Introduction
Brain metastases (BMs) are the most common
malignancies in the central nervous system, and mostly migrate from
lung cancer, melanoma and breast cancer (1). Brain metastasis is a pathological
feature associated with poor prognosis (1). Cancer genomics has expanded the
knowledge of driver mutations for various types of cancer, and has
identified potential therapeutic targets and precise therapies over
the past few decades. However, therapeutic approaches for BMs are
restricted to surgical resection, whole brain radiotherapy and
chemotherapy (2). Since traditional
therapies are insufficient to improve the prognosis of BMs
(1), there the identification of key
molecular events mediating metastasis is an urgent requirement.
Little is known regarding driver genomic alterations in BMs and to
what extent brain metastasis samples share common mutations, which
limits the mechanistic understanding and discovery of drug targets
specifically for patients with brain metastasis.
A previous study has evaluated the genetic
heterogeneity among the primary tumor site, paired normal tissue
and BMs in a limited number of patients (3). However, it is still unclear to what
extent different types of primary sites of BMs share common driver
mutations or metastasis mechanisms. Extensive heterogeneity between
primary sites and metastasis sites, and between spatially distinct
metastasis sites have been observed in other types of cancer,
including renal cell carcinoma (4).
Several small-scale genomic profiling studies have revealed genetic
alterations in patients with brain metastasis (5–7). Nearly
half of the patients with lung cancer develop brain metastasis in
the later stages of the disease (8),
and alterations in the PI3K signaling pathway have been identified
to mediate the formation of BMs in these patients (5,6). BMs
from colorectal cancer are rare; however, they can cause severe
outcomes, and genomic profiling has suggested that deficiency in
the DNA damage response is involved in the formation of BMs from
colorectal cancer (7). There remains
a requirement for comprehensive evaluation of homogeneity and
heterogeneity between primary tumor sites and BMs, as well as
between BMs from various primary tumor sites.
To gain a global view of brain metastasis
heterogeneity and potential driver genes, targeted next-generation
sequencing of 560 cancer-related genes in brain metastasis samples
from various primary sites, with an emphasis on lung cancer, was
performed in the present study. Further analysis of the mutational
profiles provided insights into the clinical outcomes associated
with genetic mutations enriched in BMs, suggesting that brain
metastasis-related gene mutations are associated with poor
prognosis.
Materials and methods
Patients, sample collection and
follow-up survey
The present study obtained the records and samples
of a total of 52 patients who underwent resection surgery for brain
metastasis at Beijing Tiantan Hospital (Beijing, China). The median
patient age was 57 years (age range, 36–73 years), 59.6% (31) were men and 40.4% (21) were women. The sequencing data were
generated using tumors resected between February 2012 and January
2016. All samples were collected and frozen in liquid nitrogen
within 5 min after resection, and were subjected to sequencing
analysis. The survival status of the patients was obtained through
phone contact every 3 months as a follow-up survey August 2018.
Library preparation and
sequencing
The sequencing library was generated using 1 µg DNA
per sample according to the guide of the Truseq Nano DNA HT Sample
Prep Kit (Illumina, Inc.) with index codes added to each sample.
The quality of genomic DNA was monitored on a 1% agarose gel, while
the concentration was measured using the Qubit® DNA
Assay Kit and Qubit® 2.0 Flurometer (Invitrogen; Thermo
Fisher Scientific, Inc.). DNA sequencing was performed for all the
exons of 559 cancer-related genes and the promoter of telomerase
reverse transcriptase (Agilent SureSelect custom kit; Agilent
Technologies, Inc.). Briefly, fragmentation was performed using a
hydrodynamic shearing system (Covaris, Inc.) to generate 180–280 bp
fragments. Extracted DNA was then amplified by ligation-mediated
PCR (LM-PCR) using Herculase II Fusion DNA Polymerase and
customized primer provided by the Agilent SureSelect custom kit
(cat. no. G9611B; Agilent Technologies, Inc.), purified and
hybridized to the probe for enrichment. The following thermocycling
conditions were used: 98°C for 2 min; 6 cycles at {98°C for 30 sec,
65°C for 30 sec and 72°C for 1 min}, and 72°C for 2 min.
Non-hybridized fragments were subsequently washed using nuclease
free water. Both non-captured and captured LM-PCR products were
subjected to quantitative PCR to estimate the magnitude of
enrichment using the KAPA Library Quantification kit (cat. no.
KK4824; Kapa Biosystems. Inc.). The primer sequences used are as
follows: Primer P1 5′-AATGATACGGCGACCACCGA-3′ and Primer P2:
5′-CAAGCAGAAGACGGCATACGA-3′. SYBR-Green I dye was used in the qPCR
analysis and library quantification DNA standards 1–6 (a 10-fold
dilution series of a linear, 452 bp template) were used as the
reference for absolute quantification. The thermocycling conditions
were as follows: 95°C for 5 min for initial
activation/denaturation, and 35 cycles denaturation, annealing and
extension at 95°C for 30 sec, and 60°C for 45 sec. The DNA
libraries were sequenced on the Illumina Hiseq 4000 platform
(Illumina, Inc.), and 150-bp paired-end reads were generated at a
depth of 1000X.
