Introduction
Breast cancer is one of the most common types of
cancer in the world. Breast cancer cases in the world have
continued to increase from 2005, with 12% of women expected to
develop this disease in their lifetime (1). Ongoing research on breast cancer is
trying to improve its treatment to assist in dealing with the issue
(2).
Ring finger protein 8 (RNF8) is a gene in the human
body that encodes the E3 ubiquitin-protein ligase RNF8 enzyme
(3). It is involved in various
activities of the body associated with immune system functions and
DNA repair (4,5). Previous studies have indicated that
RNF8 is a damage-responsive protein that integrates both protein
phosphorylation and ubiquitylation, serving an essential role in
disease treatment (6,7). Additionally, depletion of RNF8 in the
body can lead to cell cycle arrest and inhibition of cell
proliferation (8).
The vital role of RNF8 in diseases is evident in the
treatment of various types of cancer (9). It was reported that RNF8 can act as a
co-activator to promote cell proliferation of estrogen receptor
(ER)α-positive breast cancer via post-transcriptional signaling
pathways (8). Furthermore, RNF8 was
reported to promote epithelial-mesenchymal transition of breast
cancer cells and facilitate the metastasis of breast cancer cells
in vivo (10). By contrast,
another study demonstrated that mice with RNF8 deficiency have an
increased risk of mammary tumorigenesis, since RNF8 can regulate
Notch signaling by ubiquitylating the active NOTCH1 protein,
leading to its degradation (7),
while abnormal activation of Notch signaling would promote the risk
of breast cancer. Additionally, Gao et al (11) revealed that RNF8 can negatively
regulate the activation of NF-κB, of which excess activation can
lead to cancer. Therefore, there are controversial roles of RNF8 in
tumorigenesis, and the function of RNF8 in breast cancer remains
unclear.
In the present study, a series of bioinformatic
approaches were used to analyze breast cancer sequencing data to
explore the underlying mechanism of RNF8 in breast cancer.
Subsequently, further experiments were performed to verify the
bioinformatic results. The present study suggested that breast
cancer progression may be closely associated with RNF8
expression
Materials and methods
Bioinformatics analysis
RNA sequencing data from TP53-mutant mammary tumor
cells was downloaded from the GSE76075 dataset containing mRNA
sequencing data for two independent mouse TP53-mutant breast cancer
cell lines (RNF8−/− and RNF8-wild-type) (12) from the Gene Expression Omnibus (GEO)
database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76075).
The gene names and gene count files were obtained, and the GFOLD
values of each gene were calculated using the open source GFOLD
software v1.1.4 (13). A total of
500 differentially expressed genes with the largest (upward) and
minimum (downward) GFOLD values were selected, and Gene Ontology
(GO) enrichment analysis was performed on these genes using the
Database for Annotation, Visualization and Integrated Discovery
(DAVID; http://david.ncifcrf.gov/).
Furthermore, the GO enrichment results of each group were clustered
using the GOSemSim v3.11 R package (http://www.bioconductor.org/packages/release/bioc/html/GOSemSim.html)
(14), and the clustering results
between the groups were visualized using the ggtree v3.11 R package
(http://www.bioconductor.org/packages/release/bioc/html/ggtree.html)
(15). Subsequently, survival
analysis of key genes was performed using Kaplan-Meier (KM) Plotter
(http://kmplot.com/analysis/) with the
log-rank test.
Cell culture
The HCC1937 cell line was purchased from the China
Infrastructure of Cell Line Resources (Institute of Basic Medical
Sciences; Chinese Academy of Sciences). The HCC1937 cell line was
selected due to its TP53-negative status, which can eliminate the
uncertainty derived from TP53 mutations. Using the HCC1937 cell
line helps to maintain consistency between human and mouse cell
models. The cells were cultured in the RPMI 1640 (Thermo Fisher
Scientific, Inc.) medium containing 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C in the presence of 5% CO2.
Cell transfection
According to the sequence of the RNF8 gene, three
small interfering (si)RNAs (Table I)
with different target sequences of RNF8 were synthesized by
Shanghai Shenggong Biology Engineering Technology Service, Ltd., to
decrease the expression levels of the RNF8 gene in the TP53-mutant
HCC1937 cells. Untreated cells were used as negative control 1.A
non-specific scrambled siRNA (50 nM) was used as negative control 2
and specific siRNAs (siRNA1, siRNA2 and siRNA3; 50 nM), as well as
a siRNA mix (consisting of all three specific siRNAs) against RNF8
were directly transfected into HCC1937 cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Transfected cells were cultured at 37°C in the presence of 5%
CO2 for 48 h, after which subsequent experiments were
performed.
 | Table I.siRNA target sequences. |
Table I.
siRNA target sequences.
| siRNAs | Sequence |
|---|
| siRNA1 |
5′-UGCGGAGUAUGAAUAUGAAUU-3′ |
| siRNA2 |
5′-GGACAAUUAUGGACAACAAGA-3′ |
| siRNA3 |
5′-UAAGGAGAAUGCGGAGUAT-3′ |
| Control |
5′-GGUAUCGGCUUAUCAGUCCGAGUAATT-3′ |
RNA isolation, reverse
transcription-quantitative (RT-q)PCR assay and western blot
analysis
Total RNA from cells was extracted using the
MiniBEST Universal RNA Extraction kit (Takara Bio, Inc.), and RT
was performed using the PrimeScript RT reagent kit (Takara Bio,
Inc.) according to the manufacturer's protocol. The resulting cDNA
was stored at −80°C until further use. For qPCR, the TB Green Fast
qPCR mix (Takara Bio, Inc.) was used to quantify the expression
levels of the corresponding genes on the ABI 7500 (Applied
Biosystems; Thermo Fisher Scientific, Inc.), and the primer
sequences of the detected genes are shown in Table II. The thermocycling conditions
comprised an initial denaturation step at 95°C for 30 sec, followed
by 40 cycles of amplification at 95°C for 5 sec and 60°C for 30
sec. The relative transcript levels were quantified using the
2−ΔΔCq method (16) with
GAPDH used as an internal control.
 | Table II.Primer sequences of the target
genes. |
Table II.
