Introduction
Breast cancer is a heterogeneous disease, that may
be subclassified into triple-negative breast cancer (TNBC) and
non-triple-negative breast cancer (NTNBC). Compared with NTNBC,
TNBC has unique clinicopathological characteristics, such as higher
risk of recurrence, larger tumor size, lymph node metastasis and
poor prognosis, and represents a major health concern (1). TNBC accounts for 15–20% of breast
cancers and is characterized by the lack of expression of the
estrogen receptor (ER), progesterone receptor (PR) and human
epidermal growth factor receptor 2 (HER2) (2). Compared with the other subtypes of
breast cancer, TNBC has a relatively early onset and a higher
degree of malignancy (3). In
addition, patients with TNBC have a worse prognosis compared with
other breast cancer subtypes, mainly because TNBC has no specific
targets; therefore, the hormone receptors or HER2 cannot be
targeted with therapy as in other subtypes (4). Although the molecular alterations in
TNBC have been widely investigated, identifying the mechanisms that
regulate the initiation and progression of TNBC may provide further
insight into the development and progression of TNBC.
Lysine-specific demethylase 1 (LSD1), also referred
to as KDM1A and AOF2, was the first histone demethylase to be
discovered (5). LSD1 encodes a
nuclear protein containing a SWIRM domain, a FAD-binding motif and
an amine oxidase domain. The protein is a component of several
histone deacetylase complexes and may silence genes by functioning
as a histone demethylase. Notably, alternative splicing results in
multiple transcript variants. The expression levels of LSD1 in
certain subtypes of breast cancer have been investigated and LSD1
expression has been found to be frequently upregulated in several
human malignancies, including breast (6), prostate (7), lung (8)
and colon (9) cancer, neuroblastoma
(10) and hepatocellular cancer
(11). Notably, Lim et al
(12) reported a significant
positive association between LSD1 upregulation and a negative ER
status. Another previous study identified an inverse correlation
between high LSD1 expression levels and a low PR status (6). Recently, Cao et al (13) demonstrated that the overexpression of
LSD1 promoted breast cancer cell proliferation, migration and
invasion. In addition, it has been suggested that the ability of
LSD1 to promote breast cancer growth and pulmonary metastasis may
involve the resistance to immune checkpoint blockade (14). However, the expression and
significance of LSD1 in breast cancer, particularly in the most
aggressive subtype, TNBC, remain unclear.
The present study aimed to systematically
investigate the expression levels of LSD1 in normal breast tissue,
TNBC and NTNBC tissues using immunohistochemical staining, and
analyze the potential association between LSD1 expression levels
and clinicopathological characteristics of breast cancer.
Materials and methods
Bioinformatics analysis
The expression profile of LSD1 across various types
of human cancer was examined through the Broad Institute FireBrowse
portal (http://firebrowse.org). On the homepage,
‘LSD1’ was typed into the search box and ‘View Expression Profile’
was selected. The boxplots produced plotted the expression levels
of the target gene, with red bars representing tumor samples and
blue bars representing normal samples.
The mRNA expression levels of LSD1 in breast cancer
tissues were compared with their matched normal tissues using The
Cancer Genome Atlas (TCGA) datasets in the Oncomine database
(http://www.oncomine.org). The thresholds used to
obtain the most significant probes of the queried gene for each
microarray dataset included a 2-fold difference in expression
levels between the cancer and normal tissues and P<1×10-4. For
each gene, the mRNA expression levels in three independent datasets
were analyzed. The prognostic values of LSD1 in breast cancer were
analyzed using the Kaplan-Meier plotter (http://kmplot.com/analysis) and the survival rates of
patients with high and low expression levels of LSD1 were
illustrated using a Kaplan-Meier survival plot.
Tissue specimens
The participants in the present study were all newly
diagnosed patients with breast cancer who were treated at the
Department of Breast Surgery of The Affiliated People's Hospital of
Jiangsu University between December 2010 and October 2016. Samples
were collected from 238 patients with breast cancer, including 112
TNBC and 126 NTNBC tissues. In addition, 80 normal tissues adjacent
to the TNBC and 93 normal tissues adjacent to NTNBC were collected.
The pathological data of all patients were complete and included
tumor size, age, lymph node metastasis, clinical stage and
histological type. All patients were regularly followed up; the
follow-ups mainly occurred via phone or partly using an online
platform. The patients survival, living conditions and presence of
any abnormal symptoms were assessed until the follow-up deadline,
which was set as June 2019. No subjects were lost during the
follow-up period. All the tissue specimens were collected after
obtaining informed patient consent and the use of the breast cancer
specimens was approved by The Affiliated People's Hospital of
Jiangsu University Institutional Review Board.
Immunohistochemical analysis
Immunohistochemistry (IHC) was performed to
determine LSD1 expression levels in the tissues. The anti-LSD1
antibody (1:400; cat. no. 2184S) was purchased from Cell Signaling
Technology, Inc.; a rabbit two-step detection kit and DAB color
development kit were purchased from Beijing Zhongshan Jinqiao
Biotechnology, and hematoxylin and water-soluble mounting tablets
were purchased from Boster Biological Technology. All tissue
samples for the experiment were provided by The People's Hospital
of Jiangsu University and tissue chips were prepared with the
assistance of Shanghai Changzheng Hospital. Each experiment used a
known positive tissue section as the positive control and PBS
solution instead of the primary antibody as the negative
control.
