Introduction
Lung cancer is the leading cause of
cancer-associated death among men, with a 24% mortality rate, and
women, with a 23% mortality rate, in the United States (1). Based on its histology, lung
adenocarcinoma (LUAD) is classified into two main forms: Non-small
cell lung carcinoma (NSCLC) and small cell lung carcinoma, which
comprise ~85 and ~15% of all cases, respectively (2). NSCLC is further divided into three
subtypes: Squamous-cell carcinoma, adenocarcinoma and large cell
carcinoma. LUAD is the most common type of lung cancer,
representing ~40% of all lung cancer types (3). LUAD may occur due to mutations in a
variety of genes, including EGFR, ALK receptor tyrosine kinase,
KRAS, ROS proto-oncogene 1, receptor tyrosine kinase, BRAF, erb-b2
receptor tyrosine kinase 2, MET proto-oncogene, receptor tyrosine
kinase and ret proto-oncogene (4).
At present, drugs targeting mutant genes, such as EGFR (5) and KRAS (6), associated with LUAD have been
developed. However, LUAD is still one of the most aggressive and
lethal tumor types, with a 5-year survival rate <5% (7). Therefore, it is important to identify
novel target genes to improve the prognosis of patients with
LUAD.
Complement factor B (CFB), localized to the major
histocompatibility complex class III region on chromosome 6, serves
a pivotal role in the alternative pathway of the complement system
(8) and exerts a key role in
labelling target particles, resulting from the effective clearance
of the target (9). CFB is cleaved
into two fragments, non-catalytic chain Ba and the catalytic
subunit Bb, by complement factor D; the generated component 3b
(C3b) binds Bb and properdin, resulting in the formation of the C3
convertase (C3bBb) during the activation process of the alternative
pathway (10). By binding of
additional C3b to the alternative pathway, C3bBb renders it able to
cleave C5, and induces the amplification loop of the alternative
pathway (8,10). It has been reported that low
expression levels of CFB are associated with decreased expression
levels of C3bBb and facilitate the degradation of the membrane
attack complex (MAC), which leads to the inhibition of the
alternative pathway of complement activation, thereby reducing the
activation efficiency of the complement system in the whole body
(8). During the regulation of the
complex tumor microenvironment, complement proteins serve a dual
role in the tumor microenvironment, which will eventually affect
tumor progression (9). The
dysregulation of complement activation pathways serves an important
role in tumor progression (10–12).
A previous study reported that CFB is a candidate
biomarker for pancreatic cancer diagnosis (13). Furthermore, it has been revealed that
CFB expression is upregulated in patients with preeclampsia
compared with in those with normotensive pregnancies (14). CFB has also been reported to serve a
central role in mediating ultraviolet-induced immunosuppression
(15). Previous data have indicated
that CFB mRNA expression in the colonic mucosa is upregulated in
the lower parts of the crypts compared with that observed on the
luminal surface (16). Furthermore,
a previous study has identified a strong association of CFB with
molecular subtypes of breast cancer, including the luminal-A (LA),
luminal-B, triple-negative and HER2-positive subtypes (17).
As a key immunomodulatory factor in the progression
of lung cancer, complement activation induces escape from
immunosurveillance via the C3/C5-dependent signaling pathway, which
is activated by classical signaling pathways (10), thereby affecting the occurrence and
development of lung cancer (18).
Therefore, the present study aimed to determine the role of CFB in
LUAD using data from The Cancer Genome Atlas (TCGA). The present
analysis provided further evidence regarding the use of CFB as a
potential biomarker for LUAD, and CFB may become an attractive
candidate biomarker and potential prognostic target for LUAD.
Materials and methods
Data collection
RNA-sequencing (seq) expression (combining level-3
data from Illumina GA and Hi-Seq platforms) and clinical data for
patients with LUAD were downloaded from TCGA data portal
(http://cancergenome.nih.gov/). The
TCGA-LUAD dataset contained data on 514 patients with LUAD for whom
detailed clinical information was available. RNA-seq by Expectation
Maximization expression values were used for statistical
analysis.
Clinical feature analysis
LUAD samples were divided into two groups based on
the median CFB expression value of 3.64. All statistical analyses
were performed using R statistical software (version 3.4.1;
http://www.r-project.org/). The means of
the continuous variables in these two groups (high-CFB group and
low-CFB group) were compared using an independent sample t-test,
and the prevalence of categorical variables was compared using the
χ2 test. Due to the small sample size of the groups for some
variables, the comparisons were performed using Pearson's χ2 test
with Yates' continuity correction and Fisher's exact test. Violin
plots were used to visualize expression level differences for
discrete variables, such as sex, smoking history, residual tumor
and stage. Univariate logistic regression was used to investigate
the association between CFB expression (categorical) and clinical
features.
Survival analysis
Differences in overall survival and disease-free
survival between the high-CFB group and the low-CFB group were
compared using Kaplan-Meier curves, with P-values calculated via
the log-rank test using the ‘survival’ package (https://cran.r-project.org/web/packages/survival/index.html;
version 3.2–7) in R. Univariate Cox regression analysis was used to
estimate the independent effects of CFB expression and other
clinical features, including age, sex, ethnicity, smoking history,
residual tumor classification, Eastern Cooperative Oncology Group
(ECOG) score (19), Karnofsky
performance score (20) and TNM
stage (21), on overall survival and
disease-free survival. The independent effect of CFB expression on
overall survival and disease-free survival was evaluated via
multivariate Cox analysis with adjusted covariates. For the
sub-group analysis, patients were divided into three or four groups
according to the tercile or quartile expression of CFB.
Gene set enrichment analysis
(GSEA)
The JavaGSEA desktop application v3.0 was used to
perform GSEA (22) of Kyoto
Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) pathways for the following
comparisons: High-CFB vs. control, low-CFB vs. control and high-CFB
vs. low-CFB. Gene sets with <10 genes or >500 genes were
excluded. The t-statistic mean of the genes was computed for each
metabolic pathway using a permutation test with 1,000 replications.
Up- and downregulated metabolic pathways were defined as having a
normalized enrichment score (NES) >0 or <0 for patients
compared with controls, respectively. Enrichment analysis of Gene
Ontology (https://www.ebi.ac.uk/QuickGO/) biological processes
and hallmark pathways was also conducted. An absolute value of NES
>1 and a false discovery rate-corrected P-value ≤0.05 were
considered significant.
