Introduction
B-Myb, also known as MYB proto-oncogene like 2
(MYBl2), is a transcription factor that belongs to the Myb gene
family, including A-Myb and c-Myb (1). MYB was the first discovered family
member and is the mammalian homolog of the retroviral v-Myb
oncogene that causes acute leukemia in birds and can transform
hematopoietic cells (2).
A-Myb is predominantly expressed in germ cells and
primordial lymphocytes (3).
c-Myb is highly expressed in cells of the hematopoietic
system, with low expression in epithelial cells of specific
tissues, such as the colon and brain (3,4).
B-Myb is ubiquitously expressed in mammalian cells with high
proliferative ability (1,4,5), and
B-Myb is a physiological regulator of cell cycle progression, cell
survival and cell differentiation (6). However, several experimental studies
have demonstrated that B-Myb is highly expressed in
different types of human malignant tumors, including breast cancer
(7), lung cancer (8), hepatocellular carcinoma (9), endometrial cancer (10), prostate cancer (11) and ovarian cancer (12). Both in vivo and in
vitro experiments have demonstrated that high B-Myb
expression promotes colony formation, cell cycle progression,
migration and invasion of cancer cells (13). B-Myb also participates in the
occurrence of epithelial-to-mesenchymal transition (EMT) in
malignant tumor, inhibits cancer cell apoptosis and results in a
poor prognosis (14). However, the
molecular mechanisms underlying the regulation of B-Myb in
the development of malignant tumors remain unclear.
The present review highlights the association
between B-Myb proto-oncogene and the development of
malignant tumors based on available studies, aiming to provide
insight into the molecular mechanisms underlying
B-Myb-induced development of malignant tumors.
Role of B-Myb in the development of
malignant tumors
B-Myb expression in malignant
tumors
B-Myb is expressed at high levels in several
human malignant tumor tissues (Table
I). For example, there are five molecular subtypes of breast
cancer, basal-like, human epidermal growth factor receptor 2
positive (HER2+)/estrogen receptor negative (ER-), luminal A,
luminal B and normal-like (7).
Microarray analyses of B-Myb in breast cancer tissues have
demonstrated that its expression levels significantly differ among
the five subtypes, with the highest level in basal-like tumors
(7). Furthermore, overexpression of
B-Myb has been observed in 83% of primary tumors and all
cell lines of non-small cell lung cancer (NSCLC) (8), and B-Myb has also been
demonstrated to modulate cell cycle and proliferation (15). Frau et al (9) assessed B-Myb mRNA and protein
expression levels across different stages of hepatocarcinogenesis
and demonstrated that B-Myb expression levels were
substantially higher in precancerous lesions, early proliferative
nodules, advanced proliferative nodules and hepatocellular
carcinoma compared with normal liver tissues. In addition, the
highest levels of B-Myb were observed in hepatocellular
carcinoma, among the four different stages. Nakajima et al
(16) reported amplification of the
B-Myb gene copy number in 36/66 cases of primary
hepatocellular carcinoma.
 | Table I.Role of B-Myb in malignant
tumor development. |
Table I.
Role of B-Myb in malignant
tumor development.
| Tumor type | B-Myb
expression level | Patient
prognosis | B-Myb
expression treatment | In vitro
effect on cell phenotype | In vivo
effect on nude mice | (Refs.) |
|---|
| Breast cancer | High | Poor | Interference | Inhibit cell cycle
progression, cell proliferation, and cell migration and
invasion. | Inhibit tumor
formation. | (7,32) |
| Lung cancer | High | Poor | Overexpression | Promote
tumorigenesis, cell proliferation and cell cycle progression. | Promote tumor
formation. | (8,13,78) |
|
|
|
| Interference | Inhibit cell
proliferation, cell cycle progression, and cell migration and
invasion. | Inhibit tumor
formation. |
|
| Hepatocellular
carcinoma | High | Poor | Overexpression | Promote
tumorigenesis, cell proliferation and cell cycle progression. | – | (9,34) |
|
|
|
| Interference | Inhibit cell
proliferation and cell cycle progression. | – |
|
| Colorectal
cancer | High | Poor | Interference | Inhibit cell
proliferation, cell cycle progression and cell migration and
invasion. | – | (18) |
| Esophageal
squamous-cell carcinoma | High | Poor | Overexpression | Promote cell cycle
progression and cell proliferation. | – | (19,20) |
|
|
|
| Interference | Inhibit cell
proliferation. | Inhibit tumor
formation. |
|
| Gallbladder
cancer | High | Poor | Overexpression | Promote cell cycle
progression and cell proliferation. | Promote tumor
formation. | (29) |
|
|
|
| Interference | Inhibit cell cycle
progression and cell proliferation. | Inhibit tumor
formation. |
|
| Glioma | High | Poor | Overexpression | Promote cell
proliferation. |
| (25) |
|
|
|
| Interference | Inhibit cell cycle
progression and cell proliferation. |
|
|
| Prostate
cancer | High | – | – | – | – | (11,17) |
| Renal cell
carcinoma | High | – | – | – | – | (26) |
| Ovarian cancer | High | – | – | – | – | (12,31) |
| Neuroblastoma | High | Poor | – | – | – | (47) |
| Fibrosarcoma | High |
|
|
|
| (30) |
| Endometrial
cancer | High | – | – | – | – | (10) |
| Primary
leukemia | High | – | – | – | – | (28) |
In the absence of gene amplification, malignant
tumors of the prostate exhibit elevated B-Myb expression
levels, and B-Myb expression is markedly higher in
metastatic prostate cancer compared with non-metastatic prostate
cancer (11,17). Based on a study involving 180
patients with colorectal cancer, B-Myb mRNA and protein
expression levels were notably higher in cancer tissues compared
with adjacent normal tissues, and B-Myb expression was
positively associated with tumor size and clinical stage (18).