Detection and filtering of genomic
alterations
Sequencing data were mapped to the human reference
genome (UCSC hg19) using the Burrows-Wheeler Aligner software
(version 0.7.10-r789) (9). SAMtools
(version 0.1.19) was used to sort the BAM files and perform
duplicate marking, local realignment and base quality recalibration
to generate the final BAM file for computing the sequence coverage
and depth (10). To identify single
nucleotide variations (SNVs) and small insertions and deletions
(InDels) from the BM samples, GATK (https://gatk.broadinstitute.org/hc/en-us) and SAMtools
were used. In addition to default filters, polymorphisms of SNVs
and InDels referenced in the 1000 Genomes Project (11), Exome Aggregation Consortium (12) or the in-house Novozhonghua database
(not publicly available yet) with a minor allele frequency >1%
were removed. Subsequently, the variant call format result was
annotated by ANNOVAR (version 20191024) (13). The mutation frequency of the primary
lung tumor site was obtained from a previous lung pan-cancer
dataset through the cBio cancer genomics portal (https://www.cbioportal.org/) (14).
Statistical analysis
Survival analysis was performed using the R (v3.6.0)
survival package (v3.2, http://cran.r-project.org/web/packages/survival/index.html).
The overall survival rate was estimated according to the
Kaplan-Meier method using the survfit function in the R survival
package. A log-rank test was performed for comparison of survival
curves using the survdiff function. Survival analysis was performed
on 48 of the 52 patients with BM (four patients were excluded in
the survival analysis as their dates of death were not accurately
obtained). For each BM enriched mutated gene, patients were grouped
according to whether they harbored the mutation or not. Survival
analysis was performed between the two groups to identify genes
whose mutation affects the overall survival of patients with BM.
P<0.05 was set as the cutoff of significant differential overall
survival rate in the log-rant test. Survival and progression-free
survival analyses of the patients with lung cancer were performed
using data from datasets and tools in the cBio cancer genomics
portal (https://www.cbioportal.org) (14). To investigate whether BM mutation in
the primary tumor affects the PFS of patients with lung cancer, the
PFS of patients with or without BM enriched mutations in the
datasets of 1,410 patients combined in cBio cancer genomics portal
was assessed.
Results
Recurrent mutations among BMs
To identify genomic alterations associated with
brain metastasis, targeted exome sequencing of a panel of 560
cancer-related genes was performed (Table SI). Exomes of these genes were
targeted with the exception of TERT, whose promoter region was
targeted using samples from 52 patients with brain metastases from
various primary sites. Clinical characteristics of the patients are
shown in Table SII. A total of 33
patients had primary lung cancer, while the remaining 19 patients
had cancer of other primary locations. Recurrent mutations in these
brain metastasis samples were identified by comparing the
sequencing results of the targeted sites with the human reference
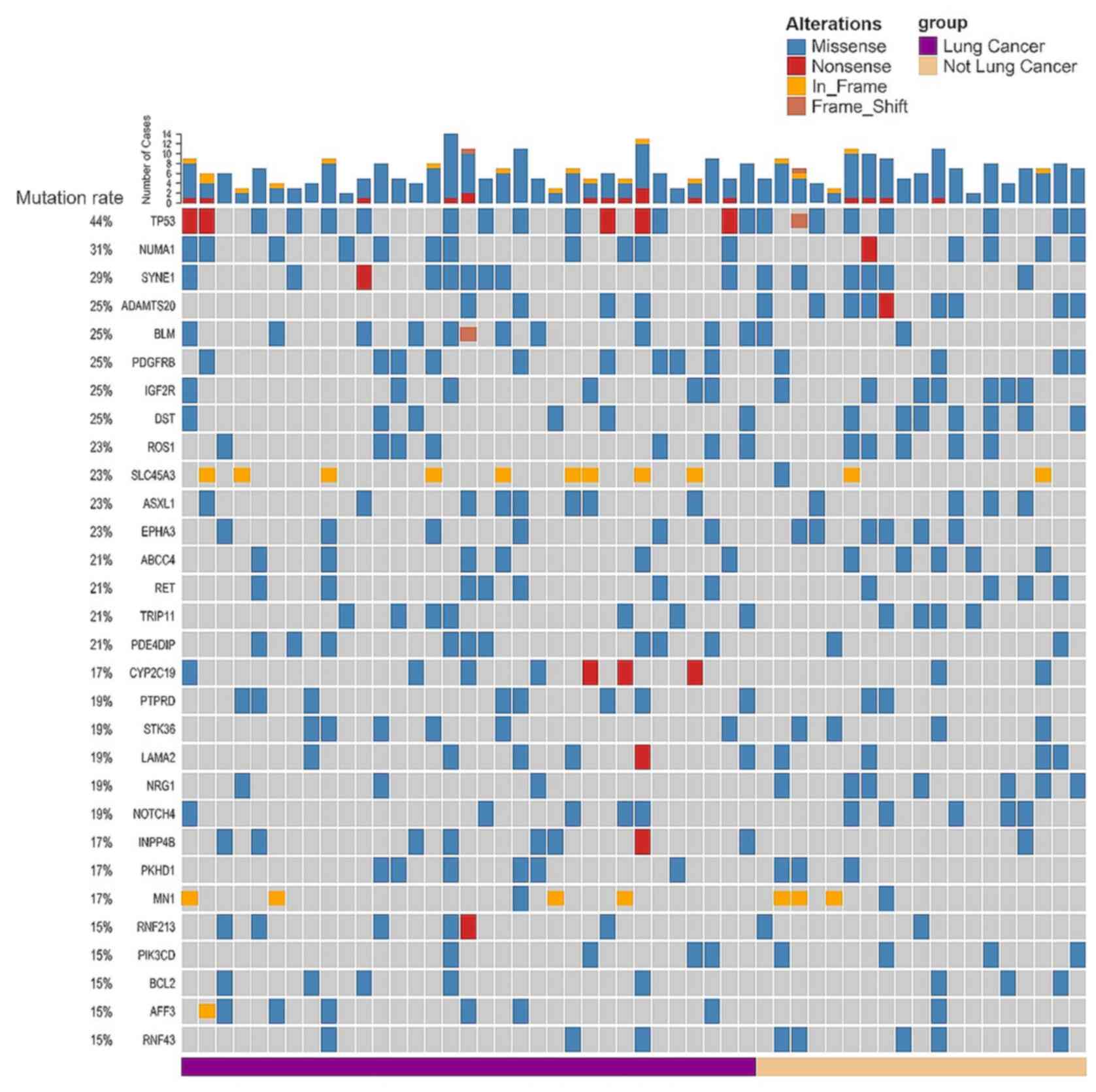
genome. TP53 was the most commonly mutated gene (44.2%)
among all the brain metastasis samples, followed by other genes
that have been frequently associated with cancer, including nuclear
mitotic apparatus protein 1 (NUMA1), SYNE1, PKHD1,
ADAMTS20, BLM, PDGFRB, IGF2R and PKHD1 (Fig. 1).
