Introduction
Malignant melanoma, which arises from melanocytes,
is lethal and common in the global population, ranking 5th for the
incidence of malignant tumors in males, and 6th in females for
mortality rate (1). Furthermore, its
mortality seriously threatens human health and imparts a grave
economic burden (2,3). Although surgical resection of malignant
tissue is recognized as the most effective therapy in the early
stages of melanoma, the 5-year survival rate in metastatic melanoma
is only 15%, with patients experiencing a very poor prognosis
(4,5). Furthermore, melanoma is characterized
by high rates of recurrence with high mortality, often diagnosed in
the last stages and resistant to current therapeutic approaches
(6,7). Therefore, further studies are needed to
identify novel biomarkers for malignant melanoma and to develop a
deeper understanding of the mechanisms that lead to melanoma
progression.
Epithelial-mesenchymal transition (EMT) is an
indispensable biological process that is closely connected to
embryogenesis and the first stage of wound healing (8). Recently, it has become widely
acknowledged that the mechanism of EMT is more complex in melanoma.
Indeed, abnormal activation of the EMT pathway alters the
microenvironment by which epithelial tumor cells that initially
undergo EMT are able to revert to epithelial phenotype by
mesenchymal-epithelial transition at the distant site, therefore
tumor cells penetrate the basement membrane and migrate (9). A variety of transcription factors act
as molecular switches that directly regulate the occurrence of the
EMT process in melanoma (10,11).
Among these, zinc finger protein E-box binding homeobox (ZEB)
proteins, especially ZEB1 and −2, participate in the initiation of
EMT in melanoma by downregulating the expression in
microphthalmia-associated transcription factor (MITF) (12).
Nuclear factor I (NFI) is a type of transcription
factor family, which is widely found in mammals and also known as
CCAAT box-binding transcription factor (CTF). This family is
characterized by a highly conserved N-terminal DNA-binding region,
and divided into four subtypes (A, B, C and X) based on the
variable C-terminus region (13,14).
NFIs are known to be involved in the regulation of DNA replication
and gene expression and to promote cell proliferation and
differentiation during the embryonic development (15–17). In
addition, NFIs are abnormally expressed in various tumors and has a
complex and diverse function in some tumors (18). For instance, human NFI type-B (NFIB)
serves a crucial role in different aspects of tumor development,
acting as an oncogene or tumor suppressor in different types of
tumor and participating in tumor-associated fusion gene formation
(19–21). Previous studies on the role of NFIB
in colorectal cancer (CRC) have indicated that NFIB triggers EMT of
CRC cells through upregulation of SNAI1 expression (22). Moreover, NFIB seems to mediate the
conversion between the two transcription factors, POU class 3
homeobox 2 and MITF, through the upregulation of enhancer of zeste
homolog 2, which increases expression of MITF and decreases
expression of BRN2, thus driving the invasive phenotype in the
melanoma (23). However, few studies
have been conducted on the specific mechanisms involved in melanoma
metastasis based on NFIB. Thus, the present study investigated the
impact of NFIB on EMT in A375 and A875 cell lines. Given that ZEB1
has two possible binding sites for the NFIB promoter (24), it was hypothesized that there was an
association between ZEB1 and NFIB. The findings of this study may
provide insight into the metastasis of malignant melanoma, with
particular emphasis on EMT.
Materials and methods
Clinical specimens
To detect the expression of NFIB by
immunohistochemistry (IHC), 15 melanoma samples, 15 benign nevus
samples and 10 normal skin samples were collected between December
2017 and January 2018 from different patients. The total number of
patients enrolled for the study was 40. Clinical samples of
melanoma and nevus in this study were provided and pathologically
diagnosed by the Department of Dermatology, Tongji Hospital
affiliated with Huazhong University of Science and Technology,
while normal skin samples were obtained from the Department of
Urology, Tongji Hospital affiliated with Huazhong University of
Science and Technology. These tissue specimens were fixed in 10%
formalin at room temperature after resection. After 24 h of
fixation, the samples were then embedded in paraffin for IHC. All
the participants were newly diagnosed and had not received any
comprehensive antitumor treatment before the surgical section. They
were informed of their rights and signed an informed consent form.
The present study was approved by the Ethics Review Committee at
Tongji Hospital affiliated with Huazhong University of Science and
Technology.
IHC analysis
The expressions of NFIB protein in the samples of
melanoma, nevus and normal skin were detected by IHC. All three
kinds of specimens were cut into uniform sections of 3-µm
thickness. The samples were treated with 10% polylysine, then fixed
on glass slides using melted paraffin at 65°C for 5 min and baked
in an oven at 65°C overnight. The next day, the prepared sections
were hydrated by dewaxing with graded alcohol and xylene at room
temperature. Thereafter, antigen retrieval was carried out using
600 ml 10 mM sodium citrate (pH 6.0) heated in a microwave oven at
100°C for 6–8 min and then cooled at room temperature. Several
drops of hydrogen peroxide were then added for incubation at room
temperature for 15 min to neutralize the excess oxygen radicals and
reduce the background. Then, a primary antibody specific for NFIB
(cat. no. ab186738; 1:100 dilution; Abcam) was added to the
sections at 4°C overnight, followed by incubation with a secondary
antibody conjugated with streptavidin-HRP at room temperature for
30 min. A freshly prepared DAB coloring solution was used to
enhance coloration with hematoxylin re-dyeing, 1% hydrochloric acid
ethanol differentiation, trypan blue pan-blue and gradient alcohol
dehydration. Subsequently, the relative intensity of NFIB
expression was evaluated using Image-Pro Plus software (IPP version
6.0; Media Cybernetics, Inc.).
Bioinformatics analysis
The present study analyzed from TCGA website
(https://xenabrowser.net/heatmap/) and
the Oncomine Cancer Microarray database (https://www.oncomine.org/resource/login.html) to
compare the expression levels of NFIB mRNA in 227 samples,
including 92 melanoma, 85 benign nevus and 50 normal skin
specimens. In each dataset, the median value of NFIB expression was
used to divide the samples into an NFIB high-expression group and
an NFIB low-expression group. The analysis was performed using
GraphPad Prism 5.0 and log-rank tests (GraphPad Software,
Inc.).
Cell lines and culture condition
The human melanoma cell lines A375, A875 and
SK-MEL-1, and the normal HaCaT keratinocyte cell line were
purchased from the China Center for Type Culture Collection. HaCaT
cells were authenticated by STR. All cell lines were resuspended in
Dulbecco's Modified Eagle Medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.) and incubated at 37°C with
5% CO2. The cell culture medium was replaced or the
cells were sub-cultured, as appropriate, every 1–2 days.
Cell transfection
A375 and A875 cells were uniformly seeded into
6-well plates at 4×105 cells/well and incubated
overnight. The cells were resuspended in Opti-MEM (Thermo Fisher
Scientific, Inc) then transfected with small interfering (si) RNA
(si-NFIB, 5′-AGGAUACUCUGAAGAACUAUU-3′; Guangzhou RiboBio Co., Ltd.)
to silence NFIB expression at 50 nM/well. The transfected cells
were screened using puromycin (5.0 µg/ml). The recombinant plasmid
pcDNA.3.1-NFIB (Guangzhou RiboBio Co., Ltd.) was transfected into
A375 and A875 cells to induce NFIB overexpression at 0.2 µg/well.
si-negative control (NC; 5′-TTCTCCGAACGTGTCACGTdTdT-3′) and empty
vectors were used as controls. Silencing and overexpression
efficiency were examined by western blot and reverse
transcription-quantitative PCR (RT-qPCR). After 10 days of
lentivirus transfection or 72 h after siRNA transfection, western
blot and RT-PCR analyses were used to detect the expression of
target genes. Plasmids pcDNA-NFIB were transfected into 293T cells
for luciferase reporter assays.
Establishment of an EMT model of
melanoma cells
The A375 and A875 cells were uniformly seeded into a
6-well plate at a density of 4×105 cells/well, and
cultured to ~75% confluence. Cells were then treated with 5 ng/ml
TGF-β1. TGF-β1 (PeproTech, Inc.) was used to stimulate EMT.
Untreated cells were used as a control. A phase-contrast microscope
(Model CKX41; magnification, ×100; Olympus Corporation) was used to
examine cell morphological changes after 24 h.
