Introduction
Ovarian serous carcinoma has the highest mortality
among the gynecological cancers (1,2). In
general, patients with malignant ovarian tumors are diagnosed at an
advanced stage (stage III or IV) because of the initial lack of
specific symptoms or biomarkers (3).
The standard treatment for advanced ovarian serous carcinoma is
maximal debulking surgery followed by platinum-taxane combination
chemotherapy (4). Despite a good
initial response to chemotherapy, many patients become resistant to
first-line drugs and the prognosis for these individuals is
particularly poor (5). The effect of
re-treatment with platinum-based chemotherapy at the time of
relapse depends on the duration of platinum non-use, which is
defined as the time between last platinum administration and
relapse (6). If the duration between
last platinum administration and recurrence is less than 6 months,
the cancer is defined as platinum-resistant, and if it is longer
than 6 months, it is defined as platinum-sensitive (7).
Sensitivity to platinum-based chemotherapy is one of
the major prognostic factors for ovarian serous carcinoma (8), and prediction of platinum sensitivity
is therefore of great importance for the development of treatment
strategies for patients with this disease. Unfortunately, however,
no effective methods currently exist to predict platinum
sensitivity in ovarian serous carcinoma. Effective prediction of
platinum-sensitivity would enable the selection of only those
patients who represent good candidates for platinum treatment,
while sparing resistant patients the adverse side-effects
associated with these drugs. Therefore, it is important to identify
specific biomarkers that predict platinum sensitivity to improve
patient prognosis in ovarian serous carcinoma.
Protein arginine methyltransferase 1 (PRMT1) is the
major arginine methyltransferase in mammalian cells and is required
for normal embryonic development, cell cycle progression, cell
viability, and signaling (9–12). PRMT1 has also been reported to be a
driver of tumorigenesis and tumor progression (13), and to contribute to chemotherapy
resistance by methylating apoptosis signal-regulated kinase 1
(ASK1) (14).
In this study, we evaluated the association between
PRMT1 expression and sensitivity to platinum-based chemotherapy in
ovarian serous carcinoma. Our aim was to identify new biomarkers
and explore new treatment options to improve the prognosis of
patients with platinum-resistant disease.
Materials and methods
Patients and samples
We reviewed 51 cases of stage III or IV serous
ovarian cancer in patients aged 75 and under treated at the Osaka
City University Hospital between January 2005 and December 2013.
Patients were assigned to one of two groups according to their
platinum sensitivity. Patients assigned to the platinum-sensitive
group (n=26) had received chemotherapy with platinum after maximal
tumor debulking surgery and had no tumor recurrence within 6 months
of the date of the last platinum dose. Patients assigned to the
platinum-resistant group (n=25) had received platinum-based
chemotherapy after maximal debulking surgery and had tumor
recurrence within 6 months of the date of the last platinum dose.
The study was approved by the Institutional Review Board (IRB) of
Osaka City University Hospital after written informed consent was
obtained from all patients prior to treatment (IRB no. 4247).
Immunohistochemistry
PRMT1 expression in ovarian serous carcinoma
specimens was determined by immunohistochemical analysis of
paraffin-embedded tissue sections using the Dako LSAB2 peroxidase
kit (cat. no. K0675; Agilent Technologies, Inc.). Four-micron-thick
tissue sections were deparaffinized, rehydrated, and soaked in 3%
hydrogen peroxide for 10 min at room temperature to block
endogenous peroxidase activity. Antigen retrieval was performed by
autoclaving sections in 10 mM citrate buffer (pH 6.0) at 110°C for
20 min. After washing with phosphate-buffered saline (PBS),
sections were incubated overnight at 4°C in a 1:100 dilution of
rabbit polyclonal antibody against PRMT1 (cat. no. ab-70724;
Abcam). Sections were washed with PBS for 15 min and incubated with
biotinylated goat IgG secondary antibody (Dako; Agilent
Technologies, Inc.) for 10 min. Additional sections were incubated
with streptavidin-peroxidase complexes and 3,3′-diaminobenzidine
was used as a chromogen. Finally, all sections were stained with
hematoxylin. As a control, primary antibodies were omitted to
examine the specificity of the immunohistochemical reaction.
