Introduction
Gastric cancer (GC) is a malignant tumor and is
associated with high morbidity and mortality (1). According to the latest statistics,
there were >1,000,000 new cases of GC and an estimated 783,000
deaths from GC worldwide in 2018 (2,3). Radical
gastrectomy and perioperative chemotherapy are the standard
treatments for GC. However, a large number of patients are
diagnosed during advanced stages, with missed opportunity for
surgery as early diagnosis is difficult due to the lack of
effective diagnostic biomarkers and most patients are asymptomatic
(3). In previous years, there has
been major progress in molecular-targeted therapy and immunotherapy
(4,5); however, the overall survival time for
patients with GC is <20%. Consequently, it is imperative to
identify effective diagnostic or therapeutic biomarkers for GC, and
to further elucidate the potential regulatory mechanisms involved
in GC progression.
Studies have identified that circular RNAs
(circRNAs) are a specific type of non-coding RNA, with a
closed-loop structure to prevent digestion by RNase R (6,7).
Research into the regulatory role of circRNAs is gaining
considerable interest. Notably, emerging evidence has shown that
circRNAs play a regulatory role in various biological processes via
sponging microRNAs (miRNAs) (7–9). Yu
et al (9) indicated that
circRNA_104718 acted as a tumor promoter gene in hepatocellular
carcinoma by regulating the miRNA-218-5p/Thioredoxin
domain-containing protein 5 signaling pathway. Besides, another
study revealed that circRNA_0084043 could sponge miR-153-3p,
upregulate Snail protein expression and promote malignant melanoma
progression (10). Nevertheless, the
biological function of circRNAs in GC progression has not been
fully elucidated.
circRNA_100395, also called circRNA_0015278, is
derived from chr1:173726114-173744981. Notably, circRNA_100395 has
been reported to play an important role in the progression of
various cancer types (11–13). Li et al (11) demonstrated that circRNA_100395 could
regulate the cellular phenotype and the malignant capacity in
ovarian cancer via regulating the miR-1228/p53 axis. Another study
showed that the upregulation of circRNA_100395 significantly
suppressed liver cancer cell proliferation (12). However, the biological role of
circRNA_100395 in GC remains unclear. Hence, the present study
aimed to explore the regulatory role of circRNA_100395 in GC
progression and further elucidate its possible mechanism in GC,
which may provide evidence for a promising novel diagnostic and
therapeutic biomarker for GC.
Materials and methods
Tissue samples
A total of 54 paired GC and normal tissue samples
were obtained from patients undergoing radical gastrectomy from
January 2013 to January 2015 at Hospital Affiliated 5 to Nantong
University, Taizhou People's Hospital (Taizhou, China). The
inclusion criteria were: i) Age 18–80 years; ii) provision of
written informed consent; iii) and pathologically confirmed GC as
assessed by veteran independent pathologists that were independent
to the present study. The exclusion criteria of the patients were:
i) Other malignant diseases; and ii) patients who had received
previous neoadjuvant chemotherapy or radiotherapy. The
clinicopathological features of the patients enrolled in the study
were assessed and recorded. The study was approved by The Ethical
Committees of Taizhou People's Hospital (Taizhou, China) (approval
no. KY2020-06511). All the patients provided written informed
consent before enrollment.
Cell culture
The 293T cells, normal epithelial cell lines (GES-1)
and human gastric cancer cell lines (HGC-27 MKN45, NCI-N87, SNU-5
and AGS) were obtained from the American Type Culture Collection.
All the cells were incubated in RPMI-1640 media and supplemented
with 10% FBS (both Gibco; Thermo Fisher Scientific, Inc.) and
incubated at 37°C in a 5% CO2 incubator.
RNA transfection
The pcDNA3.1-circRNA_100395 and pcDNA3.1 vector (NC
control) were brought from Invitrogen company (Thermo Fisher
Scientific, Inc.). HGC-27 and AGS cells were seed in a 12-well
plate and incubated overnight at 37°C with 5% CO2. At
80% confluency, these cells were transfected with 1 µg of
pcDNA3.1-circRNA_100395 or pcDNA3.1 vector respectively using
Lentiviruses (multiplicity of infection of 30) following the
manufacturer's protocol. Forty-eight hours later, these cells were
collected for subsequent experimentation.
Besides, the AGS cells were further transfected with
100 pmol miR-142-3p mimics or NC at 37°C with 5% CO2 for
48 h using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). Transfected cells were collected 48 h
after incubation for subsequent experiments.
Reverse transcription-quantitative
(RT-q)PCR
According to the manufacturer's protocols,
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) was used to extract total RNA. Subsequently, total RNA (1 µg)
was transcribed to cDNA via using PrimeScript RT Master Mix (Takara
Biotechnology Co., Ltd.) according to the manufacturer's protocol.
The temperature and duration of RT were: 37°C for 30 sec, followed
by 85°C for 5 sec and 4°C for 10 min. The cDNA (10 ng) was
subjected to qPCR (Bio-Rad Laboratories, Inc.) using SYBR Premix Ex
Taq II kit (Takara Biotechnology Co., Ltd.), and the reaction
volume was 10 µl. The thermocycling conditions used were as
follows: 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec
and 60°C for 40 sec. Analysis of circRNA_100395 and miR-142-3p
expression was performed using the 2-ΔΔCq method
(14). The primers sequences are
shown in Table I.
 | Table I.Sequences of oligomers and primers
used in the present research. |
Table I.
Sequences of oligomers and primers
used in the present research.
| Primer | Sequence, 5–3 |
|---|
| circRNA_100395
forward |
AGTGATGTGGCCCCTACAAG |
| circRNA_100395
reverse |
CCACTGGAGACCACTGGTTG |
| miR-142-3p
forward |
AGCGTGTAGTGTTTCCTACTT |
| miR-142-3p
reverse |
GTTGTGGTTGGTTGGTTTGT |
| GAPDH forward |
CCTTCCGTGTCCCCACT |
| GAPDH reverse |
GCCTGCTTCACCACCTTC |
| U6 forward |
CTCGCTTCGGCAGCACA |
| U6 reverse |
AACGCTTCACGAATTTGCGT |
| miR-142-3p
mimic |
UGUAGUGUUUCCUACUUUAUGGA |
| NC mimics |
UGUAACGUACGUUCGUACCGUGA |
In vitro cell proliferation assay
For Cell Counting Kit (CCK)-8 assay, the cells were
collected and incubated with CCK-8 kit (Dojindo Molecular
Technologies, Inc.) for 2 h, and the absorbance (OD value) at 450
nm was then determined to assess cell viability. For EdU assay, the
cells were stained using the EdU cell proliferation kit (Guangzhou
RiboBio Co., Ltd.) according to the manufacturer's instructions,
and results were captured using a fluorescence microscope.
Apoptosis
The Annexin V-FITC/PI apoptosis detection kit
(Nanjing KeyGen Biotech Co., Ltd.) was used to assess apoptosis
rate through BD FACSCelesta™ (BD Biosciences), and the total
apoptosis (early + late) rate were analyzed using FlowJo software
v.10 (Flow Jo, LLC).