RT-qPCR
Total RNA was extracted from cells 48 h following
transfection using TRIzol® solution following the
manufacturer's instructions (Thermo Fisher Scientific, Inc.). An
ultraviolet spectrophotometer with a wavelength between 260 and 280
nm was used to measure the RNA concentration. Total RNA was reverse
transcribed to cDNA with RT Master Mix (Takara Bio, Inc.) according
to the manufacturer's instructions. RT-PCR was performed with SYBR
Master Mix (Takara Bio, Inc.) using the StepOne-Plus system (Thermo
Fisher Scientific, Inc.). RT-PCR was performed with 40 cycles under
the following conditions: Denaturation at 95°C for 30 sec,
annealing at 60°C for 1 min and extension at 95°C for 5 sec. Each
experiment was conducted three times. The specific primer sequences
for evaluating the NFIB expression in melanoma cell lines were as
follows: i) NFIB forward, 5′-AAAAAGCATGAGAAGCGAATGTC-3′; ii) NFIB
reverse, 5′-ACTCCTGGCGAATATCTTTGC-3′; iii) GAPDH forward,
5′-ACAACTTTGGTATCGTGGAAGG-3′; and iv) GAPDH reverse,
5′-GCCATCACGCCACAGTTTC-3′. GAPDH served as an endogenous control to
normalize NFIB expression in each sample. The relative expression
of NFIB was calculated using the comparative Cq (2−ΔΔCq)
method (25).
Cell proliferation
The Cell Counting Kit-8 (CCK-8; Beijing Baisi
Biological Technology Co., Ltd.) was used to evaluate the rate of
cell proliferation. In brief, A375 and A875 cells were uniformly
seeded into three 96-well plates at a density of 1×103
cells/well, in culture medium volume 100 µl/well. 10 µl CCK-8
solution was added into the media. The absorbance of cells was
assessed at a fixed time every day, and the data were continuously
detected by a microplate reader (Thermo Fisher Scientific, Inc.).
The optical density of cells was measured at a 450 nm wavelength.
The cells were incubated for 24, 48 and 72 h at 37°C with 5%
CO2.
Colony formation assay
Melanoma cells in the logarithmic growth period were
seeded in a 6-well plate at a density of 5×10 cells/well containing
DMEM medium with 10% FBS and incubated at 37°C in a humidified
incubator with 5% CO2. When the colonies were visible to
the naked eye, the culture was stopped immediately and then fixed
with 4% paraformaldehyde (Google Biotechnology Co., Ltd.) and
stained by 0.1% crystal violet (Google Biotechnology Co., Ltd.).
Finally, the number of colonies (>50 cells) were analyzed under
an inverted microscope (Olympus Corporation), and the colony number
and colony formation rates were calculated. Clone formation
rate=(number of clones/number of inoculated cells) ×100%.
Wound healing assay
To assess the effects of NFIB on cell migration,
5×104 A375 and A875 cells were seeded during the
logarithmic growth period in the 6-well plate to ensure that the
cells would reach 90–100% confluency the next day. Three parallel
lines were drawn on the back of the 6-well plate with a marker pen,
and a 100-µl pipette tip was selected to draw a line on the bottom
of the cell of the 6-well plate. After this, the cells were washed
with sterile PBS repeatedly 3–5 times to remove floating cell
debris. The cells were then cultured in serum-free medium for 24
and 48 h. Migration between scratches was observed under an
inverted microscope under ×100 magnification and kept at the same
position and time under the mark.
Cell migration and invasion assay
Migration and invasion were evaluated in 24-well
Transwell chambers (Corning Inc.) in the presence or absence of
Matrigel™ ECM (Corning Inc.) coating. After transfection for 48 h,
A375 (5×104 cells/chamber) and A875 (1×105
cells/chamber) cells resuspended in 200 µl of serum-free DMEM
medium were seeded into the upper chamber. The lower chamber
received 600 µl of DMEM medium mixed with 10% FBS. Then, the cells
were incubated for an additional 24 h for migration assays or 48 h
for invasion assays at 37°C with 5% CO2. Finally, the
cells in the upper chamber were wiped off with a cotton bud, and
the penetrated cells underneath the chamber were fixed with 4%
paraformaldehyde and stained with 0.1% crystal violet staining
solution (both at room temperature for 30 min). The cells were
counted in five randomly selected fields for each membrane under an
inverted microscope and photographed at ×100 magnification.
Western blot analysis
Total protein was extracted using RIPA buffer
(Thermo Fisher Scientific, Inc.) supplemented with protease
inhibitor cocktail and 1% PMSF (Roche Diagnostics). The protein was
quantified using a BCA Protein Assay Reagent kit (Thermo Fisher
Scientific, Inc.) and were collected and mixed with loading buffer.
Equal amounts of protein (40 µg) from each sample were separated by
10% SDS-PAGE and transferred to PVDF membranes (EMD Millipore). The
membranes were blocked in 5% milk in TBST buffer for 2 h at room
temperature, then incubated with primary antibodies at the
recommended dilution overnight at 4°C. After binding with secondary
antibody conjugated HRP at room temperature for 1 h, Image J
(National Institutes of Health) was used to measure the band
density. GAPDH acted as an internal reference, and each sample was
analyzed three times. NFIB (1:1,000; cat. no. ab80835) and GAPDH
(1:1,000; cat. no. ab9485) antibodies were purchased from Abcam.
Vimentin (1:1,000; cat. no. 10366-1-AP) were purchased from
ProteinTech Group, Inc. E-cadherin (1:1,000; cat. no. sc-71007) and
N-cadherin (1:1,000; cat. no. sc-71002) were purchased from Santa
Cruz Biotechnolgy, Inc. Secondary horseradish peroxidase-goat
anti-rabbit antibodies (1:1,000; cat. no. 10285-1-AP) were
purchased from ProteinTech Group, Inc.
Statistical analysis
All data are presented as the mean ± standard
deviations (SD) from three independent experiments. All statistical
analyses were performed using GraphPad Prism (version 6.0; GraphPad
Software, Inc.). Unpaired Student's t-test was employed to compare
the difference between two groups. One-way ANOVA followed by the
Least Significant Difference was used for multigroup comparisons.
The log-rank test and Kaplan-Meier survival curves were used to
analyze the association between NFIB expression and overall
survival. Statistical analyses were performed using GraphPad Prism
5.0 (GraphPad Software, Inc.). P<0.05 was considered to indicate
a statistically significant difference.
Results
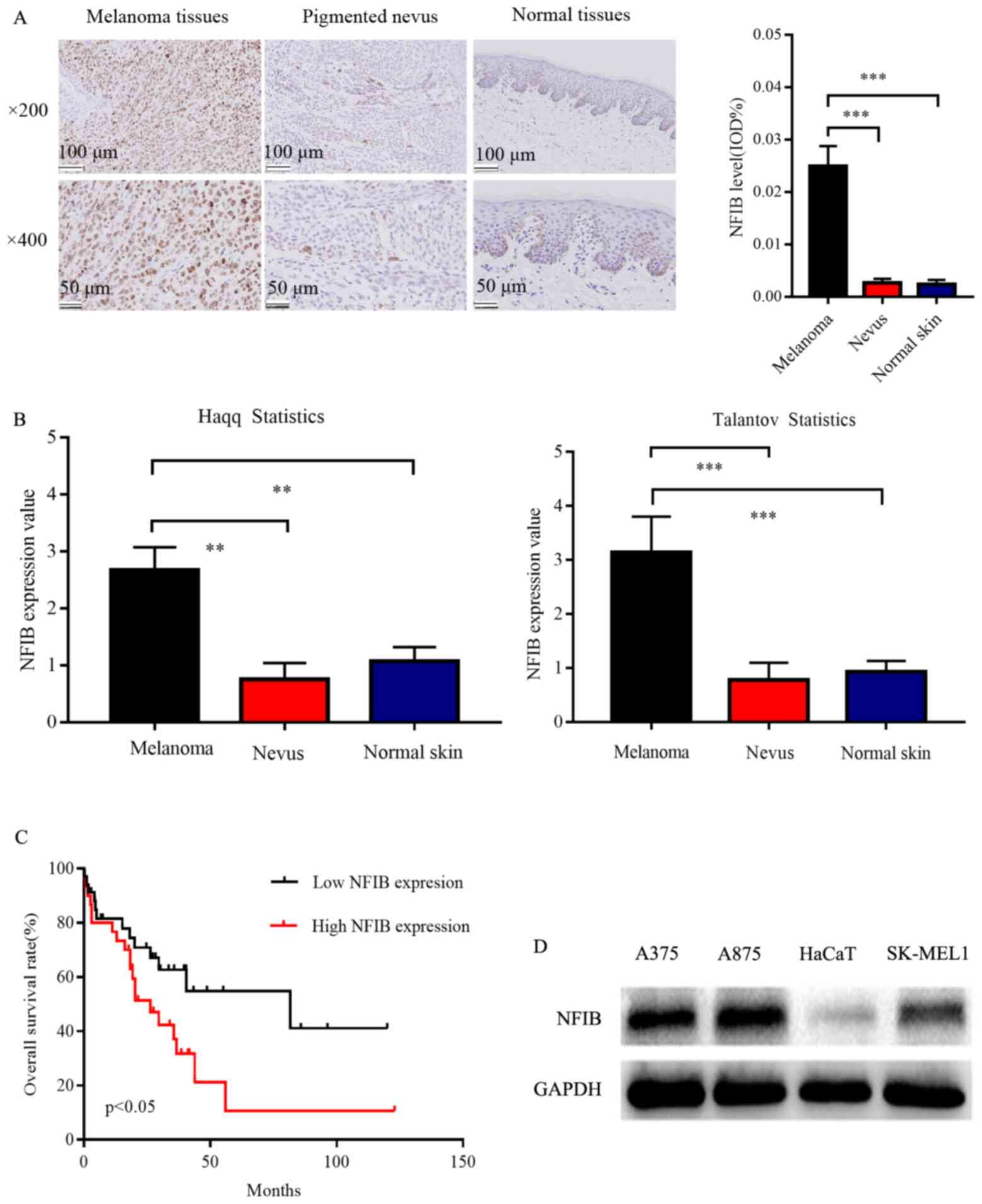
NFIB is relatively high in human
malignant melanoma and associated with poor prognosis
The expression and molecular mechanism of NIFB have
not been clearly reported in melanoma previously. Initially, the
present study analyzed the localization of NFIB in clinical
samples. NFIB expression levels were examined by IHC in 15
malignant melanoma samples, 15 benign nevus samples and in 10
normal skin non-matched samples. In total, 12 malignant melanoma
samples were positive for NFIB expression, five benign nevus
samples were positive for NFIB expression, no normal skin sample
was positive for NFIB expression. Notably, NFIB was widely
distributed in melanoma samples with a cytonuclear staining pattern
(data not shown). A significantly higher expression of NFIB was
observed in the melanoma specimens, compared with benign pigmented
nevus and normal human skin (Fig.