IHC scoring
The interpretation of the results was performed
using a double-blind reading under the guidance of a pathology
expert. The lower part that was brownish yellow referred to the
immunoreactive score, which was based on the comprehensive
evaluation of the degree of cellular staining and the percentage of
positive cells. In total, ≥10 fields of view were visualized under
a high magnification and 100 tumors were viewed in each field of
view. The percentage of positive cells was counted and the
following scoring system was used: 0 points (≤5%), 1 point (5–25%),
2 points (25–50%), 3 points (25–75%) and 4 points (≥75%). The
staining intensity was scored as follows: 0 (no staining), 1
(weakly stained), 2 (moderately stained) or 3 (strongly stained).
The LSD1 immunostaining score was calculated as (positive
percentage score) × (staining intensity score). In this experiment,
a score of ≥4 points was considered as positive.
Statistical analysis
Statistical analysis was performed using SPSS 25.0
software (IBM Corp.). The association between the expression levels
of LSD1 and clinicopathological characteristics was analyzed using
a χ2 test or Fisher exact probability method. The log-rank test and
Kaplan-Meier method were also used and the survival analysis was
depicted graphically. Following the univariate analysis, variables
with P<0.05 were used for subsequent multivariate analysis based
on the Cox proportional hazards model. P<0.05 was considered to
indicate a statistically significant difference.
Results
Upregulated LSD1 expression is not
associated with a poor prognosis in breast cancer
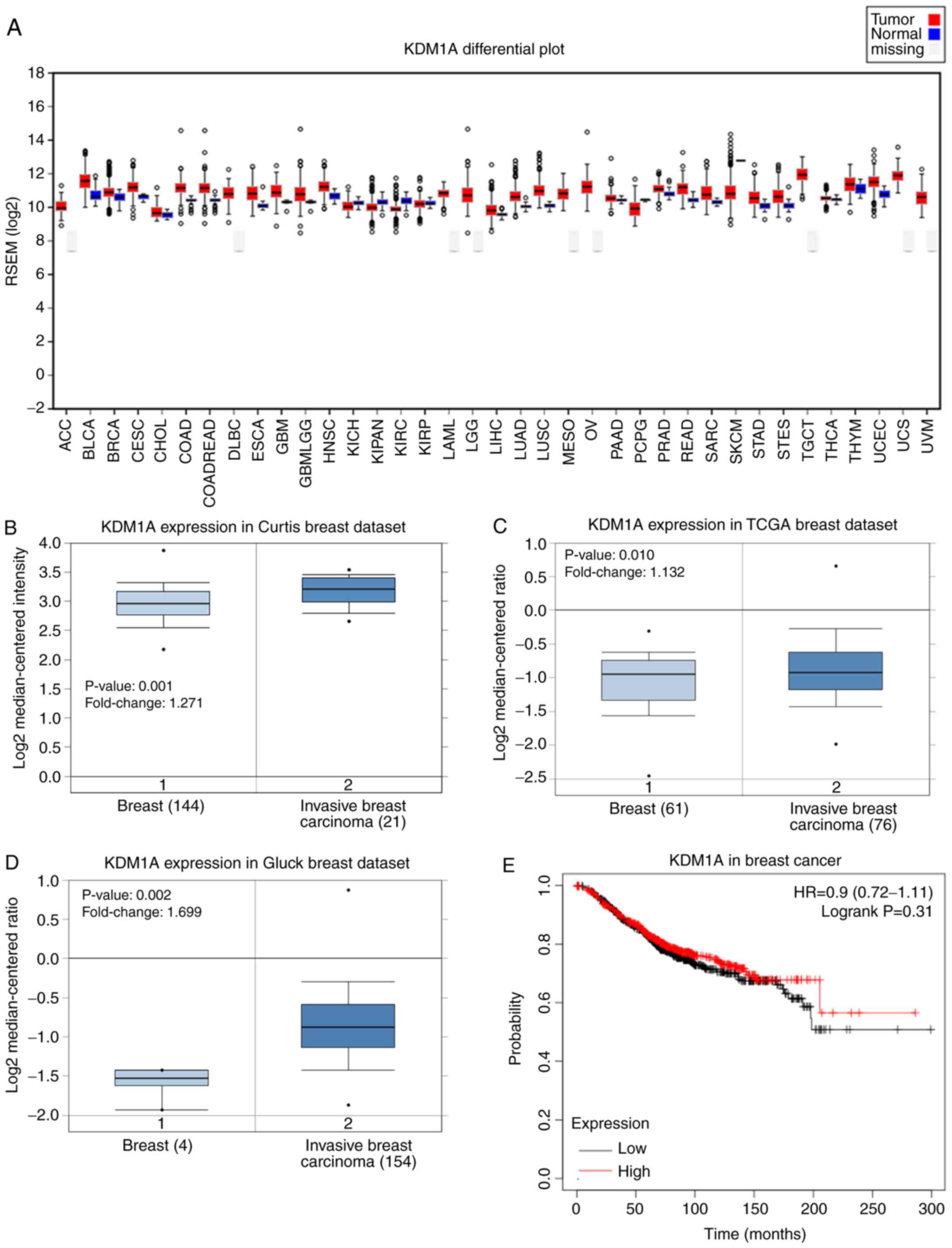
The gene expression levels of LSD1 were analyzed in
37 cases of human cancer using TCGA database. The columns in
Fig. 1A represent the accurate
quantification of the gene and isoform expression levels from the
RNA-Seq data. The results revealed that LSD1 expression levels were
upregulated in almost all cancer tissues compared with their
respective matched normal tissues. The expression levels of LSD1
were the highest in testicular germ cell tumor and the lowest in
cholangiocarcinoma. Notably, the LSD1 gene exhibited a similar
expression pattern in breast cancer (Fig. 1A). The Oncomine database analysis
comparing the cancer tissues with normal tissues also revealed that
the mRNA expression levels of LSD1 were significantly upregulated
in breast cancer tissues compared with the corresponding normal
tissues in three independent analyses (Fig. 1B-D). The results of the Kaplan-Meier
analysis revealed no significant association between the expression
levels of LSD1 and the overall survival rate of patients with
breast cancer (P=0.31; Fig. 1E).