Primer sequences of the target
genes.
| Gene name | Forward primer | Reverse primer |
|---|
| RNF8 |
5′-AAGCGACGGCAGCAGAA-3′ |
5′-AGCACCTTCACCTTCCTCAG-3′ |
| GAPDH |
5′-GCACCGTCAAGGCTGAGAAC-3′ |
5′-TGGTGAAGACGCCAGTGGA-3′ |
| EPHB2 |
5′-TGAGTGCCCTCAGATGGTCAA-3′ |
5′-AGGGCAGGGTATCACAGTGAATG-3′ |
| LHX2 |
5′-GGAAGCATCTACTGCAAGGAAG-3′ |
5′-GAGGTGATAAACCAAGTCCCG-3′ |
| BRSK1 |
5′-CGGGAACTTCATCTCCTTGGAC-3′ |
5′-ACAGCACACTGTGACTCAGGCT-3′ |
For the western blot experiments, cells were
harvested and collected via centrifugation at 800 × g for 3 min at
room temperature. Cell proteins were lysed in NP-40 Lysis-Buffer
[150 mM NaCl, 1% NP-40 and 50 mM Tris (pH 8.0)] and analyzed via
BCA protein assay (Beyotime Institute of Biotechnology). Proteins
were denatured by heating for 5 min at 85°C, separated via 12%
SDS-PAGE (50 µg protein/lane) and transferred to PVDF membranes.
Subsequently, the membranes were blocked with 5% BSA (Beyotime
Institute of Biotechnology) at room temperature for 30 min and
incubated with anti-RNF8 (1:1,000; EMD Millipore; cat. no. 09-813),
anti-GAPDH (1:5,000; BIOSS; cat. no. bsm-33033M), anti-EPH receptor
B2 (EPHB2; 1:1,000; Sino Biological, Inc.; cat. no. 100091-T08)
anti-LIM homeobox 2 (LHX2; 1:1,000; BIOSS; cat. no. bs-11200R;) and
anti-BR serine/threonine kinase 1 (BRSK1; 1:1,000; BIOSS; cat. no.
bs-7905R;) primary antibodies at 4°C overnight. After washing three
times with PBS-Tween (0.1% Tween-20), membranes were incubated with
HRP-conjugated goat anti-rabbit IgG (1:10,000; BIOSS; cat. no.
bs-0295G-HRP) and goat anti-mouse IgG secondary antibodies
(1:10,000; BIOSS; cat. no. bs-0368G-HRP) at room temperature for 2
h. ECL luminous solution (Bio-Rad Laboratories, Inc.) was used to
detect the bands.
Cell proliferation assay
Cell Counting Kit-8 (CCK-8; Takara Bio, Inc.) was
used to evaluate cell proliferation. Briefly, 5×102
cells/well were seeded into 96-well plates and cultured in medium
containing 10% FBS for 24, 48, 72 or 96 h at 37°C in the presence
of 5% CO2. After 2 h of culture with the CCK-8 reagent,
the cells were analyzed at a wavelength of 450 nm using a
microplate reader.
Flow cytometric assay
The pretreated cells were harvested by
trypsinization and fixed with 70% ethanol at 4°C for 2 h.
Subsequently, the cells were stained with 10 µl Annexin V-FITC and
5 µl propidium iodide (both Shanghai Yeasen Biotechnology Co.,
Ltd.) for 1 h at room temperature. Finally, cells were analyzed
using a FACS instrument (FACSCelesta; BD Biosciences) and
