Introduction
Cutaneous malignant melanoma is a malignancy with
one of the fastest increasing incidence rates worldwide (1,2).
Although melanoma accounts for only a limited proportion of all
skin malignancies, it causes the largest number of skin
cancer-related deaths, with ~55,500 deaths each year (3). The high mortality of melanoma mainly
occurs due to distant metastasis (4). Immunotherapy has transformed the
treatment of melanoma in the past decade, with a significant
increase in overall survival (5).
For example, an antibody against cytotoxic T-lymphocyte-associated
protein 4 (CTLA-4), ipilimumab, increased the 2-year survival rates
of patients with metastatic melanoma from 14 to 24% (6). With the application of monoclonal
antibodies against inhibitory immune checkpoints, such as CTLA-4
and anti-programmed death 1, the treatment efficacy for metastatic
melanoma has improved (5,7). However, predictive biomarkers for
melanoma are still lacking. The occurrence and progression of
melanoma is considered to result from disorders of the oncogenic
and tumor inhibitory pathway functions (8). The importance and relevance of various
biomarkers have been demonstrated in melanoma studies (9,10), and
targeted agents such as vemurafenib and dabrafenib have enhanced
the survival of patients with melanoma; however, patient prognosis
remains poor (3). Therefore, it is
crucial to study novel biomarkers that drive the occurrence and
development of melanoma, which may contribute to the development of
new diagnostic and treatment targets.
The Wnt/β-catenin pathway contributes to cancer
metastasis through regulating the epithelial-mesenchymal transition
(EMT) (11,12), which has been reported in melanoma
(13). The Wnt/β-catenin signaling
pathway is often abnormally activated and may participate in the
occurrence and development of tumors (14), leading to β-catenin nuclear
translocation and the transcription of downstream target genes,
such as MYC (15). The MYC gene is
typically expressed constitutively in malignancies, and c-Myc,
which is encoded by MYC, has been reported to regulate the gene
expression of ≤15% of the human genome (16). From a clinical perspective, high
expression levels of c-Myc protein exhibit a significant
association with distant metastasis and impaired prognosis; from a
biological perspective, the overexpression of c-Myc results in a
significant increase in cell viability, invasion and migration
(17). Furthermore, c-Myc increases
the protein expression levels of Snail, which is an EMT marker,
in vivo and in vitro (18). Thus, MYC may be a novel therapeutic
target in melanoma, and the suppression of MYC expression may be an
effective therapeutic option for patients with melanoma.
MicroRNAs (miRNAs), a type of noncoding RNAs that
are <22 nucleotides long, bind to the 3′-untranslated region
(3′-UTR) of their downstream target mRNAs, inhibiting their
expression (19,20). Depending on the extent of base
pairing, miRNAs lead to translational repression, mRNA
deadenylation or decay (21).
Changes in miRNA expression levels contribute to human tumor
occurrence and development (19). In
melanoma, deregulation of miRNA expression levels has also been
reported previously. Using C57BL/6 mice, Noori et al
(22) have demonstrated that high
levels of miRNA (miR)-30a expression in melanoma suppress
metastasis in vivo through binding to zinc finger
E-box-binding homeobox 2 and E-cadherin. A series of miRNAs, such
as miR-1908, miR-199a-5p and miR-199a-3p, have been reported as
endogenous factors promoting melanoma metastatic invasion,
angiogenesis and colonization (23).
Since distinct miRNA expression profiles have been investigated at
various stages of melanoma progression (24,25),
identifying additional miRNAs that target MYC may provide new
therapeutic options for melanoma treatment.
The aim of the present study was to identify a
miRNA/mRNA axis that may modulate the biological malignant
behaviors of melanoma cells in vitro. To achieve this, an
online tool was used to select the candidate miRNAs that could bind
to the MYC 3′UTR, and the effects of the miR-27b/MYC axis on
melanoma progression were determined.
Materials and methods
Clinical sampling
A total of 18 paired melanoma and adjacent
noncancerous tissue samples (5 cm from the edge of the tumor) were
collected from patients who received surgical treatment for
cutaneous melanoma at the Second Affiliated Hospital of Hunan
University of Chinese Medicine (Changsha, China) with the approval
of the Ethics Committee of the Second Affiliated Hospital of Hunan
University of Chinese Medicine (approval no. 2019-KY-031). All
patients provided written informed consent. The obtained clinical
samples were stored at −80°C or fixed in 10% formalin at room
temperature until further use.
Hematoxylin and eosin (H&E) and
immunohistochemical (IHC) staining
The histomorphological changes in melanoma and
adjacent noncancerous tissues were evaluated by H&E staining at
room temperature. The 5-µm sections were stained with hematoxylin
solution for 5 min, destained with 0.5% acid ethanol (pH 2.0) and
stained with eosin for 30 sec. Following dehydration with graded
ethanol (80, 90, 95% and absolute ethanol incubated for 3 min each)
and clearing with xylene, the sections were mounted with neutral
balsam and observed under an optical microscope (Olympus
Corporation) with ×100 magnification in three fields per
sample.
The protein content and distribution of c-Myc in
tissue samples were detected by IHC as previously described
(26). Briefly, 5-µm tissue sections
were deparaffinized in xylene and rehydrated using graded ethanol
(absolute ethanol, 95, 90, 80 and 75% ethanol incubated for 3 min
each) in PBS. Endogenous peroxidase was blocked with 0.3% hydrogen
peroxide for 10 min at room temperature. The sections were
incubated with 5% normal rat serum (Beijing Solarbio Science &
Technology Co., Ltd.) at room temperature for 10 min, followed by
incubation with anti-c-Myc (1:200, cat. no. ab32072; Abcam) at 4°C
overnight. Subsequently, the sections were incubated with an
HRP-polymer-conjugated anti-rabbit/mouse secondary antibody (1:500;
cat. no. SV0004; Wuhan Boster Biological Technology, Ltd) at 37°C
for 30 min and stained using a diaminobenzidine staining kit (Wuhan
Boster Biological Technology, Ltd.). The nuclei were counterstained
with hematoxylin. The sections were mounted with neutral balsam and
observed under an optical microscope (Olympus Corporation) with
×100 magnification in three fields per sample.
Bioinformatics analysis
For miRNA target gene prediction, the online tool
TargetScan was used (http://www.targetscan.org/vert_72/) (27). To analyze the differentially
expressed miRNAs (|logFC|>0.56; P<0.05) in melanoma cells,
the GEO dataset GSE77090 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE77090)
was downloaded and analyzed using the ‘Limma’ package in R
(28,29).
Cell lines and culture
Normal human melanocytes (MC) were obtained from
ATCC (cat. no. PCS-200-013) and cultured using an Adult Melanocyte
Growth kit (cat. no. PCS-200-042; ATCC). The human melanoma cell
lines A375 and A2058 were obtained from ATCC (cat. nos. CRL-1619
and CRL-11147) and cultured in DMEM (cat. no. 30-2002) supplemented
with 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher
Scientific, Inc.), 100 mg/m penicillin and 100 U/ml streptomycin.
The cells were incubated with 5% CO2 at 37°C.
Cell transfection
For miR-27b overexpression or inhibition, miR-27b
mimics, mimics-negative control (NC), miR-27b inhibitor or
inhibitor-NC were transfected into A375 and A2058 cells
(1×106 cells/ml). MYC knockdown was achieved by
transfection of small interfering (si)RNA specific to MYC (si-MYC).