PRMT1 expression scoring was calculated by
multiplying the percentage score of positive tumor cells by the
staining intensity score using the weighted scores method of
Sinicrope et al (15). The
percentage score of positive tumor cells was defined as follows: 0
(<5%), 1 (5-25%), 2 (25-50%), 3 (50-75%), and 4 (>75%). The
staining intensity was scored as follows: 0 (no staining), 1 (weak
staining), 2 (moderate staining), or 3 (strong staining).
Cell culture
The human ovarian serous carcinoma cell line OVSAHO
(cat. no. JCRB1046; National Institute of Biomedical Sciences) was
cultured in RPMI medium (Gibco; Thermo Fisher Scientific, Inc.),
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.), penicillin (100 U/ml) and streptomycin (100
U/ml), and the medium was changed every other day. Cell cultures
were maintained at 37°C in a humidified incubator containing a 5%
CO2 atmosphere. After collection, cells were stored at
−80°C for subsequent analysis by reverse transcription-quantitative
PCR (RT-qPCR).
Chemosensitivity assay and siRNA
transfection
OVSAHO cells were seeded in 96-well plates at 3,000
cells per well and divided into groups that were transfected with
or without PRMT1-specific siRNA (siPRMT1). Transfection was
performed using Lipofectamine RNAiMax (Invitrogen; Thermo Fisher
Scientific, Inc.).
The siPRMT1 sense sequence was
5′-GCAACUCCAUGUUUCAUAAtt-3′, and the antisense sequence was
5′-UUAUGAAACAUGGAGUUGCgg-3′. After 24 h of culture in the presence
or absence of siPRMT1, cell cultures received fresh medium
containing cisplatin at concentrations of 0, 5.0, 10, or 50 µM, or
carboplatin at concentrations of 0, 50, 100, or 150 µM, and then
cultured for an additional 48 h. To confirm cell viability, 10 µl
of CCK-8 and 100 µl of RPMI were added to each well using the Cell
Counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.). The
plate was then incubated at 37°C for 2 h and the absorbance
subsequently measured at 450 nm using a microplate reader (Corona
Electric Co., Ltd.). Dose-response curves were prepared to
determine the percentage of viable cells relative to the untreated
control.
RT-qPCR
Total RNA was extracted from OVSAHO cells using the
RNeasy Mini kit (QIAGEN GmbH). RNA was reverse-transcribed using
the High Capacity cDNA Reverse Transcription kit (Thermo Fisher
Scientific, Inc.) and PRMT1 mRNA expression was determined using
the TaqMan Gene Expression Assay and an ABI 7500 Fast Real-Time PCR
System (Applied Biosystems; Thermo Fisher Scientific, Inc.). mRNA
levels were normalized to those of the GAPDH house-keeping control.
The TaqMan PRMT1 (Hs01587651_g1) and GAPDH (Hs99999905_m1) assays
were used for RT-qPCR assays (Thermo Fisher Scientific, Inc.). The
2−ΔΔCq method was used to analyze the relative changes
in gene expression for RT-qPCR experiments (16).
Statistical analysis
Data are presented as means ± standard deviation in
the tables and means ± standard error in the figures. Prognosis was
assessed by Kaplan-Meier and log-rank analysis. Weighted scores
were analyzed using the Mann-Whitney test and the receiver
operating characteristic (ROC) curve. The Student's t-test was used
to assess the significance of the difference between the mean
values of the two groups and the χ2 test or Fisher's
exact test was used appropriately to identify significant
associations between the categorical variables of the two groups.
Analyses were performed with EZR (Saitama Medical Center, Jichi
Medical University, Saitama, Japan). A P value <0.05 was
considered as statistically significant.
Results
Patient characteristics
When the association between age, FIGO stage, CA125
level, and postoperative residual disease was examined, only the
size of postoperative residual disease was significantly higher in
the platinum-resistant group (P=0.005; Table I).
 | Table I.Patient characteristics for the
platinum-sensitive and platinum-resistant groups. |
Table I.