LSD1 expression is upregulated in TNBC
and NTNBC tissues
To further investigate the results obtained through
the bioinformatics analysis, the protein expression levels of LSD1
in breast cancer tissues were also investigated. For this analysis,
samples from 238 patients with breast cancer (including 112 TNBC
and 126 NTNBC cases) who were diagnosed between 2010 and 2016 were
used; >95% of the tumors were ≤5 cm, ~35% of the patients had
lymph node metastasis and ~15% of the patients were diagnosed at an
advanced TNM stage (stage III/IV). The protein expression levels of
LSD1 in both the TNBC and NTNBC subtypes were significantly
upregulated compared with those in adjacent normal tissues
(P<0.001; Table I). Subsequently,
the proportion of LSD1 expression in breast tumors was further
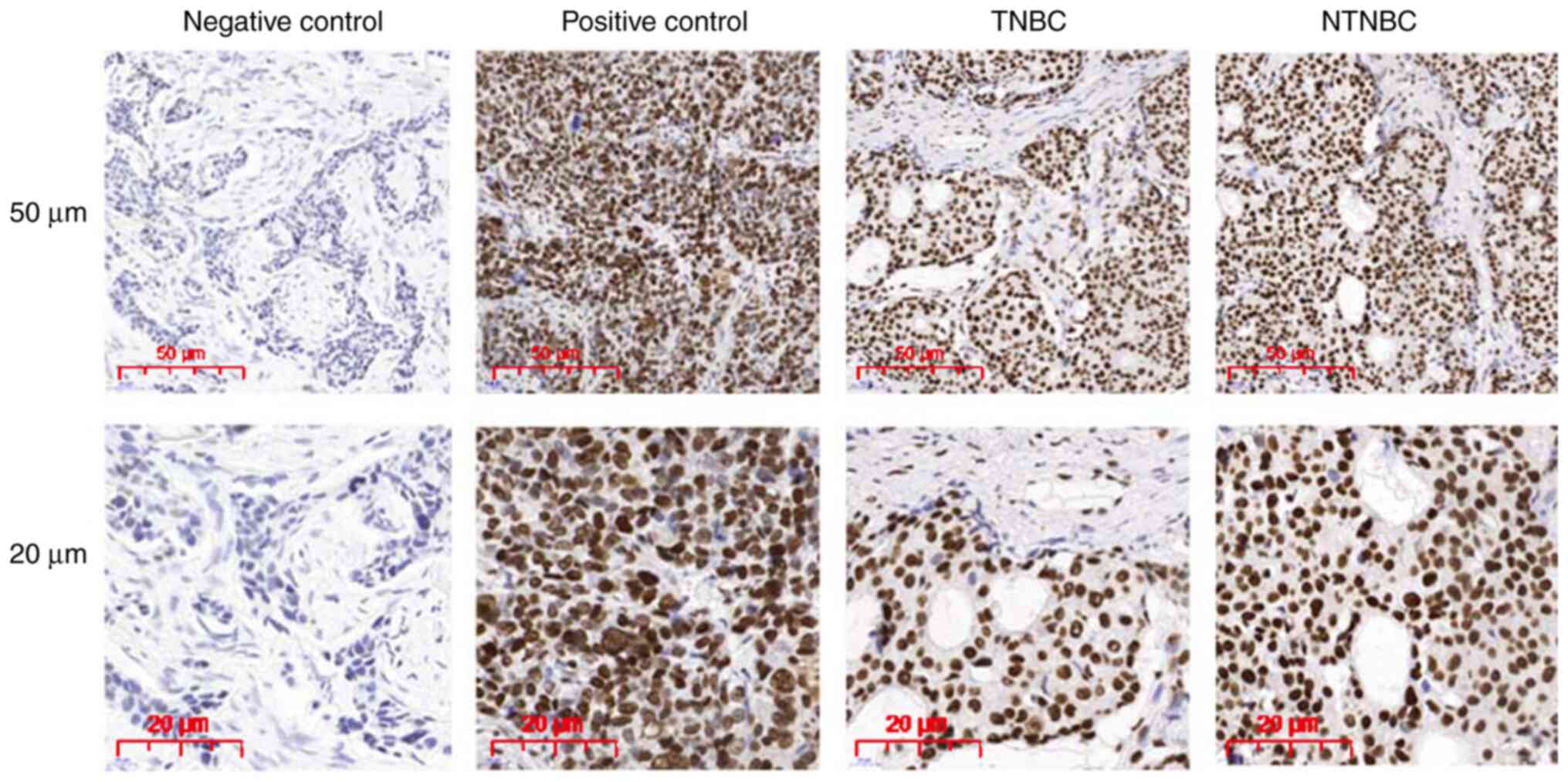
determined. IHC staining revealed that LSD1 was localized mainly to
the cell nucleus in both breast cancer subtypes (Fig. 2). Positive staining for LSD1
expression was recorded in 90 (40%) of the 231 breast cancer
samples; specifically, in 42 (40%) of the 105 TNBCs and 48 (38%) of
the 126 NTNBCs (Fig. 2). There were
no significant differences in the LSD1 expression levels between
the TNBC and NTNBC samples (P>0.05; Table SI).
 | Table I.Expression of LSD1 in breast
cancer. |
Table I.
