Introduction
Breast cancer is one of the most common malignant
tumors that endanger the health of women; its incidence rate ranked
first and its mortality rate ranked second among female
malignancies worldwide in 2018 (1).
Triple-negative breast cancer (TNBC) refers to breast cancer that
is negative for the estrogen receptor (ER), progesterone receptor
(PR) and human epidermal growth factor receptor 2 (HER2), and
accounts for 15–20% of basal cell-like breast cancer cases
(2,3). The endocrine drug tamoxifen and the
HER2 monoclonal antibody trastuzumab have been successful in the
treatment of ER+/HER2− or HER+
patients with breast cancer (4,5).
However, since patients with TNBC do not express these receptors,
endocrine and targeted drugs are not effective for these patients
(6). Understanding the underlying
molecular mechanisms may help identify biomarkers for diagnosis and
drug targets.
SRY-related high-mobility group box (SOX) family
members are a cluster of transcription factors that regulate
various cellular processes, including stemness, proliferation,
differentiation and survival (7).
Increasing evidence has demonstrated that dysregulation of SOX9
serves a pivotal role in cancer development. SOX9 expression is
upregulated in various types of cancer, including malignant
pancreatic neoplasms (8), non-small
cell lung carcinoma (9) and
esophageal squamous cell carcinoma (10). However, low SOX9 expression predicts
a poor survival in patients with gastric cancer (11). Hepatitis B virus (HBV)
transcriptionally activates SOX9 in hepatocellular carcinoma cells,
whereas SOX9 upregulation represses the replication of HBV
(12). In breast cancer, SOX9
activates FXYD domain containing ion transport regulator 3
expression, which promotes the formation of activated complex by
interacting with Src and ERα, thereby enhancing the stemness of
breast cancer cells (13). However,
the involvement of SOX9 in TNBC remains to be determined.
The present study aimed to investigate the role of
SOX9 by knocking it down in TNBC cells. Cell Counting Kit-8 (CCK-8)
and colony formation assays were performed to study the effect of
SOX9 silencing on cell proliferation. Whether SOX9-knockdown
regulated the migration and invasion of TNBC cells was also
explored. Finally, RNA sequencing was applied to explore the
downstream effectors of SOX9 in TNBC.
Materials and methods
Cell culture
Human TNBC BT-549, MDA-MB-231, MDA-MB-436 and
MDA-MB-468 cells were obtained from the American Type Culture
Collection. 293T cells were purchased from The Cell Bank of Type
Culture Collection of The Chinese Academy of Sciences. TNBC cells
were cultured in RPMI 1640 medium (HyClone; Cytiva) supplemented
with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin and streptomycin solution (Corning, Inc.). 293T cells
were cultured in DMEM (HyClone; Cytiva) supplemented with 10% FBS
and 1% penicillin and streptomycin solution. Cell culture was
maintained in a 37°C incubator with 5% CO2.
SOX9-knockdown by lentivirus
To silence SOX9 in MDA-MB-231 and MDA-MB-468 cells,
the lentivirus vector pGCSIL-GFP was constructed, which contained a
short hairpin (sh)RNA sequence targeting SOX9. shRNA sequences were
as follow: Negative control (shNC), 5′-TTCTCCGAACGTGTCACGT-3′;
shSOX9-1, 5′-GCATCCTTCAATTTCTGTATA-3′; shSOX9-2,
5′-GCGGAGGAAGTCGGTGAAGAA-3′; and shSOX9-3,
5′-CTCCACCTTCACCTACATGAA-3′. For lentivirus packaging, the
expression plasmid (pGCSIL-GFP; 20 mg; Shanghai GeneChem Co., Ltd.)
with two helper plasmids (pHelper 1, 15 mg and pHelper 2, 10 mg;
Shanghai GeneChem Co., Ltd.) were co-transfected into 293T cells
using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) in 10-cm plates. After 48 and 72 h of transfection,
lentiviruses were harvested. Following purification and titration,
the harvested virus particles were used to infect MDA-MB-231 and
MDA-MB-468 cells using a multiplicity of infection of 20. After 72
h, cells were harvested for knockdown efficiency examination when
green fluorescence was visible.