Genomic alterations in BM-related genes (e.g.
SCN7A, SCN5A, SCN2A, IKZF1, PDZRN4 and TP53) have been
identified in BMs from various primary sites (5–7,15); however, it remains unclear to what
extent BMs from different sites have common and specific mutational
signatures. Since the lungs were the primary site for 63.5% of the
brain metastasis samples (Table
SII), mutations which were common for all brain metastasis
samples were investigated, as well as those that were more frequent
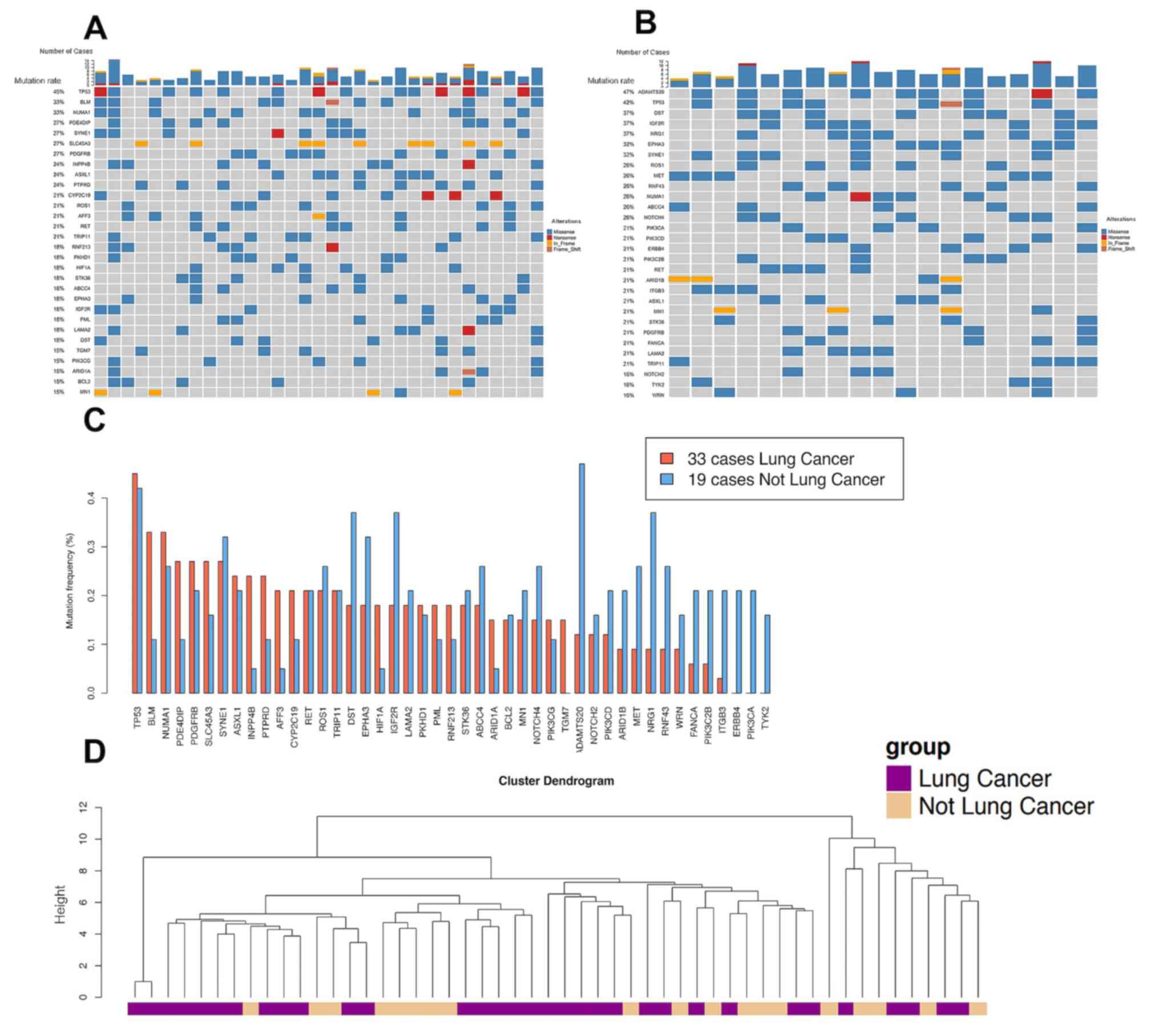
in brain metastasis originating from the lungs. The frequency of
recurrent mutations in brain metastasis from the lungs was compared
with that of other sites (Fig. 2A and
B). Among the 25 genes whose mutation frequency reached 15% in
the brain metastasis samples from the lungs, 9 genes (BLM,
PDE4DIP, INPP4B, PTRRD, AFF3, HIF1A, CYP2C19, ARID1A and TGM7;
Fig. 2C; Table SIII) harbored a >2-fold mutation
rate compared with that in brain metastasis samples from other
primary tumor sites. Recurrent mutated genes, with a similar high
mutation rate for different primary sites included TP53, NUMA1,
SYNE1, ASXL1, RET, ROS1 and TRIP11. There was no brain
metastasis mutation identified that was exclusively found in
patients with lung cancer, suggesting a limited effect of the
tissue of origin on the brain metastasis genomic signature. The
heterogeneity of brain metastasis genomic signatures was further
supported by hierarchical clustering of the mutation signature of
all brain metastasis samples (Fig.