Patients and tissue samples
Written informed consent was obtained according to
the guidelines of the Medical Ethics Committee of The First
Affiliated Hospital of Kunming Medical University (Kunming, China).
Between July and August 2020, a total of 3 patients who were
pathologically diagnosed with NSCLC at The First Affiliated
Hospital of Kunming Medical University (Kunming, China), including
2 male patients and 1 female patient (maximum age, 64 years;
minimum age, 55 years; median age, 65 years), were recruited in the
present study, and tumor types were confirmed by ≥2 experienced
pathologists. The six paired samples collected were used to
identify the expression levels of CFB in lung tumor tissues. The
enrolled patients met the following criteria: i) No patients
diagnosed as LUAD by pathology had received chemotherapy or
radiotherapy; ii) the adjacent non-tumor lung tissues were
collected ≥5 cm away from carcinoma tissues; iii) the lung tissue
samples collected during surgery were frozen in liquid nitrogen
(−196°C) within 30 min; and iv) tumor tissue samples contained ≥80%
typical tumor cellularity, while the matched adjacent non-tumor
lung tissues samples contained no cancer cells.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from tissues was isolated using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. cDNA was
synthesized using 5X All-In-One MasterMix (cat. no. G492; Applied
Biological Materials, Inc.). The mixture was incubated at 25°C for
10 min and then at 42°C for 15 min. SYBR-Green Master mix (cat. no.
MasterMix-R; Applied Biological Materials, Inc.) was used to
perform qPCR on a 7300 Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). Initially, the enzyme in the
mixture was activated at 95°C for 10 min for 1 cycle, then the
mixture was denaturalized at 95°C for 15 sec and annealed/extended
at 60°C for 60 sec for 40 cycles. The relative mRNA expression in
different samples was calculated using the 2-∆∆Cq normalization
method (23). The following primers
were used: CFB forward, 5′-AATCAAGGTCAGCGTAGGAGG-3′ and reverse,
5′-GGGAGACAAATGGGCCTGATA-3′. GAPDH was chosen as an internal
control using the following primers: GAPDH forward,
5′-GCATCCTGGGCTACACTGAG-3′ and reverse,
5′-AAGTGGTCGTTGAGGGCAAT-3′.
Western blotting
Total protein from tissues was extracted using RIPA
lysis buffer (Beijing Solarbio Science & Technology Co., Ltd.)
containing protease inhibitor (EMD Millipore), and the protein
concentration was determined using a BCA protein assay (Beijing
Solarbio Science & Technology Co., Ltd.). A total 10–15 µg
protein/lane was separated via 8–15% SDS-PAGE and transferred to
PVDF membranes, which were incubated with specific primary
antibodies overnight at 4°C after blocking with 5% non-fat milk at
room temperature for 1 h. Primary antibodies used in the present
study included anti-CFB (Abcam; cat. no. ab133765; 1:5,000) and
anti-GAPDH (Cell Signaling Technology, Inc.; cat. no. 8884;
1:1,000), which were diluted in 1X TBS-Tween (TBST; containing 0.1%
Tween-20). The membranes were washed with 1X TBST and incubated at
room temperature for 1 h with HRP-conjugated anti-rabbit IgG
secondary antibodies (Cell Signaling Technology, Inc.; cat. no.
7074; 1:10,000) diluted in 1X TBST. Finally, Protein bands were
visualized using a High-Sig ECL Western Blotting kit (Tanon Science
and Technology Co., Ltd.), and the band intensity value was
subsequently measured using ImageJ software (v1.8.0; National
Institutes of Health) with GAPDH serving as an internal
control.
Statistical analysis
All the experiments, including RT-qPCR and western
blotting, were repeated three times. All quantitative data were
presented as the mean ± standard error and analyzed using SPSS
software (version 22.0; IBM Corp) and GraphPad Prism 8 (GraphPad
Software, Inc.). A paired-sample t-test was performed to compare
the relative mRNA expression levels of the CFB gene between tumor
and matched adjacent non-tumor tissues. A Wilcoxon rank sum test
was used for comparisons of protein levels in two independent
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
Patient characteristics
The characteristics of patients with LUAD are
presented in Table I. There was no
significant difference in age, sex, ethnicity, residual tumor
classification, ECOG score or Karnofsky performance score between
the low-CFB and high-CFB groups. There was a higher percentage of
former smokers and a lower percentage of non-smokers and current
smokers in the low-CFB group compared with in the high-CFB group
(P=0.021). Furthermore, patients in the low-CFB group had a higher
stage (stage III and stage IV) compared with patients in the
high-CFB group (P=0.010). Additionally, the violin plot indicated a
negative association trend between CFB expression and stage
(Fig. 1G). Furthermore, other
clinical features showed no or an inconsistent association with CFB
expression (Fig. 1A-F).
