseline characteristics of the
patients.
|
| Group |
|
|
|---|
|
|
|
|
|
|---|
| Characteristic | Individualized
regimen | Classic
regimen |
t/χ2-value | P-value |
|---|
| Age (years) | 51.1±8.1 | 48.5±7.6 | 1.939 | 0.055 |
| BMI
(kg/m2) | 23.8±2.9 | 23.8±3.1 | 0.011 | 0.991 |
| Menstrual
status |
|
Premenopausal | 37 (52.9) | 40 (57.1) | 0.260 | 0.610 |
|
Postmenopausal | 33 (47.1) | 30 (42.9) |
|
|
| Histological
grade |
| I | 9
(12.9) | 13 (18.6) | 1.098 | 0.578 |
| II | 47 (67.1) | 46 (65.7) |
|
|
|
III | 14 (20.0) | 11 (15.7) |
|
|
| Tumor size
(cm) |
| ≤2 | 22 (31.4) | 29 (41.4) | 3.161 | 0.182 |
|
2-5 | 45 (64.3) | 35 (50.0) |
|
|
| ≥5 | 3 (4.3) | 6 (8.6) |
|
|
| Nodal status |
|
Negative | 38 (54.3) | 33 (47.1) | 0.714 | 0.398 |
|
Positive | 32 (45.7) | 37 (52.9) |
|
|
| TNM stage |
| I | 14 (20.0) | 14 (20.0) | 2.703 | 0.259 |
| II | 43 (61.4) | 35 (50.0) |
|
|
|
III | 13 (18.6) | 21 (30.0) |
|
|
| ER status |
|
Positive | 47 (67.1) | 45 (64.3) | 0.127 | 0.722 |
|
Negative | 23 (32.9) | 25 (35.7) |
|
|
| PR status |
|
Positive | 34 (48.6) | 42 (60.0) | 1.842 | 0.157 |
|
Negative | 36 (51.4) | 28 (40.0) |
|
|
| HER-2 status |
|
Positive | 32 (45.7) | 27 (38.6) | 0.732 | 0.392 |
|
Negative | 38 (54.3) | 43 (61.4) |
|
|
| Ki67 index |
|
≤14% | 15 (21.4) | 9
(12.9) | 1.810 | 0.178 |
|
>14% | 55 (78.6) | 61 (87.1) |
|
|
| Molecular type |
| Luminal
A | 6 (8.6) | 4 (5.7) | 0.541 | 0.910 |
| Luminal
B | 41 (58.6) | 43 (61.4) |
|
|
|
HER-2-enriched | 9 (12.9) | 8
(11.4) |
|
|
|
Triple-negative | 14 (20.0) | 15 (21.4) |
|
|
| Type of
surgery |
|
Modified radical
mastectomy | 64 (91.4) | 67 (95.7) | 1.844 | 0.438 |
| BCS +
SLNB/T-ALND | 4 (5.7) | 1 (1.4) |
|
|
|
Mastectomy + SLNB | 2 (2.9) | 2 (2.9) |
|
|
| Radiotherapy |
|
Yes | 48 (68.6) | 41 (58.6) | 1.511 | 0.219 |
| No | 22 (31.4) | 29 (41.4) |
|
|
| Endocrine
therapy |
|
Yes | 42 (60.0) | 35 (50.0) | 1.414 | 0.234 |
| No | 28 (40.0) | 35 (50.0) |
|
|
Gene expression
The mRNA expression levels of ERCC1, RRM1, TUBB3,
TYMS and TOP2A were detected in the individualized
chemotherapy group. Table III
shows the case distribution according to expression intensity of
the five mRNAs in the individualized group. High expression levels
of ERCC1 and RRM1 were observed in 4.3 and 5.7% of
the group, respectively, while high expression levels of
TUBB3 and TYMS were observed in 27.1 and 22.9% of the
group, respectively. A low expression level of TOP2A was
observed in 38.6% of the group.
 | Table III.Expression of five mRNAs in the
individualized group. |
Table III.
Expression of five mRNAs in the
individualized group.
| Gene | Low | Low-to-medium | Medium | Medium-to-high | High |
|---|
| ERCC1 | 32 (45.7) | 20 (28.6) | 0 (0.0) | 15 (21.4) | 3 (4.3) |
| RRM1 | 45 (64.3) | 14 (20.0) | 0 (0.0) | 7 (10.0) | 4 (5.7) |
| TUBB3 | 17 (24.3) | 19 (27.1) | 0 (0.0) | 15 (21.5) | 19 (27.1) |
| TYMS | 15 (21.4) | 21 (30.0) | 0 (0.0) | 18 (25.7) | 16 (22.9) |
| TOP2A | 27 (38.6) | 20 (28.6) | 0 (0.0) | 11 (15.7) | 12 (17.1) |
Prognosis comparison
The median follow-up time among the patients
included in the study was 67.5 months (range, 1.0–84.0 months). At
the deadline, the tumor had progressed in 24 (17.1%) patients; 17
patients in the classic group and 7 patients in the individualized
group, the latter of which included 2 patients who received TEC
(from the individualized TEC group). Moreover, 17 (12.1%) patients
had died; 13 patients in the classic group and 4 patients in the
individualized group, which included 1 patient in the
individualized TEC group. Compared with the classic group, the DFS
and OS times of the individualized group were significantly
prolonged (DFS, P=0.039; OS, P=0.031) and the OS time of the
individualized TEC group was significantly prolonged (P=0.045).