Expression of LSD1 in breast
cancer.
|
| LSD1 protein
expression |
|
|---|
|
|
|
|
|---|
| Variables | All cases | Negative | Positive | P-value |
|---|
| Breast
cancer |
|
|
| <0.001 |
|
Tumor | 231 | 141 | 90 |
|
|
Normal | 173 | 163 | 10 |
|
| TNBC |
|
|
| <0.001 |
|
Tumor | 105 | 63 | 42 |
|
|
Normal | 80 | 78 | 2 |
|
| NTNBC |
|
|
| <0.001 |
|
Tumor | 126 | 78 | 48 |
|
|
Normal | 93 | 85 | 8 |
|
Association between LSD1 expression
and clinicopathological characteristics
The present study further analyzed the association
between LSD1 expression levels and the clinicopathological
characteristics of breast tumors, including age, tumor size, lymph
node metastasis status and clinical stage. The expression levels of
LSD1 in breast cancer were not significantly associated with any of
the individual clinical indicators (P>0.05; Table II). However, in TNBC, the LSD1
expression levels were significantly associated with age (P=0.019)
and TNM stage (P=0.031), but not with tumor size or lymph node
metastasis (P>0.05; Table II).
Positive staining for LSD1 was detected in 48.9% of the patients
aged >55 years and in 44.9% of patients with TNM stage I+II
disease. In addition, positive staining for LSD1 was also detected
in 33.8% of patients aged <55 years and in 12.5% of patients
with TNM stages III+IV disease. In NTNBC, the expression levels of
LSD1 were not significantly associated with any of the
clinicopathological indicators (P>0.05; Table II). Taken together, these results
suggest that LSD1 expression levels may be associated with patient
age and TNM stage in TNBC.
 | Table II.Correlation of LSD1 expression with
clinicopathological characteristics in breast cancer. |
Table II.
Correlation of LSD1 expression with
clinicopathological characteristics in breast cancer.
|
| LSD1 protein
expression |
|
|---|
|
|
|
|
|---|
| Variables | All cases | Negative | Positive | P-value |
|---|
| Breast cancer |
| Age
(years) |
|
≤40 | 18 (missing 2) | 10 | 6 | 0.901 |
|
>40 | 220 (missing 5) | 131 | 84 |
|
|
≤50 | 93 (missing 2) | 58 | 33 | 0.498 |
|
>50 | 145 (missing 5) | 83 | 57 |
|
|
≤55 | 133 (missing 3) | 86 | 44 | 0.071 |
|
>55 | 105 (missing 4) | 55 | 46 |
|
| Tumor
size (cm) |
|
≤2 | 124 (missing
4) | 77 | 43 | 0.311 |
|
>2 | 114 (missing
3) | 64 | 47 |
|
|
≤4 | 223 (missing
7) | 130 | 86 | 0.313 |
|
>4 | 15 | 11 | 4 |
|
|
≤5 | 231 (missing
7) | 136 | 88 | 0.858 |
|
>5 | 7 | 5 | 2 |
|
| N
stage |
|
|
| 0.406 |
|
N0 | 154 (missing
5) | 88 | 61 |
|
|
N1-3 | 84 (missing 2) | 53 | 29 |
|
| TNM
stage |
|
|
| 0.179 |
|
I+II | 209 (missing
7) | 120 | 82 |
|
|
III+IV | 29 | 21 | 8 |
|
| TNBC |
| Age
(years) |
|
≤40 | 10 (missing 2) | 4 | 4 | 0.711 |
|
>40 | 102 (missing
5) | 59 | 38 |
|
|
≤50 | 38 (missing 2) | 24 | 12 | 0.314 |
|
>50 | 74 (missing 5) | 39 | 30 |
|
|
≤55 | 65 (missing 3) | 43 | 19 | 0.019 |
|
>55 | 47 (missing 4) | 20 | 23 |
|
| Tumor
size (cm) |
|
≤2 | 42 (missing 4) | 24 | 14 | 0.619 |
|
>2 | 70 (missing 3) | 39 | 28 |
|
|
≤4 | 102 (missing
7) | 57 | 38 | 1.000 |
|
>4 | 10 | 6 | 4 |
|
|
≤5 | 107 (missing
7) | 60 | 40 | 1.000 |
|
>5 | 5 | 3 | 2 |
|
| N
stage |
|
|
| 0.113 |
|
N0 | 73 (missing 5) | 37 | 31 |
|
|
N1-3 | 39 (missing 2) | 26 | 11 |
|
| TNM
stage |
|
|
| 0.031 |
|
I+II | 96 (missing 7) | 49 | 40 |
|
|
III+IV | 16 | 14 | 2 |
|
| NTNBC |
| Age
(years) |
|
≤40 | 8 | 6 | 2 | 0.680 |
|
>40 | 118 | 72 | 46 |
|
|
≤50 | 55 | 34 | 21 | 0.986 |
|
>50 | 71 | 44 | 27 |
|
|
≤55 | 68 | 43 | 25 | 0.739 |
|
>55 | 58 | 35 | 23 |
|
| Tumor
size (cm) |
|
≤2 | 82 | 53 | 29 | 0.389 |
|
>2 | 44 | 25 | 19 |
|
|
≤4 | 121 | 73 | 48 | 0.156 |
|
>4 | 5 | 5 | 0 |
|
|
≤5 | 124 | 76 | 48 | 0.525 |
|
>5 | 2 | 2 | 0 |
|
| N
stage |
|
|
| 0.743 |
|
N0 | 81 | 51 | 30 |
|
|
N1-3 | 45 | 27 | 18 |
|
| TNM
stage |
|
|
| 0.528 |
|
I+II | 113 | 71 | 42 |
|
|
III+IV | 13 | 7 | 6 |
|