2D).
Genomic alterations enriched in brain
metastasis samples compared with in tumor samples from the
lungs
To identify potential driver mutations that were
enriched in the brain metastasis samples compared with in the
primary site, the frequency of the recurrent gene mutations in the
brain metastasis samples and the primary site was compared.
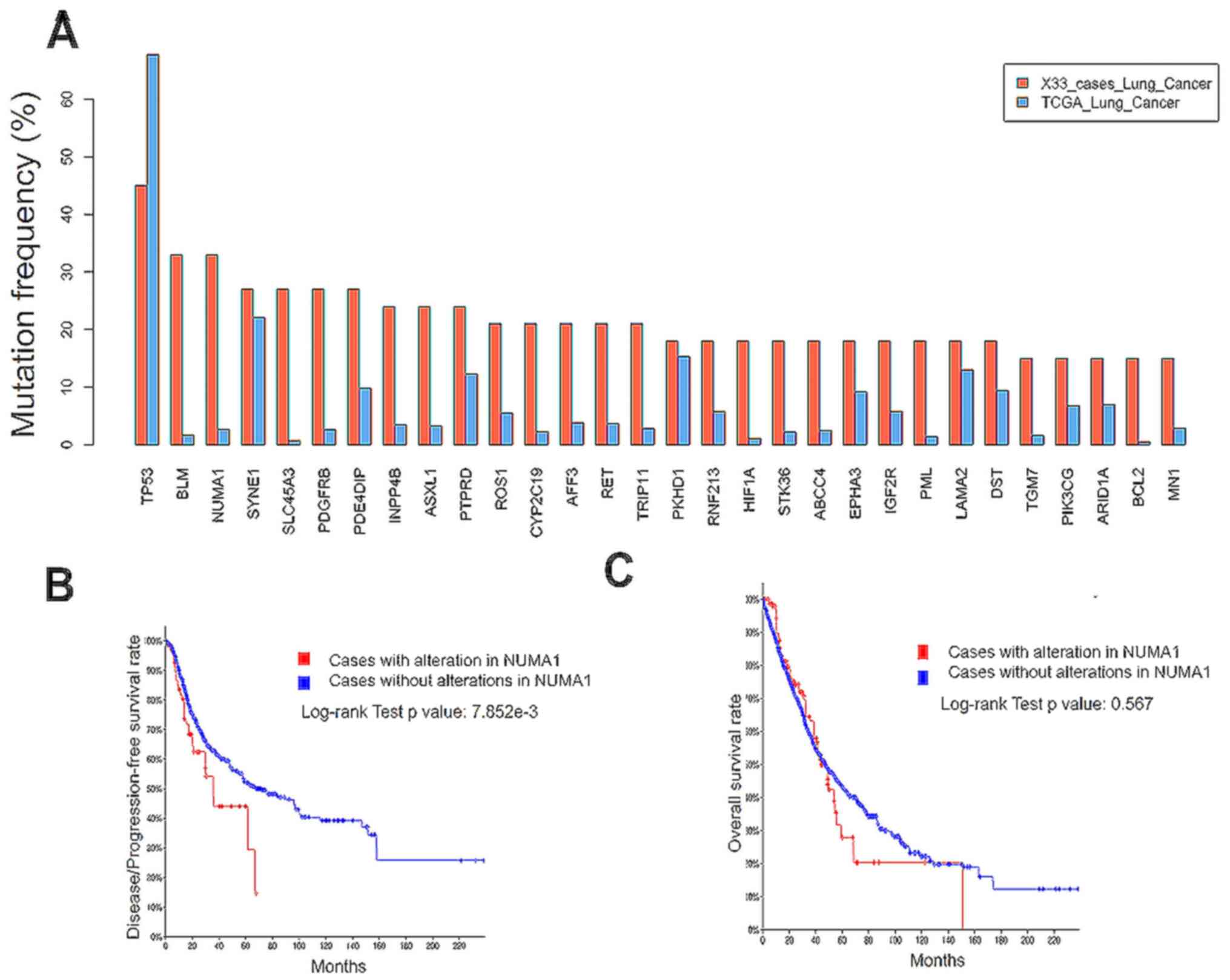
TP53 was the most frequently mutated gene in both the brain
metastasis samples and tumors in the lungs, according to data from
the present study and from a The Cancer Genome Atlas (TCGA)
dataset, respectively (Fig. 3A). A
total of two genes (BLM and NUMA1) were associated
with DNA damage response (16,17), and
had a >30% mutation rate in brain metastasis samples which had
migrated from the lungs, compared with 2–3% in the lungs, as the
primary site, suggesting that brain metastasis was associated with
dysregulated DNA damage response. Brain metastasis has been
associated with poor prognosis in patients with lung cancer
(1,5). Therefore, to examine the potential link
between brain metastasis-enriched mutated genes and clinical
outcome, a published dataset of lung cancer genome sequencing was
investigated (14,18–21). The
mutation rates for potential brain metastasis driver genes such as
BLM, NUMA1, SLC45A3 and PDGFRB were low in the primary site
(Fig. 3A); however, a mutation in
NUMA1 in the primary lung cancer site was associated with
worse progression-free survival time [35.6 months (n=69) vs. 67.2
months (n=1,341); log-rank test, P<0.01; Fig. 3B]. Furthermore, the NUMA1
mutation did not affect the overall survival rate in patients with
lung cancer (log-rank test, P=0.567; Fig. 3C). This result suggested that the
NUMA1 mutation may promote brain metastasis without
affecting the overall survival time of patients with lung cancer.