CCK-8 assay
CCK-8 reagent was purchased from Sigma-Aldrich
(Merck KGaA) and used according to the manufacturer's instructions.
MDA-MB-231 and MDA-MB-468 cells were trypsinized and seeded in
triplicate into 96-well plates at a concentration of 2,000
cells/well. Each well contained 100 µl culture medium. After 6, 12,
24, 36, 48 and 72 h, 10 µl CCK-8 reagent was added to each well.
The plates were maintained in a 37°C incubator for 3 h. The
absorbance at 450 nm was measured using a microplate reader (Thermo
Fisher Scientific, Inc.) after agitation for 30 sec.
Colony formation assay
shNC- and shSOX9-transfected MDA-MB-231 and
MDA-MB-468 cells were seeded into 6-well plates at concentrations
of 50, 100, 200, 250, 500 and 1,000 cells/well. The culture medium
was replaced by fresh medium every 3 days. The colonies were formed
after culturing for 14 days at 37°C. The colonies were washed three
times with PBS, fixed with 4% paraformaldehyde (Wuhan Servicebio
Technology Co., Ltd.) for 20 min at room temperature and stained
with Giemsa (Sigma-Aldrich; Merck KGaA) staining solution for 20
min at room temperature. A colony was defined as >50 cells. The
colony formation rate was calculated by colony number/seeding
number.
Transwell assay
Transwell assay was performed to assess migration
and invasion. For invasion, Matrigel® (Corning, Inc.)
was mixed with serum-free medium in a 1:8 ratio. A total of 100 µl
of the mixture was added onto the upper surface of the migration
chambers (Corning, Inc.; 8.0-µm filter) and incubated at 37°C for
30 min. MDA-MB-231 cells were trypsinized and washed with
serum-free medium. A total of 1×104 cells in 100 µl
serum-free medium were seeded onto the upper surface of the
chambers, while the lower chambers were filled with medium with 10%
FBS. After 24 h at 37°C, the culture medium and the cells attached
on the upper surface were removed. Cells attached on the lower
surface were fixed with methanol for 30 min at room temperature and
stained with 0.1% crystal violet for 30 min at room temperature.
The images of migratory and invasive cells were collected using a
light microscope (Olympus Corporation; magnification, ×100).
Wound-healing assay
MDA-MB-231 cells were seeded into 6-well plates and
cultured overnight. The cells were grown to ~100% confluence in the
wells. Linear scratch wounds were developed using 10-µl pipette
tips. The floating cells were washed five times with PBS.
Serum-free medium was used to maintain the cells. The images were
captured at 0, 6, 12, 18 and 24 h using an inverted light
microscope (Olympus Corporation; magnification, ×100). The
wound-healing percentage was calculated using the following
formula: (Wound area at 0 h-wound area of indicated time)/(wound
area at 0 h) ×100. Wound area was analyzed using ImageJ (v4.0;
National Institute of Health).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA in MDA-MB-231 and MDA-MB-468 cells was
extracted using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). RNA was reverse transcribed to cDNA using
the Reverse transcription system kit (Promega Corporation; cat. no.
A3500), according to the manufacturer's instructions. qPCR analysis
was performed using SYBR Master Mixture (Takara Bio, Inc.) on the
Agilent MX3000p Real Time PCR system (Agilent Technologies, Inc.)
as follows: 1 cycle at 95°C for 30 sec; 40 cycles at 95°C for 5 sec
and 60°C for 20 sec; 1 cycle at 65°C for 15 sec. qPCR primers were
as follows: SOX9 forward, 5′-AGCGAACGCACATCAAGAC-3′ and reverse,
5′-CTGTAGGCGATCTGTTGGGG-3′; and GAPDH forward,
5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′. mRNA expression was analyzed using the
2−ΔΔCq method (14).
