Introduction
Lung cancer is one of the most common primary
malignant tumors. According to the 2018 Global Cancer Statistics,
lung cancer was the leading cause of cancer-associated mortality
worldwide, with a 5-year survival rate of patients with lung cancer
of 18% (1). The most common causes
of lung cancer include smoking, air pollution, heredity factors and
activation of cancer genes, such as Ras and Raf (2). Despite the fact that the majority of
patients at early disease stages can be treated with surgery
(3), the choice of treatment for
patients at advanced stages are limited to radiotherapy and
chemotherapy (4). Furthermore, in
some patients, prolonged exposure to a single chemotherapeutic
agent such as cisplatin may lead to the development of resistance
(5). Thus, the development of novel
targeted therapeutic strategies remains critical for improved
prognosis and survival outcomes of patients with lung cancer.
Histone-lysine N-methyltransferase EZH2 (EZH2) is
the core catalytic component of polycomb repressive complex 2
(PRC2), which is involved in transcriptional repression (6). EZH2 is considered an oncogene, as it is
frequently overexpressed in several types of human cancer,
including prostate (7), gastric
(8) and breast cancer (9,10).
Furthermore, RNA interference of EZH2 has been reported to
significantly decrease cell viability in breast cancer (11,12).
However, the molecular mechanisms by which EZH2 knockdown inhibits
disease progression of lung cancer remain unclear.
The aim of the present study was to identify whether
EZH2 was associated with cell viability, migration and invasion, as
well as cell cycle progression and apoptosis, in the lung cancer
A549 cell line. By identifying whether EZH2 acts as an oncogene in
lung cancer, the present study may provide a novel diagnostic
biomarker and a therapeutic target for the treatment of lung
cancer.
Materials and methods
Bioinformatics analysis of human lung
cancer
The GSE19804 dataset (13) was downloaded from the Gene Expression
Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo) using the keywords
‘lung cancer’, ‘Homo sapiens’ and ‘Expression profiling by array or
sequencing’, which includes 60 adjacent non-tumor (within 2 cm
around tumors) and 60 tumor samples. GEO2R is an interactive online
platform, which identifies differentially expressed genes (DEGs) by
comparing samples in the GEO series (14). The cut-off criteria for the DEGs were
an adjusted P<0.05 and a |log2 fold change (FC)|
value of >2. The genes were aligned using their expression
levels, and the data are presented as a volcano plot and a heat
map. PROGgeneV2 (http://watson.compbio.iupui.edu/chirayu/proggene/database/?url=progge)
is a web-based platform used to study prognostic implications of
genes in different types of cancer. PROGgeneV2 was used to perform
the survival analysis based on EZH2 expression in the GSE30219
dataset (15) downloaded from the
GEO database.
Materials and reagents
The cell cycle kit (cat. no. 558662) and apoptosis
detection kit (cat. no. 559763) were purchased from BD Biosciences,
while the MTT reagent was purchased from Sigma-Aldrich; Merck KGaA.
The primary antibodies (all diluted 1:1,000) against EZH2 (cat. no.
4905), cyclin D1 (cat. no. 2922), p21 (cat. no. 2947),
cleaved(C)-caspase-3 (cat. no. 9661), C-caspase-9 (cat. no. 52873),
N-cadherin (cat. no. 4061), E-cadherin (cat. no. 3195), vimentin
(cat. no. 5741) and GAPDH (cat. no. 5174) were purchased from Cell
Signaling Technology, Inc., and the EZH2 inhibitor (EPZ-6438) was
obtained from Selleck Chemicals.
Cell culture
A total of five human lung cancer cell lines (H1650,
PC9, A549, H1299 and H460), and a normal lung epithelium cell line,
(BEAS-2B) were purchased from the Bank of Type Culture Collection
of the Chinese Academy of Sciences and cultured in RPMI-1640 medium
supplemented with 10% FBS and 100 U/ml penicillin (Gibco; Thermo
Fisher Scientific, Inc.) at 37°C in a humidified incubator with 5%
CO2.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from lung cancer A549 cells
using TRIzol® regent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol, and
cDNA was generated using the PrimeScript™ RT reagent kit (Takara
Biotechnology Co., Ltd.) at 37°C for 30 min, according to the
manufacturer's protocol. qPCR was subsequently performed using
SYBR® Premix ExTaq™ (Takara Biotechnology Co., Ltd.)
according to the manufacturer's protocol and detected using the
LightCycler™ 480 system (Roche Diagnostics). The reaction
conditions were as follows: Initial denaturation at 95°C for 3 min,
followed by 40 cycles of denaturation at 95°C for 10 sec, annealing
at 60°C for 40 sec and extension at 72°C for 40 sec. The following
primer sequences were used for qPCR: EZH2 forward,
5′-GAAAGCCGCCCACCTC-3′ and reverse, 5′-AAACATCGCCTACAGAAAAGC-3′;
and GAPDH forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′. Relative expression levels were
calculated using the 2−ΔΔCq method and normalized to the
internal reference gene GAPDH (16).
Western blot analysis
Total protein was extracted from A549 cells using
RIPA lysis buffer supplemented with protease inhibitors cocktail
and PMSF (1:100) (all Beyotime Institute of Biotechnology). Protein
concentration was determined using the Pierce Micro BCA protein
assay system (Pierce; Thermo Fisher Scientific, Inc.). A total of
50 µg protein/lane was separated using SDS-PAGE on a 10% gel and
subsequently transferred onto polyvinylidene difluoride membranes
(Sigma-Aldrich; Merck KGaA). The membranes were blocked with 5%
skimmed milk in TBS-Tween (1% Tween-20) for 1 h at room
temperature. After washing with TBS-Tween, the membranes were
incubated with primary antibodies against EZH2, cyclin D1, p21,
C-caspase-3, C-caspase-9, N-cadherin, E-cadherin, vimentin and
GAPDH overnight at 4°C. After washing, membranes were incubated
with goat anti-rabbit secondary antibody (1:2,000; cat. no. 7074;
Cell Signaling Technology, Inc.) at room temperature for 1 h.
Protein bands were visualized using an enhanced chemiluminescence
kit (cat. no. NCI4106; Thermo Fisher Scientific, Inc.) and scanned
using the lmageQuant 4000 system (Cytiva). The optical density of
the target bands were quantified using the Quantity One system
v4.6.2 (Bio-Rad Laboratories, Inc.).
RNA interference
A549 cells were transfected with 50 nM EZH2 small
interfering (si)RNA (Shanghai Genechem Co., Ltd.) using
Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) for 48 h before subsequent experimentation,
according to the manufacturer's protocol. The following siRNA
sequences were used: EZH2 forward, 5′-GTGCCCTTGTGTGATAGCACAA-3′ and
reverse, 5′-GGCACTTTCATTGAAGAACTAA-3′; and non-targeted negative
control (NC) forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′.