Association between LSD1 expression
and patient prognosis
To determine the role of LSD1 and the
clinicopathological indicators in predicting breast cancer
outcomes, the postoperative survival in patients with breast cancer
was tracked and analyzed. Patients were classified into
LSD1-positive and LSD1-negative expression groups according to the
IHC results. For breast cancer, the mass size (4 cm), lymph node
metastasis (N) and TNM stage were significantly inversely
associated with patient survival. The survival period of the
patients with a large tumor diameter, late clinical stage and lymph
node metastasis was significantly decreased compared with that of
patients with smaller tumors, diagnosed at an earlier clinical
stage and without metastasis to the lymph nodes (Table III). The χ2 value of the log-rank
test revealed that the cumulative survival difference between tumor
size (4 cm) was the most significant, followed by TNM stage and N
stage. In TNBC, the tumor size (4 cm) and N stage were
significantly inversely associated with patient survival (Table III). The cumulative survival rate
difference in the N stage was the most significant, followed by
tumor size. In NTNBC, the tumor size (4 cm) and TNM stage were
significantly inversely associated with patient survival (Table III). The cumulative survival rate
difference in the TNM stage was the most significant, followed by
tumor size. Taken together, these data suggested that tumor
metastasis, but not LSD1 expression levels, may be a major factor
associated with mortality in patients with TNBC and NTNBC.
 | Table III.Univariate analysis of the
association between LSD1 expression and clinicopathological
variables in patients with breast cancer (log-rank test). |
Table III.
Univariate analysis of the
association between LSD1 expression and clinicopathological
variables in patients with breast cancer (log-rank test).
| Variables | All cases | 95% CI | P-value |
|---|
| Breast cancer |
| Age
(years) |
|
≤40 | 18 | 86.864
(70.122–103.606) | 0.853 |
|
>40 | 220 | 87.143
(83.316–90.971) |
|
|
≤50 | 93 | 91.935
(85.641–98.229) | 0.571 |
|
>50 | 145 | 86.157
(81.339–90.974) |
|
|
≤55 | 133 | 91.712
(86.403–97.020) | 0.510 |
|
>55 | 105 | 85.889
(80.247–91.531) |
|
| Tumor
size (cm) |
|
≤2 | 124 | 92.546
(87.114–97.978) | 0.333 |
|
>2 | 114 | 88.493
(82.501–94.484) |
|
|
≤4 | 223 | 92.274
(88.225–96.323) | <0.001 |
|
>4 | 15 | 65.188
(48.425–81.950) |
|
|
≤5 | 231 | 90.706
(86.569–94.842) | 0.753 |
|
>5 | 7 | 88.750
(75.909–101.591) |
|
| N
stage |
|
| 0.028 |
|
N0 | 154 | 93.754
(89.110–98.399) |
|
|
N1-3 | 84 | 81.177
(74.504–87.851) |
|
| TNM
stage |
|
| 0.004 |
|
I+II | 209 | 92.601
(88.459–96.743) |
|
|
III+IV | 29 | 73.670
(61.913–85.426) |
|
| LSD1
protein expression |
|
| 0.486 |
|
Negative | 141 | 88.697
(82.812–94.582) |
|
|
Positive | 90 | 92.599
(86.766–98.432) |
|
| TNBC |
| Age
(years) |
|
≤40 | 10 | 88.200
(66.128–110.272) | 0.619 |
|
>40 | 102 | 84.273
(78.434–90.112) |
|
|
≤50 | 38 | 89.054
(78.581–99.526) | 0.748 |
|
>50 | 74 | 84.362
(77.649–91.076) |
|
|
≤55 | 65 | 87.595
(79.387–95.803) | 0.934 |
|
>55 | 47 | 85.777
(77.890–93.665) |
|
| Tumor
size (cm) |
|
≤2 | 42 | 87.073
(76.790–97.356) | 0.866 |
|
>2 | 70 | 88.353
(80.870–95.836) |
|
|
≤4 | 102 | 89.952
(83.867–96.037) | 0.027 |
|
>4 | 10 | 63.828
(41.666–85.989) |
|
|
≤5 | 107 | 87.576
(81.283–93.870) | 0.712 |
|
>5 | 5 | 89.333
(72.263–106.404) |
|
| N
stage |
|
| 0.016 |
|
N0 | 73 | 93.124
(86.437–99.812) |
|
|
N1-3 | 39 | 75.396
(65.027–85.766) |
|
| TNM
stage |
|
| 0.278 |
|
I+II | 96 | 89.218
(82.880–95.556) |
|
|
III+IV | 16 | 74.643
(57.360–91.926) |
|
| LSD1
protein expression |
|
| 0.248 |
|
Negative | 63 | 84.203
(75.189–93.217) |
|
|
Positive | 42 | 92.302
(84.161–100.444) |
|
| NTNBC |
| Age
(years) |
|
≤40 | 8 | 73.075
(52.867–93.283) | 0.411 |