Transwell assay
In brief, for cell migration assay, RPMI-1640 media
supplemented with 20% FBS (Gibco; Thermo Fisher Scientific, Inc.)
was added into the lower chamber, while 40,000 cells were suspended
in RPMI-1640 media without FBS and added into the upper chamber.
After 48 h, the cells trapped on the Transwell chamber membrane
were fixed with 4% methanol for 10 min, stained with crystal violet
for 10 min at room temperature, and finally captured with a light
microscope. However, for cell invasion assay, the filter membranes
were precoated with Matrigel (Corning, Inc.) at 37°C for 30 min,
and other steps were performed as described in the migration
assay.
Prediction of downstream molecules
regulated by circRNA-100395
A publicly available bioinformatic database Starbase
v.2.0 (15) was utilized to predict
the downstream microRNAs of circRNA-100395.
Dual-luciferase reporter assays
The test (293T) cells were co-transfected with
either the pmirGLO-circRNA_100395-wild-type or
pmirGLO-circRNA_100395-mutant plasmid along with either miR-142-3p
mimics or miR-142-3p NCs (all Shanghai GenePharma Co., Ltd.) using
Lipofectamine® 3000 Transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). A dual-luciferase reporter system
(Promega Corporation) was used 48 h later to measure the luciferase
gene activity. Firefly luciferase activity was normalized to that
of Renilla luciferase. The related sequences are shown in
Table I.
Western blotting
The transfected HGC-27 and AGS cells were collected
and lysed with RIPA buffer (Beyotime Institute of Biotechnology).
Subsequently, 40 µg/lane of proteins were loaded and resolved using
10% SDS-PAGE and transferred onto polyvinylidene fluoride
membranes. Then, the membranes were blocked with 10% BSA (Beijing
Solarbio Science & Technology Co., Ltd.) for 1 h at room
temperature. Subsequently, the membranes were incubated with
primary antibodies: PTEN (cat. no. ab267787), PI3K (cat. no.
ab154598), AKT (cat. no. ab8805), p-AKT (cat. no. ab250676) and
GAPDH (cat. no. ab9485; all 1:1,000 dilution; Abcam) overnight at
4°C. Subsequently, proteins were incubated with the goat
anti-rabbit IgG H&L (HRP; 1:2,000 dilution; cat. no. ab6721;
Abcam) for 1 h at room temperature. The membranes were washed with
0.05% Tween-20 (TBS/Tween) 3 times. Finally, the protein bands were
then analyzed with super sensitive ECL luminescence reagent (Dalian
Meilun Biology Technology Co., Ltd.). The proteins expression
levels were detected using a chemi-luminescence detection system
with Quantity One software v.3.0 (Sigma-Aldrich; Merck KGaA).
Statistical analysis
Results are shown as mean ± standard deviation. All
statistical analyses were performed using SPSS 17.0 (SPSS Inc.) and
GraphPad Prism 7.0 (GraphPad Software Inc.). Comparisons between
two groups were performed using unpaired Student's t-tests and χ2
tests. The data among data among three or more groups were analyzed
using one-way ANOVA followed by Fisher's least significant
difference post hoc test or Tukey's multiple comparisons test. The
Kaplan-Meier curve was generated and the log-rank tests were
performed using GraphPad Prism software. All experiments were
repeated at least three times. P<0.05 was considered to indicate
a statistically significant difference.
Results
circRNA_100395 is downregulated in GC
tissues
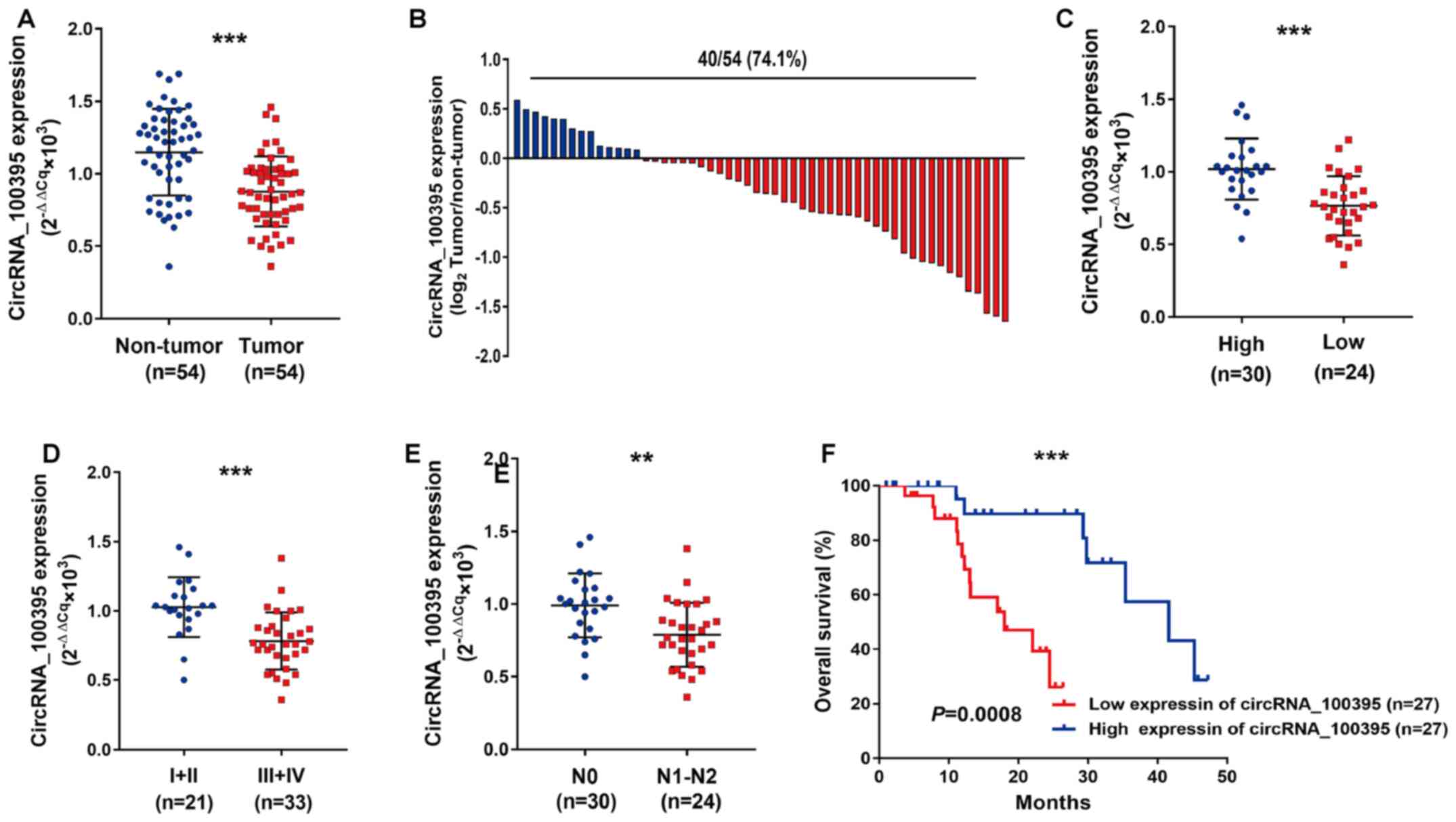
As shown in Fig. 1A,
circRNA_100395 expression was downregulated in GC tissues compared
with paired normal samples. The GC samples with low expression of
circRNA_100395 accounted for 74.1% (40/54) (Fig. 1B). Statistical analysis revealed that
circRNA_100395 expression was significantly associated with
differential status (P<0.001), vascular invasion (P=0.001),
tumor size (P=0.001), N stage (P=0.003) and TNM stage (P=0.031)
(Table II). Similarly, the patients
with GC with low expression of circRNA_100395 showed poor tumor
differentiation (P<0.0011, Fig.