Qin et al (19) demonstrated that B-Myb is
amplified in esophageal cancer, based on whole-genome sequencing
(10 pairs) and whole-exome sequencing (57 pairs) of esophageal
cancer tissues and matched adjacent normal tissues selected from
high-incidence areas of esophageal cancer in China. Additionally,
Qin et al (20) reported that
B-Myb protein was expressed at higher levels in esophageal squamous
cell carcinoma (ESCC) tissues compared with adjacent normal tissues
in a Chinese population of 107 patients with ESCC, based on
immunohistochemistry.
Among patients with neuroblastoma, individuals with
malignant metastasis and poor prognosis express B-Myb at
significantly elevated levels; this phenomenon suggests that
B-Myb expression is associated with the risk of developing
neuroblastoma (21). Notable,
inhibition of B-Myb expression prevents the proliferation of
normal human cells and cancer cells (21–24). In
addition, B-Myb expression levels are significantly higher
in glioma tissues compared with adjacent normal tissues and are
positively associated with the grade of glioma, based on the
results of reverse transcription-PCR (RT-PCR) and western blot
analyses from 79 patients with glioma (25).
In renal cell carcinoma, metastatic tumor tissues
highly express B-Myb when metastasis occurs in primary
tumors negative for B-Myb expression (26). B-Myb is also amplified in
high-grade bladder cancer (27). In
addition, high B-Myb expression has been implicated in
leukemias (28), gallbladder cancer
(29), fibrosarcoma (30), ovarian cancer (12) and aggressive T-cell lymphoma
(31).
Biological function of B-Myb in
malignant tumor development
Promotion of cancer cell
proliferation
Increasing evidence suggests that B-Myb is
overexpressed in different types of human cancer, including breast
cancer (32), cervical cancer
(33), colorectal cancer (18), liver cancer (34), leukemia cells (28) and lung cancer (35). In these types of cancer, B-Myb
promotes cell proliferation and/or cell cycle progression (18,34).
Thomas et al (36) observed a
positive correlation between B-Myb mRNA expression and Ki-67
proliferation index in breast cancer. In addition, the breast
cancer cell line, MDA-MB-231, exhibits remarkably decreased
abilities to form colonies, migrate and invade following knockdown
of B-Myb with short-hairpin RNA (37). Flow cytometric analysis demonstrated
that the cell cycle is arrested at S and G2/M phases,
while in vivo experiments indicated that both the rate of
tumor formation and the weight of tumor mass are significantly
lower in breast cancer compared with the control group (32).
Jin et al (13) demonstrated that overexpression of
B-Myb promotes the proliferation of NSCLC cells; both
extracellular regulated MAP kinase (ERK) and phosphorylated-protein
kinase B (Akt) signaling pathways participate in the modulation of
NSCLC by B-Myb. Liang et al (29) reported that B-Myb expression
is upregulated in gallbladder cancer tissues, which in turn
facilitates the proliferation of gallbladder cancer cells by
facilitating cell cycle progression through the S and
G2/M phases (30).
In ESCC, an EdU-retention assay demonstrated that
downregulation of B-Myb expression decreases the DNA
synthesis ability of EC9706 cells, while a Cell Counting Kit-8
assay demonstrated that overexpression of B-Myb also
promotes the proliferation of KYSE510 cells (20). These findings suggest that
B-Myb can promote the proliferation and DNA synthesis of
ESCC cells. Based on a colony formation assay, overexpression of
B-Myb in the low-grade glioma cell line, Hs683, considerably
increased the number of colonies, whereas knockout of B-Myb
in the high-grade glioma cell line, U251, decreased the number of
colonies (25). Subsequently, a MTT
assay demonstrated that the proliferative ability of glioma U251
cells is enhanced following transfection with small interfering
(si)RNAs (25).
Promotion of EMT
B-Myb has been demonstrated to play a role in
EMT, a process whereby epithelial cells lose their polarity,
migrate and increase polarity (38).
In breast cancer cells, downregulation of B-Myb expression
can recover the expression of the epithelial marker, E-cadherin and
promote the formation of intercellular adhesion, in addition to
inhibiting cell invasion, anchorage-dependent growth and tumor
formation (32). Conversely,
overexpression of B-Myb can decrease E-cadherin expression
and increase the expression of mesenchymal markers (14). In addition, it has been demonstrated
that B-Myb can upregulate the expression of Snail, a key
regulator of EMT, thereby mediating the promotion of EMT and cancer
cell invasion (14).
The role of B-Myb in colorectal cancer
invasion and metastasis has also been proven and is associated with
EMT (18). For example, EMT inhibits
B-Myb activity in colorectal cancer cells, thereby
upregulating the expression of the epithelial marker, E-cadherin
and downregulating the expression of the mesenchymal marker,
Vimentin and matrix metalloproteinase 9 (MMP9) (18). Based on a western blot assay on the
expression of EMT markers in glioma cells, interference with
B-Myb expression inhibits the protein expression levels of
N-cadherin, Vimentin, MMP2 and MMP9, while upregulating the protein
levels of E-cadherin and zinc finger E-box binding homeobox 1
(25). Taken together, these
findings suggest that B-Myb plays an important role in the
promotion of EMT in several malignant tumors, thereby facilitating
cancer cell infiltration and metastasis.
Inhibition of cancer cell
apoptosis
B-Myb inhibits cancer cell apoptosis
potentially through multiple pathways. First, B-Myb may
perform its anti-apoptotic function by positively regulating the
expression of the anti-apoptotic gene, Clusterin (30), also known as apolipoprotein J
(39). A study demonstrated that
inhibition of Clusterin gene expression promotes the apoptosis of
fibrosarcoma cells (30). Secondly,
B-Myb may inhibit apoptosis by positively regulating the
expression of the anti-apoptotic gene, Bcl-2 (40), a critical regulator of apoptosis
(41). In support of these pathways,
the anti-apoptotic function of B-Myb has been observed in
different cancer cell lines, including colorectal cancer (18) and liver cancer (34).
