Introduction
Multiple myeloma (MM), also known as plasma cell
myeloma, is a fatal hematological malignancy characterized by the
malignant proliferation of MM cells and an aberrant increase in
monoclonal immunoglobulin (1). In
patients with MM, this immunoglobulin excess results in platelet
dysfunction, hyperviscosity, bone loss and renal tubular damage,
which subsequently leads to bleeding, neurological impairment,
ostealgia, renal failure and other life-threatening symptoms
(2). According to a recent
epidemiological study, there were 159,985 new diagnoses and 106,105
MM-associated deaths worldwide in 2018, accounting for 0.9% of new
cancer cases and 1.1% of all cancer-associated deaths (3). Currently, the standard treatment
strategy for MM is high-dose chemotherapy (such as carfilzomib,
bortezomib and melphalan) followed by stem cell transplantation (if
accessible), which effectively prolongs patient survival time
(4). However, due to its complexity,
MM remains a challenge to treat, and its precise pathogenic
mechanisms have not been fully clarified (5,6).
Therefore, it is important to determine the underlying etiology of
MM and to identify potential new treatment targets.
lncRNA transcription factor 7 (lnc-TCF7) is a novel
lncRNA that was first identified in liver cancer cells, and
promotes the self-renewal of human liver cancer stem cells through
the activation of Wnt signaling (7).
Furthermore, lnc-TCF7 has been discovered to promote the
carcinogenesis of various malignancies, including colorectal cancer
(CRC) and non-small cell lung cancer (NSCLC) (8–10).
Moreover, the TCF7 gene promotes cellular functions, such as
self-renewal and differentiation, in hematological malignancies
(11). Furthemore, our previous
preliminary research indicated that in a small sample population,
lnc-TCF7 was upregulated in patients with MM compared with healthy
donors, and was also upregulated in MM cell lines compared with
plasma cells from healthy human primary bone marrow mononuclear
cells (BMMCs) (unpublished data). Thus it was hypothesized that
lnc-TCF7 may also facilitate MM development and progression.
However, the clinical implications of lnc-TCF7 in MM diagnosis and
prognosis, as well as its role in MM progression, remain to be
elucidated.
Therefore, the aim of the present study was to
evaluate the effects of lnc-TCF7 on the risk of developing MM, and
its influence on clinicopathological features and patient
prognosis. The study also sought to further explore the effects of
lnc-TCF7-knockdown on the regulation of cell proliferation and
apoptosis, as well as its molecular mechanism in MM.
Materials and methods
Participants
A total of 86 patients with de novo
symptomatic MM were consecutively recruited from The Affiliated
Hospital of Nantong University (Nantong, China) between July 2014
and December 2017. The inclusion criteria were as follows: i) A
diagnosis of de novo symptomatic MM based on the
International Myeloma Working Group recommendations for global
myeloma care (2013) (12), ii) age
>18 years old and iii) no history of stem cell transplantation,
chemotherapy, radiotherapy or other systematic treatments. Patients
were excluded from the study if they presented with other
malignancies, had serious infection (such as human immunodeficiency
virus), or were pregnant or lactating. In addition, 30 healthy
individuals who donated bone marrow at the hospital were enrolled
as healthy controls. The median age of the healthy controls was 55
years with a range of 31–74 years. Meanwhile, there were 19 (63.3%)
males as well as 11 (36.7%) females. The present study was approved
by the Institutional Review Board of The Affiliated Hospital of
Nantong University, and all participants or their guardians
provided written informed consent upon enrollment.
Baseline data collection
Baseline data were collected after consent for
participation was gained, which included age, sex, immunoglobulin
subtype, bone lesion, values for hemoglobin, calcium, serum
creatinine, albumin, β-2-microglobulin (β2-MG), lactate
dehydrogenase, Durie-Salmon stage (13), International Staging System (ISS)
stage (14) and cytogenetic
abnormalities. Cytogenetics abnormalities were previously
determined at the hospital using fluorescence in situ
hybridization, and the Durie-Salmon stage was evaluated according
to the Durie-Salmon Criteria (classified as stage I–III).
Samples collection, processing and
lnc-TCF7 detection
Bone marrow samples were extracted from patients
with MM before treatment initiation, and from healthy donors after
informed consent was obtained. Density gradient centrifugation (400
× g for 30 min at room temperature) was then immediately performed
to separate the mononuclear cells from the samples, and the plasma
cells were isolated using CD138-coated magnetic beads (Miltenyi
Biotec GmbH) according to the manufacturer's instructions. The
levels of plasma cell lnc-TCF7 were detected using reverse
transcription-quantitative (RT-q)PCR.
Follow up
The classification of MM was based on their
immunoglobulin subtype, which was in accordance with the National
Comprehensive Cancer Network Clinical Practice Guidelines in
Oncology: Multiple Myeloma (15).
All patients received the appropriate treatment according to these
guidelines, based on their clinical status and willingness to
receive treatment. Treatment included bortezomib/dexamethasone
(BD), bortezomib/doxorubicin/dexamethasone (BDD) or
melphalan/prednisone/bortezomib (MPB). Surveillance and follow up
were performed every 3–6 months, or as indicated, which was
performed in an out-patient clinic. The last follow-up date was
June 2018, and the median follow-up duration was 29.0 months
(range, 3.0 to 43.0 months). Complete response (CR), very good
partial response (VGPR) and partial response (PR) were assessed
based on the Revised Uniform Response Criteria by the International
Myeloma Working Group (16). Overall
response rate (ORR) was calculated as ORR=CR+VGPR+PR, and
event-free survival time (EFS) was calculated from the date of
treatment initiation to the date of disease relapse, progression or
death. Overall survival time (OS) was calculated from the date of
treatment initiation to the date of death.
Detection of lnc-TCF7 expression in MM
cell lines
Human MM cell lines [OPM2, Roswell Park Memorial
Institute (RPMI)8226, NCI-H929 and U266] were purchased from the
Leibniz Institute DSMZ-German Collection of Microorganisms and Cell
Cultures or The American Type Culture Collection, and cultured in
90% RPMI-1640 medium supplemented with 10% fetal bovine serum (both
Gibco; Thermo Fisher Scientific, Inc.). Subsequently, RT-qPCR was
used to detect lnc-TCF7 expression in these cell lines, and in
plasma cells isolated from BMMCs of health donors (as the
control).
Plasmid transfection
Empty vector negative control (NC) and lnc-TCF7
short harpin (sh)RNA (Sh-TCF7) vectors were constructed by the
Shanghai QeeJen Bio-tech Company. The vectors (1 µg) were
transfected into RPMI8226 cells with Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) for 24 h at 37°C.
After 48 h of transfection, the subsequent experiments were
performed. Transfection efficiency was confirmed 24 h later using
RT-qPCR.
Evaluation of cellular proliferation,
apoptosis and potential target miRNAs following
lnc-TCF7-knockdown
After plasmid transfection, cellular proliferation
was detected at 0, 24, 48 and 72 h using a Cell Counting Kit-8
assay (Dojindo Molecular Technologies, Inc.) according to the
manufacturer's instructions. Apoptosis rate was detected at 48 h
using the Annexin V-FITC Apoptosis Detection kit (BD Biosciences)
and a BD FACSCanto II flow cytometry (BD Biosciences) per the
manufacturer's protocol. The analysis of apoptosis rate was
performed by the FlowJo software (version 10; BD Biosciences).
Furthermore, potential targets of lnc-TCF7 were predicted using
miRanda database analysis (www.miranda.org), and microRNA (miR)-203, miR-26b and
miR-643 were selected for RT-qPCR validation and detection 24 h
post-transfection.
Compensation experiments
Compensation experiments were performed to determine
whether lnc-TCF7 shRNA influenced MM cells by targeting miR-203.
RPMI8226 cells were transfected with miR-203 inhibitor negative
control or miR-203 inhibitor, and the transfection efficacy was
evaluated. Subsequently, RPMI8226 cells were transfected with empty
vector plus miR-203 inhibitor NC (NC group), lnc-TCF7 shRNA alone
(Sh-TCF7 group) or lnc-TCF7 shRNA combined with an miR-203
inhibitor (Sh-TCF7/miR inhibitor group). The expression levels of
lnc-TCF7 and miR-203 were detected by RT-qPCR 24 h later. Cellular
proliferation and apoptosis analysis were conducted as
aforementioned.