corresponding data were analyzed using the FlowJo v10.6.2 software
(FlowJo LLC).
Statistical analysis
All experiments were performed in triplicate.
GraphPad Prism 8.0 (GraphPad Software Inc.) was used for
statistical analysis. The data are presented as the mean ± SD. The
difference between two groups was analyzed by unpaired Student's
t-test, while the comparisons among >2 groups were performed by
one-way ANOVA followed by Bonferroni's multiple comparisons test as
the post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Transcriptome analysis of
RNF8−/− TP53-mutant cells
To evaluate the signaling pathways affected by
RNF8-knockdown in TP53-mutant breast cancer cells, the GSE76075
dataset was downloaded from the GEO database, containing mRNA
sequencing data for two independent mouse TP53-mutant breast cancer
cell lines (RNF8−/− and RNF8-wild-type). The reads per
kilobase of exon per million mapped sequence reads (RPKM) values
were used to quantify the expression intensity of the genes, and
the GFOLD values were used to evaluate the degree of differential
gene expression, and a heat map was used to present the
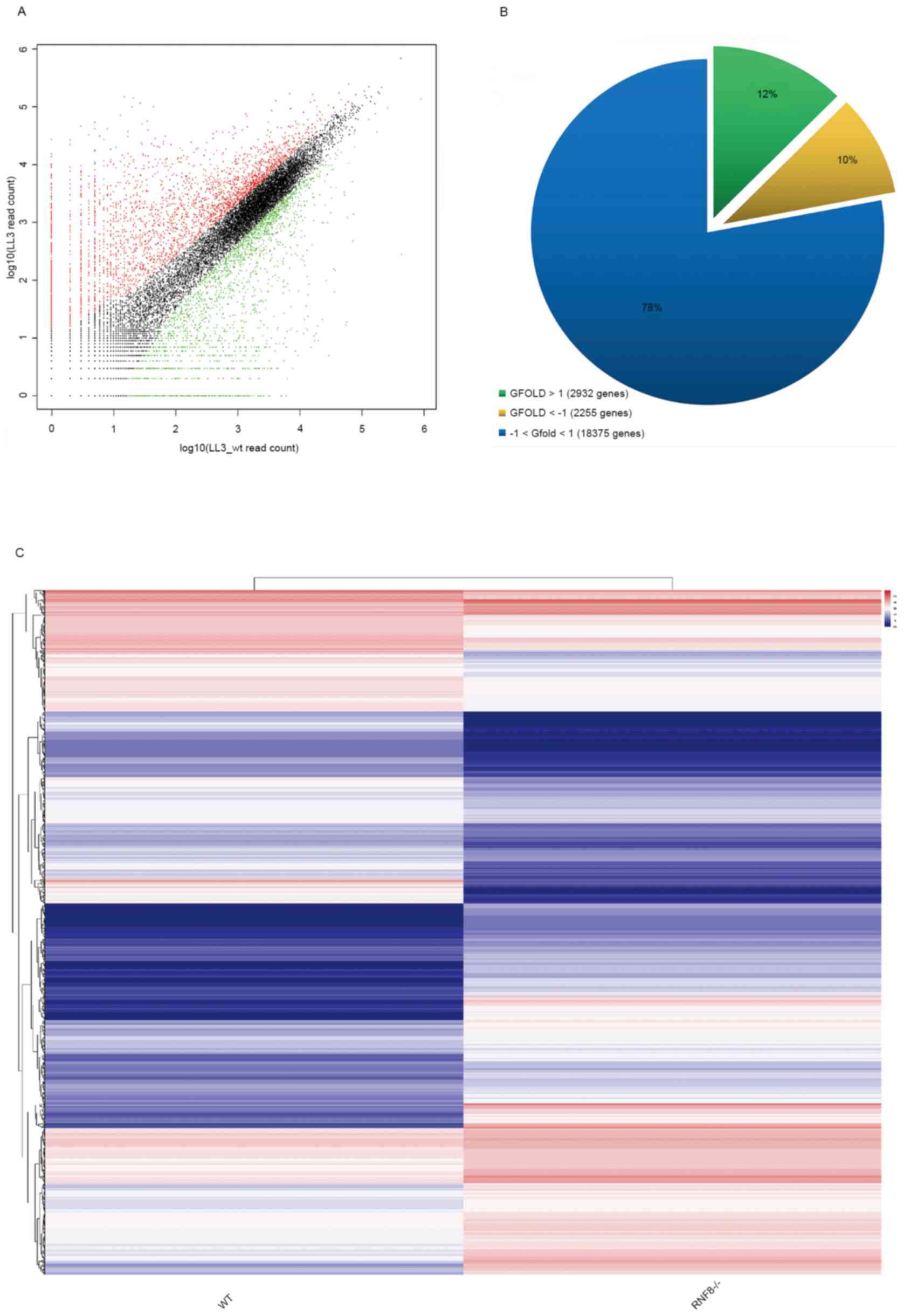
differential expression between the two cell lines. The results
revealed that the GFOLD values of most of the genes between the two
samples had different expression patterns (Fig. 1A). In addition, GFOLD values >1 or
<-1 were used as the threshold for differential expression. The
results revealed that there were 2,932 upregulated genes (~12%) and
2,255 downregulated genes (~10%) in RNF8−/− cells
compared with in wild-type cells (Fig.
1B and C).
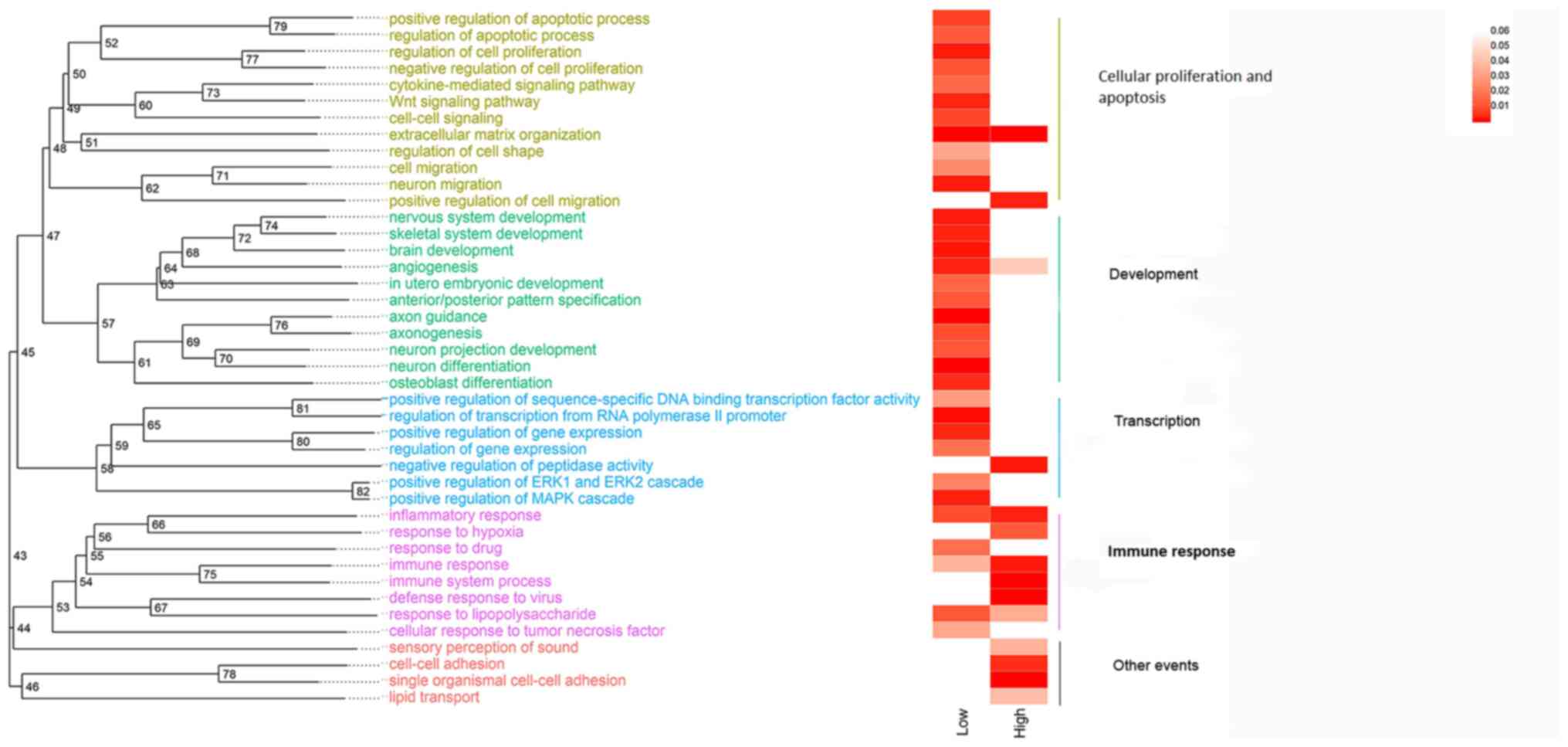
Enrichment and clustering
To further analyze the molecular mechanism of RNF8
in TP53-mutant breast cancer, a GO enrichment analysis of the top
500 downregulated and upregulated genes among the differentially
expressed genes was performed using DAVID. The GO analysis yielded
a large number of terms, making it difficult to quickly extract
effective information; therefore, statistically significant GO
terms were clustered by semantic similarity analysis. The results
revealed that in RNF8-knockout TP53-mutant breast cancer cells,
downregulated genes were significantly enriched in several pathways
involved in cell proliferation and apoptosis regulation,
development and transcription regulation, while upregulated genes
were mainly enriched in immune response-associated pathways
(Fig. 2).