MTT assay
The viability of cells was detected using the MTT
assay, according to the manufacturer's protocol. Briefly, A549
cells were trypsinized and seeded into 96-well plates at a density
of 1×104, following siRNA transfection for 48 h or
treatment with EPZ-6438 (40 µM) for 24 h, and treatment with
different concentrations (0, 2, 4, 8 or 16 µM) of cisplatin (CDDP;
Sigma-Aldrich; Merck KGaA) for 24 h. Cells were subsequently
incubated with 20 µl MTT solution (in 5 mg/ml PBS) at 37°C for 4 h.
Following which, the purple formazan crystals were dissolved using
150 µl dimethyl sulfoxide for 20 min at room temperature, and cell
viability was subsequently analyzed at a wavelength of 570 nm. All
experiments were performed in triplicate.
Colony formation assay
A total of 500 A549 cells were plated into 35 mm
dishes prior to transfection with siRNA or treatment with EPZ-6438
(40 µM) for 24 h, followed by treatment with 4 µM CDDP for 24 h,
and maintained at 37°C for 2 weeks. Cell colonies were subsequently
washed three times with PBS, fixed with ethanol for 15 min at room
temperature and stained with 1% crystal violet for 20 min at room
temperature. Cell colonies were observed using a Canon camera
(Canon, Inc.).
Cell cycle assay
Cell cycle analysis was performed using propidium
iodide (PI) staining (BD Biosciences), according to the
manufacturer's protocol. Briefly, cells were digested with
EDTA-free trypsin and centrifuged at 500 × g for 5 min at 4°C. A
total of 1×106 cells were harvested and fixed with 70%
ethanol overnight at 4°C. Subsequently, PI supplemented with RNase
A was added and the DNA content was sorted using flow cytometric
analysis (BD Biosciences). Total cell quantity at each phase was
analyzed using ModFit LT v5.0 software (Verity Software House,
Inc.).
Flow cytometric analysis of
apoptosis
The extent of apoptosis was determined using the
PE-Annexin V Apoptosis assay kit (BD Biosciences), according to the
manufacturer's protocol. Briefly, cells were digested with
EDTA-free trypsin and centrifuged at 500 × g for 5 min at 4°C.
Cells were harvested and washed three times with PBS, 48 h
post-transfection, prior to resuspension in 500 µl binding buffer.
Subsequently, cells were incubated with 5 µl PI and 5 µl Annexin
V-FITC in the dark at room temperature for 10 min and apoptotic
cells were analyzed using the FACScan flow cytometer (BD
Biosciences).
Wound healing assay
A total of 1×105 cells/well were seeded
into 24-well plates and cultured in RPMI-1640 supplemented with 1%
penicillin, 1% streptomycin and 10% FBS at 37°C in 5%
CO2 until they reached 90% confluence. Subsequently, the
cell monolayers were scratched using a 200 µl pipette tip to
produce a consistent width. Cells were washed twice with PBS to
remove non-adherent cells, while the adherent cells were
serum-starved in serum-free medium overnight. Migratory cells were
observed in five randomly selected fields under a light microscope
(magnification, ×40) at 0 and 24 h. The width of the wounds was
measured using ImageJ software 1.52 (National Institutes of
Health).
Transwell and Matrigel assays
The migration and invasion assays were performed as
previously described (17). Briefly,
4×104 cells were plated in the upper chambers of
8-µM-pore sized Transwell plates (BD Biosciences) in serum-free
medium and cultured for 37°C for both assays. A Matrigel assay was
performed to detect cell invasion. The Transwell membranes were
precoated with 50 µl Matrigel (1 mg/ml; BD Biosciences) at 37°C for
4 h. A total of 500 µl RPMI-1640 supplemented with 10% FBS was
added to the lower chambers. Following incubation at 37°C in 5%
CO2 for 24 h, both the migratory and invasive cells in
the lower chambers were washed twice with PBS and fixed with
ethanol for 10 min at room temperature. Non-invasive cells in the
upper chambers were removed using a cotton swab. Invasive cells,
and the migratory cells were stained with 1% crystal violet for 20
min at room temperature. Stained cells were observed under a light
microscope (magnification, ×100).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism software version 7.0 (GraphPad Software, Inc.). The survival
curve was calculated using the Kaplan-Meier estimator method and
the log-rank test was used to compare the two survival curves. Data
are presented as the mean ± standard deviation of three independent
experiments. Unpaired Student's t-test was used to compare
differences between two groups, while one-way ANOVA followed by the
least significant difference post hoc test was used to compare
difference between multiple groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
EZH2 expression is upregulated in
human lung cancer
Analysis of the GSE19804 dataset identified 265
DEGs, including 53 upregulated and 212 downregulated DEGs in lung
cancer compared with normal tissues (P<0.05 and
|Log2FC| >2; Fig. 1A).
The DEGs are shown in Table SI,
while the top 20 DEGs in lung cancer are presented in Fig. 1B.