1A). Furthermore, mRNA expression data from the Haqq and
Talantov Oncomine datasets supported these findings (Fig. 1B).
The Oncomine datasets were also used to explore the
relationship between NFIB expression levels and overall survival.
The log-rank test and Kaplan-Meier survival analysis indicated that
high NFIB expression was associated with lower overall survival
rate, relative to the NFIB low-expression group (Fig. 1C). This indicated that NFIB may serve
as a poor prognostic indicator for melanoma and could be associated
with the aggressiveness of melanoma cells.
In addition, western blotting suggested that NFIB
expression levels were also elevated in the A375, A875 and SK-MEL1
melanoma cell lines, compared with the normal human keratinocyte
HaCaT cell line (Fig. 1D).
NFIB expression promotes migration and
invasion in melanoma cell lines
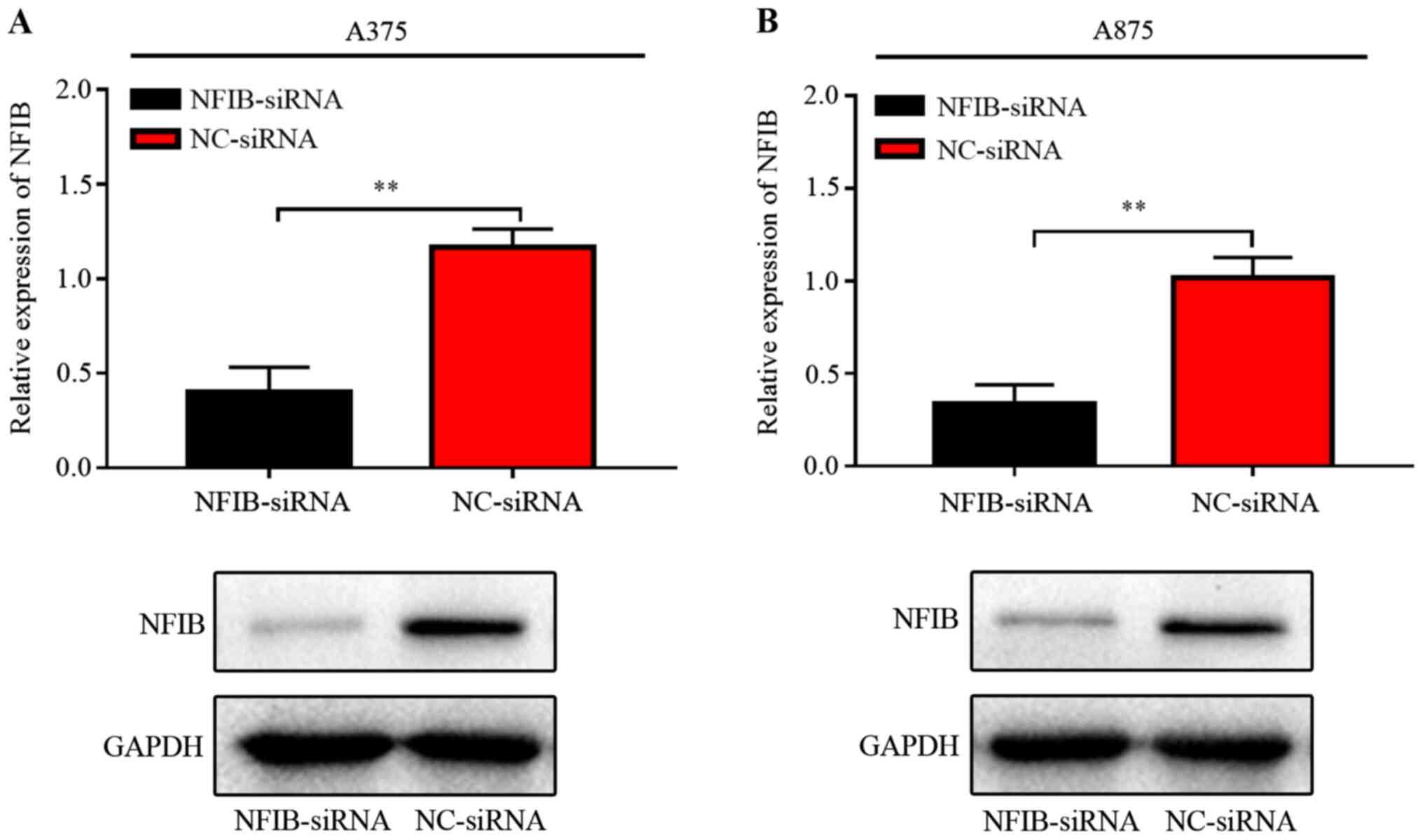
To further investigate the role of NFIB on
malignancy in melanoma, NFIB expression was silenced in A375 and
A875 cells using transfection with NFIB-siRNA. Detection of
knockdown efficiency was done by RT-qPCR and western blot analysis.