Survival analysis based on key
genes
A previous study has demonstrated that RNF8 is
involved in the reproductive development of the body (17). Therefore, the present study
investigated whether RNF8 regulated genes in development-associated
pathways in TP53-knockout breast cancer cells. A statistical
analysis of all the genes enriched in the GO ‘development’ cluster
was performed, and genes that appeared in >3 GO terms were
selected. Finally, a total of 10 genes were obtained, namely Myosin
Heavy Chain 10, Paired Box 6, LHX2, GATA Binding Protein 3,
Fibroblast Growth Factor Receptor 1 (FGFR1), EPHB2,
Sphingosine-1-Phosphate Receptor 1, Empty Spiracles Homeobox 2,
BRSK1 and GDNF family receptor α-2. Subsequently, the association
between these 10 genes and the prognosis in patients with breast
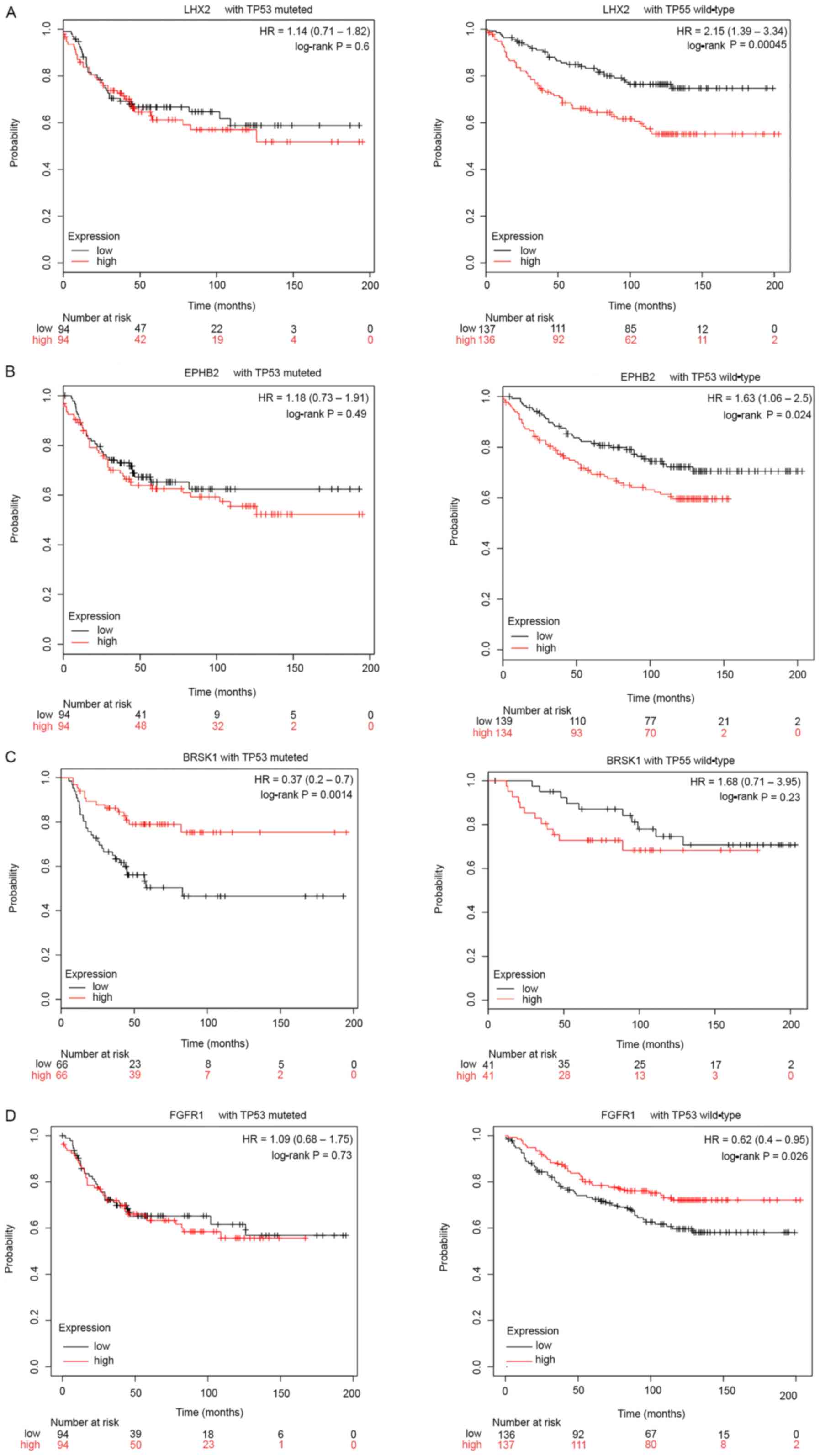
cancer was analyzed using KM Plotter (Table III). The results revealed that in
patients with TP53-wild-type breast cancer, the prognosis in
patients with low expression levels of LHX2 and EPHB2 was
significantly improved compared with that in patients with high
expression (LHX2, P=0.00045; EPHB2, P=0.024; Fig. 3A and B), while the prognosis in
patients with low expression levels of FGFR1 was significantly
decreased compared with that in patients with high expression
(FGFR1, P=0.026; Fig. 3D). In
patients with TP53-mutant breast cancer, there was no significant
difference in survival status, except for the prognosis in patients
with high BRSK1 expression, which was significantly improved
compared with that in patients with low expression (P=0.0014;
Fig. 3C).
 | Table III.Association between 10 differentially
expressed genes and the prognosis in patients with breast
cancer. |
Table III.