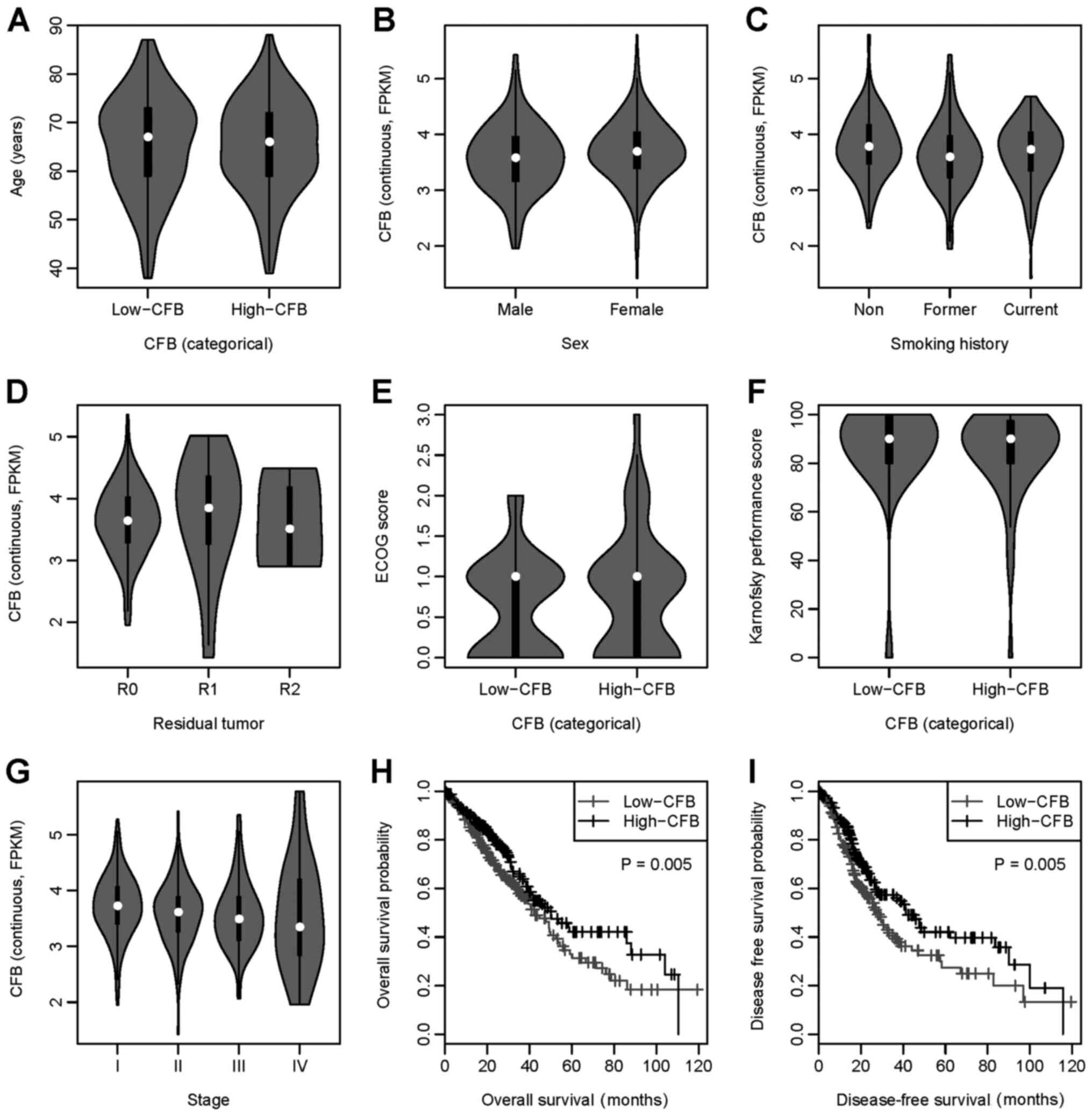 | Figure 1.CFB expression and its association
with clinical features. Association between CFB expression and (A)
age, (B) sex, (C) smoking history, (D) residual tumor
classification, (E) ECOG score, (F) Karnofsky performance score,
(G) stage, (H) overall survival and (I) disease-free survival.
Low-CFB, CFB expression <3.64; high-CFB, CFB expression ≥3.64.
CFB, complement factor B; ECOG, Eastern Cooperative Oncology
Group. |
 | Table I.Descriptive statistics stratified by
CFB expression. |
Table I.
Descriptive statistics stratified by
CFB expression.
|
Variablea | CFB <3.64
(n=257) | CFB ≥3.64
(n=256) | P-value |
|---|
| Mean age ± SD,
years | 65.44±10.25 | 65.28±9.62 | 0.855 |
| Sex, n (%) |
|
| 0.143 |
|
Male | 128 (49.81) | 110 (42.97) |
|
|
Female | 129 (50.19) | 146 (57.03) |
|
| Ethnicity, n
(%) |
|
| 0.568 |
|
Asian | 2 (0.91) | 5 (2.18) |
|
|
White | 191 (87.21) | 196 (85.59) |
|
| Black
or African American | 25 (11.42) | 28 (12.23) |
|
|
American Indian or Alaska
Native | 1 (0.46) | 0 (0.00) |
|
| Smoking history, n
(%) |
|
| 0.021 |
|
Non-smoker | 30 (12.15) | 45 (17.86) |
|
| Former
smoker | 166 (67.21) | 139 (55.16) |
|
| Current
smoker | 51 (20.65) | 68 (26.98) |
|
| Residual tumour
classification, n (%) |
|
| 0.716 |
| R0 | 172 (96.09) | 172 (94.51) |
|
| R1 | 5 (2.79) | 8 (4.40) |
|
| R2 | 2 (1.12) | 2 (1.10) |
|
| ECOG score (mean ±
SD) | 0.61±0.62 | 0.76±0.75 | 0.120 |
| Karnofsky
performance score (mean ± SD) | 86.46±20.37 | 84.40±18.53 | 0.602 |
| Stage, n (%) |
|
| 0.010 |
| Stage
I | 120 (48.00) | 155 (60.78) |
|
| Stage
II | 62 (24.80) | 59 (23.14) |
|
| Stage
III | 53 (21.20) | 31 (12.16) |
|
| Stage
IV | 15 (6.00) | 10 (3.92) |
|
CFB expression and clinical
features
The associations between CFB expression and clinical
features are shown in Table II.
There were more former smokers in the low-CFB group compared with
in the high-CFB group (P=0.026 for former smokers vs. non-smokers).
Furthermore, there were more stage III patients in the low-CFB
group compared with in the high-CFB group (P=0.002 for stage III
vs. stage I). Other clinical features (age, sex, ethnicity,
residual tumor classification, ECOG score and Karnofsky performance
score) were not observed to be associated with CFB expression.
 | Table II.Univariate logistic regression
analysis of clinical features and CFB expression. |
Table II.