Furthermore, the 5-year DFS and OS rates of the patients in the
individualized group were higher than those in the classic group
(DFS, 87.3 vs. 73.8%; OS, 94.3 vs. 84.2%). The 5-year DFS rate of
the individualized TEC group was higher than that of the classic
group (91.1 vs. 73.8%; Table IV).
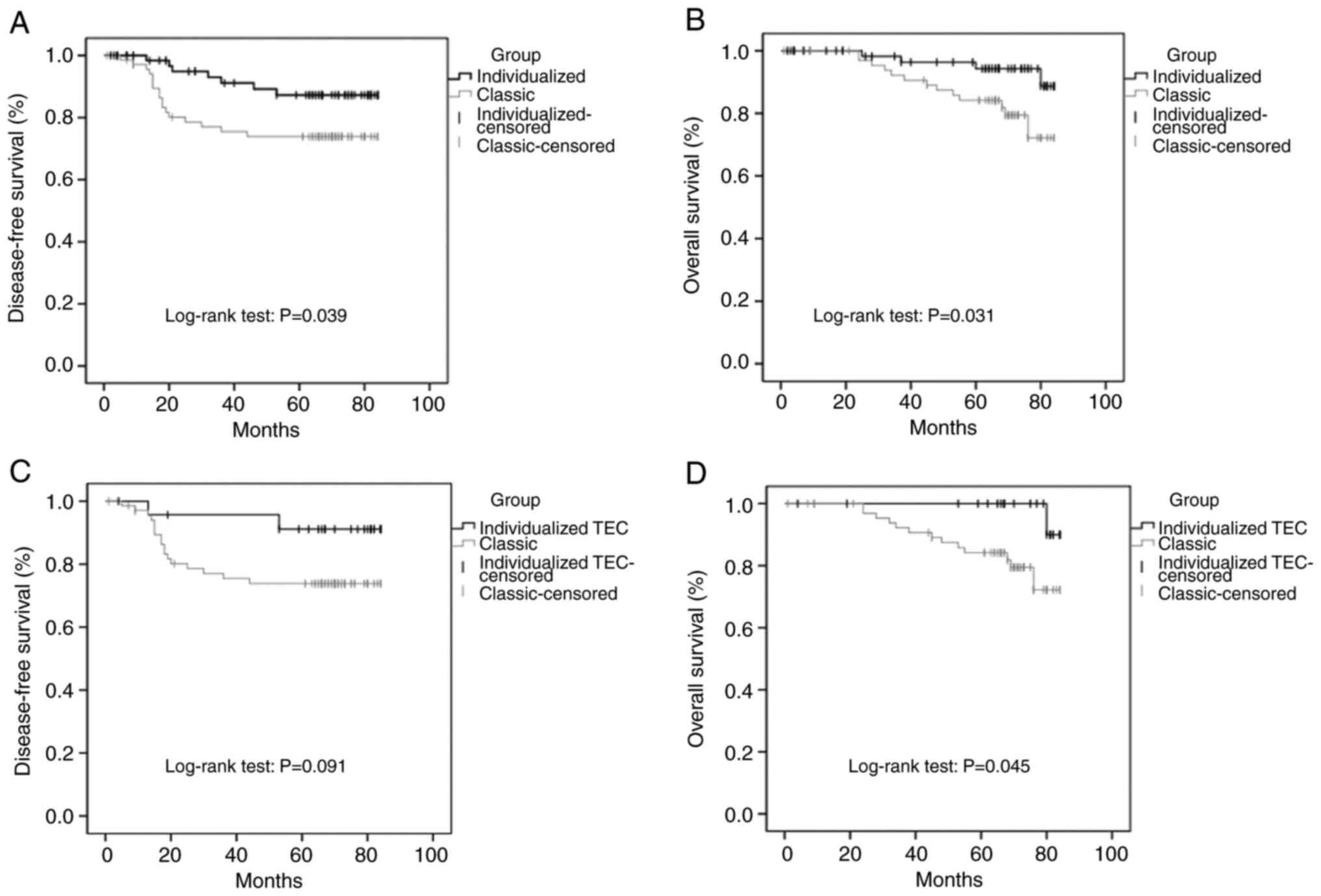
The Kaplan-Meier survival curves of the patients are shown in
Fig. 1. Compared with the classic
group, the cumulative DFS rate and cumulative OS rate of the
individualized group were significantly higher (Fig. 1A and B), and the cumulative OS rate
of the individualized TEC group was significantly higher (Fig. 1D). However, no statistically
significant difference was observed in the cumulative DFS rate
between the individualized TEC group and the classic group
(Fig. 1C).
 | Table IV.Disease-free and overall survival of
the patients. |
Table IV.