NFIB expression was notably dropped in melanoma cell lines using
transfection with NFIB-siRNA compared with NC-siRNA (Fig. 2). Subsequently, the role of NFIB on
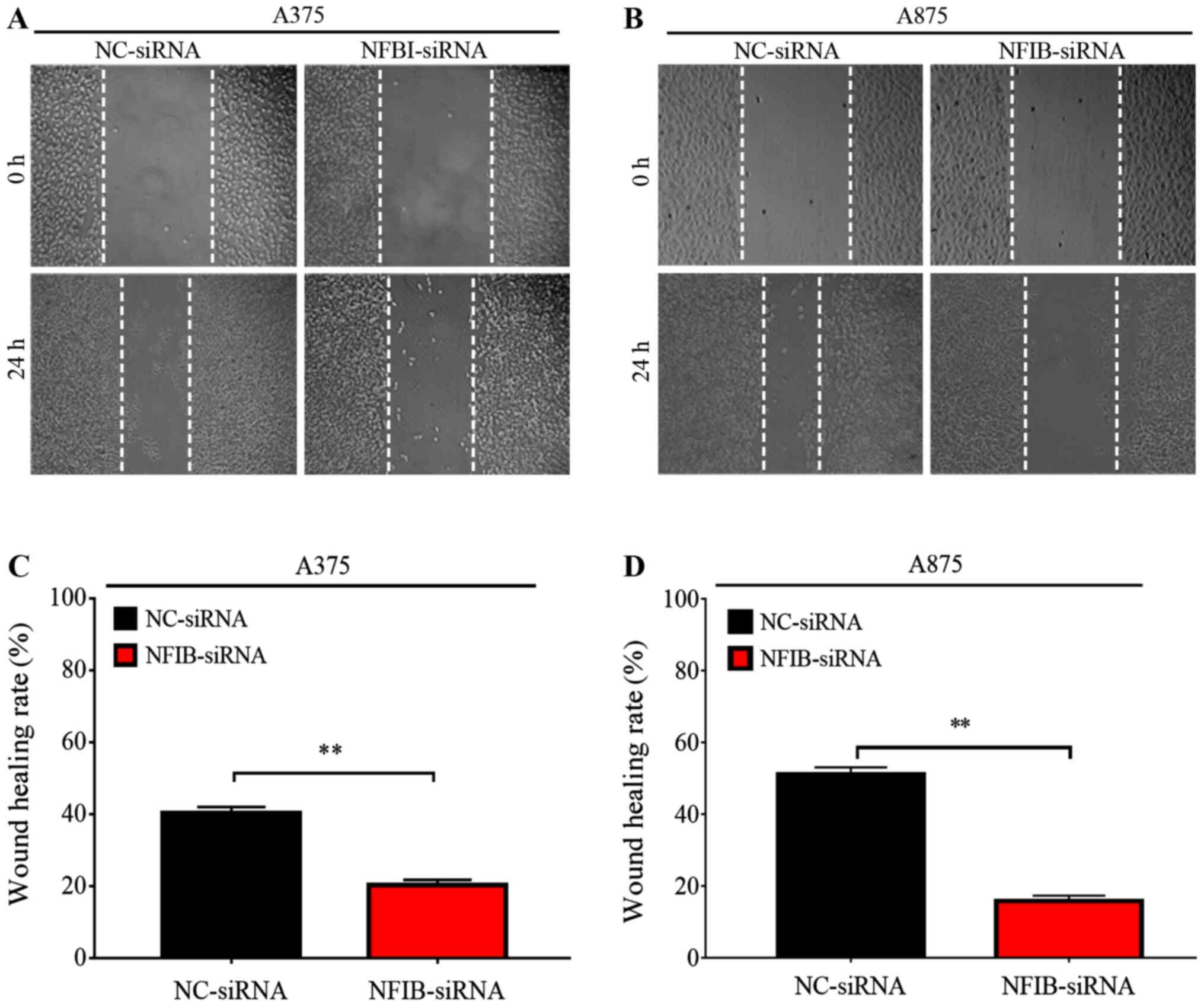
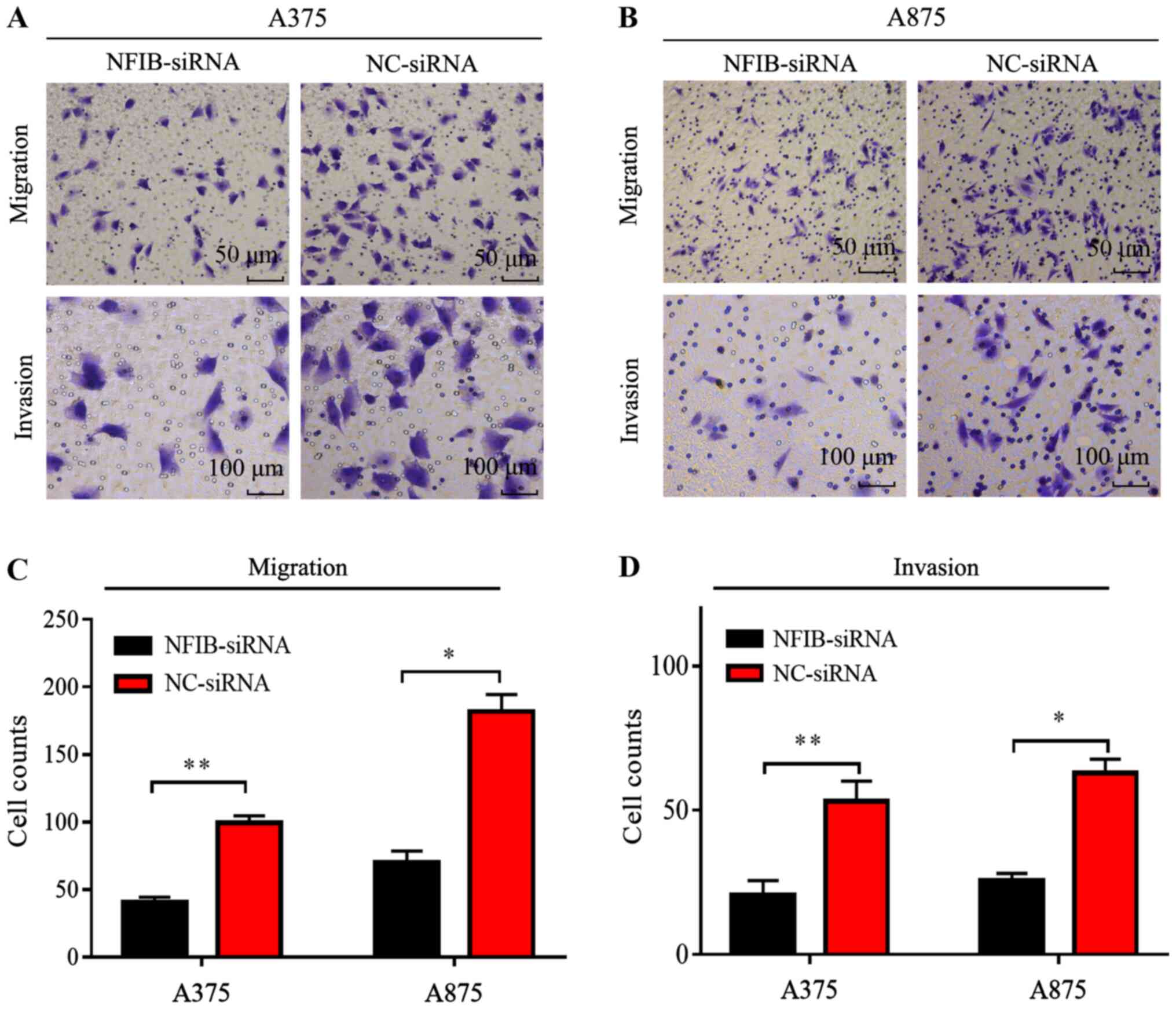
migration and invasion in melanoma cell lines was evaluated in
wound healing and Transwell assays. Transfection with NFIB-siRNA
significantly reduced the migratory capacity of both A375 and A875
cells, compared with NC-siRNA (P<0.01; Figs. 3 and 4). In addition siRNA-mediated NFIB
silencing also significantly reduced the invasion capacity of both
A375 and A875, relative to the NC-siRNA (Fig. 4).
NFIB promotes melanoma cell
proliferation and colony formation
Considering that NFIB is highly expressed in human
malignant melanoma, the present study sought to investigate whether
NFIB affects colony formation and proliferation of melanoma cells.
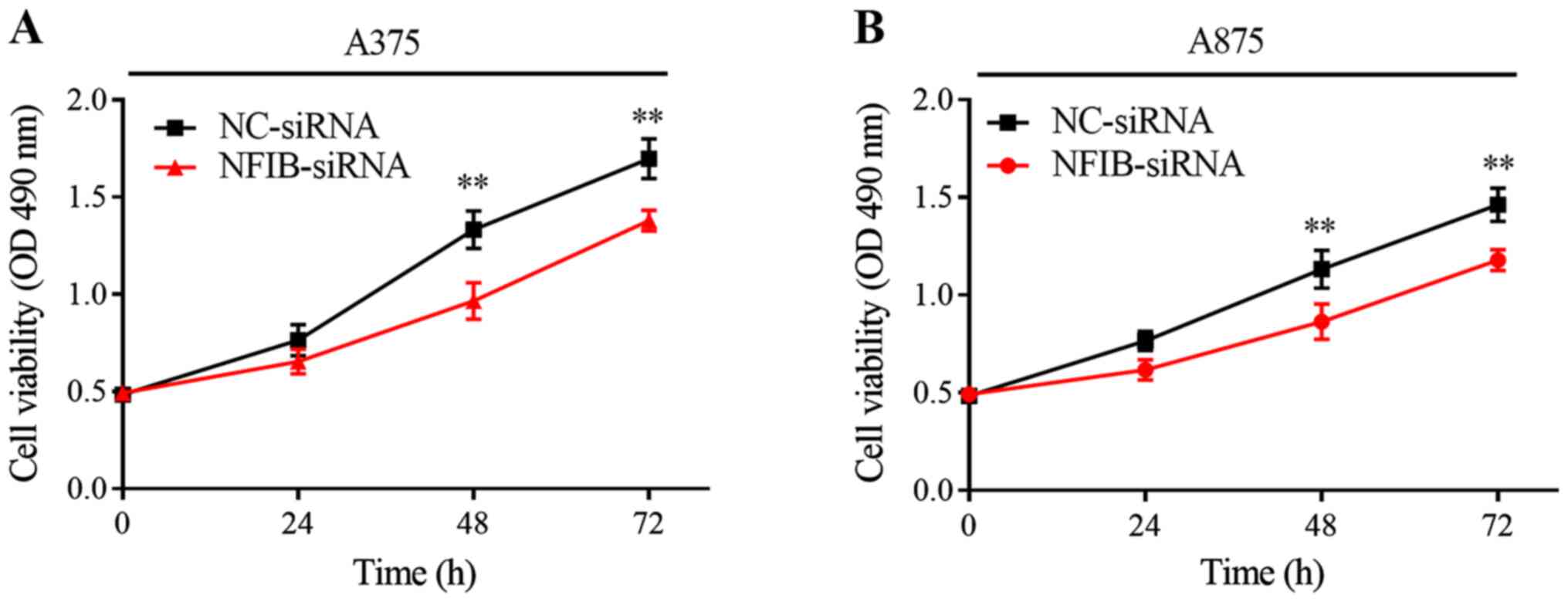
In CCK-8 assays, proliferative ability was significantly reduced in
NFIB-silenced melanoma cell lines relative to their respective
NC-siRNA controls (Fig. 5A and B).