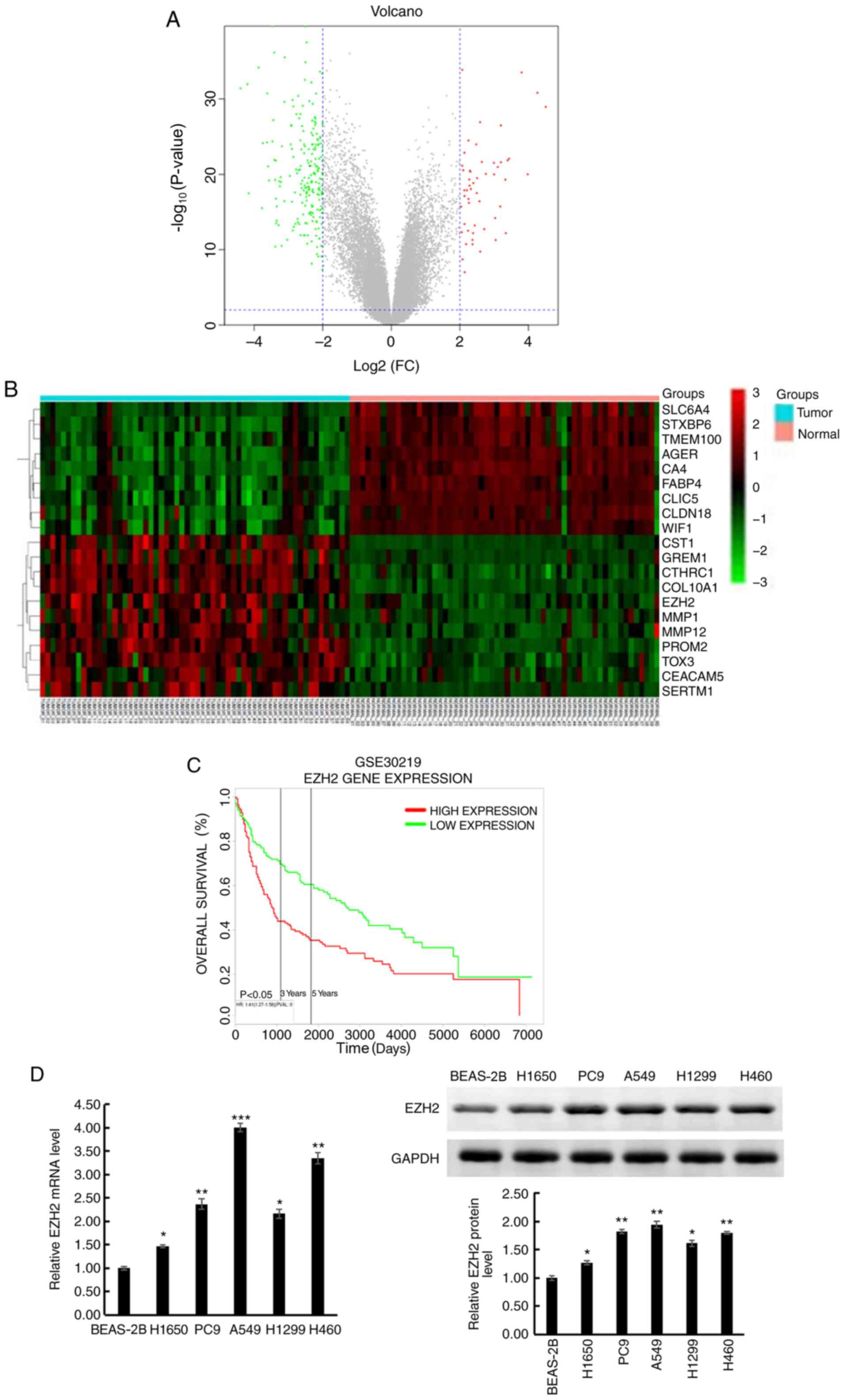 | Figure 1.EZH2 expression is upregulated in
human lung cancer tissues and cell lines. (A) Differentially
expressed genes were selected using volcano plot filtering
(P<0.05; FC >2). The red dots represent upregulated genes,
while the green dots represent downregulated genes and the grey
dots represent genes with |FC|<2. (B) Heatmap of the top 20
dysregulated genes. Red indicates upregulation and green indicates
downregulation. The values between −3 and 3 indicate the FC degree
in gene expression. The 60 GSM on the left side were collected from
tumor tissues of patients with lung cancer, while the 60 GSM on the
right side were collected from normal tissues of the same patients.
(C) Altered EZH2 expression was demonstrated to be associated with
overall survival time. (D) EZH2 expression in lung cancer cell
lines (H1650, PC9, A549, H1299 and H460) and normal human lung
epithelial cell line (BEAS-2B) was detected using reverse
transcription-quantitative PCR and western blot analyses. Data are
presented as the mean ± standard deviation of three independent
experiments. *P<0.05, **P<0.01, ***P<0.001 vs. BEAS-2B.
EZH2, histone-lysine N-methyltransferase EZH2; FC, fold change;
GSM, Gene Expression Omnibus samples. |
Survival analysis was performed for patients with
lung cancer, using the GSE30219 dataset. The results demonstrated
that high EZH2 expression was associated with a shorter overall
survival time in patients with lung cancer, thus an unfavorable
prognosis, compared with those with low EZH2 expression. The 3- and
5-year survival rates of patients with high EZH2 expression were
lower than those of patients with low EZH2 expression (Fig. 1C).
EZH2 mRNA and protein expression levels were also
determined in the six lung cancer cell lines using RT-qPCR and
western blot analyses. The results show that EZH2 was upregulated
in all the lung cancer cell lines (H1650, PC9, A549, H1299 and
H460) compared with that in the BEAS-2B cell line (Fig. 1D). As the expression level of EZH2
was highest in the A549 cell line, this was used for further
experimentations. Taken together, these results suggest that EZH2
may be associated with tumorigenesis and progression of lung
cancer.
EZH2 knockdown restricts cell
viability and induces apoptosis
To evaluate the function of EZH2 in lung cancer
cells, A549 cells were transfected with siRNA-EZH2, and
non-targeting siRNA was used as the NC. The results demonstrated
that the mRNA and protein expression levels of EZH2 were
significantly decreased in A549 cells transfected with siRNA-EZH2
compared with that in the NC cells, using RT-qPCR and western blot
analyses, respectively (Fig.
2A).