Association between 10 differentially
expressed genes and the prognosis in patients with breast
cancer.
| Gene | Count | Frequency, % | GFOLD | Rank | Kaplan-Meier
analysis P-value (TP53-mutated) | Kaplan-Meier
analysis P-value (TP53-wild-type) |
|---|
| MYH10 | 5 | 45.45 | −6.07418 | 196 | 0.57000 | 0.77000 |
| PAX6 | 4 | 36.36 | −7.30754 | 98 | 0.17000 | 0.84000 |
| LHX2 | 4 | 36.36 | −7.12856 | 116 | 0.60000 | 0.00045 |
| GATA3 | 4 | 36.36 | −4.67309 | 372 | 0.37000 | 0.62000 |
| FGFR1 | 4 | 36.36 | −4.43547 | 418 | 0.81000 | 0.05800 |
| EPHB2 | 3 | 27.27 | −7.27086 | 101 | 0.49000 | 0.02400 |
| S1PR1 | 3 | 27.27 | −7.51289 | 82 | 0.52000 | 0.74000 |
| EMX2 | 3 | 27.27 | −5.30267 | 286 | 0.86000 | 0.51000 |
| BRSK1 | 3 | 27.27 | −6.58582 | 153 | 0.00140 | 0.23000 |
| GDNF | 3 | 27.27 | −5.50851 | 254 | 0.61000 | 0.66000 |
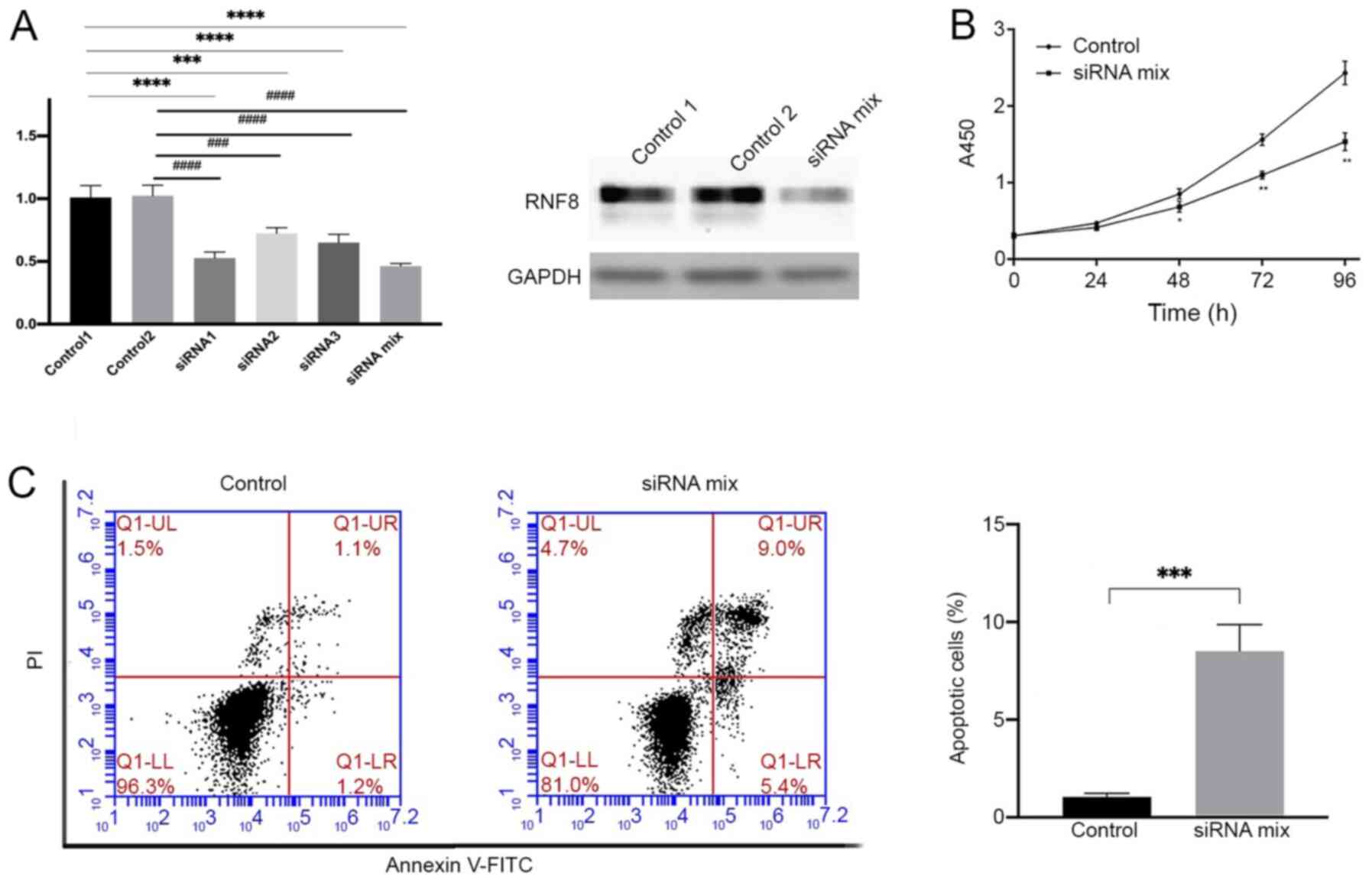
RNF8-knockdown inhibits cell
proliferation and promotes apoptosis in human TP53-mutant breast
cancer cells
To further elucidate the role of RNF8 in TP53-mutant
breast cancer, RNF8 mRNA and protein expression was knocked down in
the TP53-mutant human breast cancer HCC1937 cell line using three
different siRNAs and a siRNA mix consisting of a; three siRNAs
(Fig. 4A) In order to achieve the
best interfering effect, the siRNA mix was used in subsequent
experiments. The results demonstrated that RNF8-knockdown in
HCC1937 cells significantly decreased cell proliferation compared
with that in control cells (Fig.
4B). Since the aforementioned bioinformatics analysis revealed
that downregulated genes after RNF8-knockout were mainly enriched
in apoptosis-associated signaling pathways, the effect of
RNF8-knockdown in siRNA-treated HCC1937 cells was compared with
that in control siRNA-treated HCC1937 cells using flow cytometry.