All transfection vectors (final concentration, 20 nM) were
synthesized by Shanghai GenePharm Co., Ltd. The transfections were
performed using Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C for 6 h. Subsequently, the culture
medium was replaced with fresh culture medium. At 48 h
post-transfection, the cells were harvested for further
experiments. The sequences of the miR-27b mimics, inhibitor, si-MYC
and the corresponding NC vectors are listed in Table I.
 | Table I.Primer and microRNA sequences. |
Table I.
Primer and microRNA sequences.
| A, Primers used for
RT-quantitative PCR |
|---|
|
|---|
| Gene | Sequences
(5′→3′) |
|---|
| miR-27b-5p | RT:
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGTTCAC |
|
| F:
GCCAGAGCTTAGCTGATTG |
|
| R:
CAGTGCGTGTCGTGGA |
| MYC | F:
GGCTCCTGGCAAAAGGTCA |
|
| R:
CTGCGTAGTTGTGCTGATGT |
| GAPDH | F:
ACAGCCTCAAGATCATCAGC |
|
| R:
GGTCATGAGTCCTTCCACGAT |
| U6 | F:
CTCGCTTCGGCAGCACA |
|
| R:
AACGCTTCACGAATTTGCGT |
| MYC (RIP
assay) | F:
GCCTTGGTTCATCTGGGTCTAA |
|
| R:
TGGGGTTGATGTAGAGTTAGGGAT |
|
| B, Vectors used
for transfections |
|
|
Oligomer | Sequences
(5′→3′) |
|
| Mimics NC | S:
UUCUCCGAACGUGUCACGUTT |
|
| AS:
ACGUGACACGUUCGGAGAATT |
| miR-27b mimics | S:
AGAGCUUAGCUGAUUGGUGAAC |
|
| AS:
UCACCAAUCAGCUAAGCUCUUU |
| Inhibitor NC | S:
CAGUACUUUUGUGUAGUACAA |
| miR-27b
inhibitor | S:
GUUCACCAAUCAGCUAAGCUCU |
| Wt-MYC 3′-UTR | S:
aattctaggcgatcgctcgagAGATAATACAAAGCAGCAATCTGGAC |
|
| AS:
attttattgcggccagcggccgcTTCCCTATCAGTGAATCTTGGGC |
| Mut-MYC 3′-UTR | S:
GgcctaCTCTATTTGTGTCCCAAGCACTCCTA |
|
|
CACAAATAGAGtaggcCATTGTTATGACTTGAGTCTGTCCATT |
| Si-NC | S:
UUCUCCGAACGUGUCACGUTT |
|
| AS:
ACGUGACACGUUCGGAGAATT |
| Si-MYC | S:
AGAAUGAUUAAAAUAACCCTT |
|
| AS:
GGGUUAUUUUAAUCAUUCUTT |
5-bromo-2-deoxyuridine (BrdU)
assay
The DNA synthesis ability of the cells was examined
using BrdU assay by determining the BrdU incorporation by
proliferating cells. A375 and A2058 cells were transfected with the
miR-27b mimics, inhibitor or si-MYC for 48 h. Subsequently,
1×104 A375 and A2058 cells were cultured in 24-well
plates for 8 h and incubated with 10 µg/ml BrdU (Sigma-Aldrich;
Merck KGaA) for 24 and 48 h at 37°C with 5% CO2;
subsequently, the BrdU solution was removed. Following fixation by
4% paraformaldehyde and permeabilized by 0.1% Triton for 10 min
each at room temperature, the cells were incubated sequentially
with a BrdU antibody (1:50; cat. no. 5290; Cell Signaling
Technology, Inc.) at 4°C overnight and an HRP-goat anti-mouse
secondary antibody (1:200; cat. no. A0216; Beyotime Institute of
Biotechnology) for 1 h at room temperature. The optical density at
450 nm was measured using a microplate reader (Bio-Rad
Laboratories, Inc.).
MTT assay
The MTT assay was used to determine the cell
viability. A375 and A2058 cells transfected with the miR-27b
mimics, inhibitor or si-MYC were collected and, following 24-h
culture in 96-well plates (5×103 cells/well) at 37°C,
the cells were supplemented with 20 µl MTT (5 mg/ml; Sigma-Aldrich;
Merck KGaA) and incubated for 4 h in a humidified incubator at
37°C. Subsequently, the supernatant was removed, and 200 µl DMSO
was added to dissolve the formazan crystals. The optical density at
490 nm was measured using a microplate reader.
Transwell assay
The invasive ability of A375 and A2058 cells was
determined using a Transwell invasion assay. A375 and A2058 cells
transfected with the miR-27b mimics, inhibitor or si-MYC were
collected, suspended in FBS-free DMEM and seeded into the upper
chambers of the Transwell inserts pre-coated with Matrigel for 4 h
at 37°C. DMEM with 10% FBS was added to the lower chambers.
Following 24-h culture at 37°C, cells adhering to the upper surface
of the membrane were removed with a cotton swab, and the cells that
had penetrated to the lower surface of the membrane were fixed with
4% paraformaldehyde for 20 min, stained with crystal violet for 5
min at room temperature, and counted under an inverted optical
microscope (Olympus Corporation) with ×100 magnification in three
fields per sample.
Immunoblotting
The protein expression levels of c-Myc were
determined by western blotting. A375 and A2058 cells were
transfected with the miR-27b mimics, inhibitor or si-MYC for 48 h.
Protein samples were extracted from the cells using RIPA lysis
buffer (Beyotime Institute of Biotechnology). The protein
concentration was determined by a BCA kit (Beyotime Institute of
Biotechnology). The isolated proteins (20–100 µg) were separated by
10% SDS-PAGE. The proteins were transferred to polyvinylidene
difluoride (PVDF) membranes (Bio-Rad Laboratories, Inc.). Following
blocking with 3% skim milk for 2 h at room temperature in
Tris-buffered saline containing 0.1% Tween-20, the membranes were
incubated with an anti-c-Myc primary antibody (1:1,000; cat. no.
ab32072; Abcam) for 1 h at room temperature. Following washing with
tris-buffered saline with 0.1% Tween 20 for 5 min thrice at room
temperature, the membranes were incubated with HRP-conjugated
secondary IgG antibodies (1:5,000; cat. no. A0208; Beyotime
Institute of Biotechnology) for 1 h at room temperature. The
signals were visualized using enhanced chemiluminescence (ECL) with
a ChemiDOC XRS system (Bio-Rad Laboratories, Inc.). The
densitometry analysis was performed by ImageJ 1.52 software
(National Institutes of Health).
Reverse transcription-quantitative
(RT-q)PCR
A375 and A2058 cells were transfected with the
miR-27b mimics, inhibitor or si-MYC for 48 h. Total RNA was
extracted from the A375 and A2058 cells using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. Complementary DNA was synthesized
from the extracted RNA using the StarScript II First-strand cDNA
Synthesis Mix (cat. no. A223-2; GenStar) according to the
manufacturer's protocol (42°C for 30 min and 85°C for 5 min,
followed by storage at 4°C). The expression levels of miRNA and
mRNA were examined using a SYBR® Green PCR Master Mix
(Qiagen GmbH). An ABI7500 real-time PCR detection system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used for qPCR, and
the thermocycling conditions were as follows: Initial denaturation
at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C
for 15 sec, and annealing and extension at 60°C for 30 sec. GAPDH
(for mRNA) or RNU6B (for miRNA) expression was used as an internal
reference. The relative expression levels were calculated using the
2−ΔΔCq method (30). The
primer sequences are listed in Table
I.