Western blotting
Western blotting was used to determine the protein
expression levels in TNBC cells. Total protein (40 µg) was
extracted from MDA-MB-231 and MDA-MB-468 cells using RIPA lysis
buffer (Beyotime Institute of Biotechnology), supplemented with
protease and phosphatase inhibitors cocktail (Roche Diagnostics).
The protein concentration was detected using a BCA Protein Assay
kit. A total of 40 µg of protein was loaded and separated via 12%
SDS-PAGE, followed by transfer to PVDF membranes (EMD Millipore).
Membranes were blocked with 5% skimmed milk dissolved in PBS-Tween
(0.1% Tween) at room temperature for 1.5 h and incubated with
primary antibodies overnight at 4°C. After incubating with
peroxidase-conjugated secondary antibodies for 2 h at room
temperature, the protein abundance was analyzed using an enhanced
chemiluminescence system (EMD Millipore). Primary antibodies
against SOX9 (1:1,000; cat. no. ab185966) and GAPDH (1:1,000; cat.
no. ab8245) were purchased from Abcam. HRP-conjugated anti-mouse
(1:5,000; cat. no. sc-2005) or anti-rabbit (1:5,000; cat. no.
sc-2004) secondary antibodies were from Santa Cruz Biotechnology,
Inc.
Apoptosis analysis
Apoptosis was analyzed using an annexin V/PI
apoptosis detection kit according to the manufacturer's protocol
(cat. no. 88-8007-72; Invitrogen; Thermo Fisher Scientific, Inc.).
MDA-MB-231 and MDA-MB-468 cells were trypsinized with EDTA-free
0.25% Trypsin (Corning, Inc.). The cells and cell supernatants were
collected by centrifugation at 1,000 × g for 5 min at room
temperature. The cell pellet was washed using iced D-Hanks (pH,
7.2–7.4) buffer and 1X binding buffer. The cell pellet was
resuspended in 200 µl 1X binding buffer. After incubating with 10
µl Annexin V-APC for 10–15 min in the dark at room temperature,
cells were stained with PI staining buffer (5 µl) for 10–15 min on
ice at room temperature. A total of 400–800 µl 1X binding buffer
was added and apoptosis was analyzed on a Guava easyCyte HT system
(EMD Millipore). The data was analyzed using the guavasoft software
(v2.7; Merck KGaA).
Cell cycle analysis
PI staining was used to analyze the cell cycle. When
MDA-MB-231 and MDA-MB-468 cells reached 80% confluence, they were
trypsinized and centrifuged at 1,000 × g for 5 min at room
temperature. The cell pellet was washed using iced D-Hanks (pH
7.2–7.4) buffer and fixed with iced 75% ethanol for ≥1 h at −20°C.
The cells were centrifuged at 1,000 × g for 5 min at room
temperature and the pellet was washed twice by D-Hanks buffer.
Finally, the cell pellet was resuspended in 0.6–1 ml staining
buffer [40X PI solution (2 mg/ml):100X RNase solution (10 mg/ml):1X
D-Hanks; 25:10:1,000) at room temperature for 30 min and subjected
to cell cycle analysis using a Guava easyCyte HT system (EMD
Millipore). The data was analyzed using the guavasoft software
(v2.7; Merck KGaA).
RNA sequencing
Firstly, total RNA was extracted from shNC- and
shSOX9-transfected MDA-MB-231 cells using TRIzol reagent.