Disease-free and overall survival of
the patients.
|
| Group |
|
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| Variable | Individualized
(n=70) | Individualized TEC
(n=24) | Classic (n=70) |
χ2/Z-valuea |
P-valuea |
χ2/Z-valueb |
P-valueb |
|---|
|
Recurrence/metastasis [n (%)] | 7 (10.0) | 2 (8.3) | 17 (24.3) | 5.029 | 0.025 | 2.820 | 0.140 |
| DFS time [mean (95%
CI); months] | 77.4 (72.7,
82.1) | 79.5 (73.1,
85.9) | 67.1 (60.2,
74.1) | 4.251 | 0.039 | 2.855 | 0.091 |
| 5-year DFS rate
(%) | 87.3 | 91.1 | 73.8 | 3.609 | 0.057 | 4.518 | 0.034 |
| Mortality [n
(%)] | 4 (5.7) | 1 (4.2) | 13 (18.6) | 5.423 | 0.020 | 2.926 | 0.107 |
| OS time [mean (95%
CI); months] | 81.4 (78.6,
84.1) | 83.6 (82.9,
84.3) | 75.4 (71.1,
79.8) | 4.652 | 0.031 | 4.020 | 0.045 |
| 5-year OS rate
(%) | 94.3 | 90.0 | 84.2 | 3.249 | 0.071 | 0.302 | 0.583 |
Prognostic factors
Multivariable regression analyses were performed to
identify prognostic factors for DFS and OS (Table V). The results revealed metastasis of
axillary lymph nodes as an independent factor that increased the
risk of tumor relapse (HR=7.049, 95% CI: 1.813, 27.410, P=0.005).
Additionally, poor endocrine therapy compliance (treatment time
<5 years) was identified as an independent risk factor that
affected DFS (HR=3.378, 95% CI: 1.074, 10.624, P=0.037) and OS
(HR=8.140, 95% CI: 1.666, 39.759, P=0.010). Furthermore, the
individualized chemotherapy strategy guided by gene detection was
shown to be an independent protection factor for DFS (HR=0.389, 95%
CI: 0.153, 0.989, P=0.047) but not for OS (HR=0.340, 95% CI: 0.107,
1.078, P=0.067).
 | Table V.Multivariable Cox's regression
analysis of DFS and OS. |
Table V.
Multivariable Cox's regression
analysis of DFS and OS.
|
| DFS | OS |
|---|
|
|
|
|
|---|
| Factor | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Tumor size
(cm) |
|
|
|
|
| ≤2 | 1.00 |
|
|
|
|
2-5 | 2.700 (0.910,
8.008) | 0.073 |
|
|
| ≥5 | 1.783 (0.377,
8.443) | 0.466 |
|
|
| Nodal status |
|
|
|
|
|
Negative | 1.00 |
| 1.00 |
|
|
Positive | 7.049
(1.813, 27.410) | 0.005 | 3.360
(0.836, 13.504) | 0.088 |
| TNM stage |
|
|
|
|
| I | 1.00 |
| 1.00 |
|
| II | 0.351 (0.053,
2.330) | 0.279 | 0.704 (0.115,
4.313) | 0.704 |
|
III | 0.420 (0.051,
3.458) | 0.420 | 0.912 (0.119,
6.990) | 0.930 |
| ER status |
|
|
|
|
|
Positive | 1.00 |
| 1.00 |
|
|
Negative | 1.258 (0.225,
7.037) | 0.794 | 1.452
(0.071, 29.565) | 0.808 |
| PR status |
|
|
|
|
|
Positive | 1.00 |
| 1.00 |
|
|
Negative | 1.727 (0.321,
9.281) | 0.524 | 1.042
(0.050, 21.844) | 0.979 |
| Chemotherapy
strategy |
|
|
|
|
|
Classic | 1.00 |
| 1.00 |
|
|
Individualized | 0.389 (0.153,
0.989) | 0.047 | 0.340 (0.107,
1.078) | 0.067 |
| Endocrine therapy
compliance |
|
|
|
|
|
Good | 1.00 |
| 1.00 |
|
|
Poor | 3.378
(1.074, 10.624) | 0.037 | 8.140
(1.666, 39.759) | 0.010 |
Comparison of adverse reactions
There were no significant differences in the
incidence rate of dose reduction or reduction in the number of
chemotherapy cycles (<6 cycles) due to adverse reactions between
the individualized and classic groups (21.4 vs. 25.7%, P=0.550). In
addition, there were no mortalities associated with adverse events
in either of the treatment groups. It is noteworthy that there was
no statistically significant difference in the incidence of other
adverse events between the two groups. However, in terms of grade 2
or 3 palpitations and chest tightness, the incidence rate in the
individualized group was lower than that in the classic group (12.9
vs. 27.1%, P=0.035). Furthermore, there was no statistically
significant difference in the incidence of adverse events between
the classic group and the individualized TEC group (Table VI).
 | Table VI.Adverse events among the
patients. |
Table VI.