1C), advanced TMN stage (P=0.0001; Fig. 1D) and lymph node metastasis
(P=0.0015, Fig. 1E), suggesting that
circRNA_100395 may play a suppressor role in the development of GC.
Moreover, the Kaplan-Meier curve showed that patients with low
expression of circRNA_100395 had shorter survival times compared
with patients with high expression of circRNA_100395 (Fig. 1F). Therefore, circRNA_100395 might be
considered as a tumor suppressor in GC progression.
 | Table II.Association between the
clinicopathological data and circ_100395 expression in gastric
cancer (n=54). |
Table II.
Association between the
clinicopathological data and circ_100395 expression in gastric
cancer (n=54).
|
|
| circ_100395
expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Value, n | High | Low | P-value |
|---|
| Sex |
|
Male | 29 | 14 | 15 | 0.785 |
|
Female | 25 | 13 | 12 |
|
| Age, years |
|
<60 | 21 | 10 | 11 | 0.876 |
|
≥60 | 33 | 15 | 18 |
|
| Differential
status |
|
Moderate/well | 19 | 13 | 6 | <0.001 |
|
Undifferentiated/poorly | 35 | 7 | 28 |
|
| Vascular
invasion |
|
Negative | 22 | 14 | 8 | 0.001 |
|
Positive | 32 | 6 | 26 |
|
| Tumor size, cm |
| ≤5 | 25 | 16 | 9 | 0.001 |
|
>5 | 29 | 6 | 23 |
|
| N stage |
| N0 | 24 | 17 | 7 | 0.003 |
|
N1-N3 | 30 | 9 | 21 |
|
| TNM stage |
|
I–II | 23 | 15 | 8 | 0.031 |
|
III–IV | 31 | 11 | 20 |
|
Upregulation of circRNA_100395
inhibits GC cell proliferation
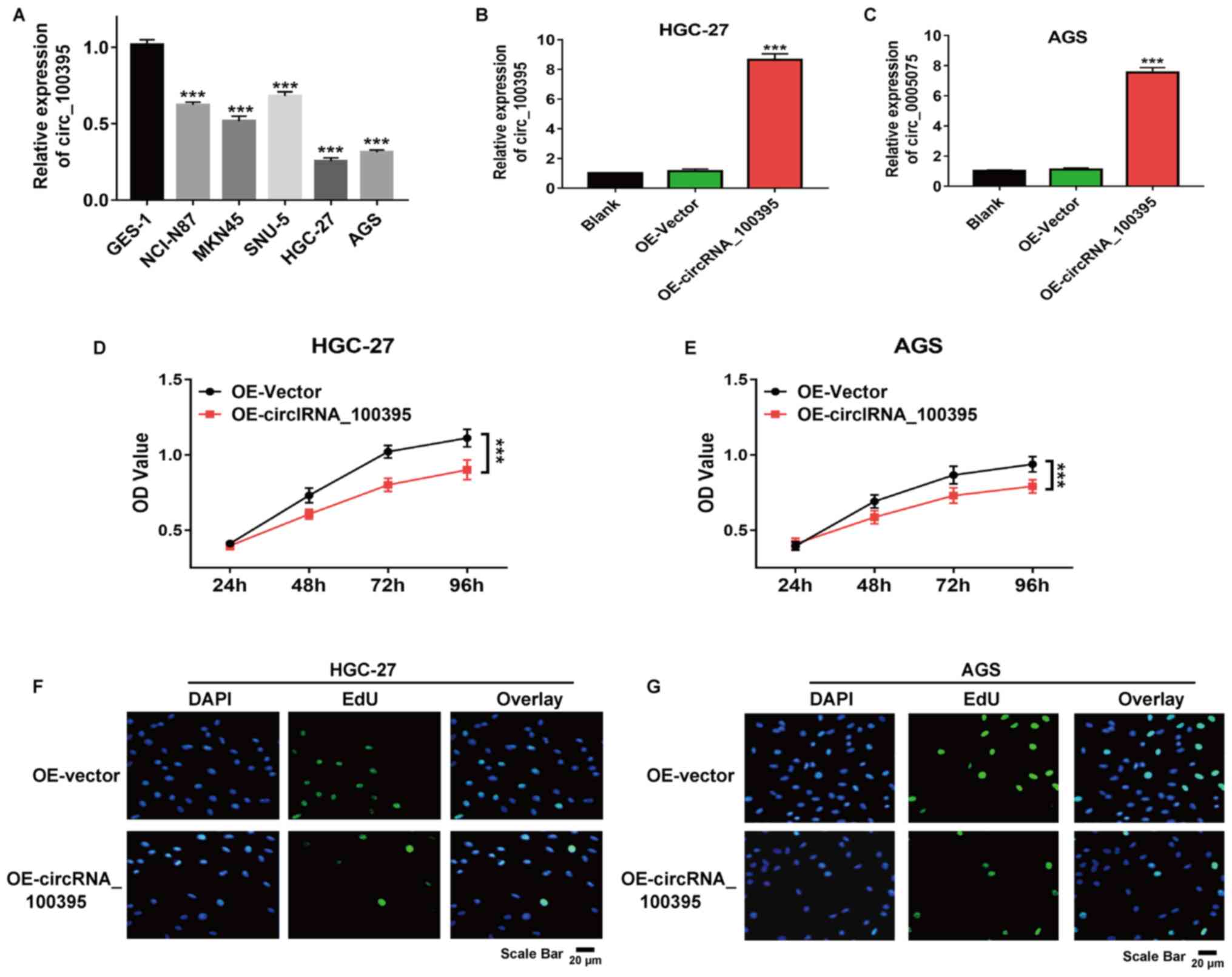
As shown in Fig. 2A,
circRNA_100395 was downregulated in GC cell lines compared with
normal epithelial cells (GES-1). HGC-27 and AGS cells showed the
lowest expression and so were selected for further experiments. The
results of RT-qPCR revealed that after transfection with
pcDNA3.1-circRNA_100395 plasmids, the circRNA_100395 expression in
HGC-27 and AGS cells were significantly increased compared with the
control group, suggesting the successful construction of the
circRNA_100395-overexpressed GC cell models (Fig. 2B and C). Besides, the CCK-8 assay
revealed that the upregulation of circRNA_100395 significantly
suppressed HGC-27 and AGS cell proliferation (both P<0.001;
Fig. 2D and E). Similarly, the EdU
incorporation assay also demonstrated that there were fewer
EdU-positive HGC-27 and AGS cells in the OE-circRNA_100395 group
compared with the OE-vector group (Fig.