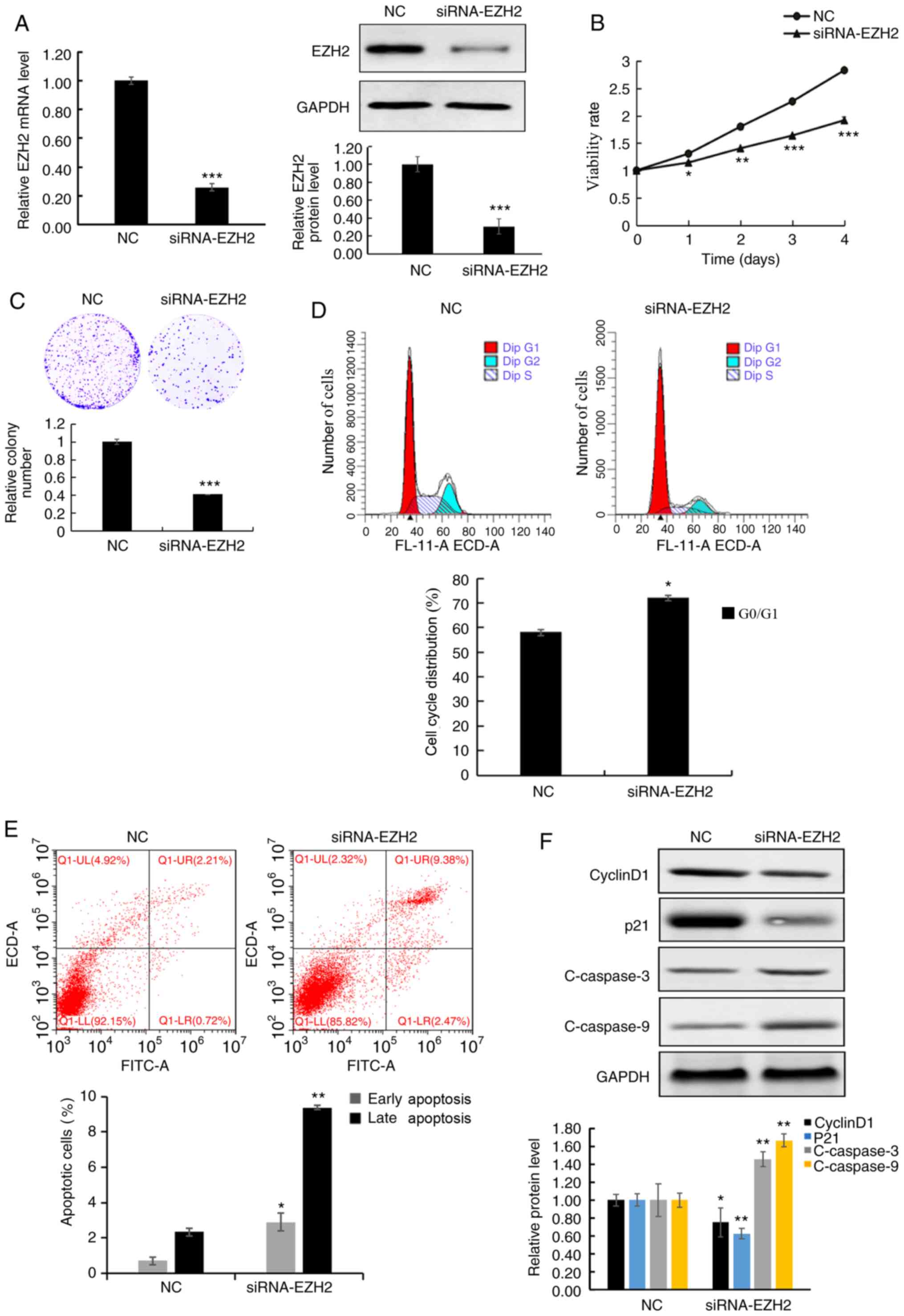 | Figure 2.EZH2 knockdown inhibits cell
viability and induces apoptosis. (A) EZH2 mRNA and protein
expression levels were significantly decreased in cells transfected
with siRNA-EZH2 compared with that in the NC. (B) MTT assay
demonstrated that the viability of A549 cells deceased following
EZH2 knockdown. (C) The colony formation assay was performed using
A549 cells, with or without EZH2 knockdown. (D) The cell cycle
distribution was analyzed using flow cytometry, with or without
EZH2 knockdown and analyzed quantitatively. (E) NC and siRNA-EZH2
transfected cells were stained with Annexin V/propidium iodide for
flow cytometric analysis and analyzed quantitatively. (F) Western
blot analysis of cyclin D1, p21, C-caspase-3 and −9 proteins in
A549 cells, treated with NC and siRNA-EZH2 and analyzed
quantitatively. Data are presented as the mean ± standard deviation
of three independent experiments. *P<0.05, **P<0.01 and
***P<0.001 vs. NC. EZH2, histone-lysine N-methyltransferase
EZH2; si, small interfering; NC, negative control; C, cleaved;
FITC, fluorescein isothiocyanate. |
To determine the biological function of EZH2 in lung
cancer cells, the viability of A549 cells following EZH2 knockdown
was analyzed using MTT and colony-formation assays. The MTT assay
results demonstrated that the cell viability was significantly
decreased in siRNA-EZH2 transfected cells compared with that in the
NC cells (Fig. 2B). Similarly,
colony formation ability was also decreased following EZH2
knockdown (Fig. 2C). In addition,
EZH2 knockdown also induced G1 phase cell cycle arrest,
while the cell percentage in the G2 phase was also
decreased in A549 cells (Fig.
2D).
Flow cytometric analysis was performed to determine
whether the inhibitory effects on cell viability were associated
with apoptosis, following EZH2 knockdown (Fig. 2E). The number of cells in early stage
apoptosis was significantly higher in the cells treated with EZH2
siRNA compared with that in the NC cells, 0.7±0.21 vs. 2.9±0.48%
cells (P<0.05). In addition, the percentage of apoptotic cells
in late stage apoptosis was also significantly increased in
siRNA-EZH2 transfected cells compared with that in the NC cells.
Furthermore, the protein expression levels of the cell cycle
proteins, cyclin D1 and p21 significantly decreased, while the
levels of the apoptosis-associated proteins, C-caspase-3 and −9
significantly increased in cells following EZH2 knockdown (Fig. 2F). These results indicate that EZH2
may play a role in regulating cell viability and apoptosis.
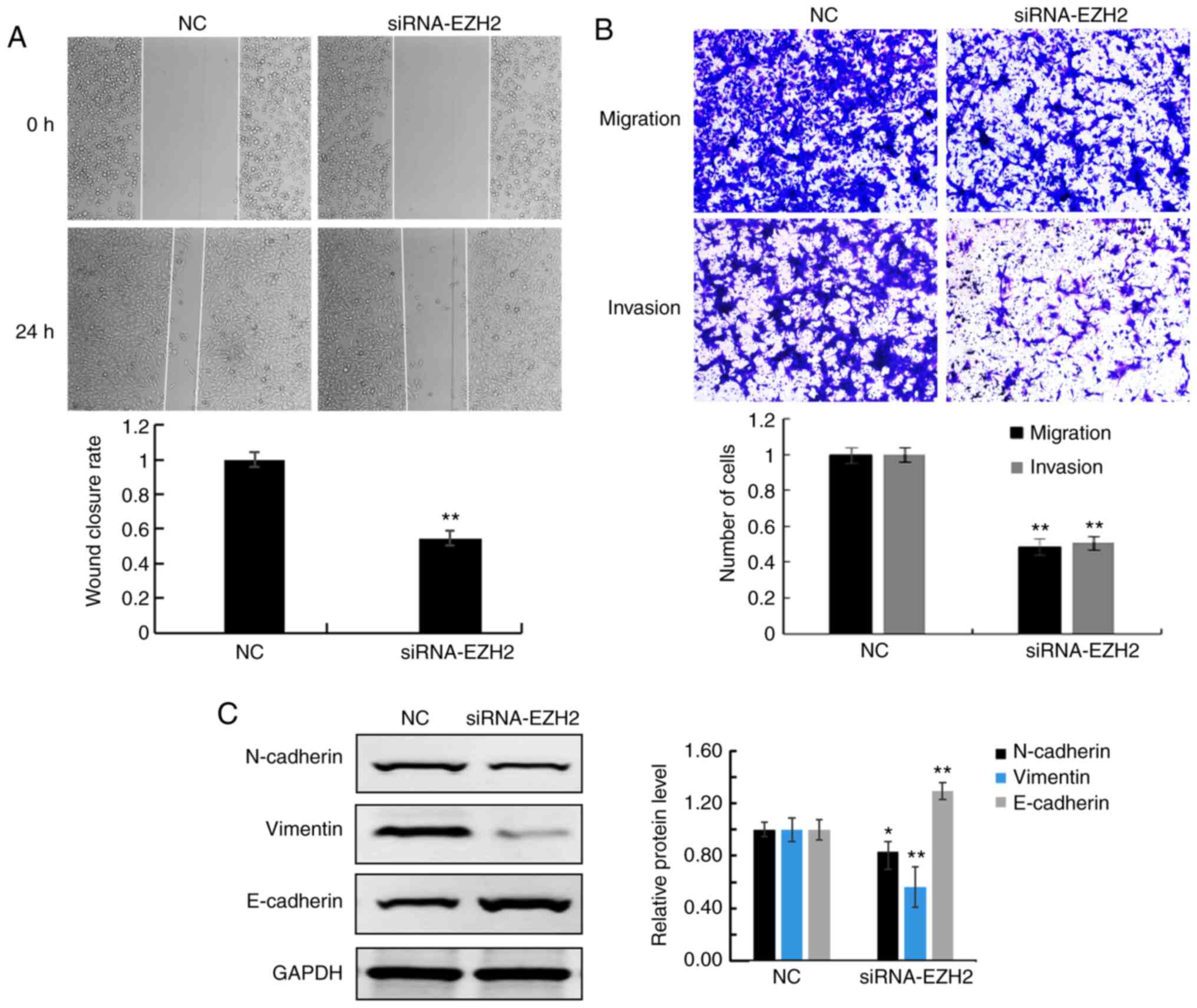
EZH2 knockdown inhibits cell migration
and invasion
The migratory ability of A549 cells was assessed
using wound healing and Transwell assays. Both assays revealed that
the migratory rate was decreased in A549 cells transfected with
EZH2 siRNA compared with that in the NC cells (Fig. 3A and B, respectively). In addition,
the Matrigel assay found that A549 invasion was also decreased in
cells transfected with EZH2 siRNA compared with that in the NC
cells (Fig. 3B). Furthermore,
decreased protein expression levels of epithelial-mesenchymal
transition (EMT)-associated proteins (N-cadherin and vimentin), and
increased levels of E-cadherin were detected in siRNA-EZH2
transfected cells compared with that in NC cells (Fig. 3C).
EZH2 knockdown enhances sensitivity of