Dual-luciferase reporter assay
The dual-luciferase reporter assay was used to
determine the potential binding between miR-27b and MYC predicted
by TargetScan. The wild-type vector (wt-MYC 3′-UTR) was generated
by amplifying and cloning the MYC 3′-UTR into the downstream region
of the Renilla psiCHECK2 vector (Promega Corporation). The
mutant reporter (mut-MYC 3′-UTR) was generated by mutating the
predicted miR-27b binding site in MYC 3′-UTR. Subsequently,
~5×104 293T cells/ml (cat. no. CRL-3216; ATCC) were
cultured in DMEM supplemented with 10% FBS and were co-transfected
with 1 µg/ml luciferase reporter vectors and 20 nM miR-27b mimics
or inhibitor using Lipofectamine® 3000 for 48 h at 37°C.
Following transfection, the luciferase activity was evaluated by
the Dual-Luciferase Reporter Assay System (Promega Corporation)
using firefly luciferase activity for normalization. The primers
used for plasmid construction are listed in Table I.
RNA immunoprecipitation (RIP)
The RIP assay was used to confirm the predicted
binding between miR-27b and MYC 3′-UTR using a Magna RIP
RNA-Binding Protein Immunoprecipitation kit (cat. no. 17-700;
MilliporeSigma) according to the manufacturer's protocol. Briefly,
A2058 cells were lysed using RIP lysis buffer (MilliporeSigma) and
centrifuged at 10,000 × g for 10 min at 4°C to collect the
supernatant. The magnetic A/G beads (50 µl) were incubated with 5
µg argonaute 2 (Ago2; cat. no. ab186733; Abcam) or IgG (cat. no.
ab109489; Abcam) antibodies. Subsequently, the bead-antibody
complex was rinsed, resuspended in 900 µl RIP Wash Buffer, and
incubated with 100 µl supernatant at 4°C overnight. Following
incubation with protease K, the RNA was extracted from the samples
for RT-qPCR detection for miR-27b and the 3′UTR of MYC.
Statistical analysis
Data from triplicate experiments were analyzed with
GraphPad Prism 6 software (GraphPad Software, Inc.) and presented
as the mean ± SD. Data normality was assessed by the
Kolmogorov-Smirnov test. Paired Student's t-test was used for
statistical comparison between tumor tissues group and adjacent
noncancerous tissues group. Differences among multiple groups were
determined using one-way ANOVA with Tukey's post hoc test. The
correlation between miR-27b and MYC mRNA expression levels was
analyzed by Pearson's correlation analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
c-Myc protein levels and distribution
in melanoma and adjacent noncancerous tissue samples
To determine the mechanism of c-Myc functions in
melanoma, melanoma and paired adjacent noncancerous tissue samples
were collected, and the histomorphological changes in these samples
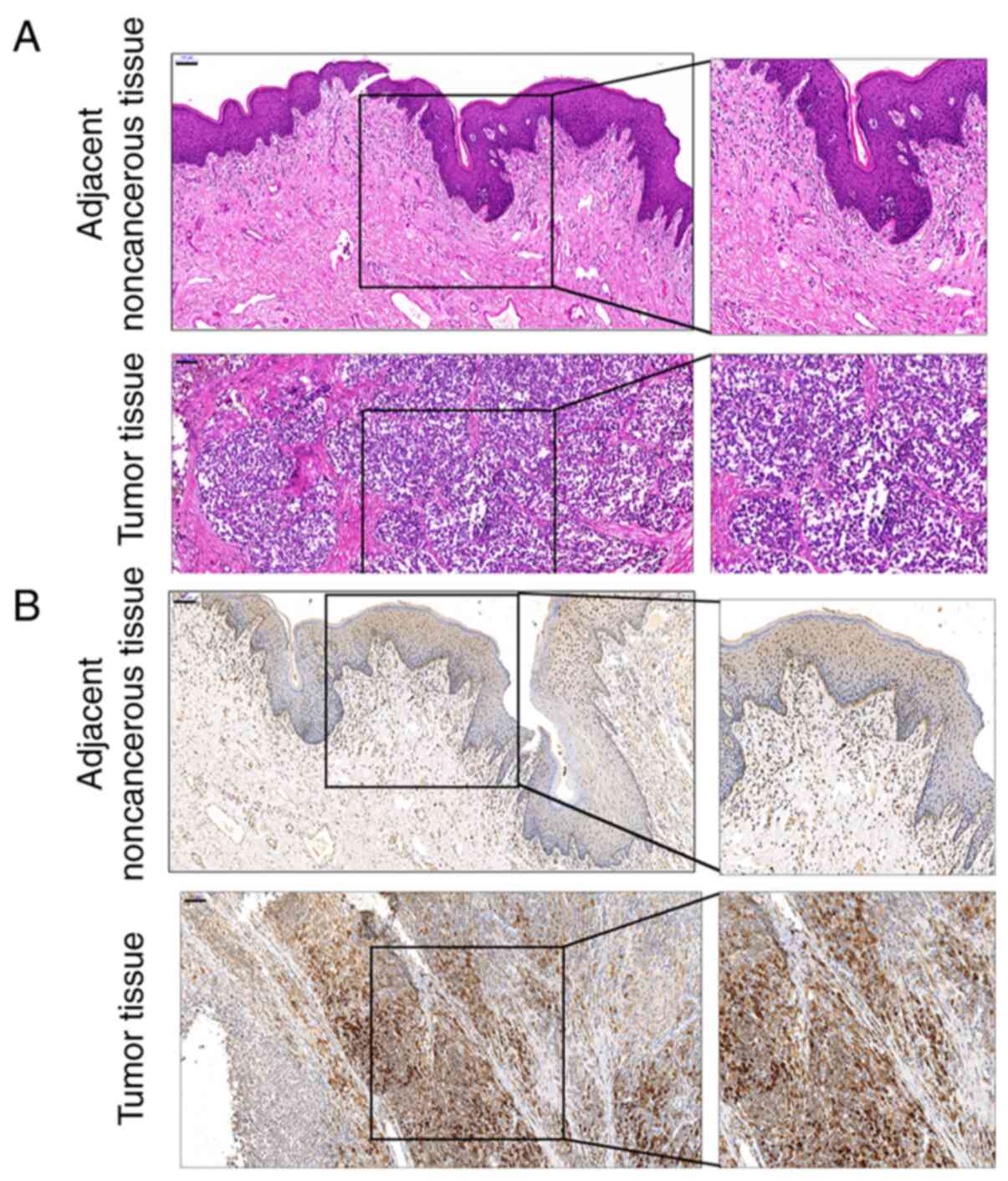
were examined by H&E staining. As demonstrated in Fig. 1A, melanoma cells were heteromorphic
and exhibited no signs of maturity. IHC staining revealed that
melanoma cells were c-Myc-positive (Fig.
1B).