|
>40 | 118 | 86.088
(81.640–90.536) |
|
|
≤50 | 55 | 85.974
(79.409–92.539) | 0.842 |
|
>50 | 71 | 85.361
(79.425–91.296) |
|
|
≤55 | 68 | 87.806
(82.385–93.226) | 0.263 |
|
>55 | 58 | 83.084
(76.003–90.165) |
|
| Tumor
size (cm) |
|
≤2 | 82 | 87.075
(81.803–92.348) | 0.268 |
|
>2 | 44 | 79.614
(72.530–86.698) |
|
|
≤4 | 121 | 86.586
(82.188–90.984) | 0.004 |
|
>4 | 5 | 68.200
(43.333–93.067) |
|
|
≤5 | 124 | 85.868
(81.437–90.299) | 0.248 |
|
>5 | 2 | 87.000
(87.000–87.000) |
|
| N
stage |
|
| 0.541 |
|
N0 | 81 | 86.275
(80.815–91.735) |
|
|
N1-3 | 45 | 84.282
(76.821–91.743) |
|
| TNM
stage |
|
| 0.002 |
|
I+II | 113 | 87.472
(83.025–91.918) |
|
|
III+IV | 13 | 70.019
(57.408–82.63) |
|
| LSD1
protein expression |
|
| 0.895 |
|
Negative | 78 | 85.406
(79.533–91.278) |
|
|
Positive | 48 | 85.399
(78.452–92.346) |
|
Cox regression analysis was subsequently used to
calculate various prognostic parameters for the survival of
patients with TNBC and NTNBC. Univariate analysis identified four
prognostic factors: TNM stage (I+II vs. III+IV), N stage (N0 vs.
N1-3), LSD1 expression levels (negative or positive) and tumor size
(≤4 vs. >4 cm). However, upon performing multivariate Cox
proportional hazard regression, increased LSD1 expression levels
were not identified as a significant independent predictor of poor
survival in patients with breast cancer (P>0.05; Table IV), which was consistent with the
results of the bioinformatics analysis. Taken together, these data
suggested that LSD1 expression levels may not be inversely
associated with a poor prognosis in breast cancer and, in fact, the
best predictor of poor prognosis in TNBC may be the N stage
(P<0.05; Table IV).
 | Table IV.Cox multivariate analysis of
prognostic factors of overall survival in breast cancer. |
Table IV.
Cox multivariate analysis of
prognostic factors of overall survival in breast cancer.
| Variables | Hazards ratio | 95% CI | P-value |
|---|
| Breast cancer |
| LSD1
protein expression (negative vs. positive) | 0.928 | 0.498–1.728 | 0.813 |
| Tumor
size, cm (≤4 vs. >4) | 2.529 | 0.998–6.411 | 0.05 |
| N stage
(N0 vs. N1-3) | 1.481 | 0.716–3.061 | 0.289 |
| TNM
stage (I+II vs. III+IV) | 1.351 | 0.522–3.495 | 0.535 |
| TNBC |
| LSD1
protein expression (negative vs. positive) | 0.699 | 0.301–1.624 | 0.405 |
| Tumor
size, cm (≤4 vs. >4) | 2.479 | 0.785–7.826 | 0.122 |
| N stage
(N0 vs. N1-3) | 2.714 | 1.112–6.622 | 0.028 |
| TNM
stage (I+II vs. III+IV) | 0.517 | 0.151–1.769 | 0.293 |
| NTNBC |
| LSD1
protein expression (negative vs. positive) | 1.145 | 0.415–3.163 | 0.794 |
| Tumor
size, cm (≤4 vs. >4) | 2.663 | 0.556–12.763 | 0.221 |
| N stage
(N0 vs. N1-3) | 0.483 | 0.107–2.185 | 0.344 |
| TNM
stage (I+II vs. III+IV) | 5.362 | 0.948–30.348 | 0.058 |
Discussion
LSD1 is a member of the monoaminoxidase enzyme
family, which play an important role in controlling gene expression
through histone modifications (15).
Consistent with the perceived role of LSD1 in cell proliferation,
overexpression of LSD1 has been reported in a diverse range of
human tumors, including breast cancer (6). For example, Serce et al
(6) reported upregulated expression
levels of LSD1 in invasive ductal breast cancer. The expression
levels of LSD1 were also found to increase with progression of
ductal carcinoma in situ (DCIS) to invasive ductal carcinoma
(6). Similarly, a study by Scoumanne
and Chen (16), which analyzed the
role of LSD1 in the human malignant breast cancer cell line MCF7,
discovered that downregulation of LSD1 expression reduced the
number of proliferating breast cancer cells. However, the
expression levels of LSD1 in TNBC and NTNBC have not been analyzed
to date. Therefore, to the best of our knowledge, the present study
was the first to systematically analyze LSD1 expression levels in
TNBC and NTNBC.