The results demonstrated that apoptosis in HCC1937 cells with
RNF8-knockdown was significantly higher compared with that in
control cells (Fig. 4C). Overall,
the present results indicated that downregulation of RNF8
expression inhibited proliferation and enhanced apoptosis in
TP53-mutant breast cancer cells.
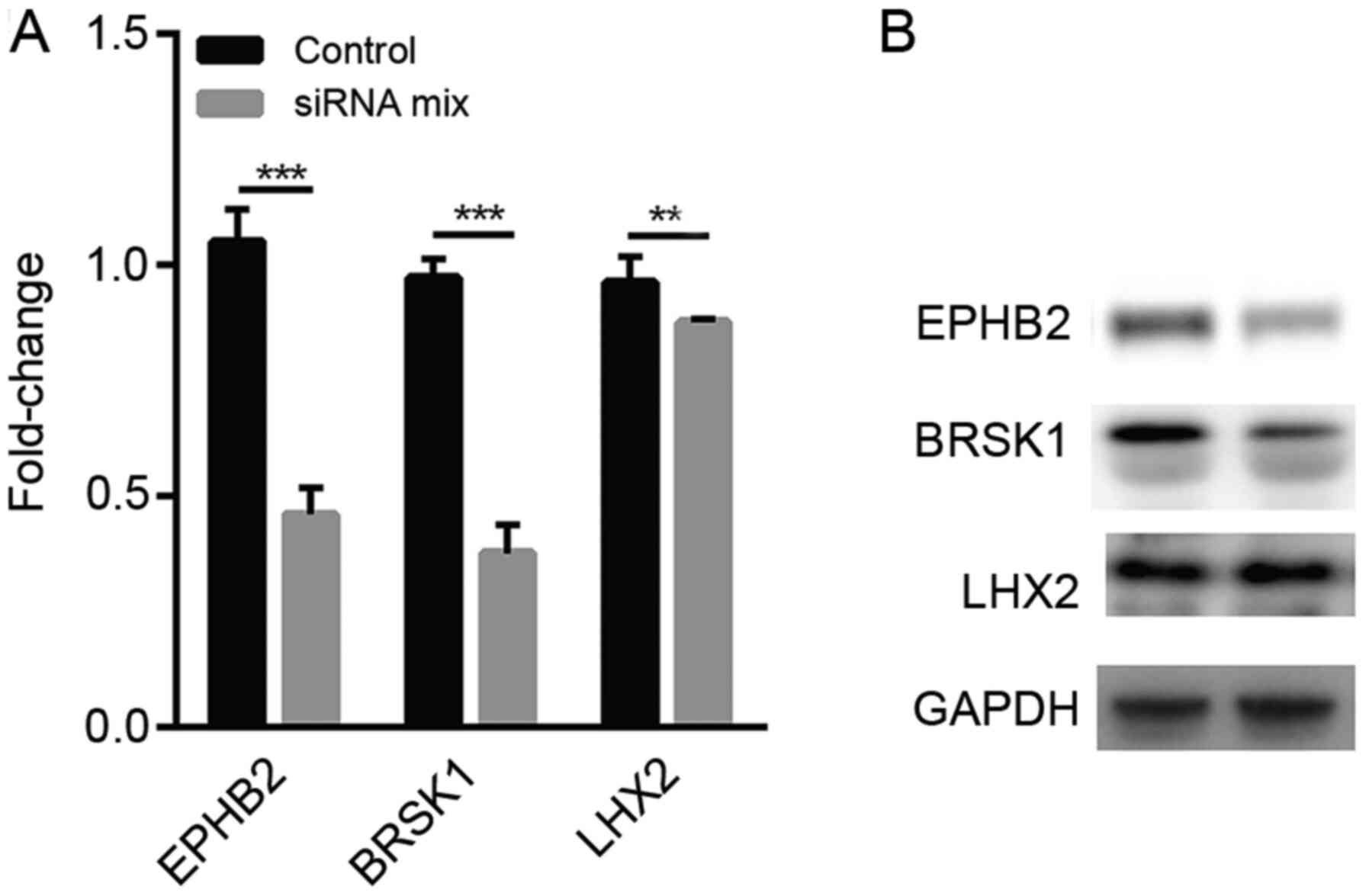
qPCR and western blot analysis of the
expression levels of LHX2, EPHB2 and BRSK1 in TP53-mutant human
breast cancer cells with low RNF8 expression
Since low FGFR1 expression was associated with a
poor prognosis, which indicated that it may have a different
function compared with LHX2 and EPHB2 in breast cancer, FGFR1 was
excluded from subsequent evaluations. The association between RNF8
and the three aforementioned genes associated with prognosis, LHX2,
EPHB2 and BRSK1, was based on sequencing data from mouse breast
cancer cells. To verify whether knockdown of RNF8 in human breast
cancer cells also caused a decrease in the expression levels of
LHX2, EPHB2 and BRSK1, qPCR was performed, revealing that the
expression levels of EPHB2, BRSK1 and LHX2 were significantly
downregulated in human breast cancer HCC1937 cells after treatment
with the siRNA mix (Fig. 5A). A
similar trend was also observed in protein expression in
RNF8-silenced cells (Fig. 5B).
Discussion
Previous studies have indicated that TP53 mutations
have important functions in multiple subtypes of breast cancer,
such as triple-negative and HER2+ breast cancer
(18–20). Similarly, some studies have reported
that RNF8 is associated with the oncogenesis of breast cancer
(10,11). For example, Kuang et al
(10) demonstrated that RNF8
promotes epithelial-mesenchymal transition in breast cancer cells
(including TP53 wild-type and mutant cells); however, the
aforementioned study did not investigate the mechanism of RNF8
promoting epithelial-mesenchymal transition in TP53-mutant breast
cancer. Other studies have tried to investigate the specific
mechanism of RNF8 in breast cancer (21,22). For
example, it has been reported that RNF8 cooperates with RNF168 to
mediate Forkhead Box M1 ubiquitination and degradation in breast
cancer (21); however, the
aforementioned study used the MCF7 cell line, which is a
TP53-wild-type breast cancer cell line. In the present study,
different datasets were used to investigate the role of RNF8 in
TP53-mutant breast cancer and to explore the specific mechanism of
RNF8 promoting TP53-mutant breast cancer through transcriptome
sequencing, and verified the co-expression of RNF8, LHX2 and EPHB2
in the HCC1937 cell line.