2F and G). Therefore, circRNA_100395-overexpression could
significantly suppress cell proliferation in GC.
Upregulation of circRNA_100395
promotes apoptosis and suppresses cell migration and invasion
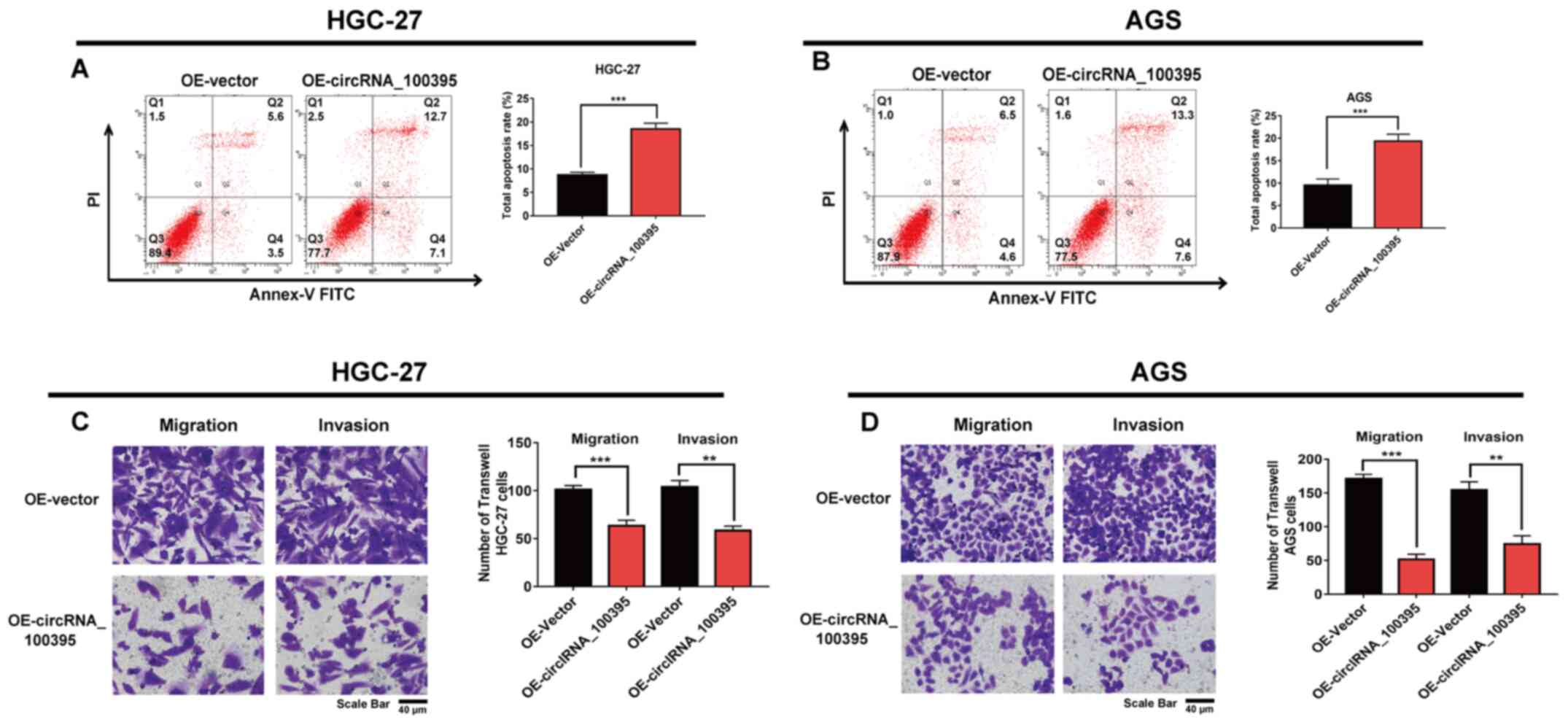
Herein, overexpression of circRNA_100395 was found
to significantly inhibit the GC cell proliferation rate. However,
it was not clear whether apoptosis contributed to the proliferation
inhibition caused by circRNA_100395-overexpression. Therefore, flow
cytometry experiments were performed to evaluate the apoptosis rate
induced by circRNA_100395-overexpression. The findings showed that
the total apoptosis rate of HGC-27 cells in the
circRNA_100395-overexpression group was higher compared with the
OE-vector group (P<0.001; Fig.
3A). Similarly, upregulating circRNA_100395 expression could
induce more apoptosis in AGS cells compared with in the OE-vector
group (Fig. 3B).
Statistical analysis revealed that circRNA_100395
expression in GC was associated with lymph node metastasis.
Therefore, a Transwell assay was performed to confirm whether
circRNA_100395 participated in regulating cell migration and
invasion in GC. Fig. 3C shows that
cell migration and invasion decreased after upregulating
circRNA_100395 expression in HGC-27 cells compared with the
OE-vector group. Similarly, Fig. 3D
also demonstrated that fewer migratory and invasive AGS cells were
observed after circRNA_100395-overexpression. Taken together, the
upregulation of circRNA_100395 could inhibit cell proliferation,
increase the apoptosis rate and suppress GC cell invasion and
migration.