From the clinical data, it may be concluded that the
survival of patients with TNBC is poor. The poor prognosis of TNBC
may be due to its biological characteristics, such as younger age
at onset, higher rate of breast cancer family history, larger tumor
size, more advanced clinical stage at diagnosis, higher rate of
lymph node metastasis, higher histological grade, earlier
recurrence and metastasis, and resistance to endocrine and targeted
therapy. The present study revealed that LSD1 expression levels
were upregulated in ~40% (90/231) of breast cancer cases.
Importantly, LSD1 was found to be upregulated in both TNBC and
NTNBC. The expression levels of LSD1 also appeared to be similar
between older and younger patients with breast cancer. In addition,
the expression levels of LSD1 were not significantly associated
with poor prognosis or the cumulative survival rate of
postoperative patients with breast cancer. Bioinformatics analysis
also revealed that LSD1 expression levels were not significantly
associated with the prognosis of breast cancer. However, Nagasawa
et al (17) concluded that
the upregulation of LSD1 was a poor prognostic factor in breast
cancer, particularly the basal-like subtype of invasive breast
cancer. This inconsistency may be due to the insufficient data on
LSD1 expression levels obtained via IHC staining in the present
study, which may lead to differences in the LSD1 prognostic impact.
In the present study, LSD1 expression levels were found to be
closely associated with the breast tumor size and distant
metastasis (TNM stage) in TNBC. Lim et al (12) demonstrated that the overexpression of
LSD1 in breast cancer was significantly positively correlated with
the absence of the ER. In addition, the knockdown of LSD1 by small
interfering RNA induced the regulation of multiple
proliferation-related genes, including p21, ERBB2 and CNA2, thereby
inhibiting the proliferation of breast cancer cells (12). However, the specific mechanisms
underlying the association between LSD1 and cancer development have
not been fully elucidated. Upregulated expression levels of LSD1
have been reported to be a hallmark of breast cancer cells
(18). However, according to the
data obtained in the present study, LSD1 was not found to play an
important role in breast cancer progression or metastasis. Thus,
LSD1 may be a secondary factor associated with breast
cancer-related mortality.
LSD1 may be involved in the carcinogenesis and
progression of breast cancer. It has been reported that the
CtBP/LSD1/COREST complex interacts with ZNF516 to participate in
the epithelial-to-mesenchymal transition (EMT) process, and
inhibits the proliferative and invasive ability of breast cancer
cells (19). LSD1 was also
discovered in another study to regulate the EGFR signaling pathway
and affect EMT (11), thereby
inhibiting breast cancer cell invasion. LSD1 appears to serve a
role in regulating the expression of oncogenic proteins in breast
cancer cells (20), and the
upregulation of LSD1 may be an early tumor-promoting event in
breast cancer. Thus, LSD1 may be involved in the occurrence and
development of TNBC, in addition to NTNBC, which may hold
therapeutic promise. The combination of a LSD1 inhibitor (21), pargyline (22) and the HDAC inhibitor SAHA
(vorinostat) (23) significantly
inhibited the growth and apoptosis of TNBC cells. Therefore, to
further elucidate the specific mechanism underlying the role of
LSD1 in breast cancer in vivo and in vitro, future
studies should explore the association between LSD1 and breast
cancer cell proliferation, migration and invasion.
In conclusion, the findings of the present study
indicated that LSD1 expression levels may be upregulated in breast
cancer and the expression levels of LSD1 may be associated with
clinical stage in TNBC. However, the detection of LSD1 was not
found to be a marker for the early diagnosis of breast cancer or a
potential target for early treatment strategies. The results of the
present study may shed some light on the complex epigenetic
regulatory mechanisms of breast cancer, which may help identify
novel therapeutic targets.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Miao Chen
(Department of Pathology, The Affiliated People's Hospital of
Jiangsu University, Zhenjiang), for reviewing the
clinicopathological data in this manuscript.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81170573)
and the Key Research and Development Program of Jiangsu Province
(grant no. BE2020678).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors contributions
GS and QL conceived and designed the study; KZ
performed the experiments; KZ and YL wrote the manuscript; YL, TH,
AS, ML and WB were involved in the conception of the study. All
authors agree to be accountable for all aspects of the research in
ensuring that the accuracy or integrity of any part of the work are
appropriately investigated and resolved. All authors have read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Human breast tissue specimens were preserved in the
Breast Cancer Tissue Bank at The Affiliated People's Hospital of
Jiangsu University (Zhenjiang, China). All the tissue specimens for
this study were collected after obtaining informed patient consent
and the use of the breast cancer specimens was approved by The
Affiliated People's Hospital of Jiangsu University Institutional
Review Board.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Redig AJ and McAllister SS: Breast cancer
as a systemic disease: A view of metastasis. J Intern Med.
274:113–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sharma P: Biology and management of
patients with triple-negative breast cancer. Oncologist.
21:1050–1062. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ho AY, Gupta G, King TA, Perez CA, Patil
SM, Rogers KH, Wen YH, Brogi E, Morrow M, Hudis CA, et al:
Favorable prognosis in patients with T1a/T1bN0 triple-negative
breast cancers treated with multimodality therapy. Cancer.
118:4944–4952. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shen M, Jiang YZ, Wei Y, Ell B, Sheng X,
Esposito M, Kang J, Hang X, Zheng H, Rowicki M, et al: Tinagl1
suppresses triple-negative breast cancer progression and metastasis
by simultaneously inhibiting integrin/FAK and EGFR signaling.