lung cancer cells to CDDP by inhibiting viability and enhancing
apoptosis
To further assess the function of EZH2 during
treatment with CDDP, the cell viability of NC and siRNA-EZH2
transfected cells following CDDP treatment was analyzed using the
MTT and colony formation assays. The viability (Fig. 4A) and colony formation ability
(Fig. 4B) of A549 cells transfected
with siRNA-EZH2 were significantly reduced compared with that in NC
cells, following CDDP treatment (0, 2, 4, 8 or 16 µM) and 4 µM CDDP
treatment, respectively. To determine whether the chemo response
effects of EZH2 on A549 cells was associated with apoptosis,
transfected cells following CDDP treatment were detected using flow
cytometry. The number of early stage apoptotic cells in the EZH2
knockdown cells (9.99±1.26%) was significantly increased compared
with that in the NC cells (6.37±0.69%), with 4 µM CDDP for 24 h
(Fig. 4C). In addition, the
percentage of late-stage apoptotic cells was significantly
increased in EZH2 knockdown cells (8.51±0.94%) compared with that
in the NC cells (3.98±0.43%), with 4 µM CDDP for 24 h. Furthermore,
the expression levels of the apoptosis-associated proteins,
C-caspase-3 and −9, increased in siRNA-EZH2 cells treated with CDDP
compared with those in NC cells (Fig.
4D).
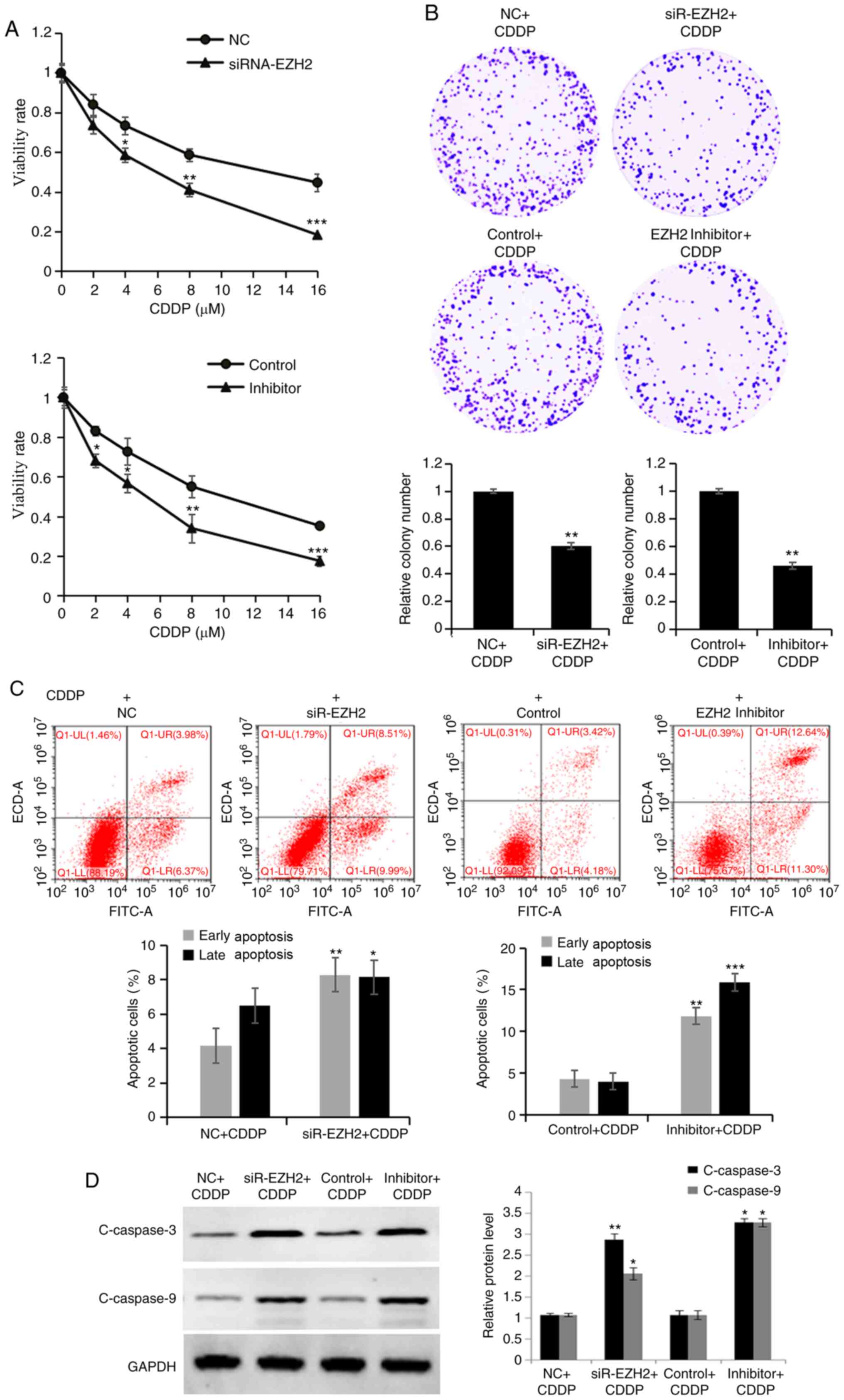 | Figure 4.Downregulation of EZH2 enhances
sensitivity to CDDP by inhibiting cell viability and promoting
apoptosis. (A) MTT and (B) colony formation assays were performed
to detect the proliferative ability of cells transfected with
siRNA-EZH2 or treated with the EZH2 inhibitor, following CDDP
treatment, respectively. (C) A higher apoptosis rate was observed
in cells transfected with siRNA-EZH2 or treated with the EZH2
inhibitor, following CDDP treatment, compared with that in the
control groups. (D) The expression levels of C-caspase-3 and −9
were detected using western blot analysis in cells transfected with
siRNA-EZH2 or treated with the EZH2 inhibitor, following CDDP
treatment. Data are presented as the mean ± standard deviation of
three independent experiments. *P<0.05, **P<0.01 and
***P<0.001 vs. NC+CDDP or Control+CDDP. EZH2, histone-lysine
N-methyltransferase EZH2; si, small interfering; NC, negative
control; CDDP, cisplatin. |
Notably, an inhibitor of EZH2 (EPZ-6438) was used to
simulate EZH2 silencing. A549 cells treated with CDDP (4 µM) alone
or in combination with EPZ-6438 (40 µM) for 24 h were subsequently
analyzed using the MTT, colony formation and apoptosis assays. The
viability (Fig. 4A) and colony
formation ability (Fig. 4B) of A549
cells treated with the EZH2 inhibitor were significantly decreased
compared with those in control cells, following CDDP treatment (0,
2, 4, 8 or 16 µM) and 4 µM CDDP treatment, respectively. The number
of early stage apoptotic cells in cells treated with the EZH2
inhibitor (11.30±1.35%) was significantly increased compared with
that in the control cells (4.18±0.53%); additionally, the
percentage of late-stage apoptotic cells was significantly
increased in cells treated with the EZH2 inhibitor (12.64±1.64%)
compared with that in the control cells (3.42±0.31%), with 4 µM
CDDP for 24 h (Fig. 4C).
Furthermore, the expression levels of the apoptosis-associated
proteins, C-caspase-3 and −9, increased in cells treated with the
EZH2 inhibitor and with CDDP compared with those in control cells