Ren et al (18) reported that Bcl-2 protein expression
is downregulated in SW480 colorectal cancer cells via interference
with B-Myb expression. Calvisi et al (34) demonstrated that B-Myb exerts
an anti-apoptotic function, and interference with B-Myb
expression via transfection with siRNA induces apoptosis in four
different hepatocellular carcinoma cell lines. In glioma U251 cells
transfected with B-Myb siRNA, Zhang et al (25) detected an increase in the percentage
of apoptotic cells by Annexin V-FITC/PI and hochest 3342 staining.
Furthermore, Zhang et al (25) assessed the effects of B-Myb
silencing on apoptosis-related proteins, including caspase-3/9,
Bcl/Bax, PTEN and P53. Western blot analysis demonstrated that
B-Myb silencing decreases Bcl-2 expression, while increasing
the expression levels of Bax, PTEN and P53, and activates
caspase-3/9 activity. Taken together, these results suggest that
downregulation of B-Myb can induce apoptosis in glioma
cells.
Enhancement of drug resistance
The role of B-Myb in tumor resistance to
therapy is associated with its pro-apoptotic function (42,43).
Overexpression of B-Myb promotes the expression of the
anti-apoptotic gene, Bcl-2 in IL2-dependent murine CTLL-2
cells, thereby increasing the therapeutic resistance to drugs,
including doxorubicin, ceramide and dexamethasone (42). Similarly, Levenson et al
(43) demonstrated that B-Myb
expression is notably upregulated in fibrosarcoma cells with
therapeutic resistance induced by chemotherapeutic drugs that
inhibit DNA synthesis, including hydroxyurea, cysteine, etoposide
and adriamycin. Furthermore, B-Myb regulates the expression
of the anti-apoptotic gene, Apolipoprotein J/Clusterin in
neuroblastoma cells to resist apoptosis caused by doxycycline
(30).
Sottile et al (44) reported that individuals with
B-Myb overexpression or MYCN amplification are more
sensitive to therapy with camptothecins (irinotecan and topotecan)
among patients with neuroblastoma. B-Myb is a downstream
target of MYCN; MYCN amplification promotes
B-Myb overexpression, while B-Myb overexpression in
turn promotes an upregulation of MYCN expression, thus these
two factors regulate each other (21). Camptothecins selectively
downregulates B-Myb and MYCN expression, while
upregulation of B-Myb decreases the killing effect of
camptothecins, suggesting that B-Myb is an important target
of camptothecins (44). In addition,
B-Myb is overexpressed in cetuximab-resistant NSCLC,
suggesting that B-Myb overexpression is associated with
cetuximab resistance (45).
Effect on patient prognosis
High B-Myb expression is associated with
tumor growth and poor prognosis of patients, making it a potential
clinical marker for poor prognosis (20,46).
Based on microRNA (miRNA/miR) prediction and RT-PCR analyses,
B-Myb may have a negative regulatory association with miR-30
family members, and biochemical relapse-free survival time is
shortened in patients with acute myeloid leukemia highly
overexpressing B-Myb; thus, B-Myb can be a predictive
marker for the prognosis of patients with acute myeloid leukemia
(46). In addition, B-Myb expression
is elevated in ESCC tissues and negatively associated with
postoperative overall survival in patients with ESCC, as revealed
by a Kaplan-Meier analysis (20).
A prognostic analysis of breast cancer and its
different subtypes revealed that the B-Myb high expression
group has a worse prognosis compared with the low expression group
(7). High B-Myb expression
also increases the risk of poor prognosis, decreases the
differentiation ability of cells, and promotes tumor development in
neuroblastoma (47). In HepG2 and
HuH7 hepatocellular carcinoma cell lines, overexpression of
B-Myb increases the cell proliferative ability and
facilitates G1-S and G2-M transitions,
whereas interference of B-Myb with siRNA results in cell
cycle arrest at G0-G1 and G2-M
phases (9).
Among patients with colorectal cancer, the 5-year
survival rate is notably lower in individuals with high
B-Myb expression than those with low B-Myb
expression; B-Myb expression and clinical stage of the tumor
can be used as independent prognostic factors of colorectal cancer
(18). With regards to the prognosis
and survival of 79 patients with glioma, Kaplan-Meier analysis and
log-rank test results indicated that high B-Myb expression
is negatively associated with survival, and is a poor prognostic
factor in patients with glioma (25).
Role of B-Myb in the diagnosis and
treatment of malignant tumors effect
B-Myb plays a role in the clinical diagnosis and
treatment of malignant tumors. B-Myb can be used as a biological
marker of cervical cancer to make up for the limitations of
conventional cytology for cervical intraepithelial neoplasia and
cervical cancer diagnosis in routine cervical cytology testing
(43,44). Astbury et al (48) measured MYBL2 expression levels in
cervical cancer cell lines, cervical intraepithelial neoplasia and
cervical glandular epithelium using genomics and proteomics
technology, and assessed the potential of B-Myb as a biomarker.
Previous studies (47) have
demonstrated that patients with neuroblastoma, with MYBL2
overexpression, are more sensitive to camptothecin therapy
(44). Camptothecin drugs can
selectively downregulate the expression of B-Myb. Conversely,
upregulating B-Myb can decrease the killing effect of camptothecin
drugs (44). Collectively, these
results suggest that B-Myb is an important target of camptothecin
drugs (44).
Regulatory mechanisms of B-Myb in
malignant tumor
B-Myb regulation by miRNAs
miRNAs are a group of small non-coding single
stranded RNAs that have been extensively observed in animals and
plants, typically 18–22 nucleotides in length (49). Both misregulation and mutation of
miRNAs can cause tumorigenesis, and they contribute to the function
of oncogenes through targeted downregulation of tumor suppressor
genes or activation of oncogene transcription factors (50,51).
B-Myb is mainly regulated by the miR-29 and miR-30 families
(52). In breast cancer cell lines,
miRNA binds to the 3′-untranslated region of B-Myb and
thereby inhibits B-Myb expression (53). Additionally, miR-29a expression is
negatively correlated with B-Myb expression, and cyclin
A2 and cyclin D1 expression is positively correlated
with B-Myb expression (54). These
results indicate that miR-29a suppresses tumor growth by
downregulating B-Myb expression (54).