BMs migrated from the breasts and the colon have been demonstrated
to be enriched with mutations and abnormal expression levels of DNA
damage repair genes (7,22,23). The
results of the present study suggested that DNA damage repair
deficiency was a common feature of BMs as genes related to DNA
damage responses (BLM and NUMA1) was frequently mutated in BMs from
various primary sties.
Common genetic alterations in BMs and
prognosis
A total of 8 genes with recurrent mutations in both
BMs from the lungs and other primary sites in at least 9 patients
were identified (Fig. 1).
Significant prognostic markers for brain metastasis samples were
rarely identified previously (1,5,6,8). To
investigate gene mutations associated with prognosis in patients
with brain metastasis, the overall survival rate of patients with
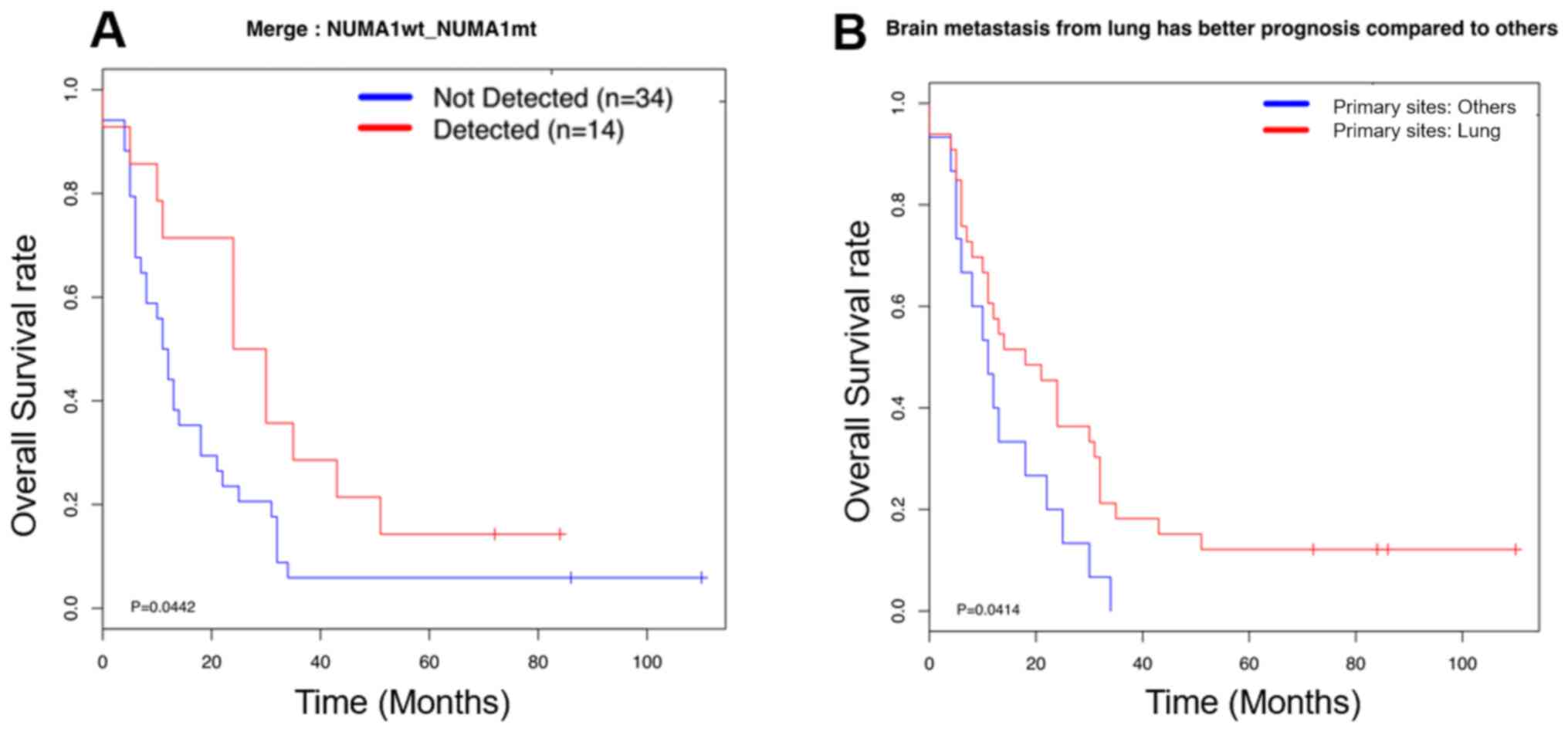
or without these mutations was investigated. Most genetic
alterations in BMs were not associated with the overall survival
rate; however, a mutation in the potential brain metastasis driver
gene, NUMA1, could predict a good prognosis in patients with
brain metastasis (Fig. 4A),
suggesting NUMA1 may be a potential prognostic marker for
brain metastasis progression. The frequency of brain metastasis for
different types of cancer varies greatly; however, the clinical
outcome of patients with brain metastasis and different primary
sites is unknown. It was identified that patients with lung cancer
were more prone to have brain metastasis; however, they had a good
prognosis compared with patients with BMs that had migrated from
other sites (Fig. 4B). These results
suggested that the clinical outcome of BMs may not be associated
with the frequency of brain metastasis formation.