Univariate logistic regression
analysis of clinical features and CFB expression.
| Variable | N1/N2a | OR (95% CI) | P-value |
|---|
| Age (years) |
| 1.00
(0.98–1.02) | 0.854 |
| Sex |
|
Male | 128/110 | Reference |
|
|
Female | 129/146 | 1.32
(0.93–1.86) | 0.121 |
| Ethnicity |
|
Asian | 2/5 | Reference |
|
|
White | 191/196 | – | – |
| Black
or African American | 25/28 | – | – |
|
American Indian or Alaska
Native | 1/0 | – | – |
| Smoking
history |
|
Non-smoker | 30/45 | Reference |
|
| Former
smoker | 166/139 | 0.56
(0.33–0.93) | 0.026 |
| Current
smoker | 51/68 | 0.89
(0.49–1.60) | 0.694 |
| Residual tumour
classification |
| R0 | 172/172 | Reference |
|
| R1 | 5/8 | 1.60
(0.51–4.99) | 0.418 |
| R2 | 2/2 | – | – |
| ECOG score |
| 1.36
(0.92–2.01) | 0.129 |
| Karnofsky
performance score |
| 0.99
(0.97–1.02) | 0.599 |
| Stage |
| Stage
I | 120/155 | Reference |
|
| Stage
II | 62/59 | 0.74
(0.48–1.13) | 0.163 |
| Stage
III | 53/31 | 0.45
(0.27–0.75) | 0.002 |
| Stage
IV | 15/10 | 0.52
(0.22–1.19) | 0.120 |
CFB expression, clinical features and
patient survival
Data on CFB expression, clinical features and
patient survival are presented in Tables III and IV. High CFB expression was significantly
associated with overall survival and disease-free survival
according to both the continuous (P=0.045 for overall survival;
P=0.006 for disease-free survival) and categorical model (P=0.005
for overall survival; P=0.002 for disease-free survival). The
Kaplan-Meier curves suggested that the low-CFB group had a
significantly decreased overall survival (Fig. 1H) and disease-free survival (Figs. 1I and S1) compared with the high-CFB group. In
the sub-group analysis of different smokers, there was no
significant association between CFB expression and overall survival
or disease-free survival (Fig. S2).
Furthermore, the residual tumor classification was significantly
associated with overall survival (P<0.001 for R1 vs. R0). The
Karnofsky performance score was significantly associated with
overall survival (P=0.031) and disease-free survival (P=0.013).
Additionally, high stage was significantly associated with overall
survival (P<0.001 for stage II vs. stage I; P<0.001 for stage
III vs. stage I; P<0.001 for stage IV vs. stage I) and
disease-free survival (P<0.001 for stage II vs. stage I;
P<0.001 for stage III vs. stage I; P=0.034 for stage IV vs.
stage I). These results indicated that the residual tumor
classification, Karnofsky performance score and cancer stage were
associated with overall survival, and that Karnofsky performance
score and stage were associated with disease-free survival.
 | Table III.Univariate Cox proportional hazards
regression analysis of CFB expression, clinical features and
overall survival. |
Table III.
Univariate Cox proportional hazards
regression analysis of CFB expression, clinical features and
overall survival.
| Variable | N | Median survival
time (Q1-Q3), months | HR (95% CI) | P-value |
|---|
| CFB
(continuous) | 504 |
| 0.77
(0.60–0.99) | 0.045 |
| CFB
(categorical) |
| CFB
<3.64 | 250 | 20.60
(11.55–35.12) | Reference |
|
| CFB
≥3.64 | 254 | 21.95
(14.67–38.68) | 0.66
(0.49–0.88) | 0.005 |
| Age (years) | 494 |
| 1.01
(0.99–1.02) | 0.324 |
| Sex |
|
Male | 235 | 20.66
(10.58–36.13) | Reference |
|
|
Female | 269 | 21.62
(14.72–37.12) | 0.96
(0.72–1.29) | 0.808 |
| Ethnicity |
|
Asian | 7 | 18.66
(7.66–24.21) | Reference |
|
|
White | 387 | 20.83
(13.63–35.35) | NA | NA |
| Black
or African American | 53 | 22.01
(16.85–37.29) | NA | NA |
|
American Indian or Alaska
Native | 1 | 15.34
(15.34–15.34) | NA | NA |
| Smoking
history |
|
Non-smoker | 72 | 23.66
(13.84–36.09) | Reference |
|
| Former
smoker | 300 | 20.02
(13.44–35.26) | 0.92
(0.60–1.41) | 0.695 |
| Current
smoker | 118 | 22.08
(14.60–36.73) | 0.84
(0.51–1.37) | 0.476 |
| Residual tumour
classification |
| R0 | 336 | 23.14
(14.59–39.77) | Reference |
|
| R1 | 13 | 21.98
(9.56–29.99) | 3.33
(1.73–6.40) | <0.001 |
| R2 | 3 | 8.02
(6.00–11.52) | NA | NA |
| ECOG score | 213 |
| 1.33
(0.97–1.82) | 0.076 |
| Karnofsky
performance score | 97 |
| 0.99
(0.97–1.00) | 0.031 |
| Stage |
| Stage
I | 271 | 22.80
(15.64–41.85) | Reference |
|
| Stage
II | 119 | 22.24
(11.11–32.75) | 2.53
(1.76–3.65) | <0.001 |
| Stage
III | 81 | 15.37
(8.80–28.88) | 3.61
(2.46–5.29) | <0.001 |
| Stage
IV | 25 | 21.55
(15.01–31.11) | 4.07
(2.33–7.11) | <0.001 |
 | Table IV.Univariate Cox proportional hazards
regression analysis of CFB expression, clinical features and
disease-free survival. |
Table IV.
Univariate Cox proportional hazards
regression analysis of CFB expression, clinical features and
disease-free survival.