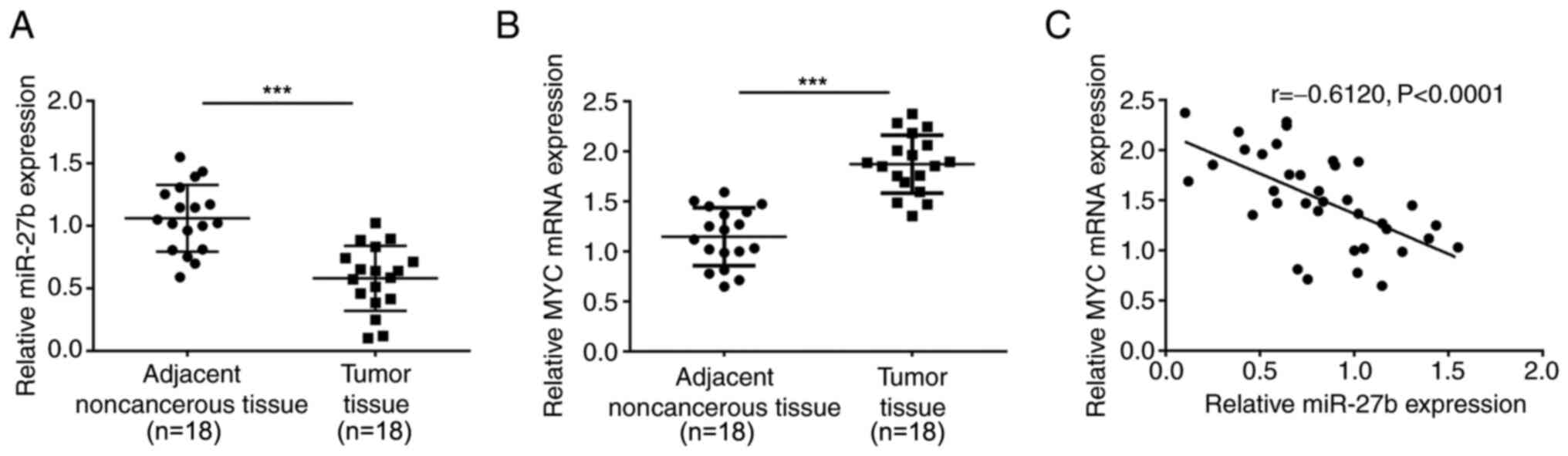
Expression levels and correlation of
miR-27b and MYC within tissues
Since the online tool TargetScan predicted that
miR-27b may target MYC, the present study determined the miR-27b
and MYC expression levels in 18 paired melanoma and noncancerous
normal tissue samples. The expression levels of miR-27b were
markedly reduced, whereas the expression levels of MYC were
increased in the melanoma tissues compared with those in the
adjacent noncancerous tissues (Fig. 2A
and B). In addition, the expression levels of miR-27b were
negatively correlated with those of MYC within the tumor and
adjacent noncancerous tissues (r=−0.6120; Fig. 2C), suggesting that miR-27b may target
MYC to negatively regulate MYC expression levels.
Effects of miR-27b on the phenotype of
melanoma cells
To validate the specific roles of miR-27b in
melanoma cell lines, miR-27b expression levels were determined in a
normal cell line, MC, and two melanoma cell lines, A375 and A2058,
by RT-qPCR; consistent with the results observed in patient tissue
samples, miR-27b expression levels were markedly lower in the two
melanoma cell lines compared with those in the MCs (Fig. 3A). The melanoma cells were
transfected with miR-27b mimics or inhibitor to achieve overexpress
or inhibit the levels of miR-27b, and the transfection efficiency
was verified by RT-qPCR; transfection with the miR-27b mimics
significantly increased the cellular miR-27b levels, whereas the
miR-27b inhibitor decreased the cellular miR-27b levels compared
with those in the corresponding NC groups (Fig. 3B). The phenotypes of the A375 and
A2058 cells transfected with the miR-27b mimics or inhibitor were
subsequently examined. The BrdU assay results demonstrated that
transfection with the miR-27b mimics significantly inhibited the
DNA synthesis ability in melanoma cells compared with that in cells
in the mimics-NC group (Fig. 3C).
The results of the MTT and Transwell assay revealed that the A375
and A2058 cell viability and invasive ability were inhibited by the
miR-27b mimics compared with those in the mimics-NC-transfected
cells (Fig. 3D-G); by contrast,
miR-27b inhibition promoted the DNA synthesis ability, viability
and invasive ability compared with those in the cells in the
inhibitor-NC group (Fig. 3D-G).
Therefore, miR-27b overexpression suppressed the biological
malignant behaviors of A375 and A2058 melanoma cells.
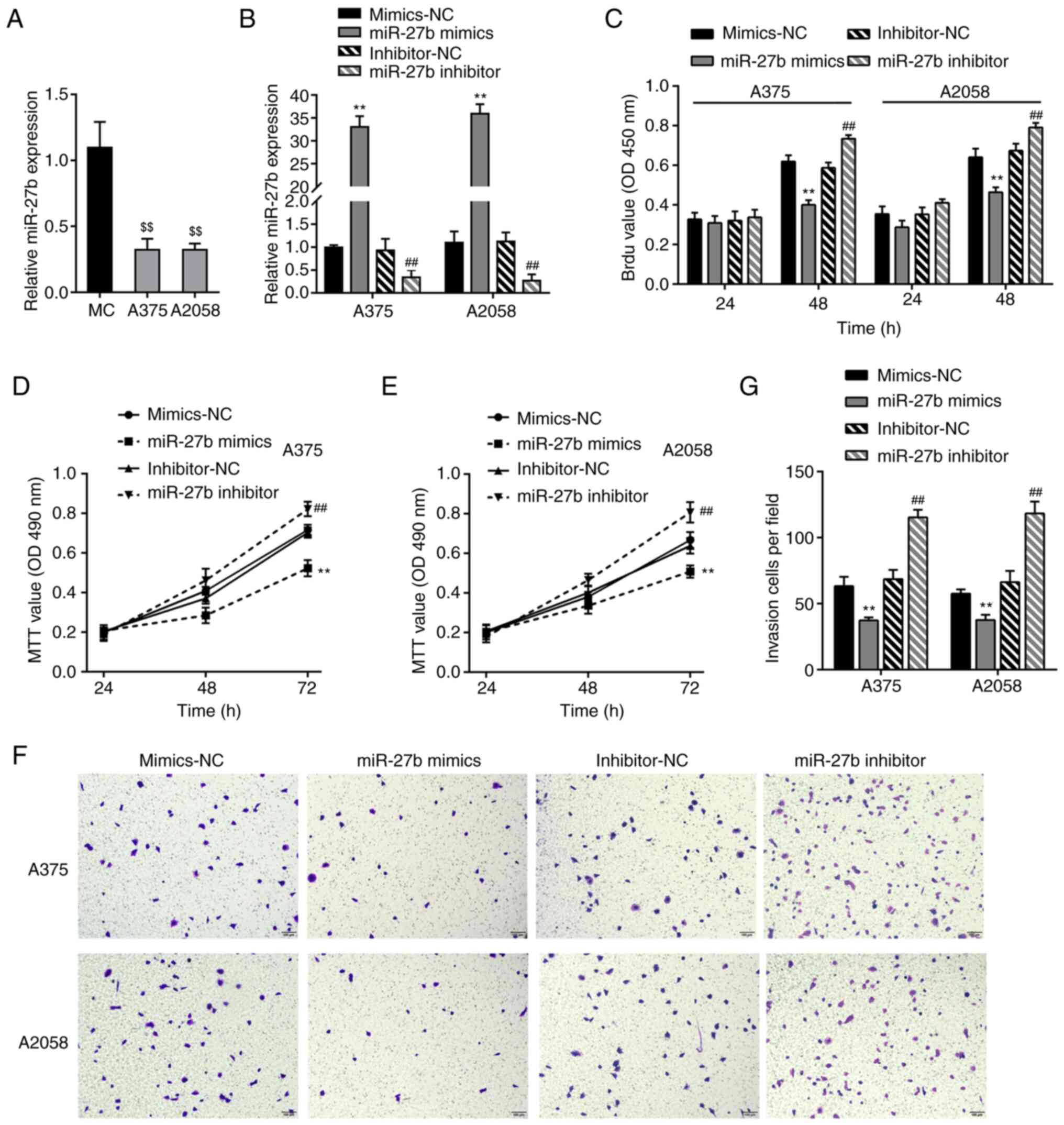 | Figure 3.Effects of miR-27b on melanoma cell
phenotype. (A) miR-27b expression was examined in a normal cell
line, MC, and two melanoma cell lines, A375 and A2058, using
RT-qPCR. miR-27b expression levels were lower in melanoma cell
lines compared with those in MC. (B) miR-27b overexpression or
miR-27b inhibition was achieved in two melanoma cell lines by the