Interestingly, the inhibition effect on the proliferation of
melanoma cells began to appear in the first 24 h, and peaked
between 24 and 48 h, whereas this effect disappeared after 48 h.
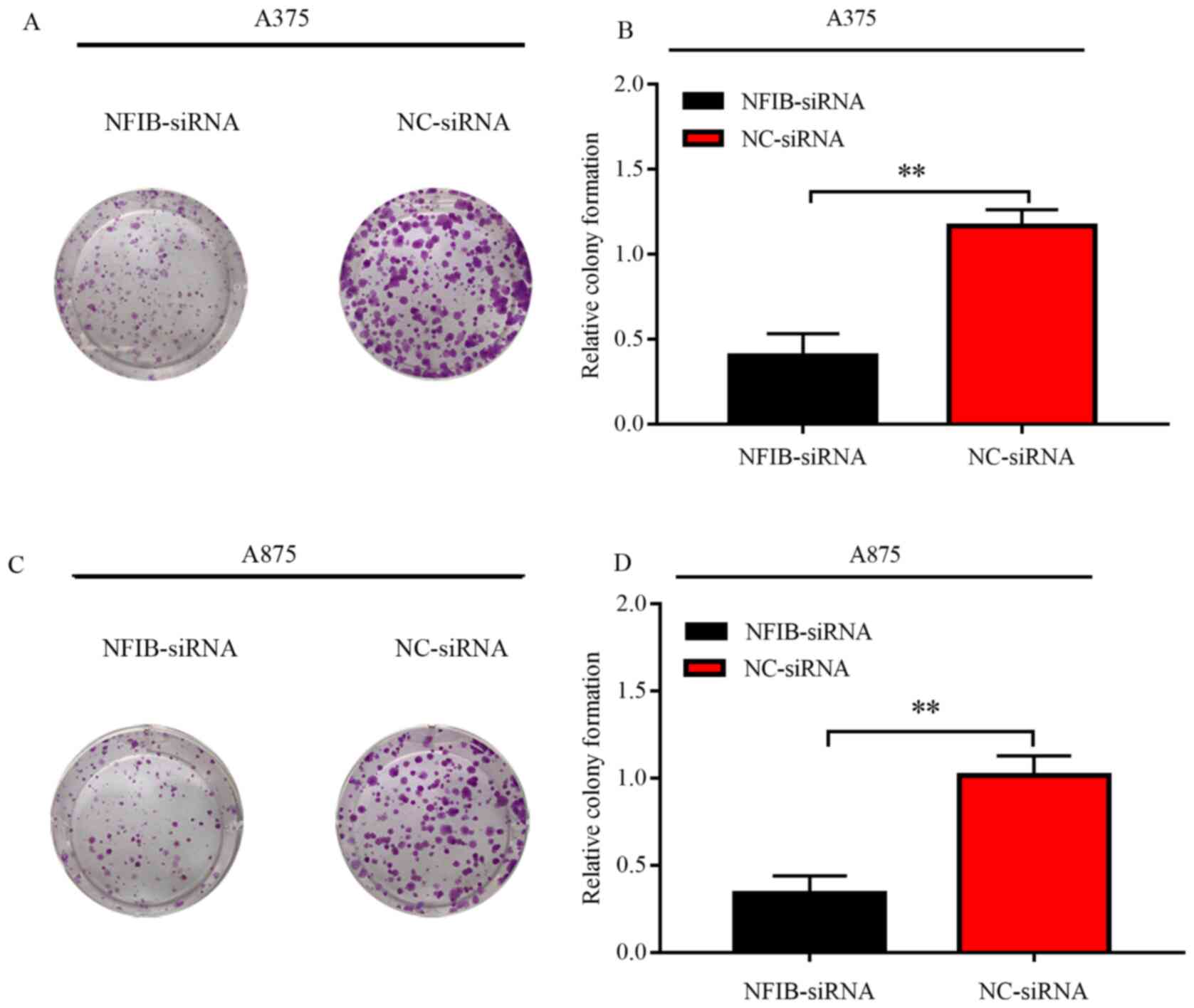
Furthermore, the colony formation rate of cells transfected with
NFIB-siRNA was significantly lower than that of the NC-siRNA group
in A375 and A875 cells (Fig. 6B and
D). Furthermore, compared with the NC-siRNA, cells transfected
with NFIB-siRNA also exhibited restricted colonies in both size and
numbers (Fig. 6A and C).
Consequently, these data suggested that NFIB may enhance the
malignancy of melanoma cells, by enhancing the proliferative
ability.
NFIB expression promotes EMT in
melanoma cell lines
A large number of studies have suggested that EMT
has an important role in malignant tumors, focusing on malignant
behaviors, such as migration and invasion. During the progression
of EMT, E-cadherin is known to be downregulated and N-cadherin is
upregulated, which reduces the polarity of epithelial cells and
weakens the connection with the basement membrane to obtain higher
invasion and migration capacities (26). Thus, the present study examined
whether NFIB had an effect on melanoma EMT. Morphological changes
of the melanoma cells were observed under TGF-β1 treatment and
NFIB-siRNA transfection. During inverted phase-contrast microscopy,
A375 and A875 cells that initially had epithelial morphology
developed an elongated fibroblast-like morphology upon exposure to
TGF-β1-induced EMT. In contrast, the NFIB-siRNA group melanoma
cells were closely packed, and the number of cells in the same
field of view was higher, compared with the NC-siRNA group,
therefore that TGF-β1 treated NFIB knockdown cells mostly retained
their primary epithelial morphology (Fig. 7A). Western blotting suggested that
the downregulation of NFIB was accompanied by relatively higher
expression levels of E-cadherin and ZO-1, compared with the control
groups. Conversely, relatively low expression levels of N-cadherin
and VIM were associated with the NFIB-siRNA transfected cells
(Fig. 7B and C). Overall, these data
indicated that NFIB can promote EMT in melanoma cells.