RT-qPCR
Total RNA was extracted from RPMI8226 cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. The extracted
RNA was then reverse transcribed into cDNA using the PrimeScript RT
reagent kit (Takara Biotechnology, Co., Ltd.) at 42°C for 2 min,
and qPCR was subsequently performed with the ABI 7500 system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) using the SYBR
Premix Ex Taq kit (Takara Biotechnology, Co., Ltd.) (95°C for 30
sec; 40 cycles of 95°C for 5 sec and 60°C for 10 sec). GAPDH was
used as the internal reference for lncRNA and mRNA, while U6 was
used as internal reference to miRNA expression analysis. Relative
RNA expression was calculated using the 2−ΔΔCq method
(17). The qPCR primer sequences are
listed in Table I.
 | Table I.Primers used for quantitative
PCR. |
Table I.
Primers used for quantitative
PCR.
| Gene | Forward primer,
5′→3′ | Reverse primer,
5′→3′ |
|---|
| lnc-TCF7 |
AATTTTGCACATCTGAACAGCC |
TTCAAAATCCCACTACGCCCA |
| Jagged1 |
GGCTTGGATCTGTTGCTTGGT |
TTGTTGGTGGTGTTGTCCTCAG |
| Notch1 |
GCAACAGCGAGGAAGAGGAG |
CGGCATCAGAGCGTGAGTAG |
| GAPDH |
GACCACAGTCCATGCCATCAC |
ACGCCTGCTTCACCACCTT |
| miR-203 |
ACACTCCAGCTGGGAGTGGTTCTTAACAGT |
TGTCGTGGAGTCGGCAATTC |
| miR-26b |
ACACTCCAGCTGGGTTCAAGTAATTCAGGA |
TGTCGTGGAGTCGGCAATTC |
| miR-643 |
ACACTCCAGCTGGGACTTGTATGCTAGCTC |
TGTCGTGGAGTCGGCAATTC |
| U6 |
CGCTTCGGCAGCACATATACTA |
ATGGAACGCTTCACGAATTTGC |
Western blotting
Total protein was isolated from cells using RIPA
Lysis and Extraction Buffer (Thermo Fisher Scientific, Inc.), and
the protein concentration was determined using the Pierce™ BCA
Protein Assay kit (Thermo Fisher Scientific, Inc.). The protein
samples (20 µg per lane) were then separated by 4–20% SDS-PAGE and
transferred to polyvinylidene fluoride membranes (EMD Millipore).
After blocking with 5% non-fat milk for 2 h at 37°C, the membranes
were incubated with the corresponding primary antibodies overnight
at 4°C. The membranes were subsequently incubated with secondary
antibody for 1 h at room temperature. The bands were visualized
using Pierce™ ECL Plus Western Blotting Substrate (Thermo Fisher
Scientific, Inc.) followed by exposure to X-ray film (Kodak). The
greyscales of the blots were analyzed using ImageJ software
(ver.1.8; National Institutes of Health). All antibodies are listed
in Table II.
 | Table II.Antibodies using in western
blotting. |
Table II.
Antibodies using in western
blotting.
| A, Primary
antibodies |
|---|
| Antibody | Company | Catalog number | Dilution |
| Jagged1
Rabbit mAb | CST | #2620 | 1:1,000 |
| Notch1
Rabbit mAb | CST | #3268 | 1:1,000 |
| GAPDH
Rabbit mAb | CST | #2118 | 1:1,000 |
|
| B, Secondary
antibody |
|
| Antibody | Company | Catalog number | Dilution |
| Goat
Anti-Rabbit | CST | #7074 | 1:3,000 |
| IgG
H&L (HRP) |
|
|
|
Statistical analysis
The study data are presented as the mean ± standard
deviation, median and interquartile range or count (percentage),
and statistical analysis was performed using SPSS 24.0 (IBM Corp).
Comparisons among three or more groups were made using the Kruskal
Wallis test, or one-way ANOVA followed by Dunnett's multiple
comparison test or Tukey's multiple comparison test. Comparisons
between two groups were made using the unpaired t-test, Wilcoxon
rank sum test or the χ2 test. Univariate and
multivariate logistic regression analyses were performed to screen
factors affecting CR. Univariate and multivariate Cox proportional
hazards regression model analyses were performed to screen factors
affecting EFS and OS. All graphs were plotted using GraphPad Prism
7.01 (GraphPad Software, Inc.). Receiver operating curve (ROC)
analysis and area under the curve (AUC) were conducted to assess
the diagnostic value of lnc-TCF7 in MM. OS and EFS are displayed as
Kaplan-Meier curves. Patients with MM were divided into
lnc-TCF7-high and -low expression groups based on the median
lnc-TCF7 level (1.91), and the survival difference between the
high- and low-lnc-TCF7 expression groups was determined using the
log-rank test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Baseline characteristics of patients
with MM
Our preliminary research (with a small sample
population) indicated that lnc-TCF7 was upregulated in patients
with MM compared with healthy controls, and in MM cells compared
with BMMCs plasma cells from health donors (data not shown). Of the
86 patients with MM, the mean age was 58.9±9.0 years, including 53
(61.6%) men and 33 (38.4%) women. For Ig subtype, the numbers of
patients positive for IgG, IgA, IgM, IgD and free light chain were
49 (57.0%), 20 (23.2%), 1 (1.2%), 1 (1.2%) and 15 (17.4%),
respectively. For Durie-Salmon stage, the numbers of patient with
stage I, II and III disease were 3 (3.5%), 40 (46.5%) and 43
(50.0%), respectively, and the numbers at ISS stage I, II and III
were 19 (22.1%), 32 (37.2%) and 35 (40.7%), respectively. Details
of the other baseline characteristics are displayed in Table III.
 | Table III.Characteristics of 86 patients with
multiple myeloma. |
Table III.
Characteristics of 86 patients with
multiple myeloma.