Geng et al (55) reported that miR-30a expression is
significantly downregulated in NSCLC tissues compared with adjacent
normal tissues, and B-Myb is a target gene of miR-30a based
on a double-luciferase reporter gene assay; miR-30a can suppress
proliferation and growth of NSCLC through targeted inhibition of
B-Myb expression. Li et al (56) demonstrated a close association
between B-Myb overexpression and low levels of miR-30a,
miR-30b and miR-30c in 291 patients with acute myeloid, based on
RT-qPCR analysis of B-Myb, miR-29 family and miR-30 family
genes. Li et al (56)
reported that the long non-coding RNA, LINC01139, upregulates
B-Myb by competitively binding to the miR-30 family, thereby
promoting the progression of hepatocellular carcinoma.
B-Myb is also regulated by other
miRNAs during tumor development
Zauli et al (28) demonstrated that the cell cycle is
arrested at G1 following downregulation of B-Myb
expression in primary leukemia cells and P53 wild-type
myeloid and lymphoblastic cells; miR-34a plays a pivotal regulatory
role in this process. Lee et al (57) reported that inhibition of miR-34a
recovers B-Myb expression, while miR-34a mimics
downregulates B-Myb expression in HCT116 colorectal cancer
cells. In addition, Li et al (56) and Yu et al (58) have concluded that the expression of
G1/S-related genes, Ezh2 and B-Myb, are
suppressed by miR-34c overexpression in the cancer cell line
cultured from a mouse model of tubal high-grade serous ovarian
carcinoma, leading to cell cycle arrest at G1 phase and
induction of apoptosis.
Wang et al (59) demonstrated that post-transcriptional
regulation of MALAT1 is regulated by miR-101 and miR-217 in
ESCC. Specifically, post-transcriptional silencing of MALAT1
significantly inhibits ESCC cell proliferation by arresting the
G2/M phase, while the migratory and invasive abilities
of ESCC cells decrease following overexpression of miR-101 and
miR-217. This may be due to MALAT1-mediated upregulation of
P21 and P27 expression and inhibition of B-Myb
expression (60). In addition, Chen
et al (61) indicated that
miR-143-3p negatively regulates B-Myb in breast cancer
cells, and modulates cancer cell proliferation and apoptosis
(62).
B-Myb regulation by E2Fs
The E2F transcription factor family plays a
crucial role in the regulation of cell cycle progression, DNA
replication and apoptosis (63). The
E2F family can be divided into two groups, E2F1-3a
are transcription factors that activate the cell cycle and mainly
encode genes that promote the progression of G1 to S
phase, while E2F3b and E2F4-8 mainly promote cells to
exit the cell replication cycle and facilitate cell differentiation
(64,65). B-Myb is a typical cell cycle
regulator that is rarely expressed in the G0 phase
(66). When external growth
factor-mediated signal pathways, such as ERK1/2, are activated,
they promotes the release and activation of E2F1-3 from pRB
through CCND1; these factors drive their target genes to
encode G1/S-related cytokines, and B-Myb
expression is also induced by E2F1-3 (4,67,68). The
induction of B-Myb expression at the end of the
G1 phase is due to a substantial increase in gene
transcription, suggesting that B-Myb may be a gene regulated
by the E2F transcription factors (62).
The promoters of both human and murine B-Myb
genes contain completely conserved E2F-binding sites, which
is key for B-Myb to participate in the transcription
regulation of the cell cycle (69,70). A
chromatin immunoprecipitation assay of NIH3T3 in fibroblasts
revealed that both E2F4/P107 and E2F4/P130 are
associated with the B-Myb promoter in the G0
phase, while E2F4/P107 is associated with the B-Myb
promoter in the early G1 phase (69). In human malignant glioma T98G cells,
E2F1 and E2F3 are associated with the B-Myb
promoter in the late G1 phase (71). B-Myb transcriptionally
activates its own promoter through the SP1-binding site
adjacent to the transcription initiation site (72). SP1 is coupled with E2F1
to promote transcriptional activation of the B-Myb promoter
(73,74). It is suggested that E2F1 and
E2F3 play a role in promoting B-Myb transcription in
at least some cells (75).
Nakajima et al (16) demonstrated that knockout of
E2F1 in JHH-5 hepatocellular carcinoma cells decreases the
expression levels of B-MYB, CCNE1, MYC, TK1 and RRM1.
The kinetics of B-Myb interaction with
G2-regulated promoters coincides with the activation of
the genes; a decrease in RNAi-regulated B-Myb expression can
inhibit cyclin B1 and cell division cycle 2 (cdc2)
expression, arrest cell cycle at G2/M, and increase
apoptosis (76). The interaction of
B-Myb with the cdc2 promoter is dependent on the
complete E2F-binding site, and the B-Myb gene is
regulated by E2F at G1/S transition, thereby
modulating the target genes associated with G1/S and
G2/M transitions, including cdc2, cyclin A2 and
cyclin B1 (77).