Discussion
The prognosis of patients with brain metastasis is
poor, with a median survival time of a few months (1). The incidence of brain metastasis is
rising, as revolutionized cancer therapy has improved the survival
of patients with advanced cancer (24). In contrast to the advancement of
treatment of primary tumors, the treatment of brain metastasis
remains a substantial challenge, primarily due to the lack of
actionable targets (1,2,8).
Previous high-throughput sequencing studies have revealed a
distinct mutational landscape of brain metastasis from primary
tumors, regional lymph nodes and extracranial metastasis (3); however, there is a lack of evaluation
regarding the magnitude of BMs from the same or different primary
sites which share common mutations. In the hierarchical clustering
analysis of brain metastasis samples in the present study, higher
genetic similarity among BMs from lungs was not observed compared
with that between BMs from lungs and other primary sites. Previous
analyses have revealed independent evolution of brain metastasis
from the primary sites (7,25,26).
These results collectively suggested that brain metastasis should
be treated by targeting genomic alterations enriched in brain
metastasis instead of the primary tumors. In the present study,
there were 15% of patients with BM that harbored mutations in the
PIKC3D gene (Fig. 1), which
was consistent with previous reports that PI3K could be a potential
brain metastasis therapeutic target (27,28).
Comparisons of the mutational landscape of brain
metastasis with that of the primary tumors revealed potential
driver mutations for brain metastasis in the KRAS, PI3K and
DNA damage response signaling pathways (7,23,29,30).
It is largely unknown whether prognosis would be affected if
patients harbor these mutations in their primary sites. Potential
mutations that could contribute to brain metastasis from the lungs
were identified by comparing the frequency of recurrent mutations
to their frequencies in the lung cancer data from TCGA. Gene
mutations associated with DNA damage response deficiency were
enriched in brain metastasis samples, and patients with the
NUMA1 mutation exhibited a shorter progression-free survival
time.
The tumor suppressor gene, TP53, has
antiproliferative effects, and somatic TP53 gene alterations
are frequent in most types of human cancer (31). It also regulates the transcription of
genes involved in processes that are essential for metastasis, such
as cell motility and adhesion (32,33). A
high mutation frequency of TP53 was identified in BMs in the
present study, and a high TP53 mutation frequency has also
been observed in samples of brain metastasis of breast cancer
(34–36). Therefore, these data collectively
suggested that the TP53 mutation not only contributed to the
development of tumors at the primary sites, but also promoted brain
metastasis.
Identifying mutations affecting the survival of
patients with brain metastasis is fundamental for developing
therapeutic approaches for brain metastasis. NUMA1 interacts
and colocalizes with the P53-binding protein 1 (P53BP1), which
prevents P53BP1 accumulation at the DNA break, and high
NUMA1 expression predicts improved patient outcomes
(17). NUMA1 also promotes
p53-dependent downstream gene transcription in cancer cells
(37,38). It was speculated that
loss-of-function of NUMA1 affects the DNA damage response and may
limit the expansion of brain metastasis subclones. NUMA1
alternative splicing has been identified to be involved in multiple
primary cancer sites (39), and has
recently been reported to be enriched in prostate cancer brain
metastasis (15). However, when
comparing the survival time of patients with brain metastasis with
or without each recurrent mutation, a missense mutation in the
structural nuclear protein, NUMA1, was associated with a
longer survival time compared with that of patients without this
mutation. Collectively, the NUMA1 mutation in the primary
sites caused more frequent brain metastasis. However, patients with
BMs and the NUMA1 mutation had a good prognosis, suggesting
that the role of the DNA damage response in the formation of brain
metastasis and the clinical outcome of brain metastasis may be
independent of each other.