| Parameters | Value |
|---|
| Age, years, mean ±
SD | 58.9±9.0 |
| Sex, n (%) |
|
|
Male | 53 (61.6) |
|
Female | 33 (38.4) |
| Immunoglobulin
subtype, n (%) |
|
|
IgG | 49 (57.0) |
|
IgA | 20 (23.2) |
|
IgM | 1 (1.2) |
|
IgD | 1 (1.2) |
| Free
light chain | 15 (17.4) |
| Bone lesion, n
(%) | 64 (74.4) |
| Hb, g/dl, median
(IQR) | 10.1
(8.6–11.5) |
| Calcium, mg/dl,
median (IQR) | 9.9 (8.9–11.4) |
| Scr, mg/dl, median
(IQR) | 1.6 (1.3–1.9) |
| ALB, mg/dl, median
(IQR) | 3.9 (3.3–4.2) |
| β2-MG, mg/l, median
(IQR) | 4.7 (2.8–8.9) |
| LDH, U/l, median
(IQR) | 193.0
(167.8–228.7) |
| Durie-Salmon stage,
n (%) |
|
| I | 3 (3.5) |
| II | 40 (46.5) |
|
III | 43 (50.0) |
| ISS stage, n
(%) |
|
| I | 19 (22.1) |
| II | 32 (37.2) |
|
III | 35 (40.7) |
| Cytogenetics, n
(%) |
|
|
t(4;14) | 7 (8.1) |
|
t(14;16) | 11 (12.8) |
|
Del(17p) | 12 (14.0) |
Relation between lnc-TCF7 expression
and risk of MM
lnc-TCF7 expression was increased in patients with
MM [median value of relative expression: 1.91 (0.98–3.75)] compared
with healthy controls [median value of relative expression: 0.64
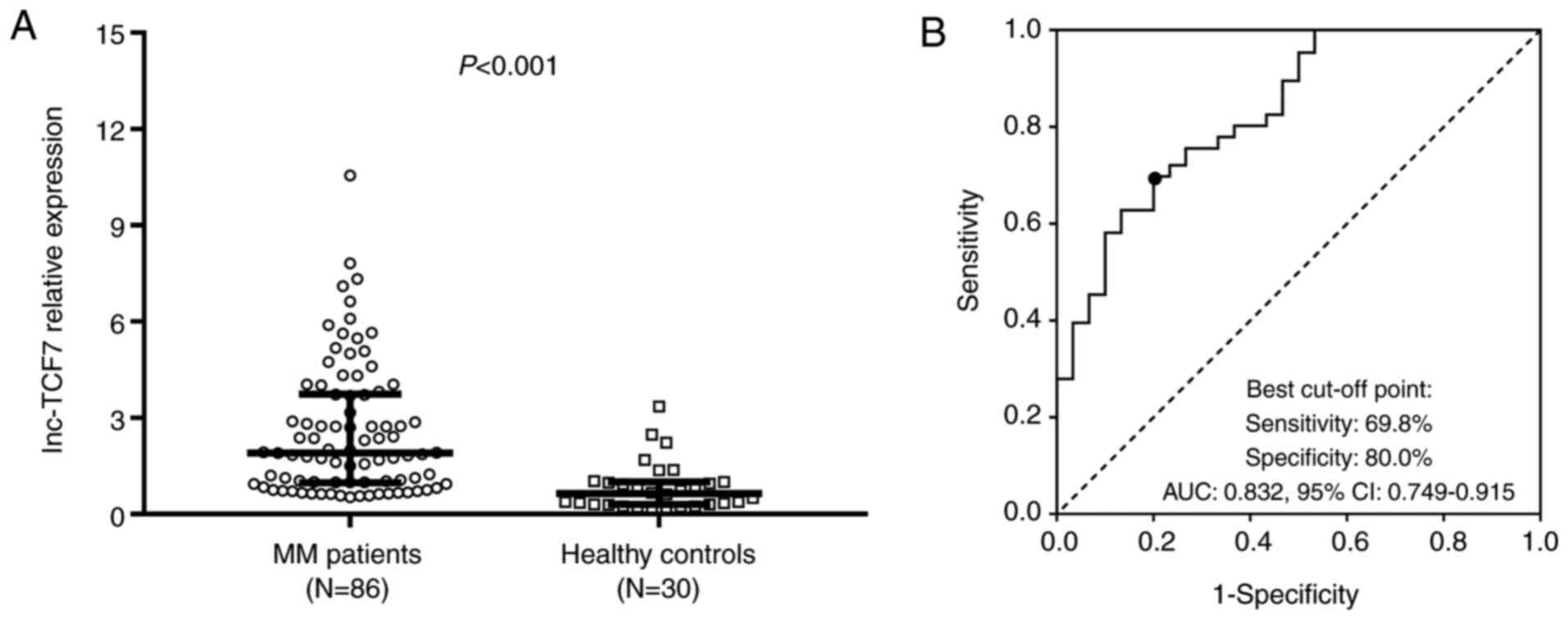
(0.30–1.01)] (P<0.001) (Fig. 1A).
ROC curve analysis revealed that lnc-TCF7 effectively discriminated
between patients and healthy controls, with an AUC of 0.832 (95%
confidence interval, 0.749–0.915; Fig.
1B), suggesting that lnc-TCF7 was related to increased risk of
MM. No differences in lnc-TCF7 expression were observed among
patients with MM who had received BD, BDD or MPB treatment
(Fig. S1).
Relation between lnc-TCF7 expression
and the clinicopathological characteristics of patients with
MM
lnc-TCF7 expression was found to be significantly
related to β2-MG expression (P=0.001) and ISS stage (P=0.001),
though no difference was observed between lnc-TCF7 expression and
other baseline clinical characteristics (Table IV).
 | Table IV.Relation between lnc-TCF7 relative
expression and clinical characteristics. |
Table IV.
Relation between lnc-TCF7 relative
expression and clinical characteristics.
| Variables | lnc-TCF7 relative
expression, median (IQR) | P-value |
|---|
| Age, years |
| 0.242 |
|
≤60 | 2.33
(0.95–4.11) |
|
|
>60 | 1.59
(1.00–3.07) |
|
| Sex |
| 0.912 |
|
Female | 1.94
(1.03–3.03) |
|
|
Male | 1.90
(0.89–4.03) |
|
| Immunoglobulin
subtype |
| 0.209 |
|
IgG | 1.84
(0.83–3.92) |
|
|
IgA | 1.72
(0.96–2.41) |
|
|
IgM | 1.99a |
|
|
IgD | 7.11a |
|
| Free
light chain | 2.74
(1.24–5.08) |
|
| Bone lesion |
| 0.138 |
| No | 2.55
(1.10–5.25) |
|
|
Yes | 1.81
(0.86–3.10) |
|
| Hb, g/dl |
| 0.349 |
|
≤10 | 1.87
(1.07–4.61) |
|
|
>10 | 2.02
(0.84–2.81) |
|
| Calcium, mg/dl |
| 0.292 |
|
≤11.5 | 1.85
(0.92–2.96) |
|
|
>11.5 | 2.57
(1.00–4.64) |
|
| Scr, mg/dl |
| 0.355 |
| ≤2 | 1.90
(0.94–3.17) |
|
|
>2 | 3.73
(1.07–5.18) |
|
| ALB, mg/dl |
| 0.878 |
|
≤3.5 | 1.96
(1.00–3.39) |
|
|
>3.5 | 1.85
(0.86–3.79) |
|
| β2-MG, mg/l |
| 0.001 |
|
≤5.5 | 1.57
(0.74–2.30) |
|
|
>5.5 | 2.87
(1.74–4.34) |
|
| LDH, U/l |
| 0.763 |
|
≤220 | 1.85
(0.96–3.96) |
|
|
>220 | 2.19
(0.98–3.75) |
|
| Durie-Salmon
stage |
| 0.297 |
| I | 0.75
(0.67–2.24) |
|
| II | 2.36
(0.84–4.02) |
|
|
III | 1.83
(1.07–3.72) |
|
| ISS stage |
| 0.001 |
| I | 1.24
(0.74–1.83) |
|
| II | 1.81
(0.73–4.24) |
|
|
III | 2.87
(1.74–4.34) |
|
| t(4;14) |
| 0.076 |
| No | 1.87
(0.94–3.72) |
|
|
Yes | 2.73
(1.79–7.33) |
|
| t(14;16) |
| 0.693 |
| No | 1.94
(0.84–3.73) |
|
|
Yes | 1.51
(1.07–5.89) |
|
| Del(17p) |
| 0.236 |
| No | 1.85
(0.92–3.70) |
|
|
Yes | 2.81
(1.15–4.76) |
|
Effects of lnc-TCF7 expression on
treatment responses and survivals in patients with MM
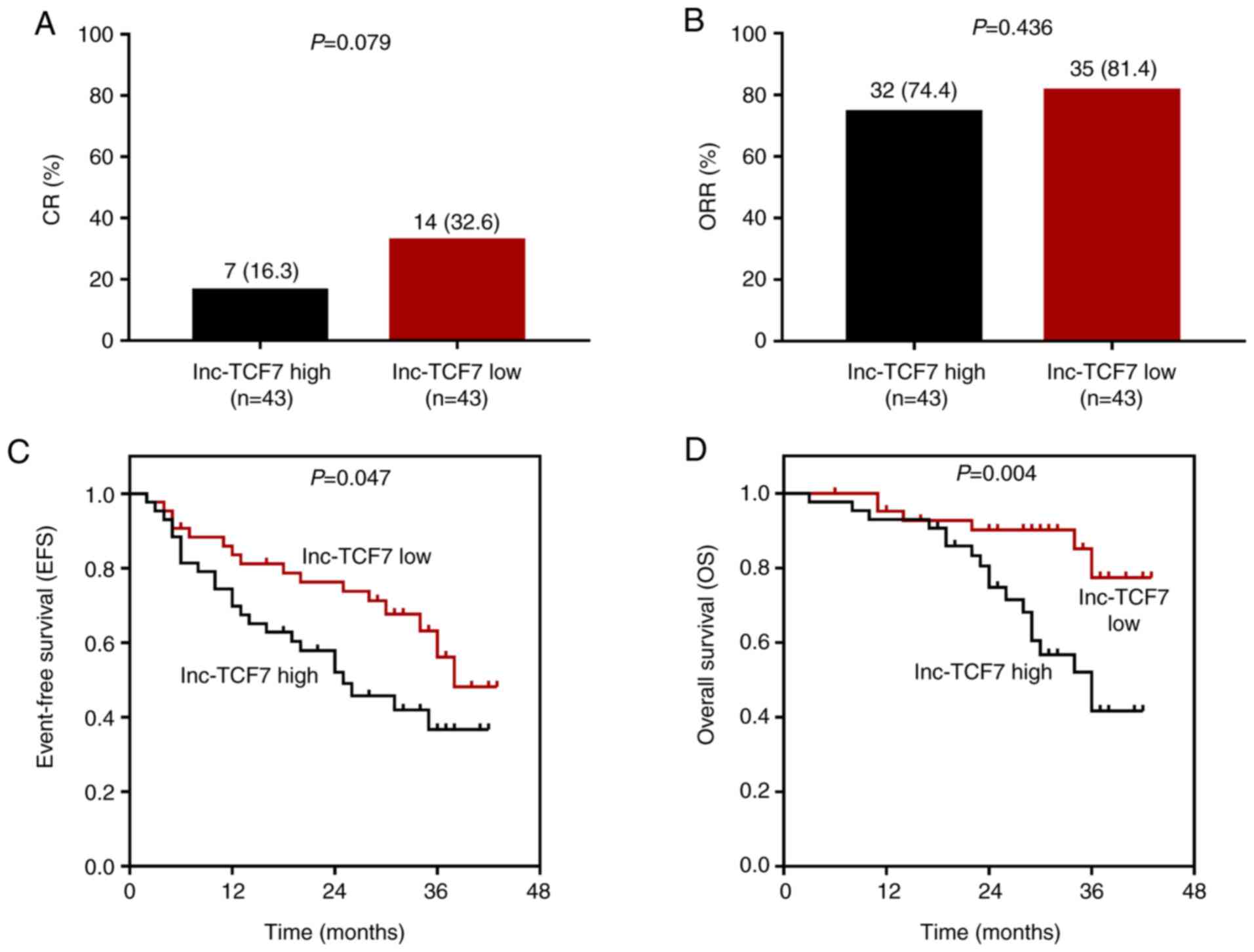
CR (P=0.079; Fig. 2A)
was lower, while ORR (P=0.436; Fig.