Patient characteristics for the
platinum-sensitive and platinum-resistant groups.
| Variable | Platinum-sensitive
group | Platinum-resistant
group | P-value |
|---|
| No. of cases | 26 | 25 |
|
| Age, years (mean ±
SD) | 60.4±11.7 | 58.5±8.6 | 0.521a |
| FIGO stage, n |
|
| 0.418b |
| IIIA | 1 | 0 |
|
| IIIB | 3 | 1 |
|
| IIIC | 20 | 18 |
|
| IVA | 1 | 3 |
|
| IVB | 1 | 3 |
|
| Tumor marker CA125,
U/ml (mean) | 3,467.6 | 2,057.4 | 0.309a |
| Postoperative
residual disease, n |
|
| 0.005b |
|
None | 5 | 0 |
|
| ≤1
cm | 8 | 2 |
|
| >1
cm | 13 | 23 |
|
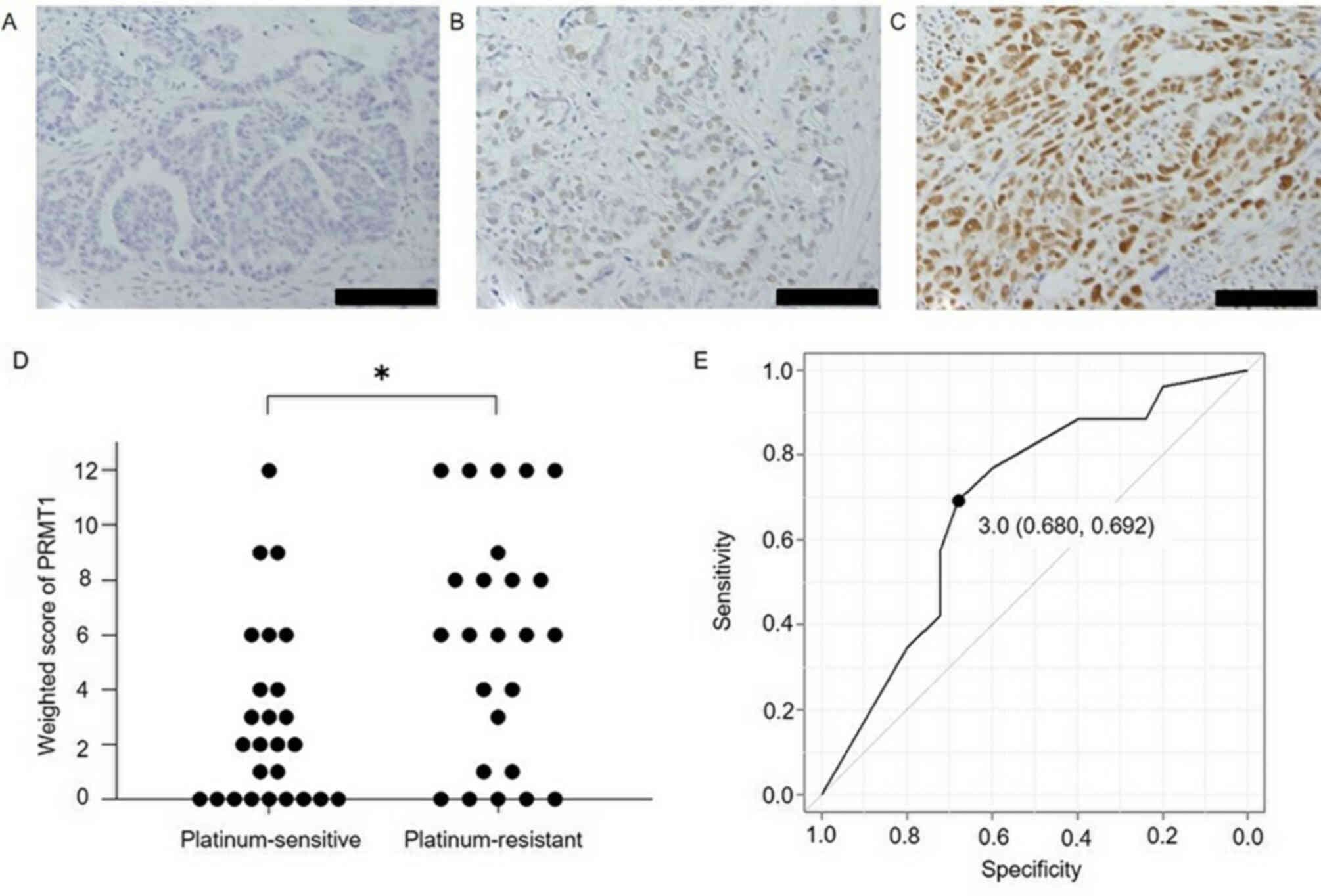
Expression of PRMT1 in ovarian serous carcinoma
tissue. Fig. 1 shows PRMT1
expression in cancer tissues, the weighted scores for the
platinum-sensitive and resistant groups and the ROC curve for
determining the PRMT1 score cut-off to evaluate the sensitivity to
platinum-based chemotherapy. The weighted score for PRMT1
expression in the platinum-resistant group was significantly higher
than that obtained for the platinum-sensitive group (P=0.019). From
the ROC curve, the cut-off value was set to 3, with a specificity
of 68.0% and a sensitivity of 69.2%. Based on this cut-off value,
cases were assigned to either a low PRMT1 expression group
(weighted score ≤3, n=26) or a high PRMT1 expression (weighted
score ≥4, n=25), according to their weighted score. There were no
significant differences in age, stage, CA125 value, and
postoperative residual lesion between the low PRMT1 expression and
high PRMT1 expression groups (Table
II).