Adverse events among the
patients.
|
| Group |
|
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| Grade | Individualized
(n=70) | Individualized TEC
(n=24) | Classic (n=70) |
χ2-valuea |
P-valuea |
χ2-valueb |
P-valueb |
|---|
| Nausea and
vomiting |
| 1 | 28 (40.0) | 8 (33.3) | 29 (41.4) | 0.478 | 0.788 | 0.674 | 0.784 |
| 2 | 37 (52.9) | 14 (58.3) | 34 (48.6) |
|
|
|
|
| 3 | 5 (7.1) | 2 (8.3) | 7 (10.0) |
|
|
|
|
| Diarrhea |
| 1 | 62 (88.6) | 20 (83.3) | 64 (91.4) | 0.317 | 0.573 | 1.232 | 0.271 |
| 2 | 8 (11.4) | 4 (16.7) | 6 (8.6) |
|
|
|
|
| Constipation |
| 1 | 63 (90.0) | 23 (95.8) | 61 (87.1) | 0.282 | 0.595 | 1.420 | 0.443 |
| 2 | 7 (10.0) | 1 (4.2) | 9 (12.9) |
|
|
|
|
| Mucositis |
| 1 | 51 (72.9) | 19 (79.2) | 56 (80.0) | 0.991 | 0.319 | 0.008 | 1.000 |
| 2 | 19 (27.1) | 5 (20.8) | 14 (20.0) |
|
|
|
|
|
Leukopenia/neutropenia |
| 1 | 23 (32.9) | 11 (45.8) | 26 (37.1) | 0.319 | 0.853 | 0.598 | 0.775 |
| 2 | 29 (41.4) | 8 (33.3) | 28 (40.0) |
|
|
|
|
|
3,4 | 18 (25.7) | 5 (20.8) | 16 (22.9) |
|
|
|
|
|
Thrombocytopenia |
| 1 | 58 (82.9) | 21 (87.5) | 59 (84.3) | 0.052 | 0.820 | 0.146 | 0.758 |
| 2 | 12 (17.1) | 3 (12.5) | 11 (15.7) |
|
|
|
|
| Anemia |
| 1 | 66 (94.3) | 22 (91.7) | 59 (84.3) | 3.659 | 0.056 | 0.817 | 0.504 |
| 2 | 4 (5.7) | 2 (8.3) | 11 (15.7) |
|
|
|
|
| Liver toxicity |
| 1 | 46 (65.7) | 16 (66.7) | 58 (82.9) | 5.351 | 0.059 | 3.189 | 0.144 |
| 2 | 19 (27.1) | 7 (29.2) | 10 (14.3) |
|
|
|
|
| 3 | 5 (7.1) | 1 (4.2) | 2 (2.9) |
|
|
|
|
| Fatigue |
| 1 | 26 (37.1) | 9 (37.5) | 20 (28.6) | 1.166 | 0.280 | 0.668 | 0.414 |
| 2 | 44 (62.9) | 15 (62.5) | 50 (71.4) |
|
|
|
|
| Palpitations and
chest tightness |
| 1 | 61 (87.1) | 19 (79.2) | 51 (72.9) | 4.464 | 0.035 | 0.374 | 0.541 |
|
2,3 | 9 (12.9) | 5 (20.8) | 19 (27.1) |
|
|
|
|
| Hand-foot
syndrome |
| 1 | 52 (74.3) | 17 (70.8) | 58 (82.9) | 1.527 | 0.271 | 1.602 | 0.243 |
| 2 | 18 (25.7) | 7 (29.2) | 12 (17.1) |
|
|
|
|
Discussion
Individualized therapy has become an intensively
pursued approach at the molecular level. Previous studies have
indicated the important roles of ERCC1, RRM1, TUBB3, TYMS
and TOP2A gene expression in the pathogenesis, diagnosis and
prognosis of various types of carcinomas. Notably, as their roles
in chemoresistance have been fully confirmed, these genes are
suitable markers to provide guidance for individualized cancer
chemotherapy. However, to the best of our knowledge, there have
been no studies on the combined detection of ERCC1, RRM1, TUBB3,
TYMS and TOP2A gene expression to guide the selection of
chemotherapy regimens for patients with breast cancer. The present
study was designed to address this issue. The results demonstrated
that individualized chemotherapy strategies can prolong DFS and OS,
and also reduce adverse cardiovascular reactions, specifically
palpitations and chest tightness, in patients with breast
cancer.
ERCC1 is a key nuclease that regulates the
nucleotide excision repair (NER) pathway, which serves an essential
role in repair of DNA damage caused by platinum compounds (20,21).