| Variable | N | Median survival
time (Q1-Q3), months | HR (95% CI) | P-value |
|---|
| CFB
(continuous) | 428 |
| 0.70
(0.55–0.90) | 0.006 |
| CFB
(categorical) |
| CFB
<3.64 FPKM | 207 | 16.79
(8.15–27.76) | Reference |
|
| CFB
≥3.64 FPKM | 221 | 19.74
(13.76–31.18) | 0.62
(0.47–0.84) | 0.002 |
| Age (years) | 418 |
| 1.01
(0.99–1.02) | 0.317 |
| Sex |
|
Male | 193 | 17.61
(8.54–29.01) | Reference |
|
|
Female | 235 | 18.17
(13.27–28.70) | 0.97
(0.72–1.29) | 0.822 |
| Ethnicity |
|
Asian | 7 | 14.49
(6.00–22.88) | Reference |
|
|
White | 329 | 17.44
(10.09–27.23) | 0.62
(0.23–1.69) | 0.354 |
| Black
or African American | 48 | 20.16
(15.66–33.78) | 0.45
(0.15–1.35) | 0.156 |
|
American Indian or Alaska
Native | 1 | 9.26
(9.26–9.26) | NA | NA |
| Smoking
history |
|
Non-smoker | 60 | 17.92
(13.12–31.47) | Reference |
|
| Former
smoker | 259 | 16.92
(9.54–26.25) | 1.07
(0.70–1.64) | 0.758 |
| Current
smoker | 97 | 20.70
(13.40–30.98) | 0.74
(0.45–1.24) | 0.253 |
| Residual tumour
classification |
| R0 | 282 | 18.99
(12.41–33.02) | Reference |
|
| R1 | 11 | 14.72
(5.18–18.84) | NA | NA |
| ECOG score | 181 |
| 1.03
(0.73–1.45) | 0.880 |
| Karnofsky
performance score | 79 |
| 0.98
(0.97–1.00) | 0.013 |
| Stage |
| Stage
I | 246 | 19.22
(13.63–31.82) | Reference |
|
| Stage
II | 102 | 16.98
(8.21–26.88) | 2.16
(1.54–3.02) | <0.001 |
| Stage
III | 59 | 13.76
(7.23–24.95) | 2.23
(1.47–3.37) | <0.001 |
| Stage
IV | 14 | 19.88
(14.02–25.67) | 2.22
(1.06–4.62) | 0.034 |
The results of the multivariate Cox regression
analysis demonstrated that high CFB expression was associated with
increased overall survival time (Table
V) and disease-free survival time (Table VI), according to both the continuous
model [hazard ratio (HR), 0.48; 95% confidence interval (95% CI),
0.25–0.93; P=0.029 for overall survival; HR, 0.29; 95% CI,
0.15–0.59; P=0.001 for disease-free survival] and the categorical
model (HR, 0.46; 95% CI, 0.22–0.93; P=0.031 for overall survival;
HR, 0.25; 95% CI, 0.12–0.55; P=0.001 for disease-free survival)
after adjusting for corresponding covariates (residual tumour
classification, Karnofsky performance score and stage).
 | Table V.Multivariate Cox proportional hazards
regression analysis of CFB expression and overall survival. |
Table V.
Multivariate Cox proportional hazards
regression analysis of CFB expression and overall survival.
| Variable | N | Median survival
time (Q1-Q3), months | HR (95% CI) | P-value |
|---|
| CFB
(continuous) | 504 |
| 0.48
(0.25–0.93) | 0.029 |
| CFB
(categorical) |
| CFB
<3.64 FPKM | 250 | 20.60
(11.55–35.12) | Reference |
|
| CFB
≥3.64 FPKM | 254 | 21.95
(14.67–38.68) | 0.46
(0.22–0.93) | 0.031 |
 | Table VI.Multivariate Cox proportional hazards
regression analysis of CFB expression and disease-free
survival. |
Table VI.
Multivariate Cox proportional hazards
regression analysis of CFB expression and disease-free
survival.
| Variable | N | Median survival
time (Q1-Q3), months | HR (95% CI) | P-value |
|---|
| CFB
(continuous) | 428 |
| 0.29
(0.15–0.59) | 0.001 |
| CFB
(categorical) |
| CFB
<3.64 FPKM | 207 | 16.79
(8.15–27.76) | Reference |
|
| CFB
≥3.64 FPKM | 221 | 19.74
(13.76–31.18) | 0.25
(0.12–0.55) | 0.001 |
CFB expression and pathway
impairment
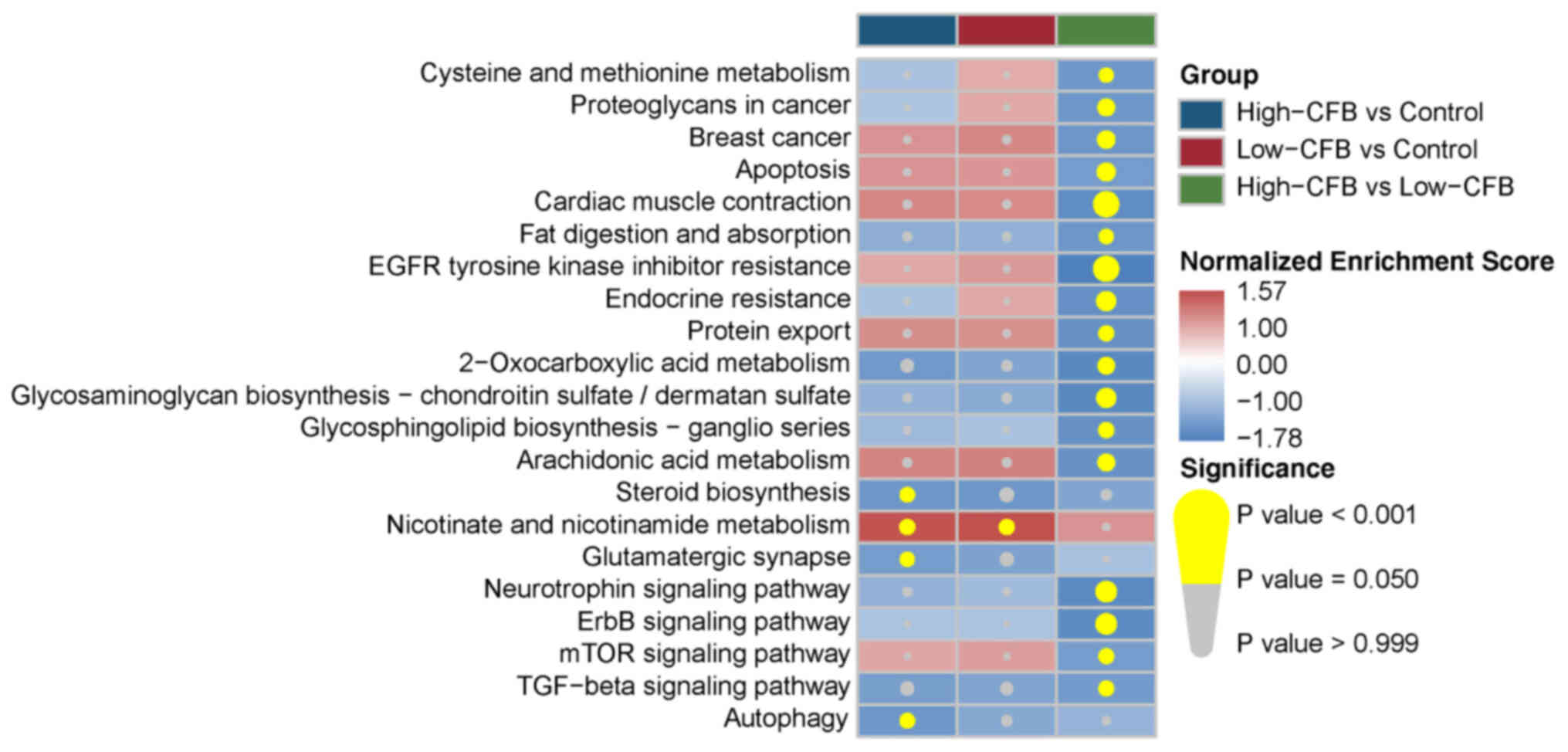
Significantly enriched KEGG pathways of the three
groups are presented in Fig. 2.