Cancer Cell. 35:64–80.e7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen Y, Yang Y, Wang F, Wan K, Yamane K,
Zhang Y and Lei M: Crystal structure of human histone
lysine-specific demethylase 1 (LSD1). Proc Natl Acad Sci USA.
103:13956–13961. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Serce N, Gnatzy A, Steiner S, Lorenzen H,
Kirfel J and Buettner R: Elevated expression of LSD1
(Lysine-specific demethylase 1) during tumour progression from
pre-invasive to invasive ductal carcinoma of the breast. BMC Clin
Pathol. 12:132012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ketscher A, Jilg CA, Willmann D, Hummel B,
Imhof A, Rüsseler V, Hölz S, Metzger E, Müller JM and Schüle R:
LSD1 controls metastasis of androgen-independent prostate cancer
cells through PXN and LPAR6. Oncogenesis. 3:e1202014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hayami S, Kelly JD, Cho HS, Yoshimatsu M,
Unoki M, Tsunoda T, Field HI, Neal DE, Yamaue H, Ponder BA, et al:
Overexpression of LSD1 contributes to human carcinogenesis through
chromatin regulation in various cancers. Int J Cancer. 128:574–586.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ding J, Zhang ZM, Xia Y, Liao GQ, Pan Y,
Liu S, Zhang Y and Yan ZS: LSD1-mediated epigenetic modification
contributes to proliferation and metastasis of colon cancer. Br J
Cancer. 109:994–1003. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Amente S, Milazzo G, Sorrentino MC,
Ambrosio S, Di Palo G, Lania L, Perini G and Majello B:
Lysine-specific demethylase (LSD1/KDM1A) and MYCN cooperatively
repress tumor suppressor genes in neuroblastoma. Oncotarget.
6:14572–14583. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shao G, Wang J, Li Y, Liu X, Xie X, Wan X,
Yan M, Jin J, Lin Q, Zhu H, et al: Lysine-specific demethylase 1
mediates epidermal growth factor signaling to promote cell
migration in ovarian cancer cells. Sci Rep. 5:153442015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lim S, Janzer A, Becker A, Zimmer A,
Schüle R, Buettner R and Kirfel J: Lysine-specific demethylase 1
(LSD1) is highly expressed in ER-negative breast cancers and a
biomarker predicting aggressive biology. Carcinogenesis.
31:512–520. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao C, Vasilatos SN, Bhargava R, Fine JL,
Oesterreich S, Davidson NE and Huang Y: Functional interaction of
histone deacetylase 5 (HDAC5) and lysine-specific demethylase 1
(LSD1) promotes breast cancer progression. Oncogene. 36:133–145.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qin Y, Vasilatos SN, Chen L, Wu H, Cao Z,
Fu Y, Huang M, Vlad AM, Lu B, Oesterreich S, et al: Inhibition of
histone lysine-specific demethylase 1 elicits breast tumor immunity
and enhances antitumor efficacy of immune checkpoint blockade.
Oncogene. 38:390–405. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Forneris F, Battaglioli E, Mattevi A and
Binda C: New roles of flavoproteins in molecular cell biology:
Histone demethylase LSD1 and chromatin. FEBS J. 276:4304–4312.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Scoumanne A and Chen X: The
lysine-specific demethylase 1 is required for cell proliferation in
both p53-dependent and -independent manners. J Biol Chem.
282:15471–15475. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nagasawa S, Sedukhina AS, Nakagawa Y,
Maeda I, Kubota M, Ohnuma S, Tsugawa K, Ohta T, Roche-Molina M,
Bernal JA, et al: LSD1 overexpression is associated with poor
prognosis in basal-like breast cancer, and sensitivity to PARP
inhibition. PLoS One. 10:e01180022015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Boulding T, McCuaig RD, Tan A, Hardy K, Wu
F, Dunn J, Kalimutho M, Sutton CR, Forwood JK, Bert AG, et al: LSD1
activation promotes inducible EMT programs and modulates the tumour
microenvironment in breast cancer. Sci Rep. 8:732018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li L, Liu X, He L, Yang J, Pei F, Li W,
Liu S, Chen Z, Xie G, Xu B, et al: ZNF516 suppresses EGFR by
targeting the CtBP/LSD1/CoREST complex to chromatin. Nat Commun.
8:6912017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hu X, Xiang D, Xie Y, Tao L, Zhang Y, Jin
Y, Pinello L, Wan Y, Yuan GC and Li Z: LSD1 suppresses invasion,
migration and metastasis of luminal breast cancer cells via
activation of GATA3 and repression of TRIM37 expression. Oncogene.
38:7017–7034. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sehrawat A, Gao L, Wang Y, Bankhead A III,
McWeeney SK, King CJ, Schwartzman J, Urrutia J, Bisson WH, Coleman
DJ, et al: LSD1 activates a lethal prostate cancer gene network
independently of its demethylase function. Proc Natl Acad Sci USA.
115:E4179–E4188. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang M, Liu X, Guo J, Weng X, Jiang G,
Wang Z and He L: Inhibition of LSD1 by Pargyline inhibited process
of EMT and delayed progression of prostate cancer in vivo. Biochem
Biophys Res Commun. 467:310–315. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu J, Sun J, Wang P, Ma X and Li S:
Pendant HDAC inhibitor SAHA derivatised polymer as a novel prodrug
micellar carrier for anticancer drugs. J Drug Target. 26:448–457.
2018. View Article : Google Scholar : PubMed/NCBI
|