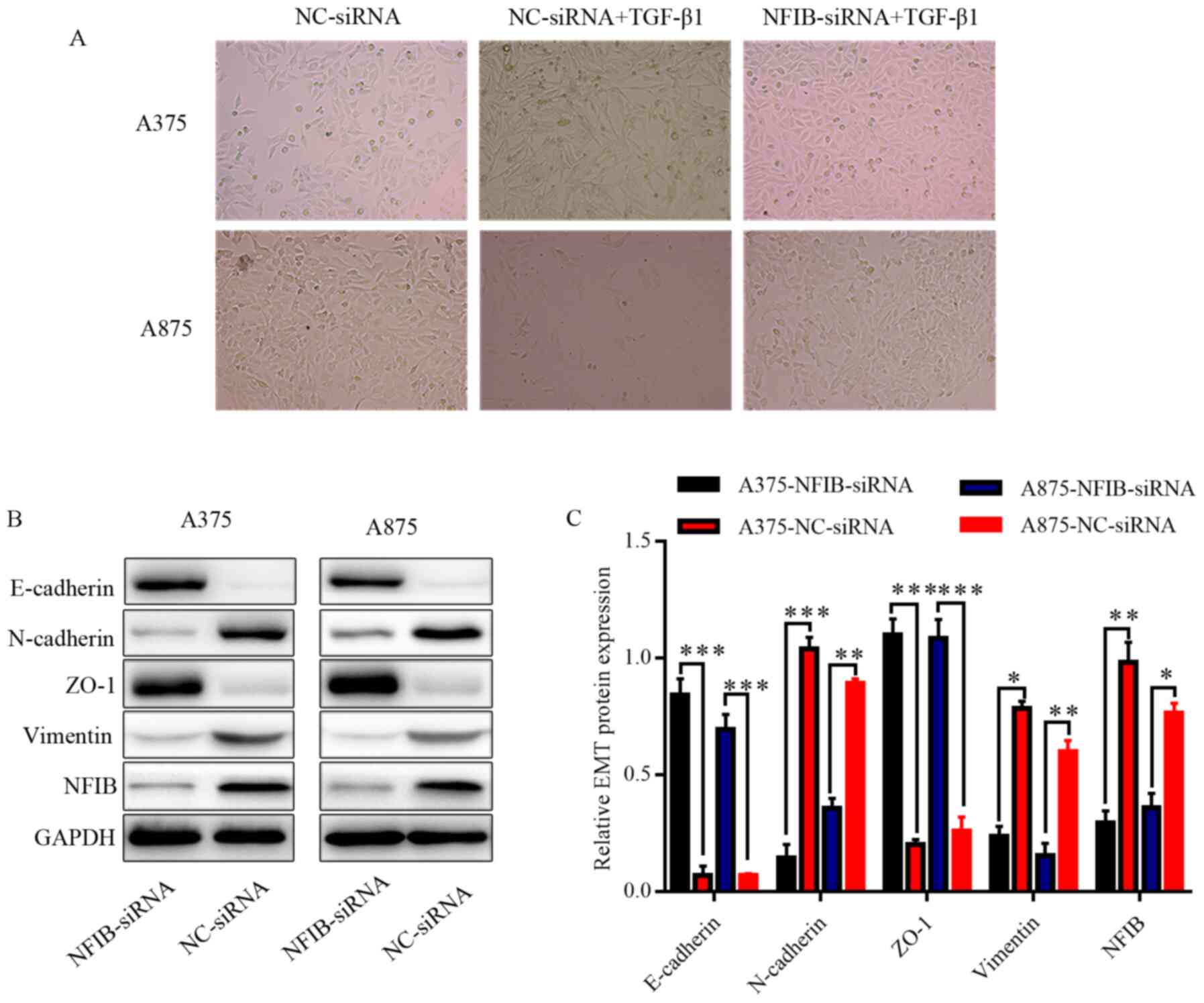 | Figure 7.NFIB promotes invasion and
proliferation of A375 and A875 cells by facilitating EMT in
vitro. (A) Inverted phase-contrast microscopy of melanoma cells
following transfection with NFIB-siRNA or NC-siRNA and TGF-β1
treatment. (B) Western blots and (C) semi-quantitative analysis of
epithelial phenotype markers E-cadherin, N-cadherin, ZO-1,
vimentin, as well as NFIB following transfection. *P<0.05,
**P<0.01, ***P<0.001. NFIB, nuclear factor I/B; siRNA, small
interfering RNA; NC, negative control; ZO-1, zona occludens-1; EMT,
epithelial-mesenchymal transition. |
NFIB positively regulates EMT by
modulating ZEB1 in melanoma cell lines
To further elucidate the specific molecular
mechanism of NFIB in the regulation of EMT in melanoma cells, the
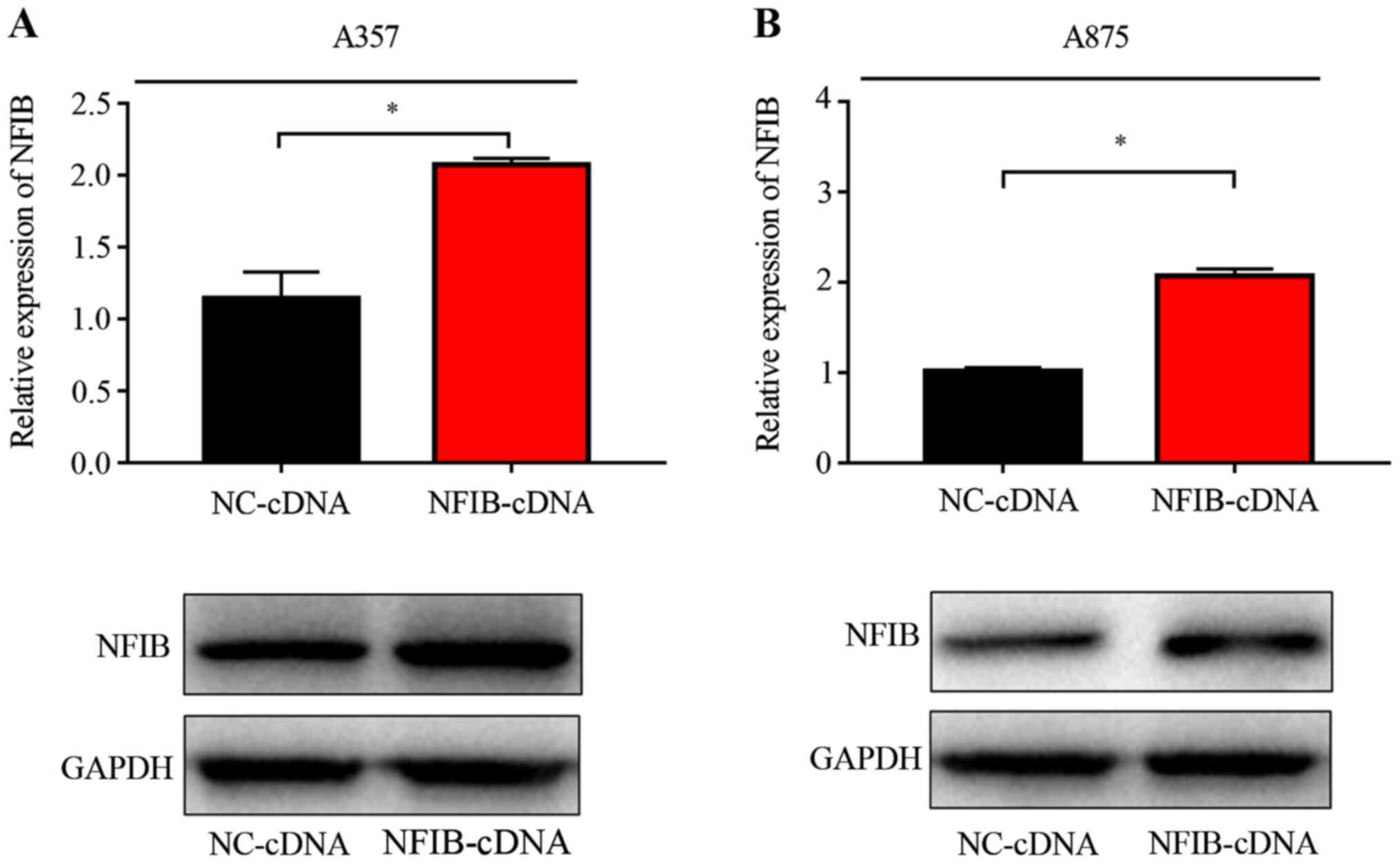
A357 and A875 cell lines were transfected with NFIB-cDNA groups or
NC-cDNA. The overexpression was detected by RT-qPCR and western
blot analysis. NFIB expression was overexpressed in A375 and A875
cells using transfection with NFIB-cDNA compared with NC-cDNA
(Fig. 8).
In gastrointestinal epithelial tumors, it has been
documented that NFIB is closely related to tumor EMT and promotes
the occurrence of EMT in tumor cells by upregulating the expression
of SNAI1. It was speculated that NFIB imbued its functions in
melanoma by modulating its downstream signaling molecules. ZEB1 has
been studied in various tumors and served an important role in EMT.