There were four deregulated KEGG pathways for the high-CFB vs.
control, including ‘steroid biosynthesis’, ‘nicotinate and
nicotinamide metabolism’, ‘glutamatergic synapse’ and ‘autophagy’,
whereas only ‘nicotinate and nicotinamide metabolism’ was
upregulated in the low-CFB vs. control group. Furthermore, there
were 17 downregulated pathways in the high-CFB vs. low-CFB group.
These findings indicated that there was a marked difference in the
pathway deregulation of individuals with different CFB expression
states. Enrichment analysis of Gene Ontology biological processes
(Table SI) and hallmark pathways
(Table SII) was also conducted. The
results demonstrated that CFB was associated with multiple
signaling pathways related to cell proliferation and tumorigenesis,
such as ‘positive regulation of G2/M transition of mitotic cell
cycle’, ‘TNFA signaling via NF-κB’ and ‘Wnt/β-catenin
signaling’.
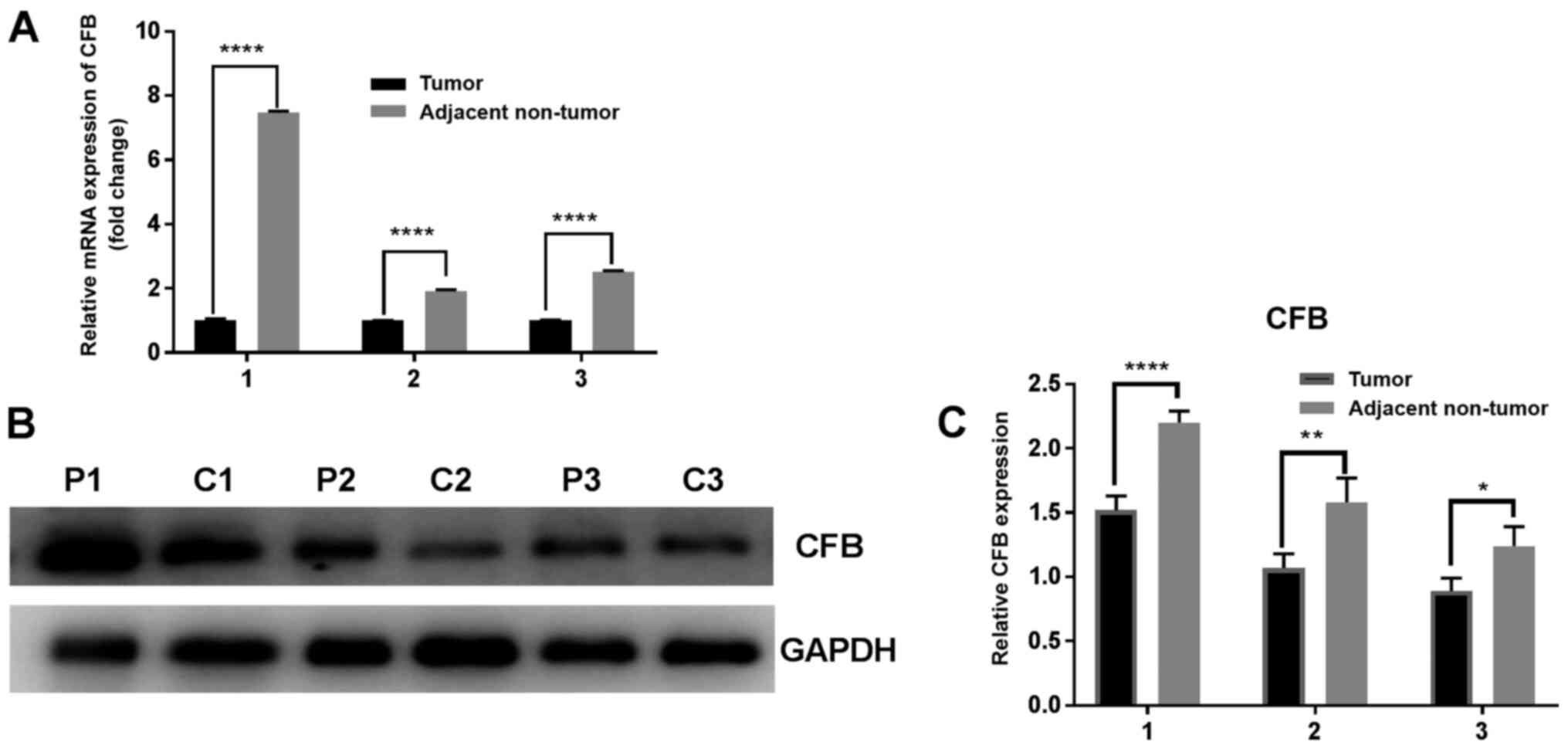
CFB expression in human lung
tissues
It was identified that both the relative mRNA and
protein expression levels of CFB were downregulated in tumor
tissues compared with in matched adjacent non-tumor tissues
(P<0.05; Fig. 3).
Discussion
Increasing attention has been paid to precise
individualized medicine in cancer treatment, which is facilitated
by the exploration and identification of biomarkers (24). The treatment of LUAD is associated
with complex factors and comprises multiple targeted therapies.
Therefore, a greater understanding of molecular biomarkers will
help to improve the diagnosis and prognosis of human LUAD.