2B) was similar in the lnc-TCF7-high expression group compared
with the low expression group. Furthermore, EFS (P=0.047; Fig. 2C) and OS (P=0.004; Fig. 2D) time were shorter in the
lnc-TCF7-high compared with lnc-TCF7-low expression group,
indicating that increased lnc-TCF7 expression was related to poorer
prognosis of patients with MM.
Analyses of factors affecting CR
The results of univariate or multivariate logistic
regression analysis revealed that lnc-TCF7 expression and any other
clinical characteristics were not related to a CR in patients with
MM (all P>0.05; Table V).
 | Table V.Univariate and multivariate logistic
regression model analyses of factors affecting complete
response. |
Table V.
Univariate and multivariate logistic
regression model analyses of factors affecting complete
response.
|
| Univariate logistic
regression | Multivariate
logistic regression |
|---|
|
|
|
|
|---|
| Variables | P-value | OR (95% CI) | P-value | OR (95% CI) |
|---|
| lnc-TCF7, high vs.
low | 0.084 | 0.403
(0.144–1.129) | 0.398 | 0.585
(0.169–2.028) |
| Age, >60 years
vs. ≤60 years | 0.923 | 1.051
(0.381–2.900) | 0.680 | 0.767
(0.217–2.712) |
| Sex, male vs.
female | 0.627 | 0.780
(0.287–2.122) | 0.352 | 0.536
(0.144–1.992) |
| Immunoglobulin
subtype, IgG vs. others | 0.986 | 1.009
(0.373–2.726) | 0.836 | 1.134
(0.345–3.725) |
| Bone lesion, yes
vs. no | 0.718 | 0.816
(0.271–2.458) | 0.892 | 0.901
(0.200–4.069) |
| Hb, >10 g/dl vs.
≤10 g/dl | 0.084 | 0.403
(0.144–1.129) | 0.262 | 0.412
(0.087–1.941) |
| Calcium, >11.5
mg/dl vs. ≤11.5 mg/dl | 0.601 | 0.721
(0.211–2.457) | 0.871 | 0.881
(0.190–4.075) |
| Scr, >2.0 mg/dl
vs. ≤2.0 mg/dl | 0.330 | 1.950
(0.509–7.461) | 0.186 | 3.238
(0.567–18.483) |
| ALB, >3.5 mg/dl
vs. ≤3.5 mg/dl | 0.486 | 1.463
(0.501–4.277) | 0.590 | 1.405
(0.407–4.850) |
| LDH, >220 U/l
vs. ≤220 U/l | 0.463 | 0.655
(0.211–2.028) | 0.820 | 0.859
(0.232–3.180) |
| Durie-Salmon stage,
III vs. II/I | 0.213 | 1.896
(0.693–5.188) | 0.967 | 0.968
(0.202–4.630) |
| ISS stage, III vs.
II/I | 0.076 | 0.365
(0.119–1.113) | 0.163 | 0.328
(0.068–1.570) |
| t(4;14), yes vs.
no | 0.523 | 0.492
(0.056–4.336) | 0.945 | 0.913
(0.067–12.354) |
| t(14;16), yes vs.
no | 0.608 | 0.655
(0.130–3.303) | 0.717 | 0.691
(0.094–5.101) |
| Del(17p), yes vs.
no | 0.192 | 0.245
(0.030–2.025) | 0.235 | 0.228
(0.020–2.619) |
Analyses of factors affecting EFS
Univariate Cox proportional hazards regression model
analysis showed that lnc-TCF7 expression (high vs. low) was related
to EFS, though this was not statistically significant (P=0.052).
However, ISS stage (III vs. II/I) was significanlty related to
shorter EFS (P<0.001; Table VI).
All factors were further analyzed using multivariate Cox analysis,
which disclosed that lnc-TCF7 expression (high vs. low) was not an
independent risk factor for EFS (P=0.307); whereas age (>60 vs.
≤60 years; P=0.029), Durie-Salmon stage (III vs. II/I; P=0.012) and
ISS stage (III vs. II/I; P<0.001) were independent risk factors
for shorter EFS; however, age and Durie-Salmon stage were
non-significant in the univariate analysis (Table VI).
 | Table VI.Univariate and multivariate Cox's
proportional hazards regression model analyses of factors affecting
event-free survival. |
Table VI.
Univariate and multivariate Cox's
proportional hazards regression model analyses of factors affecting
event-free survival.
|
| Univariate Cox's
regression | Multivariate Cox's
regression |
|---|
|
|
|
|
|---|
| Variables | P-value | HR (95% CI) | P-value | HR (95% CI) |
|---|
| lnc-TCF7, high vs.
low | 0.052 | 1.878
(0.994–3.549) | 0.307 | 1.535
(0.674–3.495) |
| Age, >60 years
vs. ≤60 years | 0.564 | 1.203
(0.642–2.254) | 0.029 | 2.561
(1.102–5.954) |
| Sex, male vs.
female | 0.111 | 0.602
(0.322–1.124) | 0.846 | 1.086
(0.472–2.503) |
| Immunoglobulin
subtype, | 0.796 | 1.087
(0.577–2.047) | 0.781 | 0.899
(0.424–1.907) |
| IgG vs. others |
|
|
|
|
| Bone lesion, yes
vs. no | 0.171 | 0.622
(0.315–1.228) | 0.208 | 0.547
(0.214–1.400) |
| Hb, >10 g/dl vs.
≤10 g/dl | 0.614 | 0.852
(0.457–1.588) | 0.754 | 1.153
(0.474–2.804) |
| Calcium, >11.5
mg/dl vs. ≤11.5 mg/dl | 0.492 | 1.287
(0.627–2.640) | 0.922 | 0.955
(0.379–2.408) |
| Scr, >2.0 mg/dl
vs. ≤2.0 mg/dl | 0.260 | 1.653
(0.690–3.959) | 0.339 | 0.575
(0.185–1.790) |
| ALB, >3.5 mg/dl
vs. ≤3.5 mg/dl | 0.643 | 1.170
(0.603–2.269) | 0.602 | 1.234
(0.560–2.718) |
| LDH, >220 U/l
vs. ≤220 U/l | 0.316 | 1.397
(0.727–2.684) | 0.151 | 1.850
(0.799–4.283) |
| Durie-Salmon stage,
III vs. II/I | 0.142 | 1.608
(0.852–3.034) | 0.012 | 3.230
(1.295–8.055) |
| ISS stage, III vs.