 | Table II.Characteristics of the patients in
the low and high PRMT1 expression groups. |
Table II.
Characteristics of the patients in
the low and high PRMT1 expression groups.
| Variable | Low PRMT1 (≤3)
group | High PRMT1 (≥4)
group | P-value |
|---|
| No. of cases | 26 | 25 |
|
| Age, years (mean ±
SD) | 62.3±9.2 | 56.5±10.7 | 0.488a |
| FIGO stage, n |
|
| 0.945b |
|
IIIA | 0 | 1 |
|
|
IIIB | 2 | 2 |
|
|
IIIC | 19 | 19 |
|
|
IVA | 2 | 2 |
|
|
IVB | 3 | 1 |
|
| Tumor marker CA125,
U/ml (mean) | 3,799.7 | 1,712.0 | 0.129a |
| Postoperative
residual disease, n |
|
| 0.068b |
|
None | 4 | 1 |
|
| ≤1
cm | 6 | 4 |
|
| >1
cm | 16 | 20 |
|
Association of platinum sensitivity
with PRMT1 expression
The proportion of patients sensitive to
platinum-based chemotherapy was significantly higher in the low
PRMT1 expression group, when compared with the high PRMT1
expression group (P=0.01, Table
III). In the low PRMT1 expression group, 18 (69.2%) cases were
platinum-sensitive and 8 (30.8%) cases were platinum-resistant; in
the high PRMT1 expression group, 8 (32.0%) cases were
platinum-sensitive and 17 (68.0%) cases were
platinum-resistant.
 | Table III.Numbers of patients with low and high
PRMT1 expression in the platinum-sensitive and platinum-resistant
groups. |
Table III.
Numbers of patients with low and high
PRMT1 expression in the platinum-sensitive and platinum-resistant
groups.
| PRMT1
expression | Platinum-sensitive,
n (%) | Platinum-resistant,
n (%) | P-value |
|---|
| Low, ≤3 | 18 (69.2) | 8