CFB is a key soluble component in the alternative
pathway (8). CFB is cleaved by
complement factor D into fragments Ba and Bb. The complement factor
D and activated component Bb are serine proteinases. Furthermore,
Bb remains attached to C3b and forms the alternative pathway
convertase, C3bBb, which is a key enzyme involved in the activation
of the alternative pathway (10). It
has been demonstrated that low CFB expression is associated with
decreased C3bBb expression and accelerates the degradation of the
MAC, which leads to inhibition of the alternative pathway of
complement activation, thereby significantly reducing the
activation efficiency of the complement system in the whole body
(8). During the regulation of the
complex tumor microenvironment, complement proteins serve a dual
role in the tumor microenvironment, which will eventually affect
tumor progression (8,9). The present study demonstrated that
there was no difference in age, sex, ethnicity, residual tumor
classification, ECOG score or Karnofsky performance score between
the low-CFB and high-CFB groups. Furthermore, there were more
former smokers and stage III patients in the high-CFB group
compared with in the low-CFB group. Notably, in both univariate and
multivariate analysis, high CFB expression was associated with
overall survival and disease-free survival, according to both the
continuous and categorical models.
CFB is a serum protein that is not only produced by
the liver (25–27) but can also be synthesized by the
choroid, retinal pigment epithelial cells and neural retina
(28). Furthermore, the CFB protein
has been found in ocular drusen and Bruch's membrane (26), and was observed to be potently
upregulated in patients with ulcerative colitis and Crohn's disease
(29). Previous data have
demonstrated that CFB may be highly expressed in association with
inflammatory bowel disease, in addition to having a possible key
role in systemic complement activation (30). A previous study has reported that
polymorphisms in CFB are associated with the risk of age-related
macular degeneration (31).
Furthermore, the study of CFB non-synonymous variants may improve
the understanding of chronic hepatitis B etiology (31). The present study first used online
public data analysis to demonstrate that low CFB expression was
associated with decreased overall and disease-free survival in
patients with LUAD. Subsequently, the prognostic value of CFB
expression in lung tumor tissues was further investigated.
Recent studies have reported that CFB is associated
with the prognosis of different types of cancer (32–34). A
previous study has demonstrated that CFB is not only identified in
all crypts in the colonic mucosa, but is also expressed in adenomas
and carcinomas (16). The expression
levels of CFB have been revealed to be increased in the plasma of
patients with pancreatic cancer, for which CFB may be a novel
biomarker (13). A previous analysis
of CFB genetic alterations and mRNA expression in breast cancer has
been performed using public TCGA invasive breast carcinoma sample
data (32). CFB exhibits a strong
association with molecular subtypes of breast cancer, particularly
the LA subtype (17). Furthermore,
CFB is a potential biomarker in pancreatic ductal adenocarcinoma
(13) and pancreatic cancer
(32) with diagnostic significance,
and is also associated with a high likelihood of relapse-free
survival (17). To the best of our
knowledge, there has been no study addressing the prognostic
significance of CFB in patients with LUAD. In the present study,
multivariate Cox regression analysis demonstrated that high CFB
expression was associated with increased overall and disease-free
survival according to a continuous model after adjusting for
corresponding covariates. Consistent with these results, both
RT-qPCR and western blotting indicated that the relative expression
levels of the CFB gene in lung tumor tissues were decreased
compared with those in adjacent non-tumor tissues.
There are several limitations to the present study.
First, the association between CFB and the occurrence and
progression of LUAD, as well as the specific pathogenesis, remains
unknown. The current results can to some extent explain an
association between CFB and LUAD; however, prospective randomized
controlled studies are required to confirm these promising results.
Second, the current data concerning drug therapy and prognosis of
LUAD are not widely available. Given the limitations of the present
study, further large-sample and in-depth studies are required to
confirm these results.
In conclusion, the present study demonstrated that
CFB expression was an independent predictor of overall and
disease-free survival in patients with LUAD. The current results
may therefore provide helpful information for the early diagnosis
and drug development of LUAD.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 81460325
and 81760384), the Applied Basic Research in Yunnan Province (grant
no. 2015FB040), and the Health Science and Technology Project in
Yunnan Province (grant no. 2017NS030). The funders had no role in
the study design, data collection and analysis, decision to publish
or preparation of the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CH and YL contributed to the design of the present
study, developed the methodology, collected the bioinformatics
data, performed the experiments, analyzed the results and wrote the
manuscript. RZ and JC contributed to the collection of patient data
and assisted with qPCR and western blotting experiments. XF
assisted in experimental design, data analysis and manuscript
editing. YD contributed to the design of the study, critically
revised the manuscript and approved the final version to be
published. CH, YL and YD confirmed the authenticity of all raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Kunming Medical
University (Kunming, China; approval no. 2020L42). Written informed
consent was obtained from patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jackman DM and Johnson BE: Small-cell lung
cancer. Lancet. 366:1385–1396. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li C and Lu H: Adenosquamous carcinoma of
the lung. OncoTargets Ther. 11:4829–4835. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Scheff RJ and Schneider BJ: Non-small-cell
lung cancer: treatment of late stage disease: chemotherapeutics and
new frontiers. Semin Intervent Radiol. 30:191–198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu HA, Paz-Ares LG, Yang JC, Lee KH,
Garrido P, Park K, Kim JH, Lee DH, Mao H, Wijayawardana SR, et al:
Phase I study of the efficacy and safety of ramucirumab in
combination with osimertinib in advanced T790M-positive EGFR-mutant
non-small cell lung cancer. Clin Cancer Res. 27:992–1002. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu Y, Gao GF, Minna JD, Williams NS and
Westover KD: Loss of wild type KRAS in KRASMUT lung adenocarcinoma
is associated with cancer mortality and confers sensitivity to FASN
inhibitors. Lung Cancer. 153:73–80. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Denisenko TV, Budkevich IN and Zhivotovsky
B: Cell death-based treatment of lung adenocarcinoma. Cell Death
Dis. 9:1172018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rutkowski MJ, Sughrue ME, Kane AJ, Mills
SA and Parsa AT: Cancer and the complement cascade. Mol Cancer Res.
8:1453–1465. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gros P, Milder FJ and Janssen BJ:
Complement driven by conformational changes. Nat Rev Immunol.