However, the association between NFIB and ZEB1 remains unclear;
thus, the present study proceeded to explore this in melanoma. In
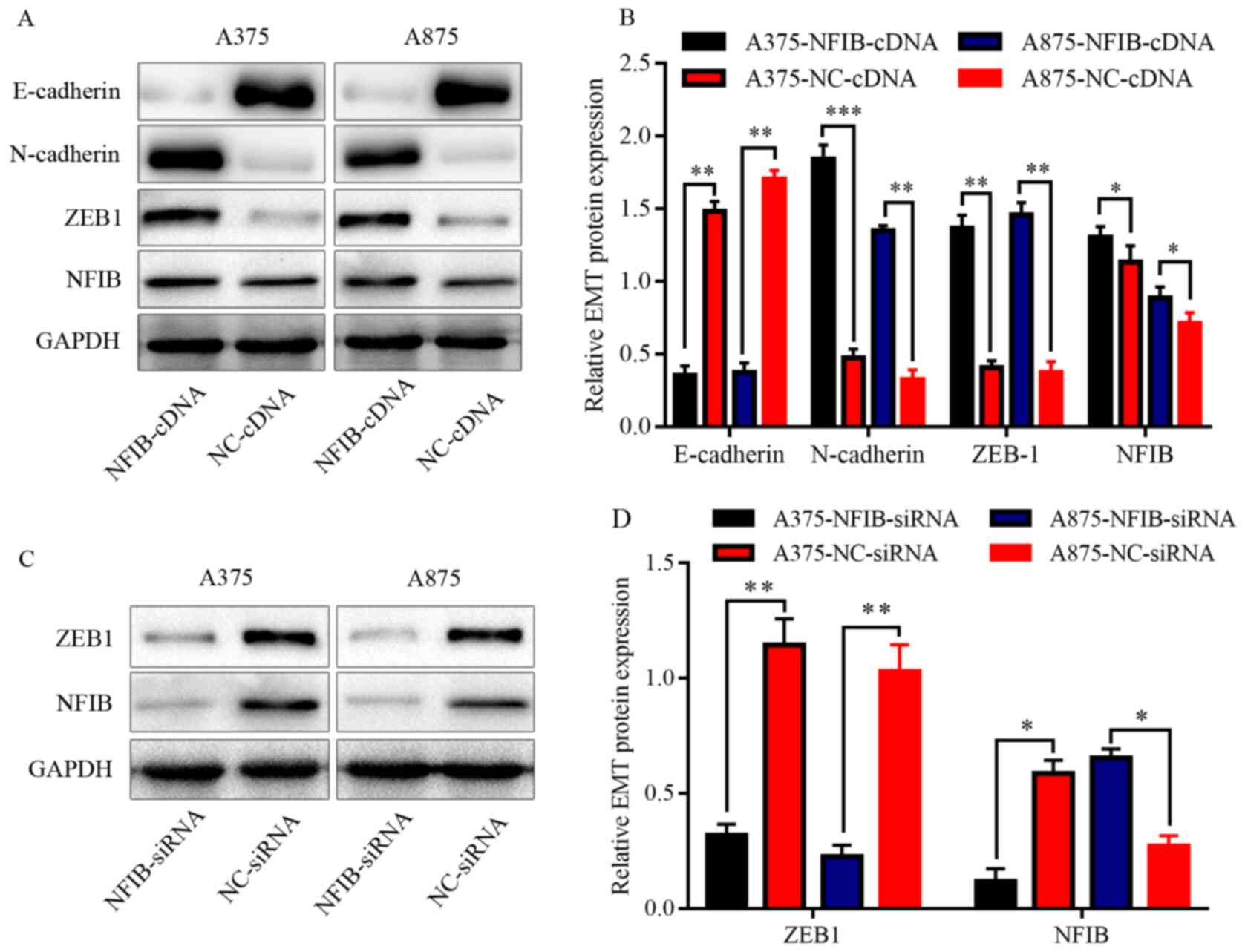
line with the data obtained from melanoma cells in western
blotting, NFIB overexpression was associated with elevated ZEB1and
N-cadherin, while the expression of E-cadherin was downregulated
(Fig. 9A and B). Supporting this,
NFIB silencing downregulated ZEB1 in A375 and A875 cells, compared
with NC-siRNA (Fig. 9C and D),
indicating that ZEB1 was associated with NFIB in the regulation of
EMT in melanoma.
Discussion
NFIB is a member of the NFI nuclear transcription
factor family. NFIB plays an important role in the development of
normal embryos and the formation and development of various organs
by participating in DNA replication and transcription and can be
detected and localized in the nucleus at the early stages of murine
embryonic development at 14.5 days (27,28).
Accumulating studies indicate that NFIB is closely related to the
occurrence of malignant tumors through gene fusion, especially in
adenoid cystic carcinoma (29).
However, some scholars have pointed out that NFIB may have
tumor-suppressive effects as rearrangement leads to NFIB gene
truncation and loss of function, together with other related genes,
such as NFIB-AIG1, NFIB-MAN1A1 and NFIB-NKAIN2 (30–32). Due
to this contradictory, double-sided effect of NFIB, which is
characterized by both oncogenic and tumor-suppressive activity, the
role of NFIB in melanoma remains unclear.
The present study demonstrated that NFIB was
upregulated in human melanoma samples, relative to nevus and normal
skin samples. Furthermore, NFIB expression was associated with a
poor prognosis in melanoma. These are consistent with previous
observational data in patients with non-small-cell lung cancer
(31). Moreover, the present study
revealed that high expression of NFIB was associated with poor
prognosis in patients with melanoma. Consistent with the
aforementioned NSCLC study, NFIB is overexpressed and regulates
cell viability and proliferation during transformation of murine
SCLC, where NFIB amplification is ~15% of primary human SCLC
(33).
Based on this evidence, and to further understand
the biological functions of NFIB in melanoma cell lines,
siRNA-mediated silencing of NFIB was employed to explore the role
of this molecule. NFIB silencing inhibited melanoma cell
proliferation, colony formation, as well as cell migration and
invasion. The cell cycle of melanoma cells was analyzed; however,
the experimental results were not satisfactory, and there is no
suggestion that there was an association between NFIB expression
and the cell cycle in melanoma (data not shown). However, in
another study, NFIB knockdown in TP53-mutated triple-negative
breast cancer cells promoted cell death, triggered cell cycle
arrest and enhanced sensitivity to docetaxel, a first-line
chemotherapeutic drug in breast cancer treatment (34). These results suggest that NFIB might
serve an important role in melanoma.
A growing number of studies have proposed the
behavior of EMT to be a reversible biological process that can
regulate migration and invasion in human melanoma (11,35,36). To
examine the underlying biological functions by which NFIB promotes
migration and invasion in melanoma cells, functional experiments
were performed, through which it was uncovered that NFIB had a
positive effect on the occurrence of EMT. A recent study revealed
that NFIB was involved in the EMT process in colorectal cancer by
encompassing various downstream effector molecules (22,37).
Western blots were not conducted on the groups treated with TGF-β1,
therefore further experimentation is needed to analyze this.
Considering that NFIB regulates numerous cancer-related pathways,
the downstream signaling pathway is of great significance for NFIB
in the development of melanoma.
ZEB1 is a well-characterized transcription factor
that facilitates tumor invasion and metastasis through an
EMT-independent mechanism in carcinoma cells (12,38–40).
Since ZEB1 belongs to the ZEB family and binds the E-box sequence
CACCT, which has two possible binding sites for NFIB, finding a
connection between NFIB would be useful to understand the oncogenic
functions of NIFB in melanoma (41).
Thus, it was hypothesized that NFIB might regulate ZEB1 to
influence the EMT of melanoma. The present data support the idea
that NFIB overexpression can increase ZEB1 expression. Conversely,
the silencing of NFIB could repress ZEB1. However, the present
conclusion only provides a new site for NFIB to regulate the
biological process in melanoma; further extensive research is
needed to verify this hypothesis and determine whether there is an
NFIB-ZEB1 axis that modulates the process of EMT. Besides,
additional in vivo, cell cycle and apoptosis assay
experiments are worth conducting to determine whether NFIB
contributes to other aspects of melanoma malignancy or if there
exist ZEB2-related or other signaling pathways that are also linked
with the function of NFIB in melanoma. Collectively, the present
findings may hold promising insights into NFIB as a novel
prognostic biomarker to predict prognosis and potential therapeutic
target in melanoma.
Acknowledgements
Not applicable.
Funding
The present study was supported by The National
Nature Science Foundation of China (grant no. 81472000).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
RC analyzed and interpreted the patient data. RC and
SG performed the western blotting and functional experiments, and
were major contributors in writing the manuscript. WH performed the
statistical analysis of the experimental data and proofread the
manuscript. YL collected the clinical data. YC contributed to the
conception of the study and designed the research plan. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics Review
Committee at Tongji Hospital affiliated with Huazhong University of
Science and Technology. All participants signed an informed consent
form.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
2
|
Siegel R, DeSantis C, Virgo K, Stein K,
Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al:
Cancer treatment and survivorship statistics, 2012. CA Cancer J
Clin. 62:220–241. 2012. View Article : Google Scholar
|
|
3
|
Boyle GM: Therapy for metastatic melanoma:
An overview and update. Expert Rev Anticancer Ther. 11:725–737.
2011. View Article : Google Scholar
|
|
4
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar
|
|
5
|
Miller AJ and Mihm MC Jr: Melanoma. N Engl
J Med. 355:51–65. 2006. View Article : Google Scholar
|
|
6
|
Mirzaei H, Gholamin S, Shahidsales S,
Sahebkar A, Jaafari MR, Mirzaei HR, Hassanian SM and Avan A:
MicroRNAs as potential diagnostic and prognostic biomarkers in
melanoma. Eur J Cancer. 53:25–32. 2016. View Article : Google Scholar
|
|
7
|
Eggermont AM and Kirkwood JM:
Re-evaluating the role of dacarbazine in metastatic melanoma: What
have we learned in 30 years? Eur J Cancer. 40:1825–1836. 2004.