II/I | 1.2×10-7 | 6.919
(3.383–14.153) | 1.9×10-7 | 12.746
(4.892–33.208) |
| t(4;14), yes vs.
no | 0.135 | 2.051
(0.800–5.254) | 0.996 | 0.997
(0.304–3.263) |
| t(14;16), yes vs.
no | 0.689 | 1.194
(0.501–2.848) | 0.977 | 0.986
(0.368–2.639) |
| Del(17p), yes vs.
no | 0.152 | 1.763
(0.811–3.832) | 0.655 | 0.792
(0.284–2.206) |
Analyses of factors affecting OS
In order to further investigate the factors
affecting OS in patients with MM, univariate Cox proportional
hazards regression model analysis was performed, which revealed
that lnc-TCF7 expression (high vs. low; P=0.008) and ISS stage (III
vs. II/I; P<0.001) were related to worse OS (Table VII). All factors were further
subjected to multivariate Cox analysis, which showed that lnc-TCF7
expression (high vs. low; P=0.123) was not an independent factor
for predicting OS, while ISS stage (III vs. II/I; P<0.001) was
an independent predictive factor for poorer OS (Table VII). These results suggested that
lnc-TCF7 may influence ISS stage rather than directly impacting OS
in patients with MM.
 | Table VII.Univariate and multivariate Cox's
proportional hazards regression model analyses of factors affecting
overall survival. |
Table VII.
Univariate and multivariate Cox's
proportional hazards regression model analyses of factors affecting
overall survival.
|
| Univariate Cox
regression | Multivariate Cox
regression |
|---|
|
|
|
|
|---|
| Variables | P-value | HR (95% CI) | P-value | HR (95% CI) |
|---|
| lnc-TCF7, high vs.
low | 0.008 | 3.513
(1.391–8.874) | 0.123 | 2.541
(0.778–8.302) |
| Age, >60 years
vs. ≤60 years | 0.874 | 1.068
(0.474–2.407) | 0.233 | 1.991
(0.642–6.178) |
| Sex, male vs.
female | 0.089 | 0.498
(0.223–1.111) | 0.471 | 0.648
(0.199–2.109) |
| Immunoglobulin
subtype, | 0.724 | 0.865
(0.387–1.934) | 0.705 | 0.832
(0.320–2.162) |
| IgG vs. others |
|
|
|
|
| Bone lesion, yes
vs. no | 0.942 | 0.964
(0.357–2.602) | 0.657 | 0.705
(0.150–3.306) |
| Hb, >10 g/dl vs.
≤10 g/dl | 0.751 | 1.139
(0.509–2.548) | 0.952 | 0.959
(0.249–3.699) |
| Calcium, >11.5
mg/dl vs. | 0.105 | 2.033
(0.863–4.790) | 0.225 | 2.301
(0.599–8.841) |
| ≤11.5 mg/dl |
|
|
|
|
| Scr, >2.0 mg/dl
vs. ≤2.0 mg/dl | 0.939 | 1.058
(0.246–4.544) | 0.616 | 0.612
(0.090–4.170) |
| ALB, >3.5 mg/dl
vs. ≤3.5 mg/dl | 0.833 | 1.096
(0.468–2.562) | 0.227 | 2.010
(0.647–6.242) |
| LDH, >220 U/l
vs. ≤220 U/l | 0.172 | 1.763
(0.782–3.975) | 0.097 | 2.634
(0.839–8.266) |
| Durie-Salmon stage,
III vs. II/I | 0.516 | 1.309
(0.581–2.952) | 0.316 | 1.921
(0.536–6.878) |
| ISS stage, III vs.
II/I | 7.7×10-7 | 11.113
(4.276–28.882) | 3.4×10-5 | 17.494
(4.519–67.727) |
| t(4;14), yes vs.
no | 0.110 | 2.408
(0.818–7.086) | 0.699 | 0.725
(0.142–3.710) |
| t(14;16), yes vs.
no | 0.471 | 0.587
(0.138–2.500) | 0.490 | 0.563
(0.110–2.875) |
| Del(17p), yes vs.
no | 0.618 | 1.315
(0.449–3.854) | 0.299 | 0.446
(0.097–2.047) |
Comparison of lnc-TCF7 expression
between MM cell lines and plasma cells from healthy BMMCs
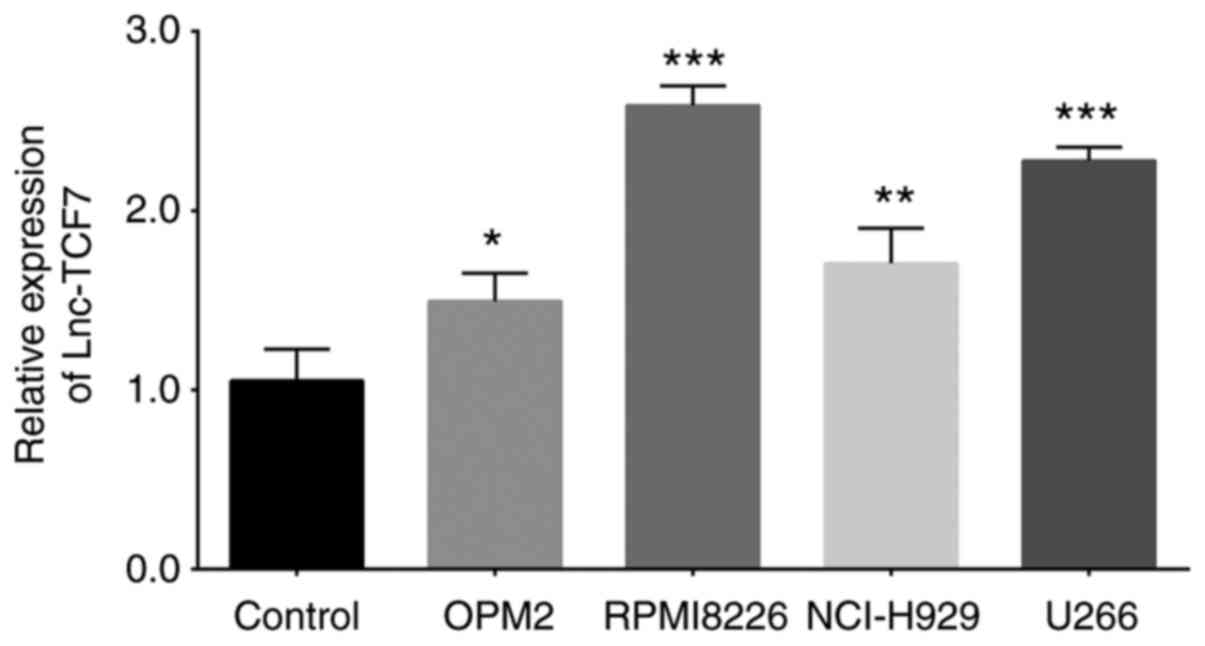
The expression levels of lnc-TCF7 were subsequently
compared between MM cell lines and BMMC from healthy donors. The
results revealed that lnc-TCF7 expression was elevated in MM cell
lines [OPM2 (P<0.05), RPMI8226 (P<0.001), NCI-H929
(P<0.01) and U266 (P<0.001)] compared with healthy donor
cells (Fig. 3).