Subsequently, RNA sequencing was performed by Beijing Aoweisen Gene
Technology Co., Ltd. Briefly, the total RNA integrity was evaluated
using an Agilent Bioanalyzer 2100 (Agilent Technologies, Inc.). The
RNA Clean XP kit (cat. no. A63987; Beckman Coulter, Inc.) and
RNase-free DNase Set (cat. no. 79254; Qiagen GmbH) were used for
RNA clean-up and DNA removing, respectively. A total of 1 µg
purified RNA was used for cDNA library generation using a VAHTSTM
mRNA-seq v2 library Prep kit (cat. no. NR612-01; Vazyme Biotech
Co., Ltd.). The quality of constructed cDNA libraries were
evaluated with an Agilent 2100 Bioanalyzer (Agilent Technologies,
Inc.) according to the manufacturer's protocol. The products were
sequenced using an Illumina NovaSeq 6000 SP Reagent kit v1.5 (300
cycles; cat. no. 20028400; Illumina, Inc.) with a paired-end read
length of 150 bp. A total of 1 nM of the final library was used for
sequencing. For data analysis, TopHat was used to map reads to a
reference genome (hg38) (15). Gene
expression was measured using the fragments per kilobase of
transcript per million mapped reads (FPKM) normalization method.
FPKM quantification was performed using Cufflinks v2.2.1
(http://cole-trapnell-lab.github.io/cufflinks/install/)