High expression of ERCC1 indicates increased NER activity that
compromises the efficacy of platinum drugs. Certain studies have
demonstrated that resistance to platinum-based chemotherapy is
associated with high ERCC1 expression levels in some
advanced cancers, including gastric cancer (22), colorectal cancer (23), urinary tract cancer (5) and non-small cell lung cancer (24). Ribonucleotide reductase consists of
two subunits, RRM1 and RRM2, and is the rate-limiting enzyme in the
DNA synthesis pathway (25). The
RRM1 subunit encoded by the RRM1 gene is the main target of
gemcitabine. Studies have shown that high RRM1 expression is
associated with gemcitabine resistance (6,7). TUBB3
is a major component of the microtubules, a constructive component
of spindles and the cytoskeleton, that control mitosis and cellular
motility (26). Upregulation of
TUBB3 expression, which may destabilize microtubules and
counteract the effects of taxanes (9,27), has
been confirmed in various cancer types, including breast (28,29),
lung, ovarian, prostate, breast, stomach and pancreatic tumors
(30). TYMS is a central enzyme in
the synthesis of pyrimidine nucleotides and a major target for
antifolate cytotoxic drugs, such as 5-fluorouracil and
capecitabine. This enzyme exerts anticancer effects by inhibiting
the synthesis of deoxythymidylate and further affecting DNA
synthesis and repair (31). In
clinical studies of breast cancer (32), colorectal cancer (33) and lung cancer (34), patients with low expression of
TYMS have exhibited improved chemotherapeutic responses to
fluorochemical drugs and a longer median survival time. TOP2A is an
essential nuclear enzyme that changes the topology of DNA and is
the primary molecular target of various cytotoxic agents, including
anthracyclines (35), which
stabilize the cleavable complex formed between DNA and
topoisomerase II. Stabilization of the DNA-topoisomerase II complex
results in increased DNA cleavage and inhibition of the rejoining
of cleaved DNA, leading to cell death. Studies of the anthracycline
chemotherapy of breast cancer showed that patients with low
TOP2A expression had a poor response to treatment and poor
prognosis (12,13,36).
These findings led to the hypothesis that the detection of the
expression of these genes will be beneficial for guiding the
selection of chemotherapeutic drugs and may improve the efficacy of
chemotherapy.
In the individualized group, the proportion of
patients with medium-to-high and high expression levels of the
genes that are negatively correlated with efficacy were as follows:
ERCC1, 25.7%; RRM1, 15.7%; TUBB3, 48.6%; and
TYMS, 48.6%. Low and low-to-medium expression levels of
TOP2A were observed in 67.2% of the individualized group. As
none of the patients received neoadjuvant therapy prior to surgery,
the results indicate that some patients had primary resistance to
certain chemotherapeutic drugs. Therefore, the regimens used for
each patient in the individualized group were selected on the basis
of their genetic report. The patients in the classic group all
received chemotherapy according to the TEC regimen.
In the present study, an analysis of the survival
data of breast cancer patients from the two groups was performed.
The results showed that the DFS time in the individualized group
was 10.3 months longer than that in the classic group (P=0.039),
and the 5-year DFS rate was higher than that in the classic group
(87.3 vs. 73.8%). The OS time in the individualized group was 6
months longer than that in the classic group (P=0.031), and the
5-year OS rate was higher than that in the classic group (94.3 vs.
84.2%). Furthermore, the Kaplan-Meier survival curves of DFS and OS
showed that the overall prognosis of the patients in the
individualized group was better than that in the classic group
(log-rank test: P=0.039 and 0.031, respectively). To investigate
the potential of selection of the individualized chemotherapy
strategy under the guidance of genetic testing as an independent
prognostic factor for breast cancer patients, the associations
between all baseline variables and survival data were initially
investigated in a univariate analysis (data not shown). Those
variables with P≤0.10 were entered into the Cox's proportional
hazards regression model for multivariable analysis. The regression
analysis revealed that this individualized chemotherapy strategy
can reduce the risk of recurrence or metastasis (HR=0.389, 95% CI:
0.153, 0.989, P=0.047). Furthermore, it was identified that
metastasis of axillary lymph nodes was an independent risk factor
for DFS, and poor endocrine therapy compliance was an independent
risk factor for DFS and OS. In terms of drug safety, the majority
of the patients tolerated and successfully completed 6–8 cycles of
chemotherapy. Although various adverse reactions did occur during
chemotherapy, they were controlled by symptomatic treatment,
reduction of drug dosage, or the interruption or termination of
chemotherapy. No grade 5 adverse events were reported in the study.
The incidence of grade 2 or 3 palpitations and chest tightness in
the individualized group was significantly lower than that in the
classic group (12.9 vs. 27.1%, P=0.035). This may be associated
with the use of anthracyclines, which were included in the classic
regimen but only used selectively in the individualized group
according to the patient's level of TOP2A gene expression.
In addition, no significant differences were detected between the
two groups in terms of the incidence of other adverse events,
namely nausea and vomiting, diarrhea, constipation, mucositis,
myelosuppression, liver toxicity, fatigue and hand-foot syndrome.