In conclusion, BM originates from distinct sites;
however, the primary tumor may have different mutational
signatures, and it was found that brain metastases from different
sites shared commonly mutated genes. In the patients with lung
cancer and brain metastasis, recurrent mutations with a higher
mutation rate in brain metastasis compared with that at the primary
site were found, indicating that these genes are potential brain
metastasis driver genes. Analysis of the TCGA lung cancer dataset
revealed that potential brain metastasis driver genes were
associated with poor progression-free survival.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Capital's
Funds for Health Improvement and Research (CFH; grant no.
2018-1-1071), The National Natural Science Fund (grant nos.
81701085 and U1804199) and the Henan Key Laboratory of the Neural
Regeneration and Repairment (grant no. HNSJXF-2018-001).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
WJ, CZ and DZ conceived and designed the present
study. DZ, SM, BM, XG and KS collected samples, performed the
experiments and recorded the clinical information. XW, WZ, JP, PL
and FX processed and analyzed the data. DZ, XW, CZ and WJ drafted
the initial manuscript with input from all authors. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of Beijing Tiantan Hospital, Capital Medical
University (Beijing, China) and performed in accordance with the
principles of the Declaration of Helsinki. Written informed consent
was provided by all patients prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BM
|
brain metastasis
|
|
TCGA
|
The Cancer Genome Atlas
|
|
SNV
|
single nucleotide variation
|
References
|
1
|
Brastianos PK, Curry WT and Oh KS:
Clinical discussion and review of the management of brain
metastases. J Natl Compr Canc Netw. 11:1153–1164. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kyritsis AP, Markoula S and Levin VA: A
systematic approach to the management of patients with brain
metastases of known or unknown primary site. Cancer Chemother
Pharmacol. 69:1–13. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brastianos PK, Carter SL, Santagata S,
Cahill DP, Taylor-Weiner A, Jones RT, Van Allen EM, Lawrence MS,
Horowitz PM, Cibulskis K, et al: Genomic characterization of brain
metastases reveals branched evolution and potential therapeutic
targets. Cancer Discov. 5:1164–1177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gerlinger M, Rowan AJ, Horswell S, Math M,
Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N,
Stewart A, et al: Intratumor heterogeneity and branched evolution
revealed by multiregion sequencing. N Engl J Med. 366:883–892.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang H, Ou Q, Li D, Qin T, Bao H, Hou X,
Wang K, Wang F, Deng Q, Liang J, et al: Genes associated with
increased brain metastasis risk in non-small cell lung cancer:
Comprehensive genomic profiling of 61 resected brain metastases
versus primary non-small cell lung cancer (Guangdong Association
Study of Thoracic Oncology 1036). Cancer. 125:3535–3544. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilson GD, Johnson MD, Ahmed S, Cardenas
PY, Grills IS and Thibodeau BJ: Targeted DNA sequencing of
non-small cell lung cancer identifies mutations associated with
brain metastases. Oncotarget. 9:25957–25970. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun J, Wang C, Zhang Y, Xu L, Fang W, Zhu
Y, Zheng Y, Chen X, Xie X, Hu X, et al: Genomic signatures reveal
DNA damage response deficiency in colorectal cancer brain
metastases. Nat Commun. 10:31902019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Steeg PS, Camphausen KA and Smith QR:
Brain metastases as preventive and therapeutic targets. Nat Rev
Cancer. 11:352–363. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li H and Durbin R: Fast and accurate
long-read alignment with Burrows-Wheeler transform. Bioinformatics.
26:589–595. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R; 1000 Genome
Project Data Processing Subgroup, : The sequence alignment/map
format and SAMtools. Bioinformatics. 25:2078–2079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
1000 Genomes Project Consortium, .
Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker
RE, Kang HM, Marth GT and McVean GA: An integrated map of genetic
variation from 1,092 human genomes. Nature. 491:562012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lek M, Karczewski KJ, Minikel EV, Samocha
KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ,
Cummings BB, et al: Analysis of protein-coding genetic variation in
60,706 humans. Nature. 536:285–291. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang K, Li M and Hakonarson H: ANNOVAR:
Functional annotation of genetic variants from high-throughput
sequencing data. Nucleic Acids Res. 38:e1642010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Campbell JD, Alexandrov A, Kim J, Wala J,
Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et
al: Distinct patterns of somatic genome alterations in lung
adenocarcinomas and squamous cell carcinomas. Nat Genet.
48:607–616. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rodriguez A, Gallon J, Akhoundova D,
Maletti S, Ferguson A, Cyrta J, Amstutz U, Garofoli A, Paradiso V,
Tomlins S, et al: The genomic landscape of prostate cancer brain
metastases. BioRxiv; 2020
|
|
16
|
Patel DS, Misenko SM, Her J and Bunting
SF: BLM helicase regulates DNA repair by counteracting RAD51
loading at DNA double-strand break sites. J Cell Biol.
216:3521–3534. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salvador Moreno N, Liu J, Haas KM, Parker
LL, Chakraborty C, Kron SJ, Hodges K, Miller LD, Langefeld C,
Robinson PJ, et al: The nuclear structural protein NuMA is a
negative regulator of 53BP1 in DNA double-strand break repair.