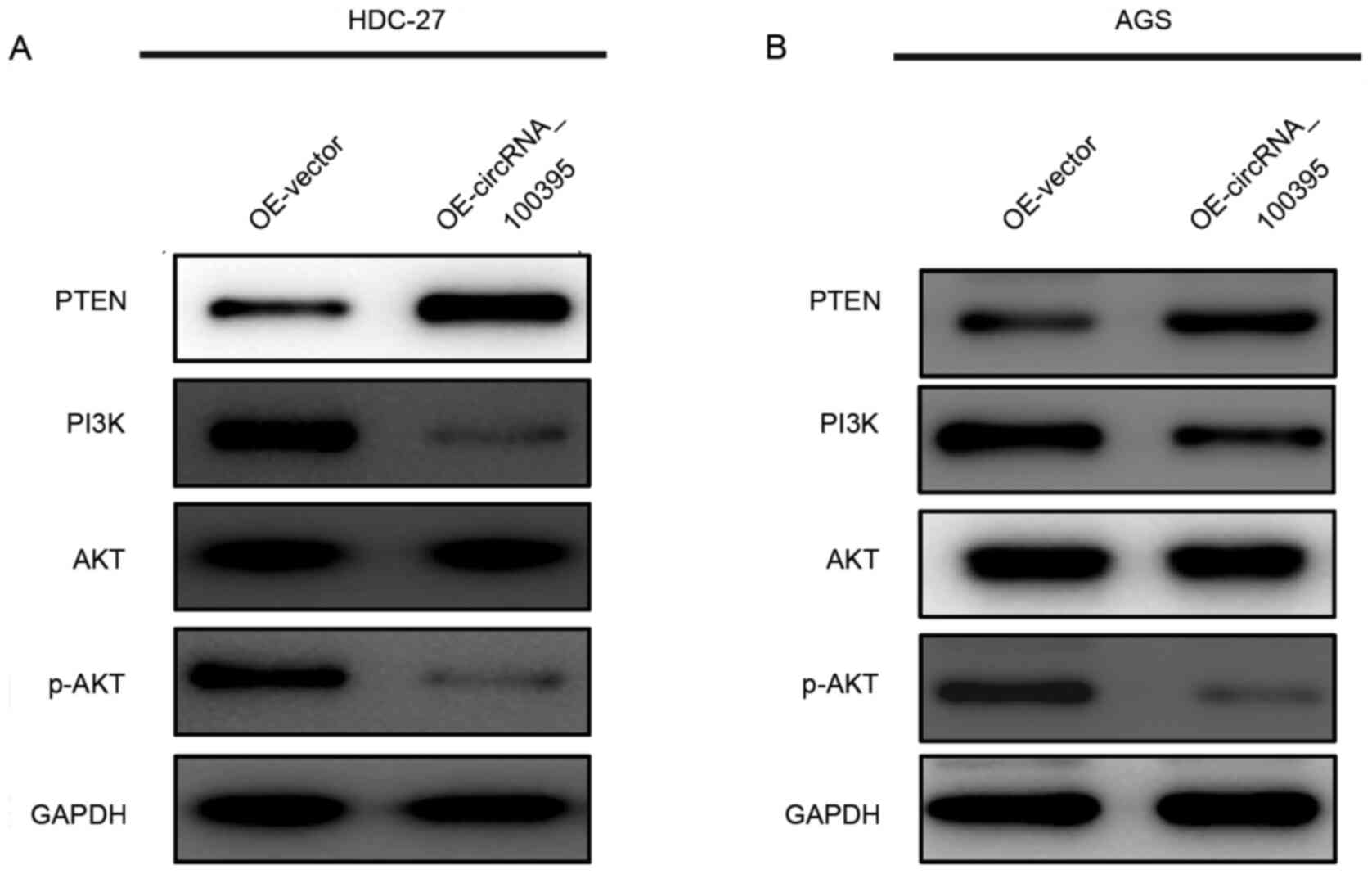
Antitumor effects of circRNA_100395 by
regulating the PTEN-PI3K/AKT signaling pathway
The PI3K/AKT signaling pathway serves an important
role in regulating cell proliferation, migration and invasion
(16–18). Therefore, in the present study, the
western blotting assay was performed to determine whether
circRNA_100395 function as a tumor inhibitor via regulating this
signaling pathway. As shown in Fig. 4A
and B, the upregulation of circRNA_100395 could lead to
increased expression of PTEN protein, accompanied with the
decreased expression of PI3K and phosphorylated (p-)AKT. Therefore,
these results provided evidence that circRNA_100395 might promote
apoptosis and suppress cell proliferation in GC by interfering with
the PTEN-PI3K/AKT signaling pathway.
miR-142-3p is a downstream regulatory
gene of circRNA_100395 in GC
The prediction results of the public database
(Starbase v.2) showed that miR-142-3p might be the downstream
target gene of circRNA_100395, and possible binding sites between
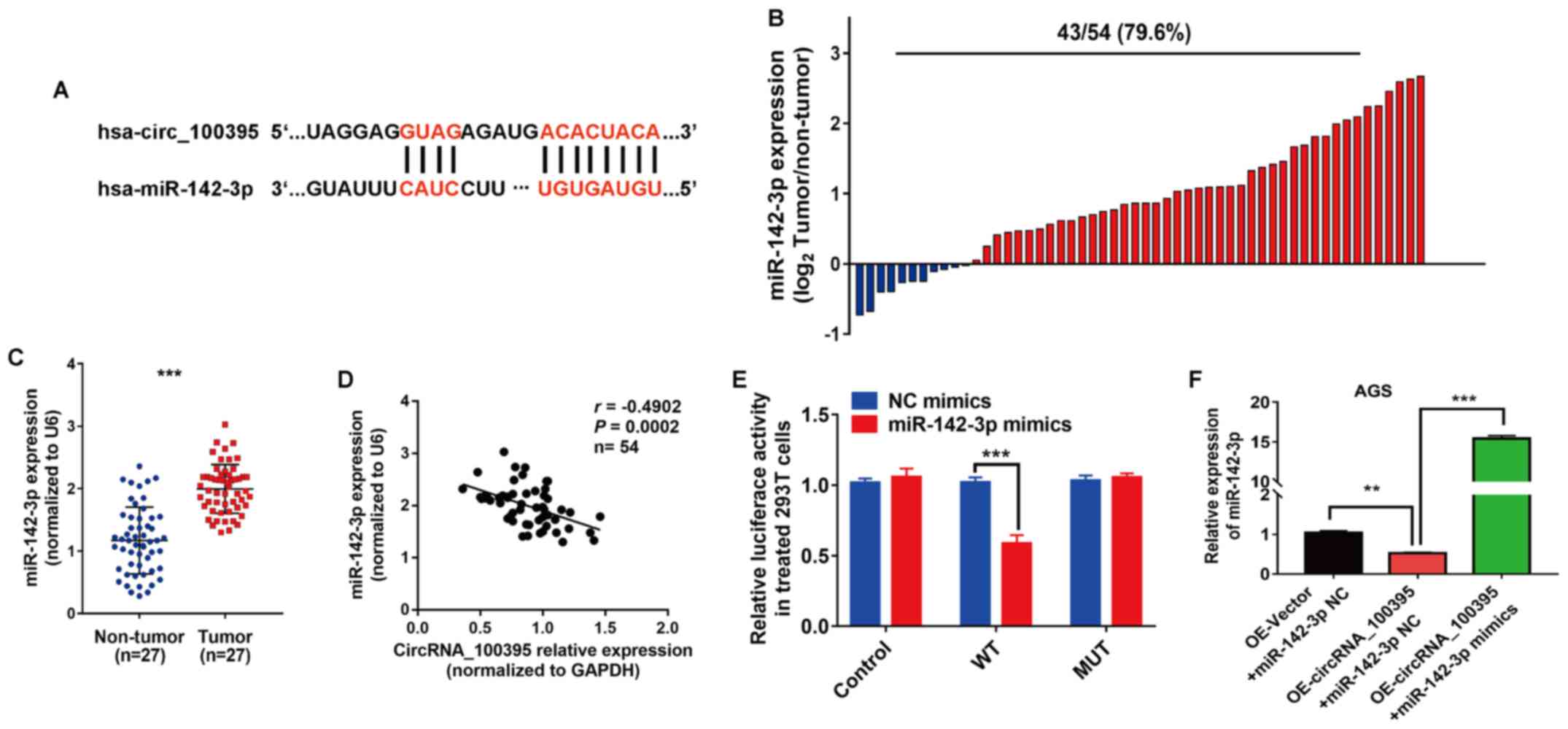
miR-142-3p and circRNA_100395 are shown in Fig. 5A. Subsequently, RT-qPCR was used to
detect miR-142-3p expression in GC tissues and cell lines. The
results revealed that miR-142-3p overexpression was found in 79.6%
(43/54) of the GC tissues (Fig. 5B).