It is noteworthy that 24 patients in the individualized group were
treated using TEC regimens. To avoid the influence of different
therapy regimens, the survival and adverse events in the classic
group were compared with those in the individualized TEC group.
Although the patients in the two groups were treated using the same
TEC regimens, the overall prognosis of the individualized TEC group
was improved compared with that of the classic group, and there was
no significant difference between these two groups in the incidence
of adverse events. These findings show that the selection of
chemotherapy regimens according to each patient's gene expression
characteristics can reduce the occurrence of drug resistance and
increase therapeutic effectiveness, as well as providing new ideas
and clinical evidence for the individualized treatment of breast
cancer patients.
Admittedly, the present study has some limitations.
First, this study used a nonrandomized patient cohort and a
relatively small sample size, which may be inconsistent with
previous studies. Second, gene expression was detected using MBL
technology, but not confirmed by other methods using normal breast
tissues or paracancerous tissue as a control. However, the
reliability of the results is supported by the use of MBL
technology, which is a mature gene detection technology that has
been widely applied for predicting the prognosis and selecting the
individualized treatment regimen for several types of tumors
(15,37–40).
Additionally, the genes investigated do not perform a single
biological function. Further research is essential to explore the
associations between the expression of these genes and other
chemotherapeutic drugs. Finally, the application of testing
technology may increase treatment costs and the benefit-cost ratio
should be evaluated for each individual patient. In summary,
large-scale, prospective studies with randomized patient cohorts,
the addition of control samples and immunohistochemical
confirmation are necessary to further investigate the guiding
significance of the expression of ERCC1, RRM1, TUBB3, TYMS,
TOP2A and other genes in the individualized therapy of breast
cancer.
In conclusion, the findings of the present study
indicate that therapeutic decision-making on the basis of ERCC1,
RRM1, TUBB3, TYMS and TOP2A gene expression can prolong
DFS and OS, improve prognosis, reduce cardiovascular adverse
reactions such as palpitations and chest tightness, enhance the
quality of life and benefit patients.
Acknowledgements
The authors would like to acknowledge Dr Yunming Li
(Department of Health Statistics at the General Hospital of Western
Theater Command, Chengdu, China), for statistical support.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
JCL contributed to the study design as well as data
analysis and interpretation, and drafted the manuscript. PS helped
conceive the present study. TH helped analyze and interpret the
data. SDH and LFL were involved in acquiring data and drafting the
manuscript. PS and TH assessed and revised the manuscript
critically for important intellectual content. GX participated in
the study conception and design, contributed to quality control of
the data and algorithms, and edited and reviewed the manuscript.
Each author participated sufficiently in the work to take public
responsibility for appropriate portions of the content, and GX
agreed to be accountable for all aspects of the work in ensuring
that questions related to the accuracy or integrity of any part of
the work are appropriately investigated and resolved. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the General Hospital of Western Theater Command
(Chengdu, China; approval no. 2011ky020). Written informed consent
was provided by all patients prior to the study start for tissue
sample retention and analysis for research purposes.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ERCC1
|
excision repair cross complementing
1
|
|
RRM1
|
ribonucleoside reductase M1
|
|
TUBB3
|
β-tubulin III
|
|
TYMS
|
thymidylate synthase
|
|
TOP2A
|
topoisomerase IIα
|
|
MBL
|
multiplex branched DNA liquidchip
|
|
T
|
docetaxel
|
|
E
|
epirubicin
|
|
C
|
cyclophosphamide
|
|
DFS
|
disease-free survival
|
|
OS
|
overall survival
|
|
P
|
cisplatin
|
|
G
|
gemcitabine
|
|
X
|
capecitabine
|
|
BMI
|
body mass index
|
|
ER
|
estrogen receptor
|
|
PR
|
progesterone receptor
|
|
HER-2
|
human epidermal growth factor receptor
2
|
|
BCS
|
breast conserving surgery
|
|
SLNB
|
sentinel lymph node biopsy
|
|
T-ALND
|
total axillary lymphadenectomy
|
|
HR
|
hazard ratio
|
|
CI
|
confidence interval
|
|
NER
|
nucleotide excision repair
|
|
RT-qPCR
|
Reverse transcription-quantitative
polymerase chain reaction
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Braunstein LZ and Taghian AG: Molecular
phenotype: Multigene assays, and the locoregional management of
breast cancer. Semin Radiat Oncol. 26:9–16. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jhan JR and Andrechek ER: Triple-negative
breast cancer and the potential for targeted therapy.
Pharmacogenomics. 18:1595–1609. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
EL Baiomy MA and El Kashef WF: ERCC1
expression in metastatic triple negative breast cancer patients
treated with platinum-based chemotherapy. Asian Pac J Cancer Pre.