Nucleic Acids Res. 47:2703–2715. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jordan EJ, Kim HR, Arcila ME, Barron D,
Chakravarty D, Gao J, Chang MT, Ni A, Kundra R, Jonsson P, et al:
Prospective comprehensive molecular characterization of lung
adenocarcinomas for efficient patient matching to approved and
emerging therapies. Cancer Discov. 7:596–609. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rizvi NA, Hellmann MD, Snyder A, Kvistborg
P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al: Cancer
immunology. Mutational landscape determines sensitivity to PD-1
blockade in non-small cell lung cancer. Science. 348:124–128. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jamal-Hanjani M, Wilson GA, McGranahan N,
Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R,
Rosenthal R, et al: Tracking the evolution of non-small-cell lung
cancer. N Engl J Med. 376:2109–2121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Woditschka S, Evans L, Duchnowska R, Reed
LT, Palmieri D, Qian Y, Badve S, Sledge G Jr, Gril B, Aladjem MI,
et al: DNA double-strand break repair genes and oxidative damage in
brain metastasis of breast cancer. J Natl Cancer Inst.
106:dju1452014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Diossy M, Reiniger L, Sztupinszki Z,
Krzystanek M, Timms KM, Neff C, Solimeno C, Pruss D, Eklund AC,
Tóth E, et al: Breast cancer brain metastases show increased levels
of genomic aberration-based homologous recombination deficiency
scores relative to their corresponding primary tumors. Ann Oncol.
29:1948–1954. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Han CH and Brastianos PK: Genetic
characterization of brain metastases in the era of targeted
therapy. Front Oncol. 7:2302017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wei Q, Ye Z, Zhong X, Li L, Wang C, Myers
RE, Palazzo JP, Fortuna D, Yan A, Waldman SA, et al: Multiregion
whole-exome sequencing of matched primary and metastatic tumors
revealed genomic heterogeneity and suggested polyclonal seeding in
colorectal cancer metastasis. Ann Oncol. 28:2135–2141. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cooper CS, Eeles R, Wedge DC, Van Loo P,
Gundem G, Alexandrov LB, Kremeyer B, Butler A, Lynch AG, Camacho N,
et al: Analysis of the genetic phylogeny of multifocal prostate
cancer identifies multiple independent clonal expansions in
neoplastic and morphologically normal prostate tissue. Nat Genet.
47:367–372. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen G, Chakravarti N, Aardalen K, Lazar
AJ, Tetzlaff MT, Wubbenhorst B, Kim SB, Kopetz S, Ledoux AA, Gopal
YN, et al: Molecular profiling of patient-matched brain and
extracranial melanoma metastases implicates the PI3K pathway as a
therapeutic target. Clin Cancer Res. 20:5537–5546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Niessner H, Schmitz J, Tabatabai G, Schmid
AM, Calaminus C, Sinnberg T, Weide B, Eigentler TK, Garbe C,
Schittek B, et al: PI3K pathway inhibition achieves potent
antitumor activity in melanoma brain metastases in vitro and in
vivo. Clin Cancer Res. 22:5818–5828. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tie J, Lipton L, Desai J, Gibbs P,
Jorissen RN, Christie M, Drummond KJ, Thomson BN, Usatoff V, Evans
PM, et al: KRAS mutation is associated with lung metastasis in
patients with curatively resected colorectal cancer. Clin Cancer
Res. 17:1122–1130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liao L, Ji X, Ge M, Zhan Q, Huang R, Liang
X and Zhou X: Characterization of genetic alterations in brain
metastases from non-small cell lung cancer. FEBS Open Bio.
8:1544–1552. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Petitjean A, Achatz MI, Borresen-Dale AL,
Hainaut P and Olivier M: TP53 mutations in human cancers:
Functional selection and impact on cancer prognosis and outcomes.
Oncogene. 26:2157–2165. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mukhopadhyay UK, Eves R, Jia L, Mooney P
and Mak AS: p53 suppresses Src-induced podosome and rosette
formation and cellular invasiveness through the upregulation of
caldesmon. Mol Cell Biol. 29:3088–3098. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Muller PA, Vousden KH and Norman JC: p53
and its mutants in tumor cell migration and invasion. J Cell Biol.
192:209–218. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lo Nigro C, Vivenza D, Monteverde M,
Lattanzio L, Gojis O, Garrone O, Comino A, Merlano M, Quinlan PR,
Syed N, et al: High frequency of complex TP53 mutations in CNS
metastases from breast cancer. Br J Cancer. 106:397–404. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee JY, Park K, Lim SH, Kim HS, Yoo KH,
Jung KS, Song HN, Hong M, Do IG, Ahn T, et al: Mutational profiling
of brain metastasis from breast cancer: Matched pair analysis of
targeted sequencing between brain metastasis and primary breast
cancer. Oncotarget. 6:43731–43742. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wei CL, Wu Q, Vega VB, Chiu KP, Ng P,
Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al: A global map of
p53 transcription-factor binding sites in the human genome. Cell.
124:207–219. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Endo A, Moyori A, Kobayashi A and Wong RW:
Nuclear mitotic apparatus protein, NuMA, modulates p53-mediated
transcription in cancer cells. Cell Death Dis. 4:e7132013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ohata H, Miyazaki M, Otomo R,
Matsushima-Hibiya Y, Otsubo C, Nagase T, Arakawa H, Yokota J,
Nakagama H, Taya Y and Enari M: NuMA is required for the selective
induction of p53 target genes. Mol Cell Biol. 33:2447–2457. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sebestyén E, Singh B, Miñana B, Mateo F,
Pujana MA, Valcárcel J and Eyras E: Large-scale analysis of genome
and transcriptome alterations in multiple tumors unveils novel
cancer-relevant splicing networks. Genome Res. 26:732–744. 2016.
View Article : Google Scholar : PubMed/NCBI
|