(30.8) | 0.012a |
| High, ≥4 | 8
(32.0) | 17 (68.0) |
|
Survival
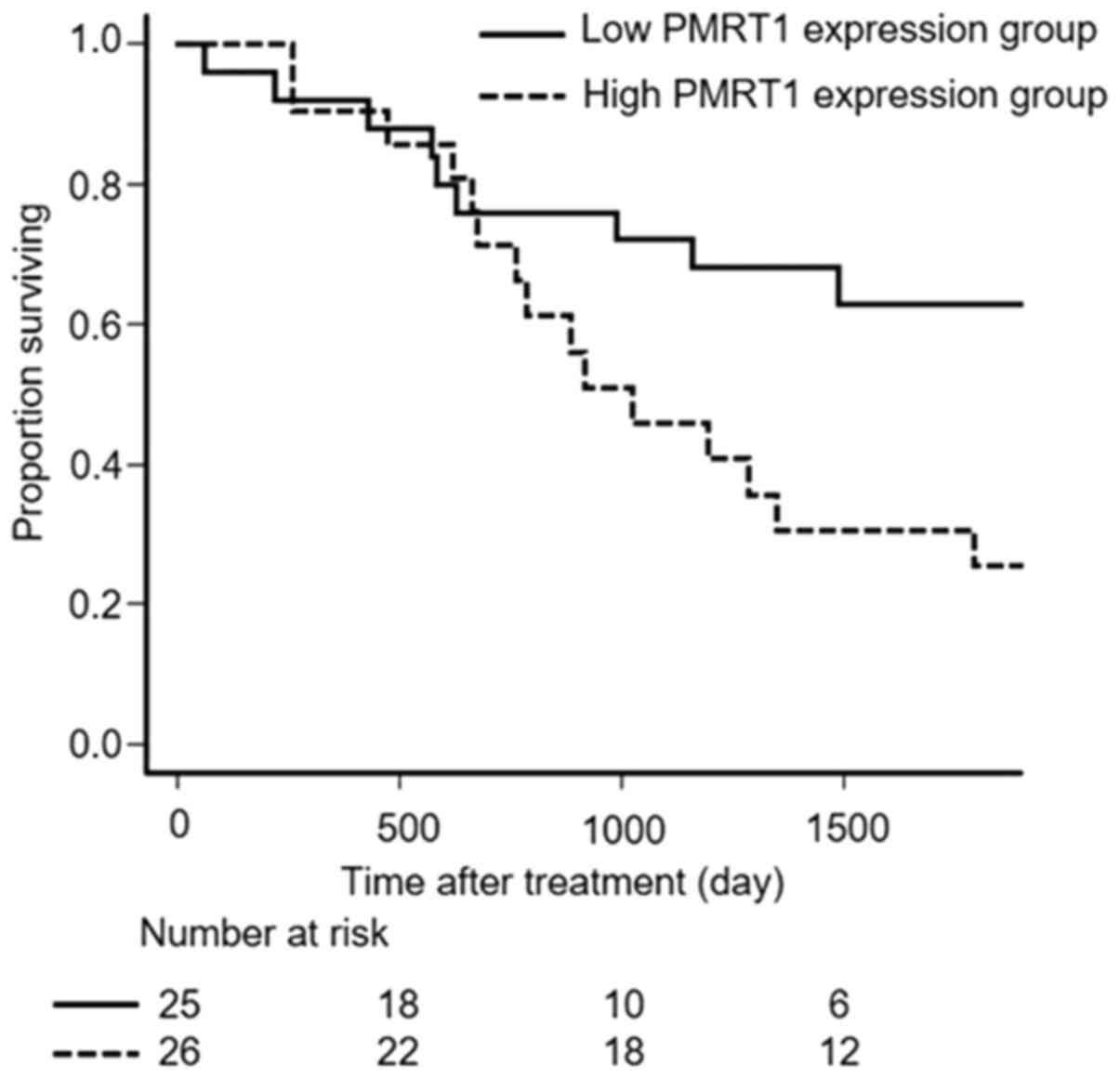
Overall survival (OS) was significantly longer for
patients of the low PRMT1 expression group than for patients of the
high PRMT1 expression group (P=0.031, Fig. 2).
siRNA-mediated silencing of PRMT1
expression enhances the sensitivity of ovarian carcinoma cells to
platinum-based therapy
PRMT1 mRNA expression in OVSAHO cells was suppressed
48 h after transfection with siPRMT1, and cells transfected with
siPRMT1 were significantly more sensitive to cisplatin or
carboplatin (Fig. 3).
Discussion
Effective treatment for serous ovarian cancer
remains a major challenge because of platinum-resistant recurrence.
It has been suggested that mechanisms of platinum-resistance may
involve decreased cellular import and increased cellular export of
the drug by transporter proteins, intracellular drug inactivation
by detoxifying enzymes, enhanced DNA damage repair, and
inactivation of cell death signaling. In platinum-resistant tumor
cells, multiple mechanisms may be involved simultaneously (17–19).
Arginine methylation is a major regulator of protein
function in mammalian cells (9).
PRMT1 represents the major arginine methyltransferase in mammalian
cells and is required for normal embryonic development, cell
division and cell viability (9–12). PRMT1
is involved in signaling pathways that regulate DNA damage, mRNA
translation, cell cycle progression, apoptosis, and genes
transcription (20–24).