transfection of the miR-27b mimics or inhibitor; mimics-NC and
inhibitor-NC were used as negative controls. The transfection
efficiency was validated by RT-qPCR. (C) DNA synthesis ability of
A375 and A2058 cells transfected with the miR-27b mimics or
inhibitor by BrdU assay; the miR-27b mimics decreased, whereas the
miR-27b inhibitor increased the DNA synthesis ability compared with
that in the corresponding NC groups. (D and E) Cell viability was
determined by MTT assay at 24, 48 and 72 h; the miR-27b mimics
decreased, whereas the miR-27b inhibitor increased cell viability
compared with that in the corresponding NC groups. (F and G) Cell
invasive ability was determined by Transwell assay; the miR-27b
mimics decreased, whereas the miR-27b inhibitor increased the
invasive ability of A375 and A2058 cells compared with that in the
corresponding NC groups. $$P<0.01 vs. MC, **P<0.01
vs. mimics-NC; ##P<0.01 vs. inhibitor-NC. miR,
microRNA; MC, melanocytes; NC, negative control; BrdU,
5-bromo-2-deoxyuridine; RT-qPCR, reverse transcription-quantitative
PCR. |
miR-27b targets MYC by binding to MYC
3′-UTR
In order to validate the binding between miR-27b and
MYC predicted by the online tool TargetScan, the A375 and A2058
cell lines were transfected with the miR-27b mimics or inhibitor,
and the c-Myc protein content was detected by immunoblotting.
Overexpression of miR-27b downregulated the protein levels of
c-Myc, whereas the inhibition of miR-27b upregulated the levels of
c-Myc compared with those in the corresponding NC groups (Fig. 4A and B).
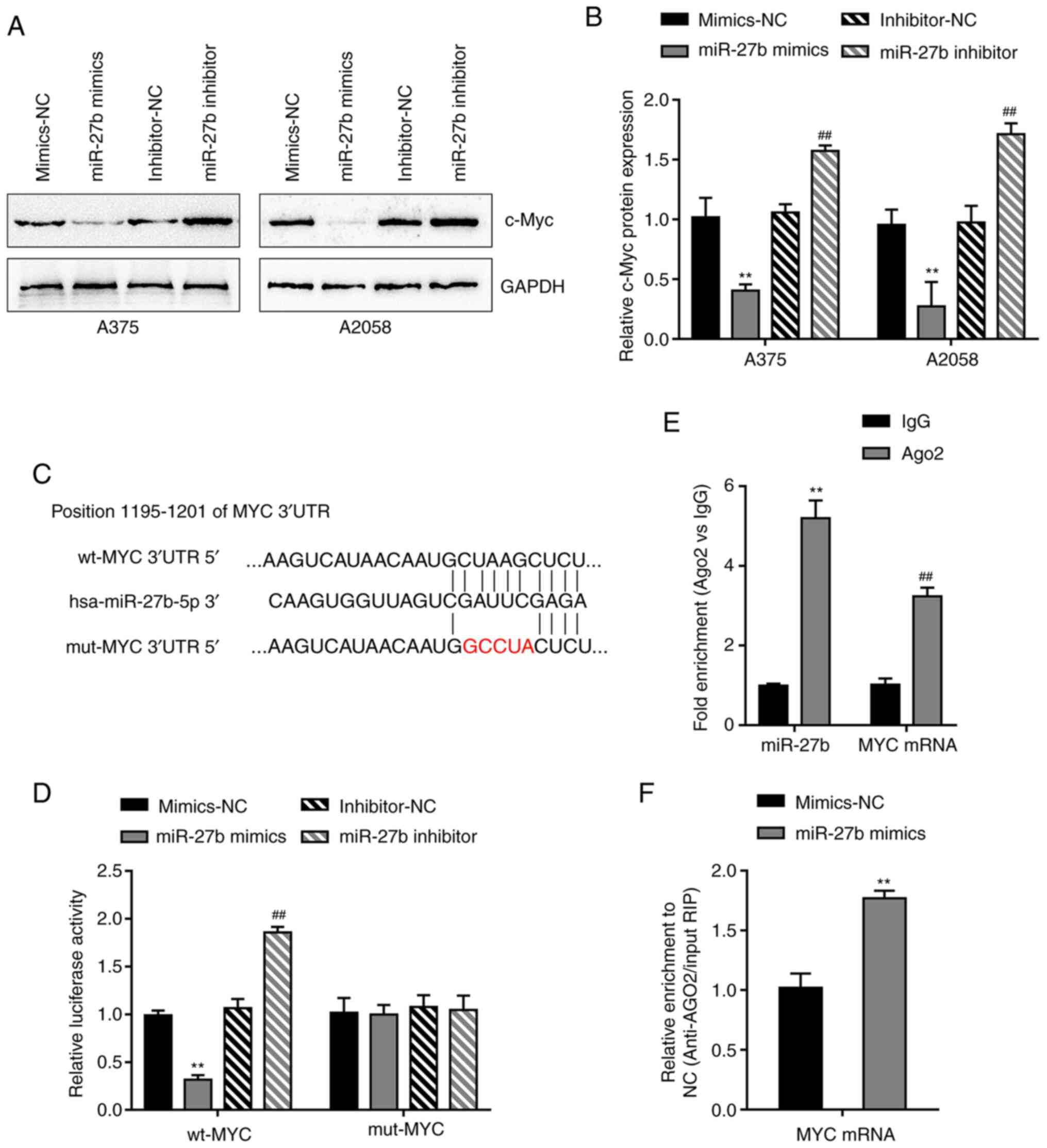 | Figure 4.miR-27b targets MYC by binding to the
MYC 3′-UTR. (A and B) A375 and A2058 cells were transfected with
the miR-27b mimics or inhibitor, and immunoblotting revealed that
the miR-27b mimics decreased, whereas the miR-27b inhibitor
increased the c-Myc protein levels compared with those in the
corresponding NC groups. (C) Wt and mut MYC 3′-UTR luciferase
reporter vectors were constructed; the mut-MYC 3′-UTR vector
contained a 5-bp mutation in the predicted miR-27b binding site.
(D) Dual-luciferase reporter assay in 293T cells demonstrated that
the miR-27b mimics reduced, whereas the miR-27b inhibitor increased
the luciferase activity in the wt-MYC group; no changes were
observed in the mut-MYC group. (E and F) RIP assay was performed to
confirm the binding of miR-27b to the MYC 3′-UTR using the Ago2
antibody; the levels of miR-27b and MYC 3′-UTR in the precipitates
were examined using reverse transcription-quantitative PCR. The
miR-27b and MYC 3′-UTR fragments were enriched in the Ago2
precipitate compared with the IgG group. In the miR-27b
mimics-transfected cells, the MYC 3′-UTR fragments were more highly
enriched compared with those in the mimics-NC transfected cells.