Statistical analysis showed that miR-142-3p was significantly
upregulated in GC compared with non-tumor tissue samples
(P<0.001; Fig. 5C). Moreover,
Pearson's correlation analysis showed that circRNA_100395
expression was negatively correlated with miR-142-3p expression in
GC (r=−0.4902, P=0.0002; Fig. 5D).
Similarly, the results of dual-luciferase reporter assays revealed
that the luciferase activity was significantly lower in wild-type
cells transfected with miR-142-3p mimics compared with NC mimics
(P<0.001; Fig. 5E). RT-qPCR
showed that the expression of miR-142-3p in
OE-circRNA_100395-overexpressing AGS cells was significantly
inhibited compared with the OE-Vector + miR142-3p NC group
(P<0.01), while the addition of miR-142-3p mimics increased
miR-142-3p expression compared with the
OE-circRNA_100395+miR-142-3p NC group (P<0.001) (Fig. 5F). Therefore, miR-142-3p was found to
be a downstream gene of circRNA_100395 in GC.
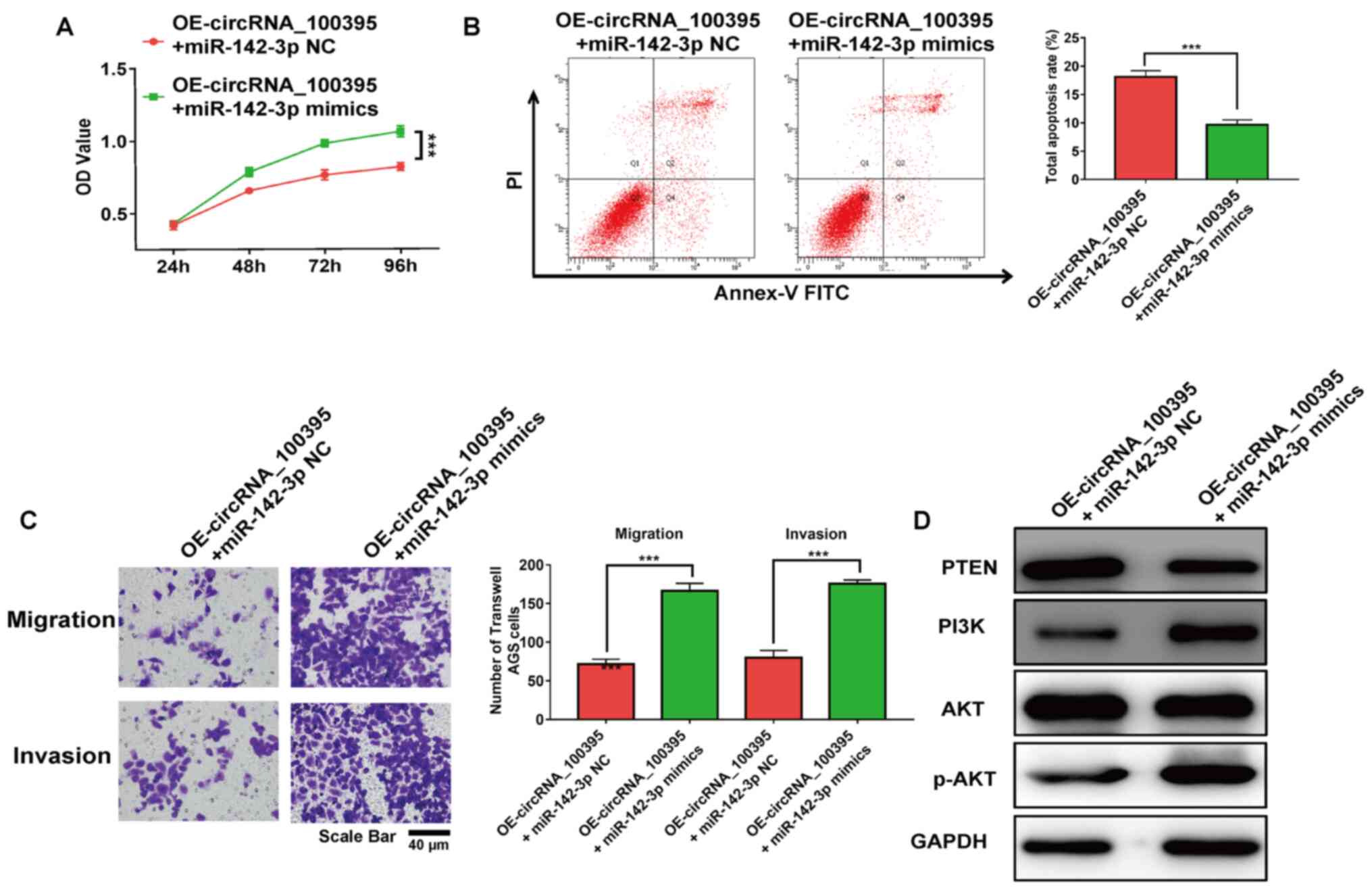
miR-142-3p rescues the antitumor
effects induced by circRNA_100395 overexpression
The results of the RT-qPCR assay revealed that the
expression level of miR-142-3p in circRNA_100395-overexpressed
cells was up-regulated with the transfection of miR-142-3p mimics.
It was reported that the addition of miR-142-3p significantly
restored the cell proliferation and apoptosis rate (both
P<0.001; Fig. 6A and B). Besides,
the addition of miR-142-3p significantly increased the migratory
and invasive AGS cell number with OE-circRNA_100395 treatment
(P<0.001; Fig. 6C). Moreover, the
PTEN-PI3K/AKT signaling pathway-related markers in GC cells were
evaluated after the upregulation of miR-142-3p expression. As shown
in Fig. 6D, the addition of
miR-142-3p reduced the expression of PTEN protein compared with the
control group. However, the expression of PI3K and p-AKT were
increased in circRNA_100395-overexpressed GC cells. Therefore,
these results showed that downregulation of circRNA_100395 could
promote apoptosis and suppress cell proliferation in GC by
regulating the miR-142-3p/PI3K/AKT axis.
Discussion
GC is one of the deadly malignant tumors globally,
causing an estimated 783,000 deaths in 2018 (2). Currently, the major therapeutic
strategies for patients with GC include surgery, chemotherapy and
molecular targeting therapy (19–21).