(16). The significantly
differentially expressed genes were obtained using a threshold of
fold-change >1.5 and P<0.05.
The Cancer Genome Atlas (TCGA)
analysis
For the analysis using TCGA data, 1,104 breast
invasion cancer tissues and 113 normal tissues were analyzed in
http://starbase.sysu.edu.cn/panCancer.php.
Statistical analysis
The data were presented as the mean ± SEM of three
technical repeats and were analyzed using GraphPad Prism 6.0
(GraphPad Software, Inc.). The difference between 2 groups was
analyzed by unpaired Student's t-test. One-way ANOVA followed by
Tukey's post-hoc test was used to compare the differences among
>2 groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
SOX9 is highly expressed in TNBC
cells
SOX9 is a pivotal modulator of stemness and
contributes to cancer development (17). To explore the relevance of SOX9 in
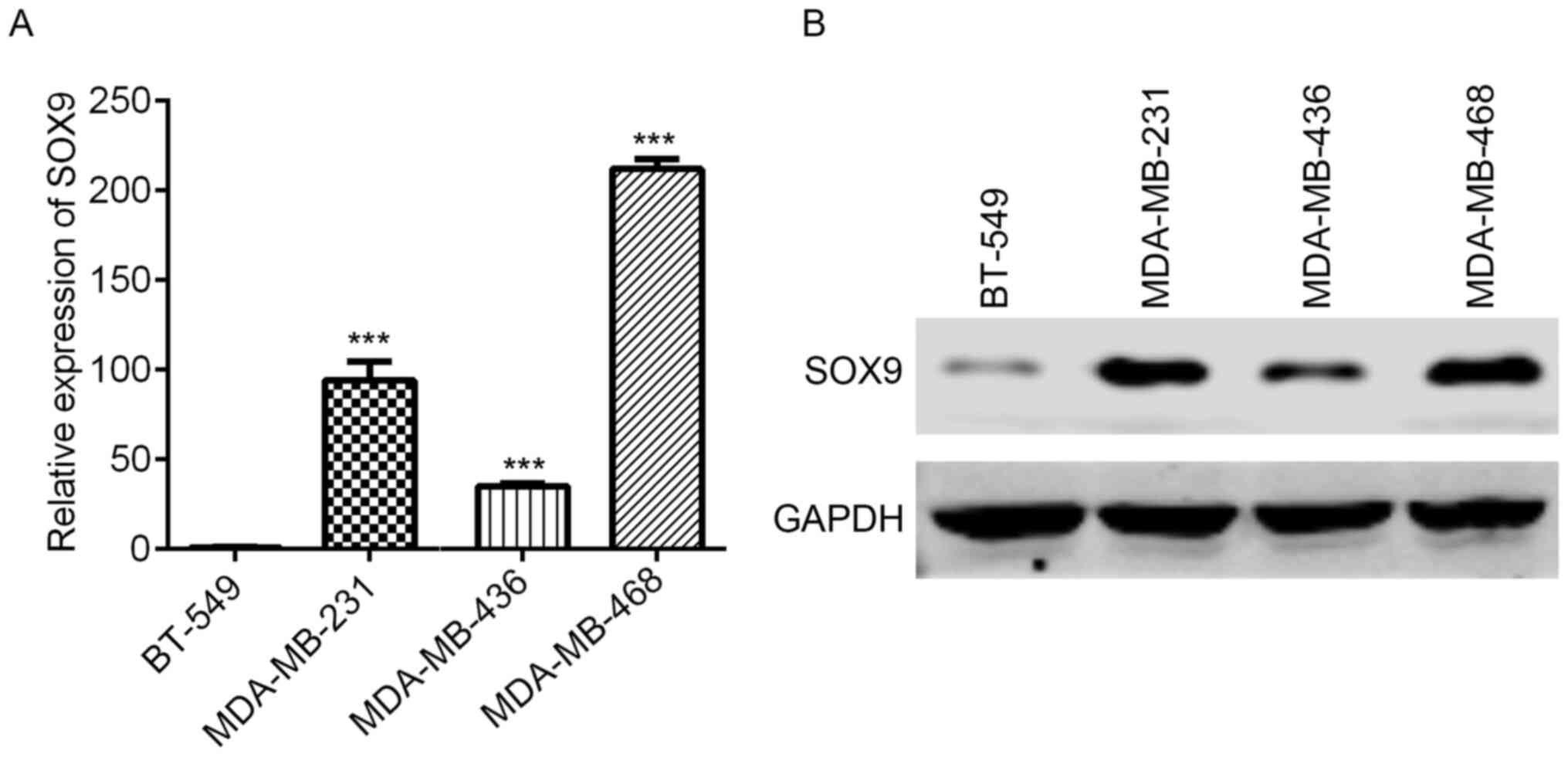
TNBC, SOX9 expression was detected in BT-549, MDA-MB-231,
MDA-MB-436 and MDA-MB-468 cells. SOX9 mRNA expression was lowest in
BT-549 cells, while MDA-MB-436 had moderate SOX9 mRNA abundance,
and MDA-MB-231 and MDA-MB-468 exhibited the highest SOX9 mRNA
expression (Fig. 1A). Western
blotting results revealed that SOX9 was highly expressed in