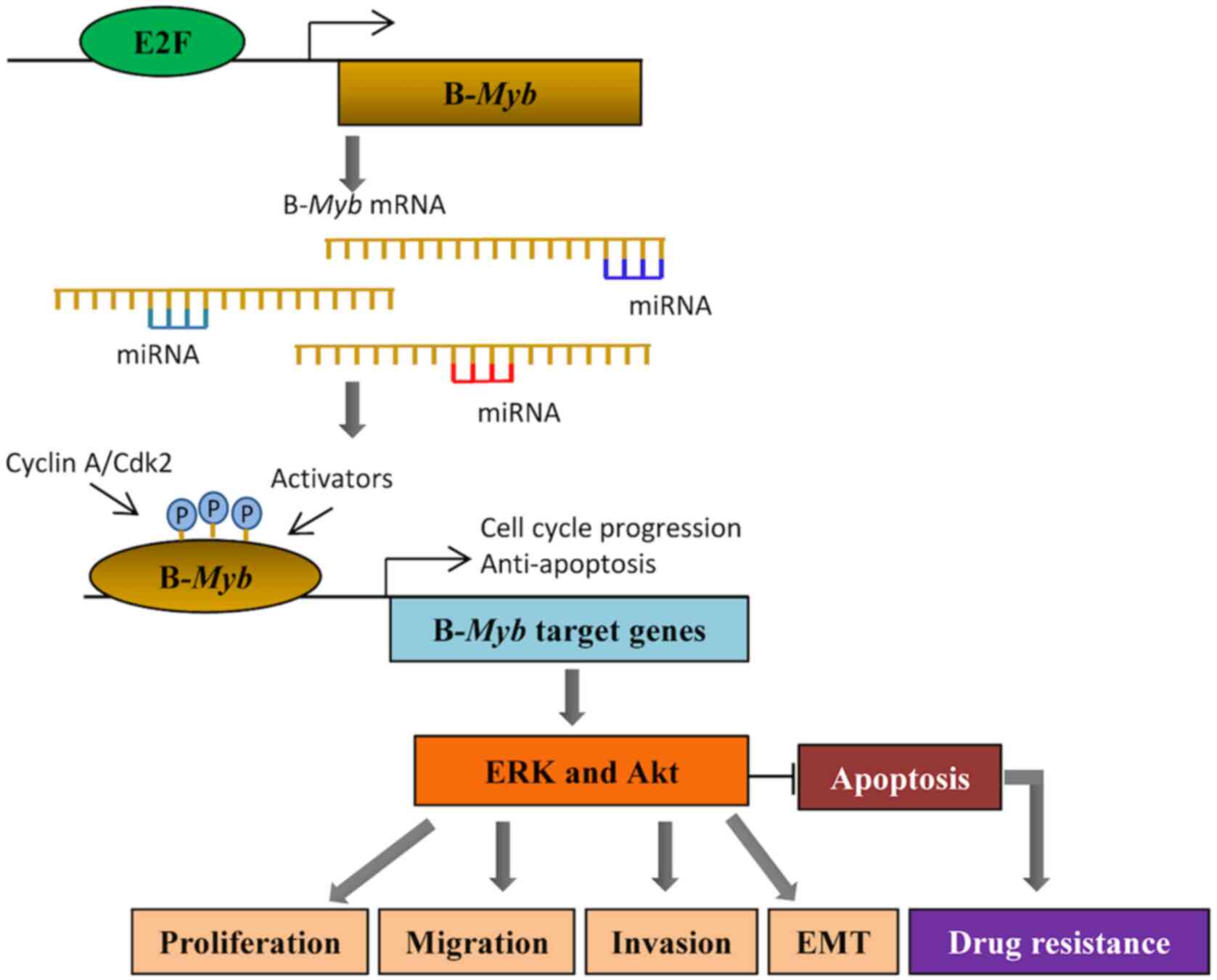
Conclusions
B-Myb, a classic oncogene, promotes the
development of malignant tumors (5).
B-Myb is highly expressed in several tumors, and its
expression is associated with the clinicopathological
characteristics of tumors (13,33,78).
High B-Myb expression severely affects the prognosis of
patients, with a relatively low 5-year survival rate (7). In vitro studies have
demonstrated that B-Myb promotes cell cycle progression,
proliferation, invasion and migration of cancer cells (35). In vivo experiments have also
reported that B-Myb facilitates tumor formation (35). Both miRNAs (46,47,51) and
E2Fs (16) contribute to the
function of B-Myb by regulating its expression. miR-29,
miR-30, miR-34, amiR-101 and miR-217 all participate in the
regulation of B-Myb, thus affecting cell functions,
including senescence, proliferation, invasion and metastasis
(Fig. 1). E2Fs interact with
B-Myb through promoter elements, which in turn activates
target genes involved in G1/S and G2/M
transitions, thereby promoting cell cycle progression (16). Following research advances on the
molecular mechanisms of malignant tumor development, it may be
possible to apply B-Myb to the diagnosis and treatment of
patients with tumors. Determining the role of B-Myb in tumor
development will provide novel tumor markers, while starting a
novel chapter of potential targeted intervention therapy of
malignant tumors.
Acknowledgements
Not applicable.
Funding
The present review was supported in part by a
grant-in-aid from the Scientific Research Fund of Zhejiang
Provincial Education Department (grant no. Y202045646 to XF), the
Scientific Research Project of Taizhou Municipal Science and
Technology Bureau (grant no. 20ywb100 to XF and grant no. 20ywb101
to YJ), and Students Science and Technology Innovation Activity
Plan of Zhejiang Province (also known as ‘New Seedling Talent
Plan’; grant no. 2020R468017 to XF). The funders had no role in the
study design, data collection and analysis, decision to publish, or
preparation of the manuscript.
Availability of data and materials
All data generated or analyzed during this study
are included in this published article.