PRMT1 has been implicated in the development and
progression of a variety of diseases, including cancer,
cardiovascular disease, diabetes, and fat production in the liver
(25–29). There are several reports regarding
PRMT1 expression or involvement in a variety of cancers, including
colorectal, breast, lung, bladder, liver, esophageal and head and
neck cancers (13,30–35).
PRMT1 has been shown to methylate cytoplasmic
proteins involved in the regulation of apoptotic signaling. It is
also associated with cell cycle arrest during the G0/G1 phase. In
addition, PRMT1 has been shown to methylate ASK1 and inhibit its
activity (36). Studies have also
shown that knockdown of PRMT1 promotes a PI3K-Akt signaling
pathway-dependent suppression of oxidative stress-induced
apoptosis, and that inhibition of PRMT1 attenuates activation of
the NF-κB pathway, making cells more susceptible to
chemotherapeutic agents (37,38).
In this study, OS was significantly longer in the
low PRMT1 expression group, when compared with the high PRMT1
expression group. siRNA-mediated suppression of PRMT1 expression
increased the sensitivity of OVSAHO cells to cisplatin and
carboplatin. Overexpression of PRMT1 may inhibit cell death by
inhibiting the activation of ASK1, thereby reducing the effects of
chemotherapy. Despite studies showing an association between PRMT1
expression and poor cancer prognosis, the mechanism by which PRMT1
induces resistance to chemotherapy remains unclear. Although our
study included only 51 cases, the clinical data suggest that PRMT1
expression may be a prognostic factor for poor outcome in serous
ovarian cancer. More studies which includes elucidating the
mechanism of ASK1 are now needed to confirm the role of PRMT1 as a
prognostic marker in serous ovarian cancer. To this end, ASK1 is
known to be methylated and inhibited the activity by PRMT1,
therefore we are planning the experiment to elucidate the enhanced
apoptosis induced by carboplatin or cisplatin when ASK1 is depleted
by RNAi resulting in higher sensitivity to those chemotherapeutic
agents.
In conclusion, our data suggest that PRMT1
expression may be a predictive marker of the efficacy of
platinum-based chemotherapy in patients with serous ovarian cancer.
To our knowledge, this is the first report of an association
between PRMT1 expression and platinum sensitivity, and these
findings should facilitate future studies that aim to improve the
prognosis of patients with serous ovarian cancer.
Acknowledgements
The authors would like to thank Dr Mary Derry and Dr
James Monypenny for editing a draft of this manuscript.
Funding
The present study was supported by The Osaka Medical
Research Foundation for Intractable Diseases (grant no.
26-2-47).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TF and TS designed the current study. TF and HM
assessed the authenticity of all the raw data. HM, YI, SN, YA, MS
and MY performed the experiments and collected the data. TF, HM, TY
and TS analyzed the data. TF and HM wrote the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of Osaka City University Hospital (IRB no. 4247;
Osaka, Japan) before initiation of the study. Written informed
consent was obtained from all patients.
Patient consent for publication
Patient consent for publication was not obtained.
However, the data included in the present study does not compromise
anonymity or confidentiality or breach local data protection
laws.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Borley J, Wilhelm-Benartzi C, Brown R and
Ghaem-Maghami S: Does tumour biology determine surgical success in
the treatment of epithelial ovarian cancer? A systematic literature
review. Br J Cancer. 107:1069–1074. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
du Bois A, Quinn M, Thigpen T, Vermorken
J, Avall-Lundqvist E, Bookman M, Bowtell D, Brady M, Casado A,
Cervantes A, et al Gynecologic Cancer Intergroup, AGO-OVAR, ANZGOG,
EORTC, GEICO, GINECO, GOG, JGOG, MRC/NCRI, NCIC-CTG, NCI-US, NSGO,
RTOG, SGCTG, IGCS, Organizational team of the two prior
International OCCC, : 2004 consensus statements on the management
of ovarian cancer: Final document of the 3rd International
Gynecologic Cancer Intergroup Ovarian Cancer Consensus Conference
(GCIG OCCC 2004). Ann Oncol. 16 (Suppl 8):viii7–viii12. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Japan Society of Gynecologic Oncology, .