8:48–58. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kleczko EK, Kwak JW, Schenk EL and
Nemenoff RA: Targeting the complement pathway as a therapeutic
strategy in lung cancer. Front Immunol. 10:9542019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Walport MJ: Complement. First of two
parts. N Engl J Med. 344:1058–1066. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Walport MJ: Complement. Second of two
parts. N Engl J Med. 344:1140–1144. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee MJ, Na K, Jeong SK, Lim JS, Kim SA,
Lee MJ, Song SY, Kim H, Hancock WS and Paik YK: Identification of
human complement factor B as a novel biomarker candidate for
pancreatic ductal adenocarcinoma. J Proteome Res. 13:4878–4888.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Low HP, Tiwari A, Janjanam J, Qiu L, Chang
CI, Strohsnitter WC, Norwitz ER, Tam SW, Evans JE, Green KM, et al:
Screening preeclamptic cord plasma for proteins associated with
decreased breast cancer susceptibility. Genomics Proteomics
Bioinformatics. 11:335–344. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Byrne SN, Hammond KJ, Chan CY, Rogers LJ,
Beaugie C, Rana S, Marsh-Wakefield F, Thurman JM and Halliday GM:
The alternative complement component factor B regulates UV-induced
oedema, systemic suppression of contact and delayed
hypersensitivity, and mast cell infiltration into the skin.
Photochem Photobiol Sci. 14:801–806. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Andoh A, Fujiyama Y, Sakumoto H, Uchihara
H, Kimura T, Koyama S and Bamba T: Detection of complement C3 and
factor B gene expression in normal colorectal mucosa, adenomas and
carcinomas. Clin Exp Immunol. 111:477–483. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Suman S, Basak T, Gupta P, Mishra S, Kumar
V, Sengupta S and Shukla Y: Corrigendum to ‘Quantitative proteomics
revealed novel proteins associated with molecular subtypes of
breast cancer’ [Journal of Proteomics 148, (2016) 183–193]. J
Proteomics. 208:1035072019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kwak JW, Laskowski J, Li HY, McSharry MV,
Sippel TR, Bullock BL, Johnson AM, Poczobutt JM, Neuwelt AJ,
Malkoski SP, et al: Complement Activation via a C3a Receptor
Pathway Alters CD4+ T Lymphocytes and Mediates Lung Cancer
Progression. Cancer Res. 78:143–156. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Azam F, Latif MF, Farooq A, Tirmazy SH,
AlShahrani S, Bashir S and Bukhari N: Performance status assessment
by using ECOG (Eastern Cooperative Oncology Group) score for cancer
patients by oncology healthcare professionals. Case Rep Oncol.
12:728–736. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Barbetta C, Allgar V, Maddocks M, Ribeiro
C, Wilcock A, Currow DC, Phillips J and Johnson MJ:
Australia-modified Karnofsky Performance Scale and physical
activity in COPD and lung cancer: an exploratory pooled data
analysis. BMJ Support Palliat Care. Jun 11–2019.(Epub ahead of
print). doi: 10.1136/bmjspcare-2019-001869. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Woodard GA, Jones KD and Jablons DM: Lung
Cancer Staging and Prognosis. Cancer Treat Res. 170:47–75. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wishart DS: Emerging applications of
metabolomics in drug discovery and precision medicine. Nat Rev Drug
Discov. 15:473–484. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gold B, Merriam JE, Zernant J, Hancox LS,
Taiber AJ, Gehrs K, Cramer K, Neel J, Bergeron J, Barile GR, et al
AMD Genetics Clinical Study Group, : Variation in factor B (BF) and
complement component 2 (C2) genes is associated with age-related
macular degeneration. Nat Genet. 38:458–462. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Colten HR: Biosynthesis of the MHC-linked
complement proteins (C2, C4 and factor B) by mononuclear
phagocytes. Mol Immunol. 19:1279–1285. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Canavese F, Botnari A, Andreacchio A,
Marengo L, Samba A, Dimeglio A, Pereira B, Mansour M and Rousset M:
Displaced tibial shaft fractures with intact fibula in children:
Nonoperative management versus operative treatment with elastic
stable intramedullary nailing. J Pediatr Orthop. 36:667–672. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen M, Muckersie E, Robertson M,
Forrester JV and Xu H: Up-regulation of complement factor B in
retinal pigment epithelial cells is accompanied by complement
activation in the aged retina. Exp Eye Res. 87:543–550. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ostvik AE, Granlund A, Gustafsson BI, Torp
SH, Espevik T, Mollnes TE, Damås JK and Sandvik AK: Mucosal
toll-like receptor 3-dependent synthesis of complement factor B and
systemic complement activation in inflammatory bowel disease.
Inflamm Bowel Dis. 20:995–1003. 2014.PubMed/NCBI
|
|
29
|
Potter BJ, Brown DJ, Watson A and Jewell
DP: Complement inhibitors and immunoconglutinins in ulcerative
colitis and Crohn's disease. Gut. 21:1030–1034. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang DK, Ma XP, Yu H, Cao G, Ding DL,
Chen H, Huang HX, Gao YZ, Wu XP, Long XD, et al: Genetic variants
in five novel loci including CFB and CD40 predispose to chronic
hepatitis B. Hepatology. 62:118–128. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ciriello G, Gatza ML, Beck AH, Wilkerson
MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et
al TCGA Research Network, : Comprehensive molecular portraits of
invasive lobular breast cancer. Cell. 163:506–519. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim SH, Lee MJ, Hwang HK, Lee SH, Kim H,
Paik YK and Kang CM: Prognostic potential of the preoperative
plasma complement factor B in resected pancreatic cancer: A pilot
study. Cancer Biomark. 24:335–342. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lim LY, Tan GH, Zainuddin ZM, Fam XI, Goh
EH, Syaris OS, Yahaya A and Singam P: Prospective evaluation of
using multiparametric magnetic resonance imaging in cognitive
fusion prostate biopsy compared to the standard systematic 12-core
biopsy in the detection of prostate cancer. Urol Ann. 12:276–282.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bauer D, Mazzio E, Hilliard A, Oriaku ET
and Soliman KF: Effect of apigenin on whole transcriptome profile
of TNFα-activated MDA-MB-468 triple negative breast cancer cells.
Oncol Lett. 19:2123–2132. 2020.PubMed/NCBI
|