Effects of lnc-TCF7-knockdown on
cellular proliferation and apoptosis, and its potential target
miRNAs in RPMI8226 cells
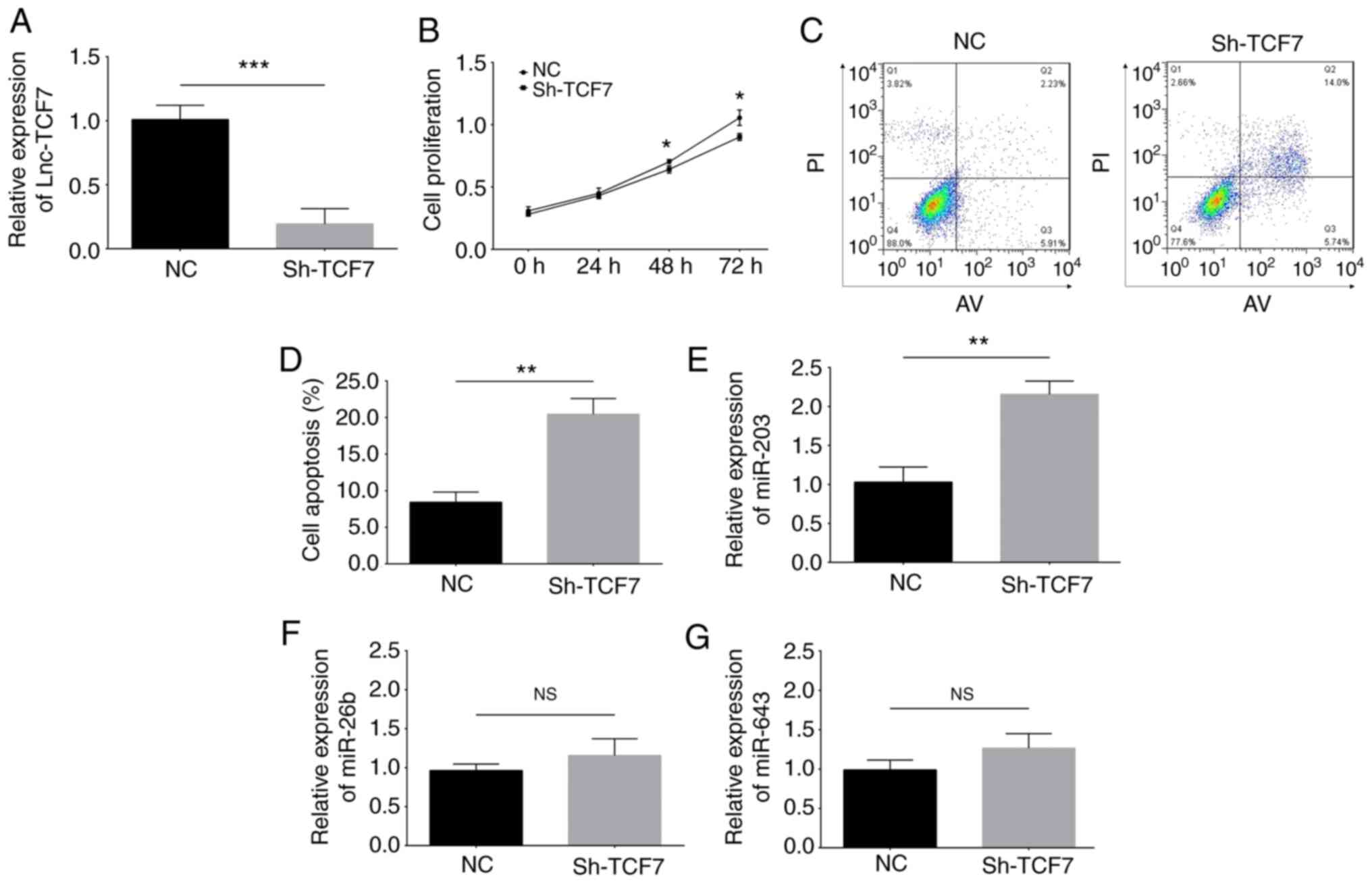
The transfection efficacy was evaluated by RT-qPCR,
which is presented in Fig. S2.
Following transfection, lnc-TCF7 expression was decreased in the
Sh-TCF7 group compared with the NC group, suggesting that the
transfection of the lnc-TCF7 shRNA vector into RPMI8226 cells was
successful (P<0.001; Fig. 4A).
RPMI8226 cell proliferation was inhibited at 48 (P<0.05) and 72
h (P<0.05) (Fig. 4B)
post-transfection, while apoptosis (P<0.01) (Fig. 4C and D) was enhanced in the Sh-TCF7
group compared with the NC group, indicating that
lnc-TCF7-knockdown suppressed cellular proliferation while
promoting apoptosis in MM cells. In addition, potential targets of
lnc-TCF7 were predicted using the miRanda database, and miR-203,
−26b and −643 were selected for validation and detection. The
results indicated that miR-203 was upregulated in the Sh-TCF7 group
compared with the NC group (P<0.01; Fig. 4E), whereas miR-26b (P>0.05;
Fig. 4F) and miR-643 (P>0.05;
Fig. 4G) levels were similar between
the two groups, illustrating that lnc-TCF7-knockdown upregulated
miR-203 expression but did not affect that of miR-26b or −643 in
RPMI8226 cells.
Interaction between lnc-TCF7 and
miR-203 mediated the Jagged1-Notch1 signaling pathway in RPMI8226
cells
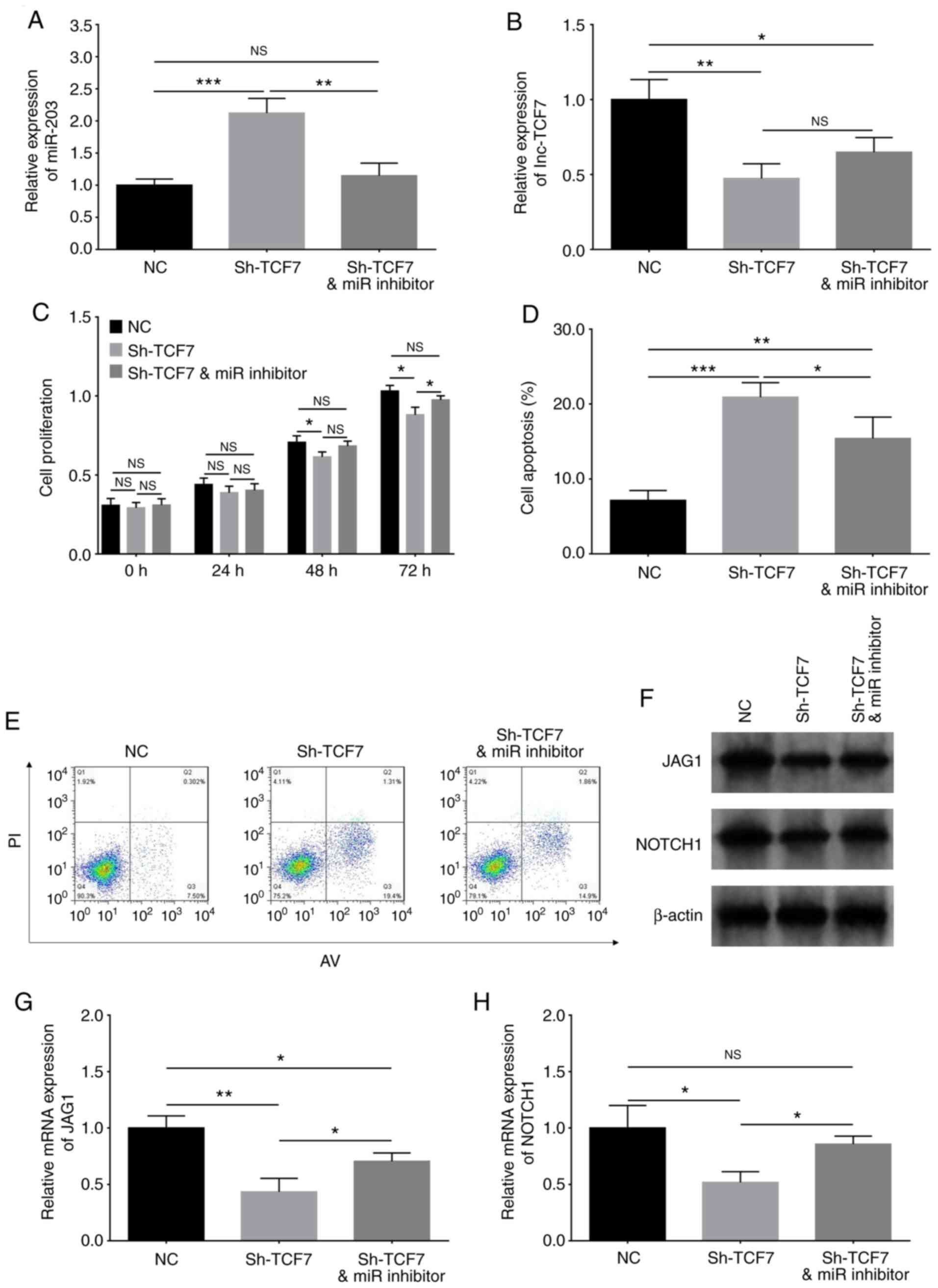
Compensation experiments were performed to determine
whether lnc-TCF7 shRNA functions in MM cells by enhancing miR-203
and regulating the downstream Jagged1-Notch1 signaling pathway. The
results indicated that miR-203 expression was decreased in the
Sh-TCF7/miR inhibitor group compared with Sh-TCF7 group (P<0.01;
Fig. 5A), whereas lnc-TCF7
expression remained consistent between the two groups (P>0.05;
Fig. 5B), indicating that lnc-TCF7
suppressed miR-203 expression, but miR-203 did not influence
lnc-TCF7 expression. In addition, cellular proliferation was
enhanced at 72 h (P<0.05; Fig.