RNF8 is an E3 ubiquitin ligase that acts primarily
on the DNA damage repair process and can rapidly accumulate at the
site of DNA damage through Fork Head Associated domain-mediated
phosphorylation of Mediator Of DNA Damage Checkpoint 1 (22–24).
Numerous proteins associated with DNA repair, such as P53 binding
protein and BRCA1, can accumulate and function through its signal
amplification (25). Therefore, RNF8
is a key factor that serves an important role in telomere
protection, maintaining genomic integrity and regulating the cell
cycle (26).
A previous study has revealed that the function of
RNF8 is highly dependent on the P53 protein, suggesting that RNF8
serves a biological role in synergy with P53 (27). However, numerous studies have
revealed a number of mutations in the P53 gene in tumor cells
(28,29); therefore, it is increasingly
important to investigate the functional role of RNF8 under the
condition of P53 mutation (27). The
present study identified the differentially expressed genes in
mouse transcriptome data from the GEO database by comparing the low
and normal expression levels of RNF8 in the TP53-mutant samples,
revealing that downregulated genes were mainly enriched in several
pathways involved in cell proliferation and apoptosis regulation,
development and transcription regulation. This is consistent with
previous studies (28,29), indicating that even in the presence
of P53 mutations, the main function of RNF8 is still focused on the
regulation of the cell cycle and transcription factors, thus
confirming that the function of RNF8 is not completely
P53-dependent and that TP53 mutations have a relatively small
effect on the physiological function of RNF8.
The role of RNF8 in tumor development and
progression exhibits a double-sided action, resulting in either
tumor inhibition or promotion. In a mouse model, the downregulation
or loss of RNF8 results in a significant increase of tumor
incidence, including the incidence of leukemia in tumorigenesis of
lymphoma (50%), thymoma (38%), mammary carcinoma (13%), skin tumor
(13%) and sarcoma (13%) (30). A
previous study has revealed that the expression levels of P53 are
significantly increased in RNF8-knockout mice (12). At the same time, RNF8 has a strong
inhibitory effect on breast cancer and can downregulate the Notch
signaling pathway by deleting the C-terminal of the NOTCH1
intracellular domain (7), thereby
limiting the spread of mammary luminal progenitors. Data from The
Cancer Genome Atlas also support a negative association between
RNF8 expression and the Notch signaling pathway (7).
However, a previous study has revealed that
decreased RNF8 expression can significantly improve radiotherapy
sensitivity in nasopharyngeal cancer, while inhibiting cell
proliferation and increasing apoptosis (31). In breast cancer MCF-7 cells,
overexpression of RNF8 can significantly increase
epithelial-mesenchymal transition and it mainly promotes
phosphorylation of GSK3β, further inhibiting its activity (10). In addition, an increase in
phosphorylation of β-catenin has been observed in
RNF8-overexpressing cells (10).
Furthermore, in the treatment of breast cancer using tamoxifen to
block ERα, tamoxifen exerts anticancer effects mainly by blocking
the ERα signaling pathway (31), but
a previous study has revealed that RNF8 can activate the ERα
signaling pathway and thus promote tumor cell proliferation
(8). The aforementioned studies did
not focus on p53 mutations. Therefore, the present study
investigated the effects of RNF8 in TP53-mutant breast cancer
cells, revealing that downregulation of RNF8 expression in a cell
model with TP53 mutation significantly inhibited the proliferation
of tumor cells. In addition, the current study hypothesized that
the tumorigenesis and development of breast cancer may be regulated
by a sophisticated network, and the role of RNF8 in the
tumorigenesis of TP53-mutant breast cancer may not be unique.
Therefore, future studies should further explore RNF8/TP53 and
immune response-associated pathways (32).
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Research Project Foundation in Higher Education of Anhui
Province in China (grant nos. KJ2019A0094 and KJ2019A0095), the 5th
‘50 Science & Technology Stars’ Innovation Research Team of
Huainan, Anhui Province in China [Huainan Talents (2018) 7], the
Key Project of Natural Science Foundation of Bengbu Medical College
in China (2019; grant no. BYKY2019318ZD) and the Science and
Technology Program of Huainan, Anhui Province in China (grant no.
2018B59).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request, and the GSE76075 dataset is available from the Gene
Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76075).
Authors' contributions
FZ performed the cell experiments, prepared the
figures and wrote the manuscript. PW and YG performed the qPCR and
western blot analysis, and data interpretation. QL and XK performed
flow cytometry. DS and HL performed the bioinformatics analysis. GL
and CL designed the study and revised the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pilevarzadeh M, Amirshahi M,
Afsargharehbagh R, Rafiemanesh H, Hashemi SM and Balouchi A: Global
prevalence of depression among breast cancer patients: A systematic
review and meta-analysis. Breast Cancer Res Treat. 176:519–533.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Greten FR and Grivennikov SI: Inflammation
and cancer: Triggers, mechanisms, and consequences. Immunity.
51:27–41. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pederiva C and Farnebo M: RNF8-The
Achilles heel of DNA repair when splicing rules. Cell Cycle.
17:137–145. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jacobs JJ: Fusing telomeres with RNF8.
Nucleus. 3:143–149. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stewart GS: Solving the RIDDLE of 53BP1
recruitment to sites of damage. Cell Cycle. 8:1532–1538. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li F, Liu B, Zhou X and Xu Q: Silencing of
E3 ubiquitin ligase RNF8 enhances ionizing radiation sensitivity of
medulloblastoma cells by promoting the deubiquitination of PCNA.
Oncol Res. 26:1365–1373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li L, Guturi KKN, Gautreau B, Patel PS,
Saad A, Morii M, Mateo F, Palomero L, Barbour H, Gomez A, et al:
Ubiquitin ligase RNF8 suppresses Notch signaling to regulate
mammary development and tumorigenesis. J Clin Invest.