**P<0.01 vs. mimics-NC; ##P<0.01 vs. inhibitor-NC.
miR, microRNA; UTR, untranslated region; NC, negative control; wt,
wild-type; mut, mutant; RIP, radioimmunoprecipitation; Ago2,
argonaute 2. |
The dual-luciferase reporter assay was subsequently
used. The predicted binding site is presented in Fig. 4C. The reporter vectors were
co-transfected into 293T cells with the miR-27b mimics or
inhibitor. The luciferase activity of wt-MYC 3′-UTR was decreased
by the overexpression of miR-27b, but increased following
inhibition of miR-27b compared with that in the corresponding NC
groups (Fig. 4D). In the presence of
mut-MYC 3′-UTR, transfection with the miR-27b mimics or inhibitor
did not affect luciferase activity, suggesting that miR-27b bound
to the predicted binding site in the MYC 3′-UTR
To confirm the binding of miR-27b to MYC, RIP assay
was performed using an Ago2 antibody. Anti-AGO2 immunoprecipitants
containing miRNAs and their interacting RNA components were
examined for the levels of miR-27b and MYC in A2058 cells; as
demonstrated in Fig. 4E, compared
with the anti-IgG group, miR-27b and MYC were more abundant in the
Ago2 group. In addition, the miR-27b mimics or mimics-NC were
transfected into A2058 cells. Higher levels of MYC mRNA were
detected in the anti-Ago2 immunoprecipitants of miR-27b
mimics-transfected cells compared with those in the
mimics-NC-transfected cells (Fig.
4F). These results confirmed that miR-27b bound to the MYC
3′-UTR.
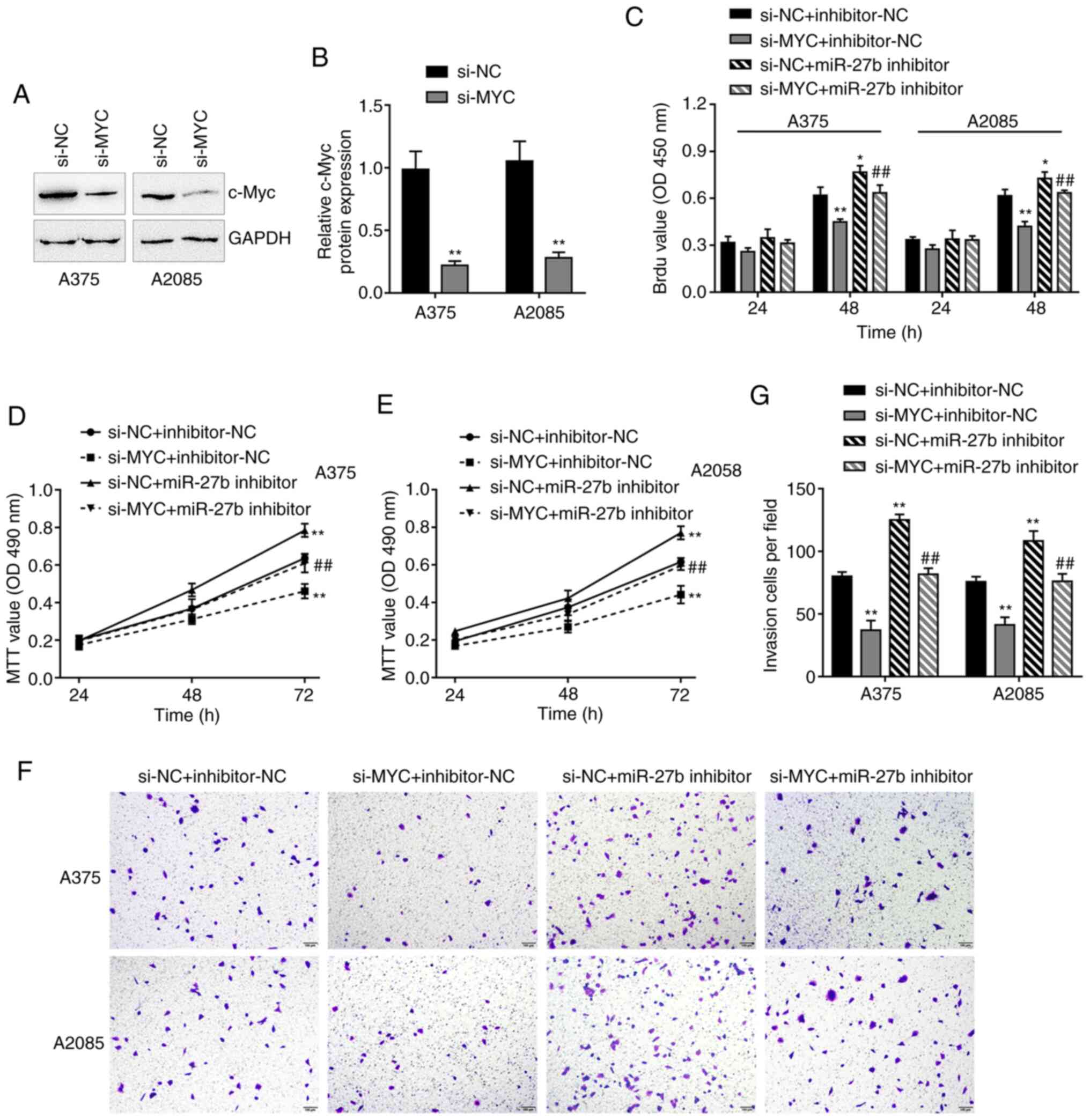
miR-27b modulates the melanoma cell
phenotype through MYC
The present study further validated the dynamic
effects of the miR-27b/MYC axis on the melanoma cell phenotype.
A375 and A2058 cells were transfected with si-MYC to achieve MYC
knockdown; the transfection efficiency was confirmed using
immunoblotting, and the results demonstrated that si-MYC
successfully reduced the c-Myc protein levels compared with those
in the cells transfected with si-NC (Fig. 5A and B). Subsequently, A375 and A2058
cells we co-transfected with the miR-27b inhibitor and si-MYC. The
results of the in vitro assays demonstrated miR-27b
inhibition promoted, whereas MYC knockdown inhibited the DNA
synthesis ability (Fig. 5C), cell
viability (Fig. 5D and E) and
invasive ability (Fig. 5F and G)
compared with those in the si-NC and inhibitor-NC group;
additionally, MYC knockdown significantly attenuated the effects of
miR-27b inhibition (Fig. 5C-G).
Thus, miR-27b may exert its effects on melanoma cells through
MYC.
Discussion
The results of the present study demonstrated that
the expression levels of miR-27b were markedly decreased in
melanoma tissue samples and cells compared with those in adjacent
normal tissues and normal cells, respectively. By contrast, MYC
expression levels were upregulated in melanoma tissues and cells
compared with those in the corresponding control groups. miR-27b
overexpression significantly inhibited melanoma cell DNA synthesis
ability, viability and invasive ability compared with those in
cells transfected with the mimic-NC. Through binding to MYC 3′-UTR,
miR-27b inhibited MYC expression. MYC knockdown in melanoma cells
exerted similar effects as miR-27b overexpression on the DNA
synthesis ability, cell viability and invasive ability; the effects
of miR-27b inhibition were significantly reversed by MYC
knockdown.
miRNAs regulate a number of biological and
pathological processes, such as the biological malignant behaviors
of cancer cells (23). For example,
miR-21 inhibits malignant biological behaviors of melanoma
(31). The abnormal regulation and
dysfunction of miRNAs have been reported in patients with melanoma
as well as in melanoma cells (32).