(Fig. 4D). Therefore, the results
were consistent to those observed with siRNA-EZH2. Overall, the
results of the present study demonstrated that EZH2 played a vital
role in CDDP response, and EZH2 inactivation increased
chemosensitivity to chemotherapeutic agents of lung cancer cells.
Thus, EZH2 inhibitor may be used to be enhance chemosensitivity of
lung cancer cells.
Discussion
According to the 2018 Global Cancer Statistics
Report, lung cancer is the world's leading malignant tumor in
morbidity (11.6%) and mortality (18.4%) rates (18). Both surgical resection or
chemotherapy have become less effective in the treatment of lung
cancer, resulting in the recurrence (19). Thus, it remains critical to research
molecular targets associated with lung cancer progression or
chemoresistance, to develop novel therapeutic strategies for cancer
treatment. Taken together, the results of the present study
demonstrated that EZH2 silencing may sensitize lung cancer cells to
the chemotherapeutic agent, CDDP. Thus, EZH2 may act as a novel
target of lung cancer progression.
EZH2 has been demonstrated to act as a
transcriptional inhibitor and is a polycomb group protein (20). Previous studies have reported that
EZH2 is overexpressed in several types of cancer (21,22). For
example, EZH2 promotes viability and migration of breast cancer
cells (23). The present study
screened the GEO database using the key words ‘lung cancer’, ‘Homo
sapiens’ and ‘Expression profiling by array or sequencing’,
identifying the GSE19804 dataset with sufficient samples for data
analysis, including 60 adjacent non-tumor and 60 tumor samples.
Both bioinformatics and western blot analyses demonstrated that the
expression level of EZH2 was increased in lung carcinoma tissues
and cell lines, which could promote carcinogenesis in lung cancer.
Furthermore, survival analysis indicated that increased levels of
EZH2 in lung carcinoma was associated with poor prognosis,
suggesting that higher levels of EZH2 may be used as a latent
molecular biomarker for the prognosis of cancer. After a large
number of clinical observations and data statistics (24,25), it
was revealed that the recurrence and metastasis of most tumor
patients (80%) occurred ~3 years after radical surgery, while those
of 10% of patients occurred ~5 years after treatment. Therefore,
the present study analyzed the 3- and 5-year survival rates of
patients with high EZH2 expression and low EZH2 expression. Similar
results have also been demonstrated in other types of human
malignant cancer, including colon (26), breast (27), lung (28), bladder (29), ovarian (30) and prostate cancer (31). Taken together, the results of the
present study highlight a novel potential role of EZH2 as a
molecular target for drug development in lung cancer.
Wang et al (32) reported that siRNA-mediated
suppression of EZH2 in bladder cancer induces apoptosis; however,
Rao et al (33) demonstrated
that siRNA-EZH2 has no effect on apoptosis in ovarian cancer. The
results of the present study are consistent with the findings by
Wee et al (34), suggesting
that the function of EZH2 varies in different types of cancer.
Furthermore, EZH2 knockdown in A549 cells suppressed viability,
whilst inducing apoptosis and producing cell cycle arrest in the
G1 phase. Taken together, these results confirm that
EZH2 was associated with both tumor cell viability and apoptosis in
lung cancer.