Nevertheless, most patients with GC are diagnosed with advanced GC,
hence their overall survival time is shorter (22). Therefore, it is important to identify
effective diagnostic and therapeutic biomarkers for early diagnosis
and treatment of GC and elucidate the possible regulatory
mechanisms of pathogenesis and progression.
Numerous studies have demonstrated that circRNAs
could serve as regulatory factors in GC progression (23–25).
Over the years, significant efforts have been made to elucidate the
regulatory function of circRNA_100395 (11–13,26).
Chen et al (13) demonstrated
that overexpression of circRNA_100395 could sponge miR-1228 and
regulate target transcription factor 21 expression, which inhibits
the cell proliferation rate in lung cancer. Another study analyzed
microarray data of 18 thyroid samples and identified circRNA_100395
as a promising biomarker for papillary thyroid carcinoma (26). However, it is still not clear whether
circRNA_100395 is involved in GC progression. In the present study,
RT-qPCR results showed that circRNA_100395 was upregulated in GC
tissues. Statistical analysis showed that patients with GC with low
expression of circRNA_100395 were more likely to have advanced TNM
stage, advanced N stage, larger tumor size, poor differential
status and shorter survival time. Therefore, circRNA_100395 might
serve as a tumor inhibitor in GC.
To confirm the biological role of circRNA_100395 in
GC development, a plasmid transfection assay was performed to
construct circRNA_100395-overexpressed GC cells. CCK-8 and EdU
assay results showed that circRNA_100395-overexpression
significantly inhibited the proliferation ability of GC cells.
Besides, the upregulation of circRNA_100395 induced apoptosis.
Since the expression of circRNA_100395 was closely associated with
advanced N stage, cell invasion and migration were analyzed using a
Transwell assay. The results revealed that the cell invasion and
migration with OE-circRNA_100395 was significantly inhibited
compared with the control group. Therefore, circRNA_100395 was
demonstrated to serve a significant role in regulating GC
progression, including increasing the apoptosis rate, inhibiting
cell proliferation, migration and invasion. However, the underlying
molecular mechanisms are not clear.
The PI3K/AKT signaling pathway has been identified
to play an important role in the progression of various cancer
types, including GC (27–29). Once the PI3K/AKT signaling pathway is
activated, phosphorylation of AKT directly regulates the expression
of a range of proteins involved in cell metabolism, proliferation,
motility, invasion and migration. Studies have also revealed that
circRNAs are significant regulatory factors in activating or
silencing the PI3K/AKT signaling pathway (30,31).
Peng et al (26) reported
that circRNA_0010882 regulates the PI3K/AKT signaling pathway, and
promotes cell proliferation in GC (32). Besides, circRNA_LARP4 has been
identified as an miR-1323 sponge and tumor inhibitor in esophageal
squamous cell carcinoma and regulates the PI3K/AKT signaling
pathway (33). Consistently, the
present results indicated that the upregulation of circRNA_100395
increased the expression of PTEN, and decreased the expression of
PI3K and p-AKT. Taken together, in the present study,
circRNA_100395 was proposed to exert antitumor effects by
suppressing the PI3K/AKT signaling pathway.
miRNA sponging is a critical function of circRNAs in
regulating cancer development (34).
Li et al (29) revealed that
the silencing of circRNA_ZNF609 suppressed cell proliferation by
regulating miR-188 expression. Another study indicated that
circRNA_CCDC66 regulated cisplatin resistance in GC via the
miR-618/BCL2 axis (35). Based on
the results of the present bioinformatics analysis, miR-142-3p was
proposed as a downstream target gene of circRNA_100395. Besides,
miR-142-3p was found to be upregulated in GC tissues, and the
expression of miR-142-3p was negatively correlated with
circRNA_100395 expression. Moreover, the relationship between the
circRNA_100395 and miR-142-3p was confirmed by dual-luciferase
reporter assays. Numerous studies have identified that miR-142-3p
plays a significant role in the progression of various cancer types
(36,37). For example, Wang et al
(33) provided evidence to show that
miR-142-3p could act as a tumor suppressor in GC progression. In
addition, miR-142-3p has been confirmed to promote prostate cancer
progression by targeting the forkhead box protein O1 pathway
(38). In the present study, the
addition of miR-142-3p rescued the antitumor effects induced by
overexpression of circRNA_100395. Therefore, these findings
suggested that circRNA_100395 acts as a tumor suppressor by
regulating the miR-143-3p/PI3K/AKT signaling pathway. However, the
present study has some limitations. Firstly, the molecules acting
downstream of the circRNA_100395/miR-142-3p axis need further
confirm via bioinformatics analysis. Secondly, in vivo
experiments are needed to further corroborate the present findings.
That is, the untransfected and transfected cells were collected and
planted in the nude mice, followed by recorded the tumor volume and
weight during an observation period.
circRNA_100395 can serve as a tumor inhibitor in GC,
and its low expression is associated with poor prognosis in
patients with GC. Besides, the upregulation of circRNA_100395
significantly inhibits cell proliferation by regulating the
miR-142-3p/PI3K/AKT axis. Overall, circRNA_100395 may be a
potential therapeutic biomarker for GC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZC and GL designed the present study. ZC, GL, CH and
XZ performed all the experiments, analyzed the data and prepared
the figures. ZC, GL and CH drafted the initial manuscript. ZC and
XZ reviewed and revised the manuscript. ZC, GL and CH confirmed the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was sought from the
participants before the samples were obtained. Authority to carry
out the present study was sought from the Ethical Committees of
Taizhou People's Hospital (Taizhou, China) (approval no.
KY2020-06511).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu W, Yang Z and Lu N: Molecular targeted
therapy for the treatment of gastric cancer. J Exp Clin Cancer Res.
35:12016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dolcetti R, De Re V and Canzonieri V:
Immunotherapy for gastric cancer: Time for a personalized approach?
Int J Mol Sci. 19:E16022018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang
Y, Li X, Wu Z, Yang D, Zhou Y, et al: Circular RNAs in Cancer:
Emerging functions in hallmarks, stemness, resistance and roles as
potential biomarkers. Mol Cancer. 18:902019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheng J, Zhuo H, Xu M, Wang L, Xu H, Peng
J, Hou J, Lin L and Cai J: Regulatory network of circRNA-miRNA-mRNA
contributes to the histological classification and disease
progression in gastric cancer. J Transl Med. 16:2162018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu J, Yang M, Zhou B, Luo J, Zhang Z,
Zhang W and Yan Z: CircRNA-104718 acts as competing endogenous RNA
and promotes hepatocellular carcinoma progression through
microRNA-218-5p/TXNDC5 signaling pathway. Clin Science (Lond.).