5C), while apoptosis was repressed (P<0.05) (Fig. 5D and E) in the Sh-TCF7/miR inhibitor
compared with the Sh-TCF7 groups. These findings suggested that
miR-203 inhibition accelerated cellular proliferation while
reducing inhibition in lnc-TCF7-knockdown cells. Furthermore,
miRanda database analysis predicted Jagged1 to be a target gene of
miR-203, and the Jagged1-Notch1 signaling pathway has been reported
to be directly regulated by miR-203 (18). Thus, in the present study, Jagged1
and Notch1 expression were detected using western blotting and
RT-qPCR. Jagged1 (P<0.05) (Fig. 5F
and G) and Notch1 (P<0.05) (Fig.
5F and H) expression were detected using western blot and
RT-qPCR analyses, which revealed that both were upregulated in the
Sh-TCF7/miR inhibitor group compared with the Sh-TCF7 group. These
data indicated that lnc-TCF7-knockdown suppressed cellular
proliferation while promoting apoptosis by regulating the
miR-203-mediated Jagged1-Notch1 signaling pathway in RPMI8226
cells.
Discussion
lnc-TCF7, a newly identified lncRNA, has been
reported to be upregulated in various types of cancer (8–10). For
example, Wang et al (7)
reported that, in 39 patients with liver cancer, lnc-TCF7 was
upregulated in tumor tissues compared with paired-adjacent normal
tissues. Furthermore, Li et al (10) revealed that lnc-TCF7 expression was
increased in tumor tissues compared with adjacent normal tissues in
patients with CRC, and that increased lnc-TCF7 levels were
associated with larger tumors and a higher Tumor-Node-Metastasis
stage. These studies reveal that lnc-TCF7 expression is elevated in
several types of solid tumor, and is associated with deteriorating
clinical features. Another previous study demonstrated that the
associated lnc-TCF7 gene (TCF7) regulates the functions of
malignant hematological cells (11).
Our preliminary research (with a small sample population) indicated
that lnc-TCF7 was upregulated in patients with MM compared with
healthy controls, and in MM cells compared with BMMCs plasma cells
from health donors (unpublished data). Based on these findings, it
was hypothesized that lnc-TCF7 may also be upregulated in patients
with MM, which may be used to predict the clinical features of MM
progression. However, limited information could be obtained. In the
current study, lnc-TCF7 expression was found to be elevated in
patients with MM compared with healthy controls, which effectively
predicted an increased risk of MM. In addition, lnc-TCF7 expression
was related to β2-MG level and the ISS stage. There are a number of
possible reasons for these results. Firstly, lnc-TCF7 may act as a
competing endogenous RNA (ceRNA) that sponges specific tumor
suppressor genes (including miR-203) and stimulates MM
tumorigenesis. This potentially increases the risk of MM, as
increased β2-MG level may be related to more advanced ISS stage.
Secondly, lncRNAs may directly activate specific signaling pathways
associated with carcinogenesis (such as the Wnt pathway), thus
increasing the risk of MM, as well as patient β2-MG levels and ISS
stage (7). Finally, lnc-TCF7 may
serve as a gene pool for TCF7, while the loss of TCF7 diminishes
hematopoietic stem/progenitor cell functions. Hence, lnc-TCF7 may
promote TCF7 expression and stimulate the malignant proliferation
of MM cells, thus increasing risk of MM, β2-MG levels and ISS stage
(11). The predictive value of
lnc-TCF7 for the prognosis of patients with MM was subsequently
evaluated, and lnc-TCF7 expression was revealed to be negatively
related to EFS and OS. These results align with those of a previous
study, which indicated that lnc-TCF7 is associated with poorer OS
in patients with CRC (10). The
negative relation between lnc-TCF7, EFS and OS in MM may be the
result of lnc-TCF7 promoting MM tumorigenesis, which exacerbates
clinical features and therefore promotes poorer EFS and OS.
Alternatively, lnc-TCF7 may facilitate disease relapse via the
activation of mesenchymal stem cells, thus promoting poorer EFS and
OS in patients with MM. In addition, univariate analysis indicated
that lnc-TCF7 expression level was related to reduced EFS and OS
times, while multivariate analysis revealed that lnc-TCF7 was not
an independent risk factor for shorter EFS or OS. These findings
may be explained by lnc-TCF7 indirectly influencing survival by
affecting other malignant features (such as ISS stage). Also, it
was found that older age, Durie-Salmon stage and ISS stage were
independent risk factors for shorter EFS, while age and
Durie-Salmon stage were non-significant in the univariate analysis.
Further studies should be conducted to verify these findings.
A number of in vitro studies have sought to
clarify the effects and molecular mechanisms of lnc-TCF7 in cancer
pathology (8–10). For instance, Wang et al
(7) discovered that lnc-TCF7 is
highly expressed in liver cancer stem cells (CSCs; Hep3B and Huh7
cell lines), and that its overexpression enhances the tumorigenic
capacity of liver CSCs by targeting the Wnt signaling pathway
(7). Similarly, Li et al
(10) revealed that
lnc-TCF7-knockdown decreases the migration and invasiveness of DLD1
and LoVo CRC cell lines by regulating the Wnt/β-catenin signaling
pathway, while its overexpression has the opposite effect. In
addition, lnc-TCF7 promotes the invasiveness and self-renewal of
NSCLC cells (A549 and 95D cell lines) by upregulating Slug and
epithelial cell adhesion molecule expression (9). However, few studies had previously
investigated the effects of lnc-TCF7 on the activities of MM cells.
The results of the present study revealed that lnc-TCF7-knockdown
inhibited MM cell proliferation while promoting apoptosis,
potentially by acting as a ceRNA and sponging tumor suppressor
miRNAs, such as miR-203. Alternatively, lncTCF7 may directly
recruit the SWItch/sucrose non-fermentable complex to the TCF7
promoter, activating tumorigenesis-related pathways (such as the
Wnt signaling pathway), and then stimulating cellular proliferation
while repressing apoptosis (7).
Finally, lnc-TCF7 may serve as the gene pool of TCF7, inducing the
upregulation of TCF7 (which may stimulate the activities of
malignant hematological cells), thereby promoting proliferation and
suppressing apoptosis (11).
However, further studies are required to clarify the effects of
lnc-TCF7 on the migration and invasion abilities of MM cells.