Metastasis is a complex process and can cause
difficulties in the treatment of lung cancer (35), which has been defined as a multi-step
process by which cell invasion contributes to metastasis (36). Previous studies have demonstrated
that high expression levels of EZH2 was associated with cancer
recurrence (37), distant metastasis
(38), invasion (39) and angiogenesis (40) in numerous types of cancer, including
melanoma and breast cancer. EZH2 is an adhesion protein expressed
in breast and gastric cancer that promotes cancer metastasis by
promoting ribosome synthesis; ribosomes are the cellular components
that produce proteins, and their increased synthesis provides the
conditions for cell metastasis (41). The results of the present study
indicated that transfection with siRNA-EZH2 in lung cancer cells
significantly inhibited invasion and migration. Furthermore, the
expression levels of the EMT pathway proteins verified the
molecular mechanisms of metastasis. Thus, EZH2 is a protein
involved in the migration and invasion of lung cancer cells,
whereby increased EZH2 expression may have a selective advantage on
the migratory and invasive abilities of lung cancer cells. The
detection of EZH2 expression can be performed as an additional tool
to identify patients with lung cancer, with tumor progression and
metastatic risk.
EPZ-6438 is a novel EZH2-specific inhibitor, which
has demonstrated efficacy in different types of cancer, such as
non-Hodgkin's lymphoma (42,43). In the present study, EPZ-6438 was
used in combination with CDDP, which was demonstrated to have an
additive effect on lung cancer cells. Treatment with the EZH2
inhibitor enhanced the CDDP-induced inhibition of cell viability
and promoted apoptosis. However, further investigations into the
molecular mechanism underlying EZH2 in the response of CDDP are
required. Notably, the results of the present study highlight the
potential applications of EZH2 inhibitors in future clinical
treatment interventions and anti-chemotherapy resistance. Further
studies should perform additional downstream experiments to reveal
the underlying molecular mechanism and should verify the present
results using animal models, which are the limitations of the
present study.
In conclusion, the results of the present study
demonstrated that EZH2 played a vital role in molecular diagnosis
and in the CDDP response of lung cancer. Suppression of EZH2
inhibited tumorigenesis and enhanced chemosensitivity, thus
suggesting that EZH2 may be used as a novel molecular therapeutic
target for lung carcinoma.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the Shanghai Sailing
Program (grant no. 17YF1415800) and the National Natural Science
Foundation of China (grant no. 81802803).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article. The datasets generated and/or
analyzed during the current study (GSE19804 and GSE30219) are
available in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/).
Authors' contributions
ZC, WW and ZD designed the study and analyzed the
data. WW, HW, WZ and YH performed the experiments. ZC drafted the
initial manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pietrzak S, Wójcik J, Scott RJ, Kashyap A,
Grodzki T, Baszuk P, Bielewicz M, Marciniak W, Wójcik N, Dębniak T,
et al: Influence of the selenium level on overall survival in lung
cancer. J Trace Elem Med Biol. 56:46–51. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang L, Pu D, Liu D, Wang Y, Luo W, Tang
H, Huang Y and Li W: Identification and validation of novel
circulating biomarkers for early diagnosis of lung cancer. Lung
Cancer. 135:130–137. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Peng A, Li G, Xiong M, Xie S and Wang C:
Role of surgery in patients with early stage small-cell lung
cancer. Cancer Manag Res. 11:7089–7101. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nix MG, Rowbottom CG, Vivekanandan S,
Hawkins MA and Fenwick JD: Chemoradiotherapy of locally-advanced
non-small cell lung cancer: Analysis of radiation dose-response,
chemotherapy and survival-limiting toxicity effects indicates a low
α/β ratio. Radiother Oncol. 143:58–65. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Naghizadeh S, Mohammadi A, Baradaran B and
Mansoori B: Overcoming multiple drug resistance in lung cancer
using siRNA targeted therapy. Gene. 714:1439722019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang X, Hua Y, Xu G, Deng S, Yang D and
Gao X: Targeting EZH2 for glioma therapy with a novel
nanoparticle-siRNA complex. Int J Nanomedicine. 14:2637–2653. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sellers WR and Loda M: The EZH2 polycomb
transcriptional repressor-a marker or mover of metastatic prostate
cancer? Cancer Cell. 2:349–350. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu J, Wang Z, Lu W, Jiang H, Lu J, Qiu J
and Ye G: EZH2 promotes gastric cancer cells proliferation by
repressing p21 expression. Pathol Res Pract. 215:1523742019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chien YC, Liu LC, Ye HY, Wu JY and Yu YL:
EZH2 promotes migration and invasion of triple-negative breast
cancer cells via regulating TIMP2-MMP-2/-9 pathway. Am J Cancer
Res. 8:422–434. 2018.PubMed/NCBI
|
|
10
|
Guo S, Li X, Rohr J, Wang Y, Ma S, Chen P
and Wang Z: EZH2 overexpression in different immunophenotypes of
breast carcinoma and association with clinicopathologic features.
Diagn Pathol. 11:412016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han L, Zhang HC, Li L, Li CX, Di X and Qu
X: Downregulation of long noncoding RNA HOTAIR and EZH2 induces
apoptosis and inhibits proliferation, invasion, and migration of
human breast cancer cells. Cancer Biother Radiopharm. 33:241–251.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mu Z, Li H, Fernandez SV, Alpaugh KR,
Zhang R and Cristofanilli M: EZH2 knockdown suppresses the growth
and invasion of human inflammatory breast cancer cells. J Exp Clin
Cancer Res. 32:702013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC,
Lin CW, Shih JY, Yang PC, Hsiao CK, Lai LC and Chuang EY:
Identification of a novel biomarker, SEMA5A, for non-small cell
lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers
Prev. 19:2590–2597. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dong S, Men W, Yang S and Xu S:
Identification of lung adenocarcinoma biomarkers based on
bioinformatic analysis and human samples. Oncol Rep. 43:1437–1450.
2020.PubMed/NCBI
|
|
15
|
Rousseaux S, Debernardi A, Jacquiau B,
Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY,
Lantuejoul S, Hainaut P, et al: Ectopic activation of germline and
placental genes identifies aggressive metastasis-prone lung
cancers. Sci Transl Med. 5:186ra662013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Singh C and Roy-Chowdhuri S: Quantitative
real-time PCR: Recent advances. Methods Mol Biol. 1392:161–176.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kramer N, Walzl A, Unger C, Rosner M,
Krupitza G, Hengstschläger M and Dolznig H: In vitro cell migration
and invasion assays. Mutat Res. 752:10–24. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Khorrami M, Jain P, Bera K, Alilou M,
Thawani R, Patil P, Ahmad U, Murthy S, Stephans K, Fu P, et al:
Predicting pathologic response to neoadjuvant chemoradiation in
resectable stage III non-small cell lung cancer patients using
computed tomography radiomic features. Lung Cancer. 135:1–9. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Isla D, De Las Peñas R, Insa A, Marsé R,
Martínez-Banaclocha N, Mut P, Morán T, Sala MA, Massuti B, Ortega
AL, et al: Oral vinorelbine versus etoposide with cisplatin and
chemo-radiation as treatment in patients with stage III non-small
cell lung cancer: A randomized phase II (RENO study). Lung Cancer.