133:1487–1503. 2019. View Article : Google Scholar
|
|
10
|
Luan W, Shi Y, Zhou Z, Xia Y and Wang J:
circRNA_0084043 promote malignant melanoma progression via
miR-153-3p/Snail axis. Biochem Biophys Res Commun. 502:22–29. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li X, Lin S, Mo Z, Jiang J, Tang H, Wu C
and Song J: CircRNA_100395 inhibits cell proliferation and
metastasis in ovarian cancer via regulating
miR-1228/p53/epithelial-mesenchymal transition (EMT) axis. J
Cancer. 11:599–609. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen Q, Chen Z, Cao S, Guo B, Chen Y, Feng
Z, Wang J, Guo G, Chen X and Huang X: Role of CircRNAs_100395 in
proliferation and metastases of liver cancer. Med Sci Monit.
25:6181–6192. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen D, Ma W, Ke Z and Xie F: CircRNA
hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung
cancer progression. Cell Cycle. 17:2080–2090. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Feng W, Li B, Wang J, Zhang H, Liu Y, Xu
D, Cheng K and Zhuang J: Long Non-coding RNA LINC00115 contributes
to the progression of colorectal cancer by targeting miR-489-3p via
the PI3K/AKT/mTOR pathway. Front Genet. 11:5676302020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu M, Zhu S, Xiong S, Xue X and Zhou X:
MicroRNAs and the PTEN/PI3K/Akt pathway in gastric cancer (Review).
Oncol Rep. 41:1439–1454. 2019.PubMed/NCBI
|
|
17
|
Huang Y, Zhang J, Hou L, Wang G, Liu H,
Zhang R, Chen X and Zhu J: LncRNA AK023391 promotes tumorigenesis
and invasion of gastric cancer through activation of the PI3K/Akt
signaling pathway. J Exp Clin Cancer Res. 36:1942017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu L, Chen J, Jia L, Chen X, Awaleh Moumin
F and Cai J: SLC1A3 promotes gastric cancer progression via the
PI3K/AKT signalling pathway. J Cell Mol Med. 24:14392–14404. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu J, Huang C, Sun Y, Su X, Cao H, Hu J,
Wang K, Suo J, Tao K, He X, et al Chinese laparoscopic
gastrointestinal surgery study (CLASS) group, : Effect of
laparoscopic vs. open distal gastrectomy on 3-year disease-free
survival in patients with locally advanced gastric cancer: The
CLASS-01 randomized clinical trial. JAMA. 321:1983–1992. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song Z, Wu Y, Yang J, Yang D and Fang X:
Progress in the treatment of advanced gastric cancer. Tumour Biol.
Jul 3–2017.doi: org/10.1177/1010428317714626. View Article : Google Scholar
|
|
21
|
Ilson DH: Advances in the treatment of
gastric cancer. Curr Opin Gastroenterol. 34:465–468. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu Y, Huang C, Sun Y, Su X, Cao H, Hu J,
Xue Y, Suo J, Tao K, He X, et al: Morbidity and mortality of
laparoscopic versus open D2 distal gastrectomy for advanced gastric
cancer: A Randomized Controlled Trial. J Clin Oncol. 34:1350–1357.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tang W, Fu K, Sun H, Rong D, Wang H and
Cao H: CircRNA microarray profiling identifies a novel circulating
biomarker for detection of gastric cancer. Mol Cancer. 17:1372018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gu W, Sun Y, Zheng X, Ma J, Hu XY, Gao T
and Hu MJ: Identification of gastric cancer-related circular RNA
through microarray analysis and bioinformatics analysis. BioMed Res
Int. 2018:23816802018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Peng N, Shi L, Zhang Q, Hu Y, Wang N and
Ye H: Microarray profiling of circular RNAs in human papillary
thyroid carcinoma. PLoS One. 12:e01702872017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qian Y, Yan Y, Lu H, Zhou T, Lv M, Fang C,
Hou J, Li W, Chen X, Sun H, et al: Celastrus orbiculatus extracts
inhibit the metastasis through attenuating PI3K/Akt/mTOR signaling
pathway in human gastric cancer. Anticancer Agents Med Chem.
19:1754–1761. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang H, Pan YZ, Cheung M, Cao M, Yu C,
Chen L, Zhan L, He ZW and Sun CY: LAMB3 mediates apoptotic,
proliferative, invasive, and metastatic behaviors in pancreatic
cancer by regulating the PI3K/Akt signaling pathway. Cell Death
Dis. 10:2302019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li M, Li Y and Yu M: CircRNA ZNF609
Knockdown Suppresses Cell Growth via Modulating miR-188/ELF2 Axis
in Nasopharyngeal Carcinoma. Onco Targets Ther. 13:2399–2409. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xin J, Zhang XY, Sun DK, Tian LQ and Xu P:
Up-regulated circular RNA hsa_circ_0067934 contributes to
glioblastoma progression through activating PI3K-AKT pathway. Eur
Rev Med Pharmacol Sci. 23:3447–3454. 2019.PubMed/NCBI
|
|
31
|
Chen T, Shao S, Li W, Liu Y and Cao Y: The
circular RNA hsa-circ-0072309 plays anti-tumour roles by sponging
miR-100 through the deactivation of PI3K/AKT and mTOR pathways in
the renal carcinoma cell lines. Artif Cells Nanomed Biotechnol.
47:3638–3648. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Peng YK, Pu K, Su HX, Zhang J, Zheng Y, Ji
R, Guo QH, Wang YP, Guan QL and Zhou YN: Circular RNA
hsa_circ_0010882 promotes the progression of gastric cancer via
regulation of the PI3K/Akt/mTOR signaling pathway. Eur Rev Med
Pharmacol Sci. 24:1142–1151. 2020.PubMed/NCBI
|
|
33
|
Wang Y, Cao Z, Wang L, Liu S and Cai J:
Downregulation of microRNA-142-3p and its tumor suppressor role in
gastric cancer. Oncol Lett. 15:8172–8180. 2018.PubMed/NCBI
|
|
34
|
Ebert MS and Sharp PA: MicroRNA sponges:
Progress and possibilities. RNA. 16:2043–2050. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Q, Miao Y, Fu Q, Hu H, Chen H, Zeng
A, Jin Y, Jiang Y, Qian L, Wu L, et al: CircRNACCDC66 regulates
cisplatin resistance in gastric cancer via the miR-618/BCL2 axis.
Biochem Biophys Res Commun. 526:713–720. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Troschel FM, Böhly N, Borrmann K, Braun T,
Schwickert A, Kiesel L, Eich HT, Götte M and Greve B: miR-142-3p
attenuates breast cancer stem cell characteristics and decreases
radioresistance in vitro. Tumour Biol. Aug 9–2018.(Epub ahead of
print). doi: 10.1177/1010428318791887. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhu X, Ma SP, Yang D, Liu Y, Wang YP, Lin
T, Li Y, Yang S, Zhang W and Wang X: miR-142-3p suppresses cell
growth by targeting CDK4 in colorectal cancer. Cell Physiol
Biochem. 51:1969–1981. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tan YF, Chen ZY, Wang L, Wang M and Liu
XH: MiR-142-3p functions as an oncogene in prostate cancer by
targeting FOXO1. J Cancer. 11:1614–1624. 2020. View Article : Google Scholar : PubMed/NCBI
|