18:507–513. 2017.
|
|
5
|
Kim KH, Do IG, Kim HS, Chang MH, Kim HS,
Jun HJ, Uhm J, Yi SY, Lim DH, Ji SH, et al: Excision repair
cross-complementation group 1 (ERCC1) expression in advanced
urothelial carcinoma patients receiving cisplatin-based
chemotherapy. APMIS. 118:941–948. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bepler G, Williams C, Schell MJ, Chen W,
Zheng Z, Simon G, Gadgeel S, Zhao X, Schreiber F, Brahmer J, et al:
Randomized international phase III trial of ERCC1 and RRM1
expression-based chemotherapy versus gemcitabine/carboplatin in
advanced non-small-cell lung cancer. J Clin Oncol. 31:2404–2412.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gong W, Zhang X, Wu J, Chen L, Li L, Sun
J, Lv Y, Wei X, Du Y, Jin H and Dong J: RRM1 expression and
clinical outcome of gemcitabine-containing chemotherapy for
advanced non-small-cell lung cancer: A meta-analysis. Lung Cancer.
75:374–380. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kamath K, Wilson L, Cabral F and Jordan
MA: BetaIII-tubulin induces paclitaxel resistance in association
with reduced effects on microtubule dynamic instability. J Biol
Chem. 280:12902–12907. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Narvi E, Jaakkola K, Winsel S,
Oetken-Lindholm C, Halonen P, Kallio L and Kallio MJ: Altered TUBB3
expression contributes to the epothilone response of mitotic cells.
Br J Cancer. 15:82–90. 2013. View Article : Google Scholar
|
|
10
|
Shan F, Liu YL, Wang Q and Shi YL:
Thymidylate synthase predicts poor response to pemetrexed
chemotherapy in patients with advanced breast cancer. Oncol Lett.
16:3274–3280. 2018.PubMed/NCBI
|
|
11
|
Gao Y, Cui J, Xi H, Cai A, Shen W, Li J,
Zhang K, Wei B and Chen L: Association of thymidylate synthase
expression and clinical outcomes of gastric cancer patients treated
with fluoropyrimidine-based chemotherapy: A meta-analysis. Onco
Targets Ther. 9:1339–1350. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brase JC, Schmidt M, Fischbach T, Sültmann
H, Bojar H, Koelbl H, Hellwig B, Rahnenführer J, Hengstler JG and
Gehrmann MC: ERBB2 and TOP2A in breast cancer: A comprehensive
analysis of gene amplification, RNA levels, and protein expression
and their influence on prognosis and prediction. Clin Cancer Res.
16:2391–2401. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
O'Malley FP, Chia S, Tu D, Shepherd LE,
Levine MN, Huntsman D, Bramwell VH, Andrulis IL and Pritchard KI:
Topoisomerase II alpha protein and responsiveness of breast cancer
to adjuvant chemotherapy with CEF compared to CMF in the NCIC CTG
randomized MA.5 adjuvant trial. Breast Cancer Res Treat.
128:401–409. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang C, Zhou Q, He J, Yang H, Luo X and Xu
J: Application of multiplex branched DNA liquidchip technology
(mbl) for optimal selection of chemotherapy in elderly patients. J
Geriatr Oncol. 5:S142014. View Article : Google Scholar
|
|
15
|
Han Y, Li G, Su C, Ren H, Chu X, Zhao Q,
Zhu Y, Wang Z, Hu B, An G, et al: Exploratory study on the
correlation between 14 lung cancer-related gene expression and
specific clinical characteristics of NSCLC patients. Mol Clin
Oncol. 1:887–893. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ren GJ, Zhao YY, Zhu YJ, Xiao Y, Xu JS,
Shan B and Zhang L: Tumor gene mutations and messenger RNA
expression: Correlation with clinical response to icotinib
hydrochloride in non-small cell lung cancer. Chin Med J (Engl).
124:19–25. 2011.PubMed/NCBI
|
|
17
|
Sun S, Shi W, Wu Z, Zhang G, Yang BO and
Jiao S: Prognostic significance of the mRNA expression of ERCC1,
RRM1, TUBB3 and TYMS genes in patients with non-small cell lung
cancer. Exp Ther Med. 10:937–941. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chinese Anti-Cancer Association, Committee
of Breast Cancer Society. Guideline for diagnosis and treatment of
breast cancer (version 2011). China Oncol. 27:695–759. 2011.(In
Chinese).
|
|
20
|
Altaha R, Liang X, Yu JJ and Reed E:
Excision repair cross complementing-group 1: Gene expression and
platinum resistance. Int J Mol Med. 14:959–970. 2004.PubMed/NCBI
|
|
21
|
Croteau DL, Peng Y and Van Houten B: DNA
repair gets physical: Mapping an XPA-binding site on ERCC1. DNA
Repair (Amst). 7:819–826. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kwon HC, Roh MS, Oh SY, Kim SH, Kim MC,
Kim JS and Kim HJ: Prognostic value of expression of ERCC1,
thymidylate synthase, and glutathione S-transferase P1 for
5-fluorouracil/oxaliplatin chemotherapy in advanced gastric cancer.