In addition, miR-137, miR-148 and miR-182 have been demonstrated to
regulate the levels of microphthalmia-associated transcription
factor in melanoma (33). miR-26a
induces apoptosis in melanoma cells by modulating SMAD1 and
silencer of death domains (34).
miRNAs have also been implicated in the development of melanoma
drug resistance and organ-specific metastasis (35). For example, a miR-150-5p, miR-15b-5p,
miR-16-5p and miR-374b-3p prognostic signature distinguishes brain
metastatic melanomas and non-brain metastatic primary melanomas
(36). miR-200c inhibits melanoma
progression and drug resistance through downregulation of BMI-1
(37). According to the online
expression profiles GSE77090, a total of 58 miRNAs are
downregulated in melanoma (data not shown); among them, miR-27b may
target MYC, which is a proto-oncogene (17). As previously reported, miR-27b
suppresses the capacity of a number of cancer cell types to
proliferate, invade and migrate. For example, miR-27b inhibits the
capacity of aggressive prostate cancer cells to migrate and invade,
but does not affect their proliferative ability (38). By targeting peroxisome
proliferator-activated receptor γ, miR-27b suppresses neuroblastoma
cell proliferation, tumor development and inflammatory response
(39). In colorectal cancer, miR-27b
binds to vascular endothelial growth factor C to suppress tumor
development and the formation of new blood vessels (40). In the present study, consistent with
its abnormal downregulation in melanoma, miR-27b inhibition
significantly promoted melanoma cell DNA synthesis ability,
viability and invasive ability; by contrast, miR-27b overexpression
markedly suppressed these biological malignant behaviors of
melanoma cells, suggesting that miR-27b may exert a
tumor-suppressive effect on melanoma.
Regarding the mechanisms by which miRNAs exert their
biological functions, miRNAs are considered to target various
downstream mRNAs, leading to translation inhibition, mRNA
deadenylation or decay (21). Based
on the TargetScan analysis performed in the preliminary experiments
of the present study, it was determined that miR-27b may target the
3′-UTR of MYC, which was confirmed by experimental analyses. The
c-Myc proto-oncogene is located on chromosome 8, and a variety of
anomalies related to its activated expression occur during the
development of numerous types of malignant tumors (41). c-Myc is considered to modulate the
expression of ≤15% of all human genes (16). A number of genes modulated by c-Myc
are involved in cell proliferation, migration, invasion and
apoptosis, thus promoting the occurrence and development of tumors
(42–45). A previous study supports the
suggestion that oncogenic c-Myc affects the presence of
determinants of immunological importance on melanoma cells
(46). c-Myc overexpression promotes
melanoma metastasis by promoting vasculogenic mimicry (18). By contrast, MYC depletion results in
the repression of the expression of a number of genes encoding
enzymes that are rate-limiting for dNTP metabolism, including
phosphoribosyl pyrophosphate synthetase 2 and inosine monophosphate
dehydrogenase 2, resulting in the inhibition of melanoma
proliferation (47). In the present
study, MYC mRNA expression levels were demonstrated to be
upregulated in melanoma tissue samples compared with those in
paired adjacent noncancerous tissues. MYC knockdown in melanoma
cells significantly inhibited the DNA synthesis ability, viability
and invasive ability compared with those in the cells transfected
with si-NC. In addition, miR-27b served as a tumor suppressor in
melanoma cells, as miR-27b overexpression inhibited melanoma cell
proliferation and invasive capability compared with those in the NC
group. By contrast, miR-27b inhibition promoted cell proliferation
and invasion compared with those in the inhibitor-NC-transfected
cells. Additionally, MYC expression levels were reduced by miR-27b
overexpression and increased by miR-27b inhibition compared with
those in the corresponding NC groups. MYC knockdown significantly
reversed the effects of miR-27b inhibition, indicating that miR-27b
may serve as a tumor suppressor in melanoma by targeting MYC. Due
to the crucial role of c-Myc in melanoma progression, targeting
c-Myc is considered a potential therapeutic strategy for melanoma.
In a previous study, nanodelivery of the c-Myc inhibitor 10058-F4
effectively inhibited human and mouse melanoma cell proliferation
(10). Nanoparticle-delivered
si-c-Myc sensitizes melanoma cells to paclitaxel and inhibits tumor
growth (48).
In conclusion, the results of the present study
demonstrated that the miR-27b/MYC axis modulated melanoma cell
biological malignant behaviors, suggesting that it may be a
potential target for melanoma treatment.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81774325) and the project of
the Hunan Province Health Committee (grant no. 20201683).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YT and JZ performed the experiments and wrote the
manuscript. ZY supervised the research. JZ revised the manuscript.
ZY designed the study. YT, ZY and JZ confirm the authenticity of
all the raw data. All authors read and approve the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the Second Affiliated Hospital of Hunan University of Chinese
Medicine (approval No. 2019-KY-031; Changsha, China). All patients
provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dimitriou F, Krattinger R, Ramelyte E,
Barysch MJ, Micaletto S, Dummer R and Goldinger SM: The world of
melanoma: Epidemiologic, genetic, and anatomic differences of
melanoma across the globe. Curr Oncol Rep. 20:872018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carr S, Smith C and Wernberg J:
Epidemiology and risk factors of melanoma. Surg Clin North Am.
100:1–12. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schadendorf D, van Akkooi ACJ, Berking C,
Griewank KG, Gutzmer R, Hauschild A, Stang A, Roesch A and Ugurel
S: Melanoma. Lancet. 392:971–984. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lopez-Soto A, Gonzalez S, Smyth MJ and
Galluzzi L: Control of metastasis by NK cells. Cancer Cell.
32:135–154. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhandaru M and Rotte A: Monoclonal
antibodies for the treatment of melanoma: Present and future
strategies. Methods Mol Biol. 1904:83–108. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
McDermott D, Haanen J, Chen TT, Lorigan P
and O'Day S; MDX010-20 investigators, : Efficacy and safety of
ipilimumab in metastatic melanoma patients surviving more than 2
years following treatment in a phase III trial (MDX010-20). Ann
Oncol. 24:2694–2698. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Weiss SA, Wolchok JD and Sznol M:
Immunotherapy of melanoma: Facts and hopes. Clin Cancer Res.
25:5191–5201. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Paluncic J, Kovacevic Z, Jansson PJ,
Kalinowski D, Merlot AM, Huang MLH, Lok HC, Sahni S, Lane DJR and
Richardson DR: Roads to melanoma: Key pathways and emerging players
in melanoma progression and oncogenic signaling. Biochim Biophys
Acta. 1863:770–784. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ankeny JS, Labadie B, Luke J, Hsueh E,
Messina J and Zager JS: Review of diagnostic, prognostic, and
predictive biomarkers in melanoma. Clin Exp Metastasis. 35:487–493.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Karagiannis P, Fittall M and Karagiannis
SN: Evaluating biomarkers in melanoma. Front Oncol.
4:3832015.PubMed/NCBI
|
|
11
|
Sun J, Zhang D, Bae DH, Sahni S, Jansson
P, Zheng Y, Zhao Q, Yue F, Zheng M, Kovacevic Z and Richardson DR:
Metastasis suppressor, NDRG1, mediates its activity through
signaling pathways and molecular motors. Carcinogenesis.