View Article : Google Scholar
|
|
8
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar
|
|
9
|
Pastushenko I and Blanpain C: EMT
transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar
|
|
10
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar
|
|
11
|
Caramel J, Papadogeorgakis E, Hill L,
Browne GJ, Richard G, Wierinckx A, Saldanha G, Osborne J,
Hutchinson P, Tse G, et al: A switch in the expression of embryonic
EMT-inducers drives the development of malignant melanoma. Cancer
Cell. 24:466–480. 2013. View Article : Google Scholar
|
|
12
|
Denecker G, Vandamme N, Akay O, Koludrovic
D, Taminau J, Lemeire K, Gheldof A, De Craene B, Van Gele M,
Brochez L, et al: Identification of a ZEB2-MITF-ZEB1
transcriptional network that controls melanogenesis and melanoma
progression. Cell Death Differ. 21:1250–1261. 2014. View Article : Google Scholar
|
|
13
|
Blomquist P, Belikov S and Wrange O:
Increased nuclear factor 1 binding to its nucleosomal site mediated
by sequence-dependent DNA structure. Nucleic Acids Res. 27:517–525.
1999. View Article : Google Scholar
|
|
14
|
Gronostajski RM: Roles of the NFI/CTF gene
family in transcription and development. Gene. 249:31–45. 2000.
View Article : Google Scholar
|
|
15
|
Chaudhry AZ, Lyons GE and Gronostajski RM:
Expression patterns of the four nuclear factor I genes during mouse
embryogenesis indicate a potential role in development. Dev Dyn.
208:313–325. 1997. View Article : Google Scholar
|
|
16
|
Li S and Rosen JM: Nuclear factor I and
mammary gland factor (STAT5) play a critical role in regulating rat
whey acidic protein gene expression in transgenic mice. Mol Cell
Biol. 15:2063–2070. 1995. View Article : Google Scholar
|
|
17
|
Kane R, Finlay D, Lamb T and Martin F:
Transcription factor NF 1 expression in involuting mammary gland.
Adv Exp Med Biol. 480:117–122. 2000. View Article : Google Scholar
|
|
18
|
Chen KS, Lim JWC, Richards LJ and Bunt J:
The convergent roles of the nuclear factor I transcription factors
in development and cancer. Cancer Lett. 410:124–138. 2017.
View Article : Google Scholar
|
|
19
|
Stringer BW, Bunt J, Day BW, Barry G,
Jamieson PR, Ensbey KS, Bruce ZC, Goasdoué K, Vidal H, Charmsaz S,
et al: Nuclear factor one B (NFIB) encodes a subtype-specific
tumour suppressor in glioblastoma. Oncotarget. 7:29306–29320. 2016.
View Article : Google Scholar
|
|
20
|
Moon HG, Hwang KT, Kim JA, Kim HS, Lee MJ,
Jung EM, Ko E, Han W and Noh DY: NFIB is a potential target for
estrogen receptor-negative breast cancers. Mol Oncol. 5:538–544.
2011. View Article : Google Scholar
|
|
21
|
Xing D, Bakhsh S, Melnyk N, Isacson C, Ho
J, Huntsman DG, Gilks CB, Ronnett BM and Horlings HM: Frequent
NFIB-associated gene rearrangement in adenoid cystic carcinoma of
the vulva. Int J Gynecol Pathol. 36:289–293. 2017. View Article : Google Scholar
|
|
22
|
Liu Z, Chen J, Yuan W, Ruan H, Shu Y, Ji
J, Wu L, Tang Q, Zhou Z, Zhang X, et al: Nuclear factor I/B
promotes colorectal cancer cell proliferation,
epithelial-mesenchymal transition and 5-fluorouracil resistance.
Cancer Sci. 110:86–98. 2019. View Article : Google Scholar
|
|
23
|
Fane ME, Chhabra Y, Hollingsworth DEJ,
Simmons JL, Spoerri L, Oh TG, Chauhan J, Chin T, Harris L, Harvey
TJ, et al: NFIB Mediates BRN2 driven melanoma cell migration and
invasion through regulation of EZH2 and MITF. Ebiomedicine.
16:63–75. 2017. View Article : Google Scholar
|
|
24
|
Hebert SL, Simmons C, Thompson AL, Zorc
CS, Blalock EM and Kraner SD: Basic helix-loop-helix factors
recruit nuclear factor I to enhance expression of the NaV 1.4
Na+ channel gene. Biochim Biophys Acta. 1769:649–658.
2007. View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Chaffer CL, San Juan BP, Lim E and
Weinberg RA: EMT, cell plasticity and metastasis. Cancer Metastasis
Rev. 35:645–654. 2016. View Article : Google Scholar
|
|
27
|
Barry G, Piper M, Lindwall C, Moldrich R,
Mason S, Little E, Sarkar A, Tole S, Gronostajski RM and Richards
LJ: Specific glial populations regulate hippocampal morphogenesis.
J Neurosci. 28:12328–12340. 2008. View Article : Google Scholar
|
|
28
|
Piper M, Barry G, Harvey TJ, McLeay R,
Smith AG, Harris L, Mason S, Stringer BW, Day BW, Wray NR, et al:
NFIB-mediated repression of the epigenetic factor Ezh2 regulates
cortical development. J Neurosci. 34:2921–2930. 2014. View Article : Google Scholar
|
|
29
|
Becker-Santos DD, Lonergan KM,
Gronostajski RM and Lam WL: Nuclear factor I/B: A master regulator
of cell differentiation with paradoxical roles in cancer.
EBioMedicine. 22:2–9. 2017. View Article : Google Scholar
|
|
30
|
Suzuki H, Aoki K, Chiba K, Sato Y,
Shiozawa Y, Shiraishi Y, Shimamura T, Niida A, Motomura K, Ohka F,
et al: Mutational landscape and clonal architecture in grade II and
III gliomas. Nat Genet. 47:458–468. 2015. View Article : Google Scholar
|
|
31
|
Dooley AL, Winslow MM, Chiang DY, Banerji
S, Stransky N, Dayton TL, Snyder EL, Senna S, Whittaker CA, Bronson
RT, et al: Nuclear factor I/B is an oncogene in small cell lung
cancer. Genes Dev. 25:1470–1475. 2011. View Article : Google Scholar
|
|
32
|
Brayer KJ, Frerich CA, Kang H and Ness SA:
Recurrent fusions in MYB and MYBL1 define a common, transcription
factor-driven oncogenic pathway in salivary gland adenoid cystic
carcinoma. Cancer Discov. 6:176–187. 2016. View Article : Google Scholar
|
|
33
|
Denny SK, Yang D, Chuang CH, Brady JJ, Lim
JS, Grüner BM, Chiou SH, Schep AN, Baral J, Hamard C, et al: Nfib
promotes metastasis through a widespread increase in chromatin
accessibility. Cell. 166:328–342. 2016. View Article : Google Scholar
|
|
34
|
Liu RZ, Vo TM, Jain S, Choi WS, Garcia E,
Monckton EA, Mackey JR and Godbout R: NFIB promotes cell survival
by directly suppressing p21 transcription in TP53-mutated
triple-negative breast cancer. J Pathol. 247:186–198. 2019.
View Article : Google Scholar
|
|
35
|
Lin K, Baritaki S, Militello L, Malaponte
G, Bevelacqua Y and Bonavida B: The role of B-RAF mutations in
melanoma and the induction of EMT via dysregulation of the
NF-ΚB/Snail/RKIP/PTEN circuit. Genes Cancer. 1:409–420. 2010.
View Article : Google Scholar
|
|
36
|
Perrot CY, Javelaud D and Mauviel A:
Insights into the transforming growth factor-beta signaling pathway
in cutaneous melanoma. Ann Dermatol. 25:135–144. 2013. View Article : Google Scholar
|
|
37
|
Wu C, Zhu X, Liu W, Ruan T, Wan W and Tao
K: NFIB promotes cell growth, aggressiveness, metastasis and EMT of
gastric cancer through the Akt/Stat3 signaling pathway. Oncol Rep.
40:1565–1573. 2018.
|
|
38
|
Ren J, Chen Y, Song H, Chen L and Wang R:
Inhibition of ZEB1 reverses EMT and chemoresistance in
docetaxel-resistant human lung adenocarcinoma cell line. J Cell
Biochem. 114:1395–1403. 2013. View Article : Google Scholar
|
|
39
|
Zhang P, Sun Y and Ma L: ZEB1: At the
crossroads of epithelial-mesenchymal transition, metastasis and
therapy resistance. Cell Cycle. 14:481–487. 2015. View Article : Google Scholar
|
|
40
|
Spaderna S, Schmalhofer O, Wahlbuhl M,
Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D, Eger A,
et al: The transcriptional repressor ZEB1 promotes metastasis and
loss of cell polarity in cancer. Cancer Res. 68:537–544. 2008.
View Article : Google Scholar
|
|
41
|
Gheldof A, Hulpiau P, van Roy F, De Craene
B and Berx G: Evolutionary functional analysis and molecular
regulation of the ZEB transcription factors. Cell Mol Life Sci.
69:2527–2541. 2012. View Article : Google Scholar
|