Authors' contributions
YJ, GQ, GC, CW and XF made substantial
contributions to conception and design, or acquisition of data, or
analysis and interpretation of data. XF and YJ drafted the initial
manuscript and critically revised it for important intellectual
content. GC and CW designed the study, and critically revised the
article. XF approved the final version to be published.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Oh IH and Reddy EP: The myb gene family in
cell growth, differentiation and apoptosis. Oncogene. 18:3017–3033.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lipsick JS and Wang DM: Transformation by
v-Myb. Oncogene. 18:3047–3055. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Toscani A, Mettus RV, Coupland R, Simpkins
H, Litvin J, Orth J, Hatton KS and Reddy EP: Arrest of
spermatogenesis and defective breast development in mice lacking
A-myb. Nature. 386:713–717. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sala A: B-MYB, a transcription factor
implicated in regulating cell cycle, apoptosis and cancer. Eur J
Cancer. 41:2479–2484. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Iness AN, Felthousen J, Ananthapadmanabhan
V, Sesay F, Saini S, Guiley KZ, Rubin SM, Dozmorov M and Litovchick
L: The cell cycle regulatory DREAM complex is disrupted by high
expression of oncogenic B-Myb. Oncogene. 38:1080–1092. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Musa J, Aynaud MM, Mirabeau O, Delattre O
and Grünewald TG: MYBL2 (B-Myb): A central regulator of cell
proliferation, cell survival and differentiation involved in
tumorigenesis. Cell Death Dis. 8:e28952017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thorner AR, Hoadley KA, Parker JS, Winkel
S, Millikan RC and Perou CM: In vitro and in vivo analysis of B-Myb
in basal-like breast cancer. Oncogene. 28:742–751. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Long EM, Long MA, Tsirigotis M and Gray
DA: Stimulation of the murine Uchl1 gene promoter by the B-Myb
transcription factor. Lung Cancer. 42:9–21. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Frau M, Ladu S, Calvisi DF, Simile MM,
Bonelli P, Daino L, Tomasi ML, Seddaiu MA, Feo F and Pascale RM:
Mybl2 expression is under genetic control and contributes to
determine a hepatocellular carcinoma susceptible phenotype. J
Hepatol. 55:111–119. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Krakstad C, Tangen IL, Hoivik EA, Halle
MK, Berg A, Werner HM, Ræder MB, Kusonmano K, Zou JX, Øyan AM, et
al: ATAD2 overexpression links to enrichment of B-MYB-translational
signatures and development of aggressive endometrial carcinoma.
Oncotarget. 6:28440–28452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bar-Shira A, Pinthus JH, Rozovsky U,
Goldstein M, Sellers WR, Yaron Y, Eshhar Z and Orr-Urtreger A:
Multiple genes in human 20q13 chromosomal region are involved in an
advanced prostate cancer xenograft. Cancer Res. 62:6803–6807.
2002.PubMed/NCBI
|
|
12
|
Tanner MM, Grenman S, Koul A, Johannsson
O, Meltzer P, Pejovic T, Borg A and Isola JJ: Frequent
amplification of chromosomal region 20q12-q13 in ovarian cancer.
Clin Cancer Res. 6:1833–1839. 2000.PubMed/NCBI
|
|
13
|
Jin Y, Zhu H, Cai W, Fan X, Wang Y, Niu Y,
Song F and Bu Y: B-Myb is up-regulated and promotes cell growth and
motility in non-small cell lung cancer. Int J Mol Sci. 18:8602017.
View Article : Google Scholar
|
|
14
|
Tao D, Pan Y, Jiang G, Lu H, Zheng S, Lin
H and Cao F: B-Myb regulates snail expression to promote
epithelial-to-mesenchymal transition and invasion of breast cancer
cell. Med Oncol. 32:4122015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Borczuk AC, Gorenstein L, Walter KL,
Assaad AA, Wang L and Powell CA: Non-small-cell lung cancer
molecular signatures recapitulate lung developmental pathways. Am J
Pathol. 163:1949–1960. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakajima T, Yasui K, Zen K, Inagaki Y,
Fujii H, Minami M, Tanaka S, Taniwaki M, Itoh Y, Arii S, et al:
Activation of B-Myb by E2F1 in hepatocellular carcinoma. Hepatol
Res. 38:886–895. 2008.PubMed/NCBI
|
|
17
|
Skotheim RI, Monni O, Mousses S, Fossa SD,
Kallioniemi OP, Lothe RA and Kallioniemi A: New insights into
testicular germ cell tumorigenesis from gene expression profiling.
Cancer Res. 62:2359–2364. 2002.PubMed/NCBI
|
|
18
|
Ren F, Wang L, Shen X, Xiao X, Liu Z, Wei
P, Wang Y, Qi P, Shen C, Sheng W and Du X: MYBL2 is an independent
prognostic marker that has tumor-promoting functions in colorectal
cancer. Am J Cancer Res. 5:1542–1552. 2015.PubMed/NCBI
|
|
19
|
Qin HD, Liao XY, Chen YB, Huang SY, Xue
WQ, Li FF, Ge XS, Liu DQ, Cai Q, Long J, et al: Genomic
characterization of esophageal squamous cell carcinoma reveals
critical genes underlying tumorigenesis and poor prognosis. Am J
Hum Genet. 98:709–727. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qin H, Li Y, Zhang H, Wang F, He H, Bai X
and Li S: Prognostic implications and oncogenic roles of MYBL2
protein expression in esophageal squamous-cell carcinoma.
OncoTargets Ther. 12:1917–1927. 2019. View Article : Google Scholar
|
|
21
|
Gualdrini F, Corvetta D, Cantilena S,
Chayka O, Tanno B, Raschellà G and Sala A: Addiction of MYCN
amplified tumours to B-MYB underscores a reciprocal regulatory
loop. Oncotarget. 1:278–288. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sala A, Casella I, Bellon T, Calabretta B,
Watson RJ and Peschle C: B-myb promotes S phase and is a downstream
target of the negative regulator p107 in human cells. J Biol Chem.
271:9363–9367. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Arsura M, Introna M, Passerini F,
Mantovani A and Golay J: B-myb antisense oligonucleotides inhibit
proliferation of human hematopoietic cell lines. Blood.
79:2708–2716. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sala A and Calabretta B: Regulation of
BALB/c 3T3 fibroblast proliferation by B-myb is accompanied by
selective activation of cdc2 and cyclin D1 expression. Proc Natl
Acad Sci USA. 89:10415–10419. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang X, Lv QL, Huang YT, Zhang LH and
Zhou HH: Akt/FoxM1 signaling pathway-mediated upregulation of MYBL2
promotes progression of human glioma. J Exp Clin Cancer Res.
36:1052017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sakai N, Kubota Y and Shuin T:
Statistically significant expression of B-myb in clinically
advanced human renal-cell carcinomas. Int J Oncol. 2:419–423.
1993.PubMed/NCBI
|
|
27
|
Nord H, Segersten U, Sandgren J, Wester K,
Busch C, Menzel U, Komorowski J, Dumanski JP, Malmström PU and Díaz
de Ståhl T: Focal amplifications are associated with high grade and
recurrences in stage Ta bladder carcinoma. Int J Cancer.