34:1943–1954. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Smalley MJ and Dale TC: Wnt signalling in
mammalian development and cancer. Cancer Metastasis Rev.
18:215–230. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zheng L and Pan J: The anti-malarial drug
artesunate blocks wnt/beta-catenin pathway and inhibits growth,
migration and invasion of uveal melanoma cells. Curr Cancer Drug
Targets. 18:988–998. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sinnberg T, Levesque MP, Krochmann J,
Cheng PF, Ikenberg K, Meraz-Torres F, Niessner H, Garbe C and Busch
C: Wnt-signaling enhances neural crest migration of melanoma cells
and induces an invasive phenotype. Mol Cancer. 17:592018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Clevers H: Wnt/beta-catenin signaling in
development and disease. Cell. 127:469–480. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gearhart J, Pashos EE and Prasad MK:
Pluripotency redux-advances in stem-cell research. N Engl J Med.
357:1469–1472. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Meškytė EM, Keskas S and Ciribilli Y: MYC
as a multifaceted regulator of tumor microenvironment leading to
metastasis. Int J Mol Sci. 21:77102020. View Article : Google Scholar
|
|
18
|
Lin X, Sun R, Zhao X, Zhu D, Zhao X, Gu Q,
Dong X, Zhang D, Zhang Y, Li Y and Sun B: C-myc overexpression
drives melanoma metastasis by promoting vasculogenic mimicry via
c-myc/snail/Bax signaling. J Mol Med (Berl). 95:53–67. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nishikawa S, Ishii H, Haraguchi N, Kano Y,
Fukusumi T, Ohta K, Ozaki M, Dewi DL, Sakai D, Satoh T, et al:
microRNA-based cancer cell reprogramming technology. Exp Ther Med.
4:8–14. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Noori J, Sharifi M and Haghjooy Javanmard
S: miR-30a inhibits melanoma tumor metastasis by targeting the
E-cadherin and zinc finger E-box binding homeobox 2. Adv Biomed
Res. 7:1432018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pencheva N, Tran H, Buss C, Huh D,
Drobnjak M, Busam K and Tavazoie SF: Convergent multi-miRNA
targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis
and angiogenesis. Cell. 151:1068–1082. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gajos-Michniewicz A and Czyz M: Role of
miRNAs in melanoma metastasis. Cancers (Basel). 11:3262019.
View Article : Google Scholar
|
|
25
|
Sole C, Tramonti D, Schramm M, Goicoechea
I, Armesto M, Hernandez LI, Manterola L, Fernandez-Mercado M,
Mujika K, Tuneu A, et al: The circulating transcriptome as a source
of biomarkers for melanoma. Cancers (Basel). 11:702019. View Article : Google Scholar
|
|
26
|
Wang X, Zou M, Li J, Wang B, Zhang Q, Liu
F and Lü G: LncRNA H19 targets miR-22 to modulate H2O2-induced
deregulation in nucleus pulposus cell senescence, proliferation,
and ECM synthesis through Wnt signaling. J Cell Biochem.
119:4990–5002. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
ELife. 4:e050052015. View Article : Google Scholar
|
|
28
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Team RC: R: A language and environment for
statistical computing. R Foundation for Statistical Computing;
Vienna: 2013
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Javanmard SH, Vaseghi G, Ghasemi A, Rafiee
L, Ferns GA, Esfahani HN and Nedaeinia R: Therapeutic inhibition of
microRNA-21 (miR-21) using locked-nucleic acid (LNA)-anti-miR and
its effects on the biological behaviors of melanoma cancer cells in
preclinical studies. Cancer Cell Int. 20:3842020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lorusso C, De Summa S, Pinto R, Danza K
and Tommasi S: miRNAs as key players in the management of cutaneous
melanoma. Cells. 9:4152020. View Article : Google Scholar
|
|
33
|
Kunz M: MicroRNAs in melanoma biology. Adv
Exp Med Biol. 774:103–120. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qian H, Yang C and Yang Y: MicroRNA-26a
inhibits the growth and invasiveness of malignant melanoma and
directly targets on MITF gene. Cell Death Discov. 3:170282017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ryu B, Hwang S and Alani RM: MicroRNAs as
an emerging target for melanoma therapy. J Invest Dermatol.
133:1137–1139. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hanniford D, Zhong J, Koetz L,
Gaziel-Sovran A, Lackaye DJ, Shang S, Pavlick A, Shapiro R, Berman
R, Darvishian F, et al: A miRNA-based signature detected in primary
melanoma tissue predicts development of brain metastasis. Clin
Cancer Res. 21:4903–4912. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu S, Tetzlaff MT, Cui R and Xu X:
miR-200c inhibits melanoma progression and drug resistance through
down-regulation of BMI-1. Am J Pathol. 181:1823–1835. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ishteiwy RA, Ward TM, Dykxhoorn DM and
Burnstein KL: The microRNA-23b/-27b cluster suppresses the
metastatic phenotype of castration-resistant prostate cancer cells.
PLoS One. 7:e521062012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lee JJ, Drakaki A, Iliopoulos D and Struhl
K: MiR-27b targets PPARg to inhibit growth, tumor progression and
the inflammatory response in neuroblastoma cells. Oncogene.
31:3818–3825. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ye J, Wu X, Wu D, Wu P, Ni C, Zhang Z,
Chen Z, Qiu F, Xu J and Huang J: miRNA-27b targets vascular
endothelial growth factor C to inhibit tumor progression and
angiogenesis in colorectal cancer. PLoS One. 8:e606872013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Stine ZE, Walton ZE, Altman BJ, Hsieh AL
and Dang CV: MYC, metabolism, and cancer. Cancer Discov.
5:1024–1039. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Campbell KJ and White RJ: MYC regulation
of cell growth through control of transcription by RNA polymerases
I and III. Cold Spring Harb Perspect Med. 4:a0184082014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang Q, Spears E, Boone DN, Li Z, Gregory
MA and Hann SR: Domain-specific c-Myc ubiquitylation controls c-Myc
transcriptional and apoptotic activity. Proc Natl Acad Sci USA.
110:978–983. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kong LM, Liao CG, Zhang Y, Xu J, Li Y,
Huang W, Zhang Y, Bian H and Chen ZN: A regulatory loop involving
miR-22, Sp1, and c-Myc modulates CD147 expression in breast cancer
invasion and metastasis. Cancer Res. 74:3764–3778. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang L, Hou Y, Ashktorab H, Gao L, Xu Y,
Wu K, Zhai J and Zhang L: The impact of C-MYC gene expression on
gastric cancer cell. Mol Cell Biochem. 344:125–135. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Poźniak J, Nsengimana J, Laye JP, O'Shea
SJ, Diaz JMS, Droop AP, Filia A, Harland M, Davies JR, Mell T, et
al: Genetic and environmental determinants of immune response to
cutaneous melanoma. Cancer Res. 79:2684–2696. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Mannava S, Grachtchouk V, Wheeler LJ, Im
M, Zhuang D, Slavina EG, Mathews CK, Shewach DS and Nikiforov MA:
Direct role of nucleotide metabolism in C-MYC-dependent
proliferation of melanoma cells. Cell Cycle. 7:2392–2400. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen Y, Bathula SR, Yang Q and Huang L:
Targeted nanoparticles deliver siRNA to melanoma. J Invest
Dermatol. 130:2790–2798. 2010. View Article : Google Scholar : PubMed/NCBI
|