The miRNAs are a class of non-coding RNAs of ~20
nucleotides in length that are abundantly expressed in mammalian
cell types (19). In the past few
decades, a large number of miRNAs have been reported to act as
modulators of cancer pathogenesis (in malignancies such as lung and
ovarian cancer) (20,21). In MM, several miRNAs have been
reported to exert anti-cancerous functions (22,23). Wu
et al (22) observed that
miR-203 suppressed MM cell proliferation and regulated
G1/S transition by targeting Bmi-1. Jia et al
(23) demonstrated that miR-26b
inhibited cellular proliferation and induced apoptosis by targeting
Jagged1 in MM cells. These studies indicate that miRNAs serve
crucial roles in inhibiting the progression of MM. Considering that
miR-203 and miR-26b play supressive roles in MM development, and in
addition to miR-643, are predicted targets of lnc-TCF7 (22,23), the
present study aimed to determine whether lnc-TCF7 reversely
mediated these three miRNAs by evaluating their expression
following lnc-TCF-knockdown. The data showed that
lnc-TCF7-knockdown stimulated miR-203 expression but did not affect
that of miR-26b or miR-643, suggesting that lnc-TCF7 reversely
modulated miR-203 expression in MM cells. To further investigate
the effects of lnc-TCF7-knockdown on MM cell proliferation and
apoptosis via miR-203, the miRanda database was used to predict
critical genes regulated by miR-203, which revealed Jagged1 as a
potential target gene. A previous study has also reported that the
Jagged1-Notch1 signaling pathway is directly regulated by miR-203
(18), thus Jagged1 and Notch1 were
selected as candidate genes in further miR-203 compensation
experiments in the present study. The expeiments revealed that the
effects of lnc-TCF7-knockdown on MM cell proliferation and
apoptosis may result from its regulation of the miR-203-mediated
Jagged1-Notch1 signaling pathway. However, further rescue
experiments could be conducted to investigate the regulatory role
of lnc-TCF7 and miR-203 on the Jagged1-Notch1 signaling pathway. In
brief, the present study provides an alternative perspective for
understanding the molecular mechanisms of MM etiopathogenesis, and
may facilitate the identification of novel therapeutic targets for
MM treatment.
In conclusion, lnc-TCF7 is clinically valuable for
predicting increased MM risk, and its elevated expression level is
related to deteriorating clinical features and poor prognosis.
Furthermore, lnc-TCF7-knockdown inhibits cellular proliferation
while stimulating apoptosis by regulating the miR-203-mediated
Jagged1-Notch1 signaling pathway in MM.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by The Key Medical Subject
of Qiang Wei Project in Jiangsu Province (grant no. ZDXKB2016009),
The Nantong Hematological Clinical Medical Research Center (grant
no. HS2015004), The National Natural Science Foundation of China
Grants (grant no. 81070400) and The Medical Innovation Team and the
Leading Talent Project of Jiangsu (grant no. LJ201136).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HL, HH and YJ conceived and designed the experiments
and confirm the authenticity of all raw data. HYL, YS and YX
performed the experiments. LW, CZ and LH made substantial
contributions to the acquisition, analysis and interpretation of
data. YJ was involved in drafting the manuscript. LH revised the
manuscript critically for important intellectual content. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by The Institutional Review
Board of The Affiliated Hospital of Nantong University (Nantong,
China). All participants or their guardians provided written
informed consent upon enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
MM
|
multiple myeloma
|
|
lncRNAs
|
long non-coding RNAs
|
|
lnc-TCF7
|
lncRNA transcription factor 7
|
|
CRC
|
colorectal cancer
|
|
NSCLC
|
non-small cell lung cancer
|
|
BMMCs
|
bone marrow mononuclear cells
|
|
β2-MG
|
β-2-microglobulin
|
|
ISS
|
International Staging System
|
|
qPCR
|
quantitative polymerase chain
reaction
|
|
CR
|
complete response
|
|
VGPR
|
very good partial response
|
|
ORR
|
overall response rate
|
|
EFS
|
event-free survival
|
|
OS
|
overall survival
|
|
RPMI
|
Roswell Park Memorial Institute
|
|
AUC
|
area under the curve
|
|
ROC
|
receiver operating characteristic
|
|
ceRNA
|
competing endogenous RNA
|
|
miRNAs
|
microRNAs
|
References
|
1
|
Chan HSH, Chen CI and Reece DE: Current
review on high-risk multiple myeloma. Curr Hematol Malig Rep.
12:96–108. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weaver CJ and Tariman JD: Multiple myeloma
genomics: A systematic review. Semin Oncol Nurs. 33:237–253. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rafei H, Haroun F and Tabbara IA: Novel
immunotherapeutic agents for the treatment of multiple myeloma. Am
J Clin Oncol. 42:317–329. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Riccomi G, Fornaciari G and Giuffra V:
Multiple myeloma in paleopathology: A critical review. Int J
Paleopathol. 24:201–212. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vo MC, Lakshmi TJ, Jung SH, Cho D, Park
HS, Chu TH, Lee HJ, Kim HJ, Kim SK and Lee JJ: Cellular
immunotherapy in multiple myeloma. Korean J Intern Med. 34:954–965.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, He L, Du Y, Zhu P, Huang G, Luo J,
Yan X, Ye B, Li C, Xia P, et al: The long noncoding RNA lncTCF7
promotes self-renewal of human liver cancer stem cells through
activation of Wnt signaling. Cell Stem Cell. 16:413–425. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu B, Chen M, Gao M, Cong Y, Jiang L, Wei
J and Huang J: Down-regulation of lncTCF7 inhibits cell migration
and invasion in colorectal cancer via inhibiting TCF7 expression.
Hum Cell. 32:31–40. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu J and Wang D: Long noncoding RNA TCF7
promotes invasiveness and self-renewal of human non-small cell lung
cancer cells. Hum Cell. 30:23–29. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li T, Zhu J, Wang X, Chen G, Sun L, Zuo S,
Zhang J, Chen S, Ma J, Yao Z, et al: Long non-coding RNA lncTCF7
activates the Wnt/β-catenin pathway to promote metastasis and
invasion in colorectal cancer. Oncol Lett. 14:7384–7390.
2017.PubMed/NCBI
|
|
11
|
Huls G, van Es J, Clevers H, de Haan G and
van Os R: Loss of Tcf7 diminishes hematopoietic stem/progenitor
cell function. Leukemia. 27:1613–1614. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ludwig H, Miguel JS, Dimopoulos MA,
Palumbo A, Garcia Sanz R, Powles R, Lentzsch S, Ming Chen W, Hou J,
Jurczyszyn A, et al: International Myeloma Working Group
recommendations for global myeloma care. Leukemia. 28:981–992.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Durie BG and Salmon SE: A clinical staging
system for multiple myeloma. Correlation of measured myeloma cell
mass with presenting clinical features, response to treatment, and
survival. Cancer. 36:842–854. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Greipp PR, San Miguel J, Durie BG, Crowley
JJ, Barlogie B, Bladé J, Boccadoro M, Child JA, Avet-Loiseau H,
Kyle RA, et al: International staging system for multiple myeloma.
J Clin Oncol. 23:3412–3420. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Anderson KC, Alsina M, Atanackovic D,
Biermann JS, Chandler JC, Costello C, Djulbegovic B, Fung HC,
Gasparetto C, Godby K, et al: Multiple myeloma, version 2.2016:
Clinical practice guidelines in oncology. J Natl Compr Canc Netw.
13:1398–1435. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kumar S, Paiva B, Anderson KC, Durie B,
Landgren O, Moreau P, Munshi N, Lonial S, Bladé J, Mateos MV, et
al: International myeloma working group consensus criteria for
response and minimal residual disease assessment in multiple
myeloma. Lancet Oncol. 17:e328–e346. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou Z, Shu B, Xu Y, Liu J, Wang P, Chen
L, Zhao J, Liu X, Qi S, Xiong K, et al: microRNA-203 modulates
wound healing and scar formation via suppressing Hes1 expression in
epidermal stem cells. Cell Physiol Biochem. 49:2333–2347. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Duchaine TF and Fabian MR: Mechanistic
Insights into MicroRNA-Mediated gene silencing. Cold Spring Harb
Perspect Biol. 11:a0327712019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mihanfar A, Fattahi A and Nejabati HR:
MicroRNA-mediated drug resistance in ovarian cancer. J Cell
Physiol. 234:3180–3191. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu SG, Chang TH, Liu YN and Shih JY:
MicroRNA in lung cancer metastasis. Cancers (Basel). 11:2652019.
View Article : Google Scholar
|
|
22
|
Wu SQ, Niu WY, Li YP, Huang HB and Zhan R:
miR-203 inhibits cell growth and regulates G1/S transition by
targeting Bmi-1 in myeloma cells. Mol Med Rep. 14:4795–4801. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jia CM, Tian YY, Quan LN, Jiang L and Liu
AC: miR-26b-5p suppresses proliferation and promotes apoptosis in
multiple myeloma cells by targeting JAG1. Pathol Res Pract.
214:1388–1394. 2018. View Article : Google Scholar : PubMed/NCBI
|