128:4525–4542. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang S, Luo H, Wang C, Sun H, Sun G, Sun
N, Zeng K, Song H, Zou R, Zhou T, et al: RNF8 identified as a
Co-activator of estrogen receptor α promotes cell growth in breast
cancer. Biochim Biophys Acta Mol Basis Dis. 1863:1615–1628. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamamoto T, Taira Nihira N, Yogosawa S,
Aoki K, Takeda H, Sawasaki T and Yoshida K: Interaction between
RNF8 and DYRK2 is required for the recruitment of DNA repair
molecules to DNA double-strand breaks. FEBS Lett. 591:842–853.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kuang J, Li L, Guo L, Su Y, Wang Y, Xu Y,
Wang X, Meng S, Lei L, Xu L and Shao G: RNF8 promotes
epithelial-mesenchymal transition of breast cancer cells. J Exp
Clin Cancer Res. 35:882016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gao S, Wu J, Liang L and Xu R: RNF8
negatively regulates NF-kappaB signaling by targeting IkappaB
kinase: Implications for the regulation of inflammation signaling.
Biochem Biophys Res Commun. 488:189–195. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu G, Chapman JR, Brandsma I, Yuan J,
Mistrik M, Bouwman P, Bartkova J, Gogola E, Warmerdam D, Barazas M,
et al: REV7 counteracts DNA double-strand break resection and
affects PARP inhibition. Nature. 521:541–544. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Feng J, Meyer CA, Wang Q, Liu JS, Shirley
Liu X and Zhang Y: GFOLD: A generalized fold change for ranking
differentially expressed genes from RNA-seq data. Bioinformatics.
28:2782–2788. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu G: Gene Ontology Semantic Similarity
Analysis Using GOSemSim. Kidder B: Stem Cell Transcriptional
Networks. Methods Mol Biol. 2117:207–215. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu G, Lam TT, Zhu H and Guan Y: Two
methods for mapping and visualizing associated data on phylogeny
using ggtree. Mol Biol Evol. 35:3041–3043. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li L, Halaby MJ, Hakem A, Cardoso R, El
Ghamrasni S, Harding S, Chan N, Bristow R, Sanchez O, Durocher D
and Hakem R: Rnf8 deficiency impairs class switch recombination,
spermatogenesis, and genomic integrity and predisposes for cancer.
J Exp Med. 207:983–997. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Giordano A, Liu Y, Armeson K, Park Y,
Ridinger M, Erlander M, Reuben J, Britten C, Kappler C, Yeh E and
Ethier S: Polo-like kinase 1 (Plk1) inhibition synergizes with
taxanes in triple negative breast cancer. PLoS One.
14:e02244202019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Petry V, Bonadio RC, Cagnacci AQC, Senna
LAL, Campos RDNG, Cotti GC, Hoff PM, Fragoso MCBV and Estevez-Diz
MDP: Radiotherapy-induced malignancies in breast cancer patients
with TP53 pathogenic germline variants (Li-Fraumeni syndrome). Fam
Cancer. 19:47–53. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Arturo JF, Chobrutskiy BI, Yeagley M,
Patel DN, Falasiri S, Patel JS and Blanck G: Electrostatic
complementarity of B-cell receptor CDR3s and TP53-mutant amino
acids in breast cancer is associated with increased disease-free
survival rates. Cell Mol Immunol. 17:776–778. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kongsema M, Zona S, Karunarathna U,
Cabrera E, Man EP, Yao S, Shibakawa A, Khoo US, Medema RH, Freire R
and Lam EW: RNF168 cooperates with RNF8 to mediate FOXM1
ubiquitination and degradation in breast cancer epirubicin
treatment. Oncogenesis. 5:e2522016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee HJ, Li CF, Ruan D, Powers S, Thompson
PA, Frohman MA and Chan CH: The DNA damage transducer RNF8
facilitates cancer chemoresistance and progression through twist
activation. Mol Cell. 63:1021–1033. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mandemaker IK, van Cuijk L, Janssens RC,
Lans H, Bezstarosti K, Hoeijmakers JH, Demmers JA, Vermeulen W and
Marteijn JA: DNA damage-induced histone H1 ubiquitylation is
mediated by HUWE1 and stimulates the RNF8-RNF168 pathway. Sci Rep.
7:153532017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Paul A and Wang B: RNF8-and
Ube2S-dependent ubiquitin lysine 11-linkage modification in
response to DNA damage. Mol Cell. 66:458–472.e5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yoshida K and Miki Y: Role of BRCA1 and
BRCA2 as regulators of DNA repair, transcription, and cell cycle in
response to DNA damage. Cancer Sci. 95:866–871. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rai R, Li JM, Zheng H, Lok GT, Deng Y,
Huen MS, Chen J, Jin J and Chang S: The E3 ubiquitin ligase Rnf8
stabilizes Tpp1 to promote telomere end protection. Nat Struct Mol
Biol. 18:1400–1407. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Halaby MJ, Hakem A, Li L, El Ghamrasni S,
Venkatesan S, Hande PM, Sanchez O and Hakem R: Synergistic
interaction of Rnf8 and p53 in the protection against genomic
instability and tumorigenesis. PLoS Genet. 9:e10032592013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kaur RP, Vasudeva K, Kumar R and Munshi A:
Role of p53 Gene in breast cancer: Focus on mutation spectrum and
therapeutic strategies. Curr Pharm Des. 24:3566–3575. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Duffy MJ, Synnott NC and Crown J: Mutant
p53 in breast cancer: Potential as a therapeutic target and
biomarker. Breast Cancer Res Treat. 170:213–219. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou T, Yi F, Wang Z, Guo Q, Liu J, Bai N,
Li X, Dong X, Ren L, Cao L and Song X: The functions of DNA damage
factor RNF8 in the pathogenesis and progression of cancer. Int J
Biol Sci. 15:909–918. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang M, Chen X, Chen H, Zhang X, Li J,
Gong H, Shiyan C and Yang F: RNF8 plays an important role in the
radioresistance of human nasopharyngeal cancer cells in
vitro. Oncol Rep. 34:341–349. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fu R, Han CF, Ni T, Di L, Liu LJ, Lv WC,
Bi YR, Jiang N, He Y, Li HM, et al: A ZEB1/p53 signaling axis in
stromal fibroblasts promotes mammary epithelial tumors. Nat Commun.
10:32102019. View Article : Google Scholar : PubMed/NCBI
|