126:1390–1402. 2010.PubMed/NCBI
|
|
28
|
Zauli G, Voltan R, di Iasio MG, Bosco R,
Melloni E, Sana ME and Secchiero P: miR-34a induces the
downregulation of both E2F1 and B-Myb oncogenes in leukemic cells.
Clin Cancer Res. 17:2712–2724. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liang HB, Cao Y, Ma Q, Shu YJ, Wang Z,
Zhang F, Ye YY, Li HF, Xiang SS, Song XL, et al: MYBL2 is a
potential prognostic marker that promotes cell proliferation in
gallbladder cancer. Cell Physiol Biochem. 41:2117–2131. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cervellera M, Raschella G, Santilli G,
Tanno B, Ventura A, Mancini C, Sevignani C, Calabretta B and Sala
A: Direct transactivation of the anti-apoptotic gene apolipoprotein
J (clusterin) by B-MYB. J Biol Chem. 275:21055–21060. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mao X, Orchard G, Lillington DM,
Russell-Jones R, Young BD and Whittaker SJ: Amplification and
overexpression of JUNB is associated with primary cutaneous T-cell
lymphomas. Blood. 101:1513–1519. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tao D, Pan Y, Lu H, Zheng S, Lin H, Fang H
and Cao F: B-myb is a gene implicated in cell cycle and
proliferation of breast cancer. Int J Clin Exp Pathol. 7:5819–5827.
2014.PubMed/NCBI
|
|
33
|
Martin CM, Astbury K, Kehoe L, O'Crowley
JB, O'Toole S and O'Leary JJ: The use of MYBL2 as a novel candidate
biomarker of cervical cancer. Methods Mol Biol. 1249:241–251. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Calvisi DF, Simile MM, Ladu S, Frau M,
Evert M, Tomasi ML, Demartis MI, Daino L, Seddaiu MA, Brozzetti S,
et al: Activation of v-Myb avian myeloblastosis viral oncogene
homolog-like2 (MYBL2)-LIN9 complex contributes to human
hepatocarcinogenesis and identifies a subset of hepatocellular
carcinoma with mutant p53. Hepatology. 53:1226–1236. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Iltzsche F, Simon K, Stopp S, Pattschull
G, Francke S, Wolter P, Hauser S, Murphy DJ, Garcia P, Rosenwald A
and Gaubatz S: An important role for Myb-MuvB and its target gene
KIF23 in a mouse model of lung adenocarcinoma. Oncogene.
36:110–1121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Thomas C, Robinson C, Dessauvagie B, Wood
B, Sterrett G, Harvey J and Amanuel B: Expression of proliferation
genes in formalin-fixed paraffin-embedded (FFPE) tissue from breast
carcinomas. Feasibility and relevance for a routine histopathology
laboratory. J Clin Pathol. 70:25–32. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wolter P, Hanselmann S, Pattschull G,
Schruf E and Gaubatz S: Central spindle proteins and mitotic
kinesins are direct transcriptional targets of MuvB, B-MYB and
FOXM1 in breast cancer cell lines and are potential targets for
therapy. Oncotarget. 8:11160–11172. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Diepenbruck M and Christofori G:
Epithelial-mesenchymal transition (EMT) and metastasis: Yes, no,
maybe? Curr Opin Cell Biol. 43:7–13. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang H, Kim JK, Edwards CA, Xu Z,
Taichman R and Wang CY: Clusterin inhibits apoptosis by interacting
with activated Bax. Nat Cell Biol. 7:909–915. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lang G, Gombert WM and Gould HJ: A
transcriptional regulatory element in the coding sequence of the
human Bcl-2 gene. Immunology. 114:25–36. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee HJ, Lee EK, Seo YE, Shin YH, Kim HS,
Chun YH, Yoon JS, Kim HH, Han MY, Kim CK, et al: Roles of Bcl-2 and
caspase-9 and −3 in CD30-induced human eosinophil apoptosis. J
Microbiol Immunol Infect. 50:145–152. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Grassilli E, Salomoni P, Perrotti D,
Franceschi C and Calabretta B: Resistance to apoptosis in CTLL-2
cells overexpressing B-Myb is associated with B-Myb-dependent bcl-2
induction. Cancer Res. 59:2451–2456. 1999.PubMed/NCBI
|
|
43
|
Levenson VV, Davidovich IA and Roninson
IB: Pleiotropic resistance to DNA-interactive drugs is associated
with increased expression of genes involved in DNA replication,
repair, and stress response. Cancer Res. 60:5027–5030.
2000.PubMed/NCBI
|
|
44
|
Sottile F, Gnemmi I, Cantilena S, D'Acunto
WC and Sala A: A chemical screen identifies the chemotherapeutic
drug topotecan as a specific inhibitor of the B-MYB/MYCN axis in
neuroblastoma. Oncotarget. 3:535–545. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Iida M, Brand TM, Campbell DA, Li C and
Wheeler DL: Yes and Lyn play a role in nuclear translocation of the
epidermal growth factor receptor. Oncogene. 32:759–767. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fuster O, Llop M, Dolz S, García P, Such
E, Ibáñez M, Luna I, Gómez I, López M, Cervera J, et al: Adverse
prognostic value of MYBL2 overexpression and association with
microRNA-30 family in acute myeloid leukemia patients. Leuk Res.
37:1690–1696. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Raschella G, Cesi V, Amendola R, Negroni
A, Tanno B, Altavista P, Tonini GP, De Bernardi B and Calabretta B:
Expression of B-myb in neuroblastoma tumors is a poor prognostic
factor independent from MYCN amplification. Cancer Res.
59:3365–338. 1999.PubMed/NCBI
|
|
48
|
Astbury K, McEvoy L, Brian H, Spillane C,
Sheils O, Martin C and O'Leary JJ: MYBL2 (B-MYB) in cervical
cancer: putative biomarker. Int J Gynecologic Cancer. 21:206–212.