Formulation committee of the treatment guidelines for ovarian.
simplehttps://jsgo.or.jp/guideline/ransou2015.htmlSeptember
1–2017
|
|
5
|
Ozols RF, Bundy BN, Greer BE, Fowler JM,
Clarke-Pearson D, Burger RA, Mannel RS, DeGeest K, Hartenbach EM
and Baergen R; Gynecologic Oncology Group, : Phase III trial of
carboplatin and paclitaxel compared with cisplatin and paclitaxel
in patients with optimally resected stage III ovarian cancer: A
Gynecologic Oncology Group study. J Clin Oncol. 21:3194–3200. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Friedlander M, Trimble E, Tinker A,
Alberts D, Avall-Lundqvist E, Brady M, Harter P, Pignata S,
Pujade-Lauraine E, Sehouli J, et al Gynecologic Cancer InterGroup,
: Clinical trials in recurrent ovarian cancer. Int J Gynecol
Cancer. 21:771–775. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Markman M, Rothman R, Hakes T, Reichman B,
Hoskins W, Rubin S, Jones W, Almadrones L and Lewis JL Jr:
Second-line platinum therapy in patients with ovarian cancer
previously treated with cisplatin. J Clin Oncol. 9:389–393. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kyrgiou M, Salanti G, Pavlidis N,
Paraskevaidis E and Ioannidis JP: Survival benefits with diverse
chemotherapy regimens for ovarian cancer: Meta-analysis of multiple
treatments. J Natl Cancer Inst. 98:1655–1663. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Spriggs KA, Bushell M and Willis AE:
Translational regulation of gene expression during conditions of
cell stress. Mol Cell. 40:228–237. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Truitt ML and Ruggero D: New frontiers in
translational control of the cancer genome. Nat Rev Cancer.
16:288–304. 2016.Erratum in: Nat Rev Cancer 17: 332, 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Marcel V, Catez F and Diaz JJ: p53, a
translational regulator: Contribution to its tumour-suppressor
activity. Oncogene. 34:5513–5523. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wooderchak WL, Zang T, Zhou ZS, Acuña M,
Tahara SM and Hevel JM: Substrate profiling of PRMT1 reveals amino
acid sequences that extend beyond the ‘RGG’ paradigm. Biochemistry.
47:9456–9466. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yoshimatsu M, Toyokawa G, Hayami S, Unoki
M, Tsunoda T, Field HI, Kelly JD, Neal DE, Maehara Y, Ponder BA, et
al: Dysregulation of PRMT1 and PRMT6, type I arginine
methyltransferases, is involved in various types of human cancers.
Int J Cancer. 128:562–573. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hirata Y, Katagiri K, Nagaoka K, Morishita
T, Kudoh Y, Hatta T, Naguro I, Kano K, Udagawa T, Natsume T, et al:
TRIM48 promotes ASK1 activation and cell death through
ubiquitination-dependent degradation of the ASK1-negative regulator
PRMT1. Cell Rep. 21:2447–2457. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sinicrope FA, Ruan SB, Cleary KR, Stephens
LC, Lee JJ and Levin B: bcl-2 and p53 oncoprotein expression during
colorectal tumorigenesis. Cancer Res. 55:237–241. 1995.PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dasari S and Tchounwou PB: Cisplatin in
cancer therapy: Molecular mechanisms of action. Eur J Pharmacol.
740:364–378. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ghosh Cisplatin S.: The first metal based
anticancer drug. Bioorg Chem. 88:1029252019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Amable L: Cisplatin resistance and
opportunities for precision medicine. Pharmacol Res. 106:27–36.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Infantino S, Benz B, Waldmann T, Jung M,
Schneider R and Reth M: Arginine methylation of the B cell antigen
receptor promotes differentiation. J Exp Med. 207:711–719. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sakamaki J, Daitoku H, Ueno K, Hagiwara A,
Yamagata K and Fukamizu A: Arginine methylation of BCL-2 antagonist
of cell death (BAD) counteracts its phosphorylation and
inactivation by Akt. Proc Natl Acad Sci USA. 108:6085–6090. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu Z, Chen T, Hébert J, Li E and Richard
S: A mouse PRMT1 null allele defines an essential role for arginine
methylation in genome maintenance and cell proliferation. Mol Cell
Biol. 29:2982–2996. 2009.Erratum in: Mol Cell Biol 37: e00298-17,
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dominguez-Sola D, Kung J, Holmes AB, Wells
VA, Mo T, Basso K and Dalla-Favera R: The FOXO1 transcription
factor instructs the germinal center dark zone program. Immunity.