Ann Oncol. 18:504–509. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim SH, Kwon HC, Oh SY, Lee DM, Lee S, Lee
JH, Roh MS, Kim DC, Park KJ, Choi HJ and Kim HJ: Prognostic value
of ERCC1, thymidylate synthase, and glutathione S-transferase pi
for 5-FU/oxaliplatin chemotherapy in advanced colorectal cancer. Am
J Clin Oncol. 32:38–43. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hwang IG, Ahn MJ, Park BB, Ahn YC, Han J,
Lee S, Kim J, Shim YM, Ahn JS and Park K: ERCC1 expression as a
prognostic marker in N2(+) nonsmall-cell lung cancer patients
treated with platinum-based neoadjuvant concurrent
chemoradiotherapy. Cancer. 113:1379–1386. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Herrick J and Sclavi B: Ribonucleotide
reductase and the regulation of DNA replication: An old story and
an ancient heritage. Mol Microbiol. 63:22–34. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Katsetos CD, Herman MM and Mörk SJ: Class
III beta-tubulin in human development and cancer. Cell Motil
Cytoskeleton. 55:77–96. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kavallaris M: Microtubules and resistance
to tubulin-binding agents. Nat Rev Cancer. 10:194–204. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen X, Wu J, Lu H, Huang O and Shen K:
Measuring β-tubulin III, Bcl-2, and ERCC1 improves pathological
complete remission predictive accuracy in breast cancer. Cancer
Sci. 103:262–268. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Y, Sparano JA, Fineberg S, Stead L,
Sunkara J, Horwitz SB and McDaid HM: High expression of class III
β-tubulin predicts good response to neoadjuvant taxane and
doxorubicin/cyclophosphamide-based chemotherapy in estrogen
receptor-negative breast cancer. Clin Breast Cancer. 13:103–108.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sève P and Dumontet C: Is class III
beta-tubulin a predictive factor in patients receiving
tubulin-binding agents? Lancet Oncol. 9:168–175. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Longley DB, Harkin DP and Johnston PG:
5-fluorouracil: Mechanisms of action and clinical strategies. Nat
Rev Cancer. 3:330–338. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kakimoto M, Uetake H, Osanai T, Shirota Y,
Takagi Y, Takeshita E, Toriya Y, Danenberg K, Danenberg PV and
Sugihara K: Thymidylate synthase and dihydropyrimidine
dehydrogenase gene expression in breast cancer predicts 5-FU
sensitivity by a histocultural drug sensitivity test. Cancer Lett.
223:103–111. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Soong R, Shah N, Salto-Tellez M, Tai BC,
Soo RA, Han HC, Ng SS, Tan WL, Zeps N, Joseph D, et al: Prognostic
significance of thymidylate synthase, dihydropyrimidine
dehydrogenase and thymidine phosphorylase protein expression in
colorectal cancer patients treated with or without
5-fluorouracil-based chemotherapy. Ann Oncol. 19:915–919. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shintani Y, Ohta M, Hirabayashi H, Tanaka
H, Iuchi K, Nakagawa K, Maeda H, Kido T, Miyoshi S and Matsuda H:
Thymidylate synthase and dihydropyrimidine dehydrogenase mRNA
levels in tumor tissues and the efficacy of 5-fluorouracil in
patients with non-small-cell lung cancer. Lung Cancer. 45:189–196.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jacot W, Fiche M, Zaman K, Wolfer A and
Lamy PJ: The HER2 amplicon in breast cancer: Topoisomerase IIA and
beyond. Biochim Biophys Acta. 1836:146–157. 2013.PubMed/NCBI
|
|
36
|
Moretti E, Desmedt C, Biagioni C, Regan
MM, Oakman C, Larsimont D, Galardi F, Piccart-Gebhart M, Sotiriou
C, Rimm DL and Di Leo A: TOP2A protein by quantitative
immunofluorescence as a predictor of response to epirubicin in the
neoadjuvant treatment of breast cancer. Future Oncol. 9:1477–1487.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Han Y, Wang XB, Xiao N and Liu ZD: mRNA
expression and clinical significance of ERCC1, BRCA1, RRM1, TYMS
and TUBB3 in postoperative patients with non-small cell lung
cancer. Asian Pac J Cancer Prev. 14:2987–2990. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yu Y, Ding S, Liang Y, Zheng Y, Li W, Yang
L, Zheng X and Jiang J: Expression of ERCC1, TYMS, TUBB3, RRM1 and
TOP2A in patients with esophageal squamous cell carcinoma: A
hierarchical clustering analysis. Exp Ther Med. 7:1578–1582. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Su C, Zhou S, Zhang L, Ren S, Xu J, Zhang
J, Lv M, Zhang J and Zhou C: ERCC1, RRM1 and BRCA1 mRNA expression
levels and clinical outcome of advanced non-small cell lung cancer.
Med Oncol. 28:1411–1417. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhao H, Zhang H, Du Y and Gu X: Prognostic
significance of BRCA1, ERCC1, RRM1, and RRM2 in patients with
advanced non-small cell lung cancer receiving chemotherapy. Tumour
Biol. 35:12679–12688. 2014. View Article : Google Scholar : PubMed/NCBI
|