2011. View Article : Google Scholar
|
|
49
|
Komiya R: Biogenesis of diverse plant
phasiRNAs involves an miRNA-trigger and Dicer-processing. J Plant
Res. 130:17–23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro- RNA genes miR15
and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad
Sci USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Martinez I, Cazalla D, Almstead LL, Steitz
JA and DiMaio D: miR-29 and miR-30 regulate B-Myb expression during
cellular senescence. Proc Natl Acad Sci USA. 108:522–527. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Martinez I and Dimaio D: B-Myb, cancer,
senescence, and microRNAs. Cancer Res. 71:5370–5373. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wu Z, Huang X, Huang X, Zou Q and Guo Y:
The inhibitory role of Mir-29 in growth of breast cancer cells. J
Exp Clin Cancer Res. 32:982013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Geng GJ, Yang YT, Jiang J, Yu XY and Fa
XE: MicroRNA-30a suppresses non-small-cell lung cancer by targeting
Myb-related protein B. Exp Ther Med. 15:1633–1639. 2018.PubMed/NCBI
|
|
56
|
Li ZB, Chu HT, Jia M and Li L: Long
noncoding RNA LINC01139 promotes the progression of hepatocellular
carcinoma by upregulating MYBL2 via competitively binding to miR-30
family. Biochem Biophys Res Commun. 525:581–588. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lee YJ, Kang YR, Lee SY, Jin Y, Han DC and
Kwon BM: Ginkgetin induces G2-phase arrest in HCT116 colon cancer
cells through the modulation of bMyb and miRNA34a expression. Int J
Oncol. 51:1331–1342. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yu Z, Kim J, He L, Creighton CJ, Gunaratne
PH, Hawkins SM and Matzuk MM: Functional analysis of miR-34c as a
putative tumor suppressor in high-grade serous ovarian cancer. Biol
Reprod. 91:1132014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang X, Li M, Wang Z, Han S, Tang X, Ge Y,
Zhou L, Zhou C, Yuan Q and Yang M: Silencing of long noncoding RNA
MALAT1 by miR-101 and miR-217 inhibits proliferation, migration,
and invasion of esophageal squamous cell carcinoma cells. J Biol
Chem. 290:3925–3935. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang Y, Tang X, Shi M, Wen C and Shen B:
MiR-216a decreases MALAT1 expression, induces G2/M arrest and
apoptosis in pancreatic cancer cells. Biochem Biophys Res Commun.
483:816–822. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chen J and Chen X: MYBL2 is targeted by
miR-143-3p and regulates breast cancer cell proliferation and
apoptosis. Oncol Res. 26:913–922. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lam EW, Robinson C and Watson RJ:
Characterization and cell cycle-regulated expression of mouse
B-myb. Oncogene. 7:1885–1890. 1992.PubMed/NCBI
|
|
63
|
de Bruin A, Maiti B, Jakoi L, Timmers C,
Buerki R and Leone G: Identification and characterization of E2F7,
a novel mammalian E2F family member capable of blocking cellular
proliferation. J Biol Chem. 278:42041–42049. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wong JV, Dong P, Nevins JR, Mathey-Prevot
B and You L: Network calisthenics: Control of E2F dynamics in cell
cycle entry. Cell Cycle. 10:3086–3094. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhu W, Giangrande PH and Nevins JR:
Temporal control of cell cycle gene expression mediated by E2F
transcription factors. Cell Cycle. 4:633–636. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Robinson C, Light Y, Groves R, Mann D,
Marias R and Watson R: Cell-cycle regulation of B-Myb protein
expression: Specific phosphorylation during the S phase of the cell
cycle. Oncogene. 12:1855–1864. 1996.PubMed/NCBI
|
|
67
|
Mowla SN, Lam EW and Jat PS: Cellular
senescence and aging: The role of B-MYB. Aging Cell. 13:773–779.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Inoue K and Fry EA: Novel molecular
markers for breast cancer. Biomark Cancer. 8:25–42. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lam EW, Bennett JD and Watson RJ:
Cell-cycle regulation of human B-myb transcription. Gene.
160:277–281. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Lam EW and Watson RJ: An E2F-binding site
mediates cell-cycle regulated repression of mouse B-myb
transcription. EMBO J. 12:2705–2713. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Lukas J, Petersen BO, Holm K, Bartek J and
Helin K: Deregulated expression of E2F family members induces
S-phase entry and overcomes p16INK4A-mediated growth suppression.
Mol Cell Biol. 16:1047–1057. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Sala A, Saitta B, De Luca P, Cervellera
MN, Casella I, Lewis RE, Watson R and Peschle C: B-MYB
transactivates its own promoter through SP1-binding sites.
Oncogene. 18:1333–1339. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Müller H, Bracken AP, Vernell R, Moroni
MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD and
Helin K: E2Fs regulate the expression of genes involved in
differentiation, development, proliferation, and apoptosis. Genes
Dev. 15:267–285. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Infante A, Laresgoiti U, Fernandez-Rueda
J, Fullaondo A, Galán J, Díaz-Uriarte R, Malumbres M, Field SJ and
Zubiaga AM: E2F2 represses cell cycle regulators to maintain
quiescence. Cell Cycle. 7:3915–3927. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Joaquin M and Watson RJ: Cell cycle
regulation by the B-Myb transcription factor. Cell Mol Life Sci.
60:2389–2401. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Santilli G, Schwab R, Watson R, Ebert C,
Aronow BJ and Sala A: Temperature-dependent modification and
activation of B-MYB: Implications for cell survival. J Biol Chem.
280:15628–15634. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zhu W, Giangrande PH and Nevins JR: E2Fs
link the control of G1/S and G2/M transcription. EMBO J.
23:4615–4626. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Fan X, Wang Y, Jiang T, Cai W, Jin Y, Niu
Y, Zhu H and Bu Y: B-Myb mediates proliferation and migration of
non-small-cell lung cancer via suppressing IGFBP3. Int J Mol Sci.
19:14792018. View Article : Google Scholar
|