135:161–168. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ang PP, Tan GC, Karim N and Wong YP:
Diagnostic value of the EZH2 immunomarker in malignant effusion
cytology. Acta Cytol. 64:248–255. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Drelon C, Berthon A, Mathieu M, Ragazzon
B, Kuick R, Tabbal H, Septier A, Rodriguez S, Batisse-Lignier M,
Sahut-Barnola I, et al: EZH2 is overexpressed in adrenocortical
carcinoma and is associated with disease progression. Hum Mol
Genet. 25:2789–2800. 2016.PubMed/NCBI
|
|
22
|
Herviou L, Jourdan M, Martinez AM, Cavalli
G and Moreaux J: EZH2 is overexpressed in transitional
preplasmablasts and is involved in human plasma cell
differentiation. Leukemia. 33:2047–2060. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Puppe J, Opdam M, Schouten PC, Jóźwiak K,
Lips E, Severson T, van de Ven M, Brambillasca C, Bouwman P, van
Tellingen O, et al: EZH2 Is overexpressed in BRCA1-like breast
tumors and predictive for sensitivity to high-dose platinum-based
chemotherapy. Clin Cancer Res. 25:4351–4362. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yan X, Jiao SC, Zhang GQ, Guan Y and Wang
JL: Tumor-associated immune factors are associated with recurrence
and metastasis in non-small cell lung cancer. Cancer Gene Ther.
24:57–63. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fang D, Zhang D and Zhang R: Study of
recurrence and metastasis after radical resection of carcinoma of
the lung. Zhonghua Zhong Liu Za Zhi. 21:284–286. 1999.(In Chinese).
PubMed/NCBI
|
|
26
|
Katona BW, Liu Y, Ma A, Jin J and Hua X:
EZH2 inhibition enhances the efficacy of an EGFR inhibitor in
suppressing colon cancer cells. Cancer Biol Ther. 15:1677–1687.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pourakbar S, Pluard TJ, Accurso AD and
Farassati F: Ezh2, a novel target in detection and therapy of
breast cancer. Onco Targets Ther. 10:2685–2687. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Geng J, Li X, Zhou Z, Wu CL, Dai M and Bai
X: EZH2 promotes tumor progression via regulating VEGF-A/AKT
signaling in non-small cell lung cancer. Cancer Lett. 359:275–287.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Martínez-Fernández M, Rubio C, Segovia C,
López-Calderón FF, Dueñas M and Paramio JM: EZH2 in bladder cancer,
a promising therapeutic target. Int J Mol Sci. 16:27107–27132.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yi X, Guo J, Guo J, Sun S, Yang P, Wang J,
Li Y, Xie L, Cai J and Wang Z: EZH2-mediated epigenetic silencing
of TIMP2 promotes ovarian cancer migration and invasion. Sci Re.
7:35682017.
|
|
31
|
Chinaranagari S, Sharma P and Chaudhary J:
EZH2 dependent H3K27me3 is involved in epigenetic silencing of ID4
in prostate cancer. Oncotarget. 5:7172–7182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Y, Xiang W, Wang M, Huang T, Xiao X,
Wang L, Tao D, Dong L, Zeng F and Jiang G: Methyl jasmonate
sensitizes human bladder cancer cells to gambogic acid-induced
apoptosis through down-regulation of EZH2 expression by miR-101. Br
J Pharmacol. 171:618–635. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rao ZY, Cai MY, Yang GF, He LR, Mai SJ,
Hua WF, Liao YJ, Deng HX, Chen YC, Guan XY, et al: EZH2 supports
ovarian carcinoma cell invasion and/or metastasis via regulation of
TGF-beta1 and is a predictor of outcome in ovarian carcinoma
patients. Carcinogenesis. 31:1576–1583. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wee ZN, Li Z, Lee PL, Lee ST, Lim YP and
Yu Q: EZH2-mediated inactivation of IFN-γ-JAK-STAT1 signaling is an
effective therapeutic target in MYC-driven prostate cancer. Cell
Rep. 8:204–216. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dar WR and Mir MH: Pontine metastasis as
an initial presentation of lung cancer. Neurol India. 67:918–920.
2019.PubMed/NCBI
|
|
36
|
Aminorroaya A, Khoshniatnikoo M,
Farrokhpour H, Vafaeimanesh J and Bagherzadeh M: Squamous cell
carcinoma of the lung and pulmonary metastasis of papillary thyroid
carcinoma: A case report. J Med Case Rep. 13:2592019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nakagawa S, Okabe H, Ouchi M, Tokunaga R,
Umezaki N, Higashi T, Kaida T, Arima K, Kitano Y, Kuroki H, et al:
Enhancer of zeste homolog 2 (EZH2) regulates tumor angiogenesis and
predicts recurrence and prognosis of intrahepatic
cholangiocarcinoma. HPB (Oxford). 20:939–948. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Alford SH, Toy K, Merajver SD and Kleer
CG: Increased risk for distant metastasis in patients with familial
early-stage breast cancer and high EZH2 expression. Breast Cancer
Res Treat. 132:429–437. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Manning CS, Hooper S and Sahai EA:
Intravital imaging of SRF and notch signalling identifies a key
role for EZH2 in invasive melanoma cells. Oncogene. 34:4320–4332.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Crea F, Fornaro L, Bocci G, Sun L, Farrar
WL, Falcone A and Danesi R: EZH2 inhibition: Targeting the
crossroad of tumor invasion and angiogenesis. Cancer Metastasis
Rev. 31:753–761. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu Z, Sun Y, Guo Y, Qin G, Mu S, Fan R,
Wang B, Gao W, Wu H, Wang G and Zhang Z: NF-YA promotes invasion
and angiogenesis by upregulating EZH2-STAT3 signaling in human
melanoma cells. Oncol Rep. 35:3630–3638. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen J, Chen X, Yao J, Li M and Yang X:
The combination of decitabine and EPZ-6438 effectively facilitate
adipogenic differentiation of induced pluripotent stem cell-derived
mesenchymal stem cells. Biochem Biophys Res Commun. 516:307–312.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Knutson SK, Kawano S, Minoshima Y,
Warholic NM, Huang KC, Xiao Y, Kadowaki T, Uesugi M, Kuznetsov G,
Kumar N, et al: Selective inhibition of EZH2 by EPZ-6438 leads to
potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol
Cancer Ther. 13:842–854. 2014. View Article : Google Scholar : PubMed/NCBI
|