43:1064–1074. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kuhn P, Chumanov R, Wang Y, Ge Y, Burgess
RR and Xu W: Automethylation of CARM1 allows coupling of
transcription and mRNA splicing. Nucleic Acids Res. 39:2717–2726.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim DI, Park MJ, Choi JH, Kim IS, Han HJ,
Yoon KC, Park SW, Lee MY, Oh KS and Park SH: PRMT1 and PRMT4
regulate oxidative stress-induced retinal pigment epithelial cell
damage in SIRT1-dependent and SIRT1-independent manners. Oxid Med
Cell Longev. 2015:6179192015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mathioudaki K, Papadokostopoulou A,
Scorilas A, Xynopoulos D, Agnanti N and Talieri M: The PRMT1 gene
expression pattern in colon cancer. Br J Cancer. 99:2094–2099.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Park MJ, Kim DI, Lim SK, Choi JH, Kim JC,
Yoon KC, Lee JB, Lee JH, Han HJ, Choi IP, et al:
Thioredoxin-interacting protein mediates hepatic lipogenesis and
inflammation via PRMT1 and PGC-1α regulation in vitro and in vivo.
J Hepatol. 61:1151–1157. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Siroen MP, Teerlink T, Nijveldt RJ, Prins
HA, Richir MC and van Leeuwen PA: The clinical significance of
asymmetric dimethylarginine. Annu Rev Nutr. 26:203–228. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sydow K, Mondon CE and Cooke JP: Insulin
resistance: Potential role of the endogenous nitric oxide synthase
inhibitor ADMA. Vasc Med. 10 (Suppl 1):S35–S43. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li B, Liu L, Li X and Wu L: miR-503
suppresses metastasis of hepatocellular carcinoma cell by targeting
PRMT1. Biochem Biophys Res Commun. 464:982–987. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou W, Yue H, Li C, Chen H and Yuan Y:
Protein arginine methyltransferase 1 promoted the growth and
migration of cancer cells in esophageal squamous cell carcinoma.
Tumour Biol. 37:2613–2619. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chuang CY, Chang CP, Lee YJ, Lin WL, Chang
WW, Wu JS, Cheng YW, Lee H and Li C: PRMT1 expression is elevated
in head and neck cancer and inhibition of protein arginine
methylation by adenosine dialdehyde or PRMT1 knockdown
downregulates proliferation and migration of oral cancer cells.
Oncol Rep. 38:1115–1123. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Avasarala S, Van Scoyk M, Karuppusamy
Rathinam MK, Zerayesus S, Zhao X, Zhang W, Pergande MR, Borgia JA,
DeGregori J, Port JD, et al: PRMT1 is a novel regulator of
epithelial-mesenchymal-transition in non-small cell lung cancer. J
Biol Chem. 290:13479–13489. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Baldwin RM, Bejide M, Trinkle-Mulcahy L
and Côté J: Identification of the PRMT1v1 and PRMT1v2 specific
interactomes by quantitative mass spectrometry in breast cancer
cells. Proteomics. 15:2187–2197. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Papadokostopoulou A, Mathioudaki K,
Scorilas A, Xynopoulos D, Ardavanis A, Kouroumalis E and Talieri M:
Colon cancer and protein arginine methyltransferase 1 gene
expression. Anticancer Res. 29:1361–1366. 2009.PubMed/NCBI
|
|
36
|
Cho JH, Lee MK, Yoon KW, Lee J, Cho SG and
Choi EJ: Arginine methylation-dependent regulation of ASK1
signaling by PRMT1. Cell Death Differ. 19:859–870. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Baldwin RM, Morettin A and Côté J: Role of
PRMTs in cancer: Could minor isoforms be leaving a mark? World J
Biol Chem. 5:115–129. 2014.PubMed/NCBI
|
|
38
|
Musiani D, Giambruno R, Massignani E,
Ippolito MR, Maniaci M, Jammula S, Manganaro D, Cuomo A, Nicosia L,
Pasini D and Bonaldi T: PRMT1 is recruited via DNA-PK to chromatin
where it sustains the senescence-associated secretory phenotype in
response to cisplatin. Cell Rep. 30:1208–1222.e9. 2020. View Article : Google Scholar : PubMed/NCBI
|