MDA-MB-231, MDA-MB-468 and MDA-MB-436 cells, while a marginal
signal band was observed in BT-549 cells (Fig. 1B).
Establishment of SOX9-knockdown in
MDA-MB-231 and MDA-MB-468 cells
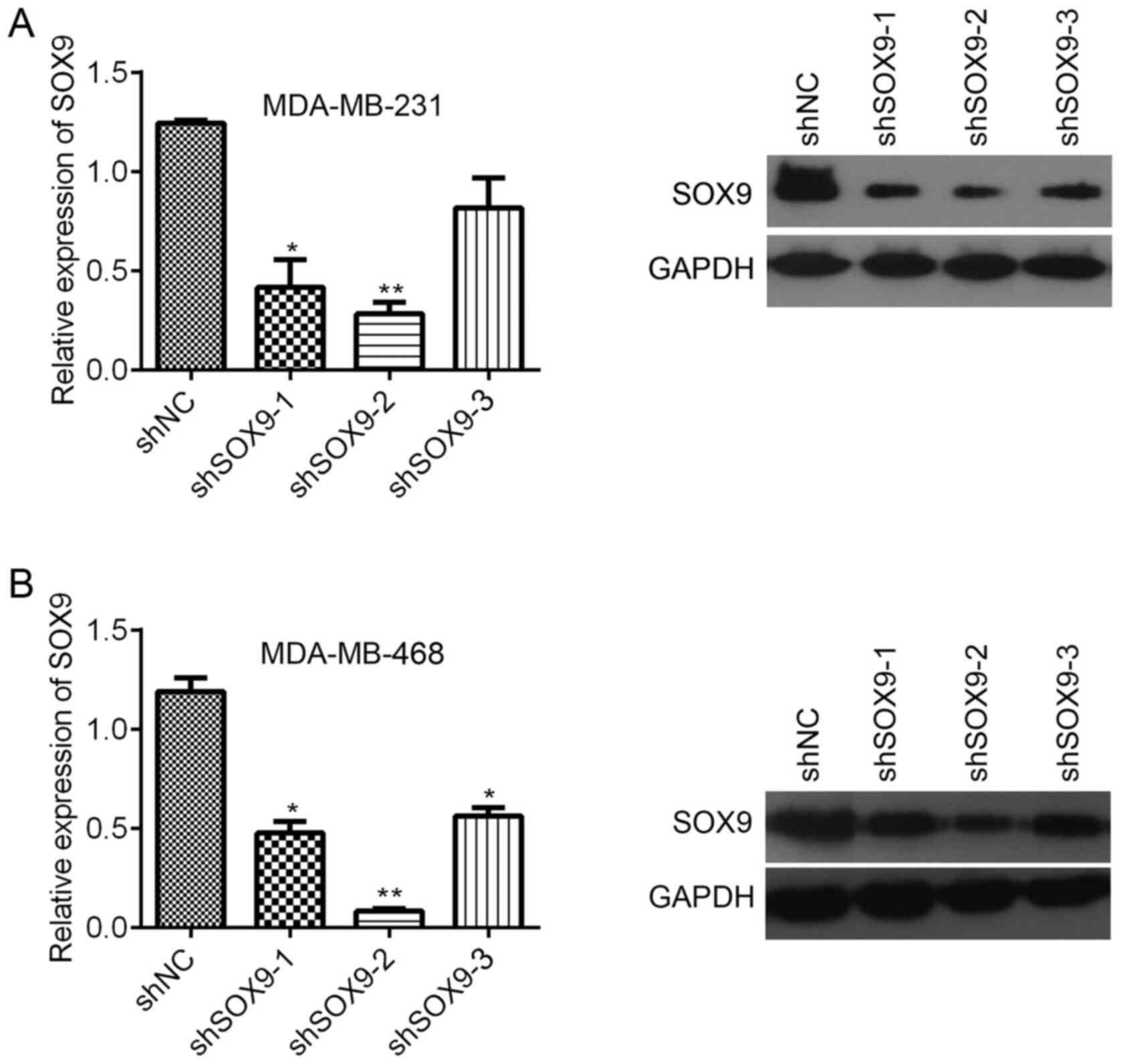
Due to SOX9 was higher expression in both MDA-MB-231
and MDA-MB-468 cells than other TNBC cells MDA-MB-436 and BT-549.
MDA-MB-231 and MDA-MB-468 cells were chosen for subsequent
experiments to study the role of SOX9 in TNBC. Three shRNA
sequences were used to inhibit SOX9 expression in the cells.
Compared with shNC, shSOX9-1 and shSOX9-2 significantly decreased
the SOX9 mRNA expression in MDA-MB-231 cells, while shSOX9-3 did
not significantly inhibit SOX9 mRNA expression (Fig. 2A). Western blotting results revealed
that all of these three shRNAs downregulated SOX9 expression
(Fig. 2A). In MDA-MB-468 cells, all
three shRNAs significantly suppressed SOX9 mRNA expression, with
shSOX9-2 being the most efficient (Fig.
2B). Western blotting results were consistent with the RT-qPCR
results (Fig. 2B). Since shSOX9-2
exhibited the highest knockdown efficiency, it was used for
subsequent experiments.
SOX9-knockdown suppresses the
proliferation and colony formation in TNBC cells
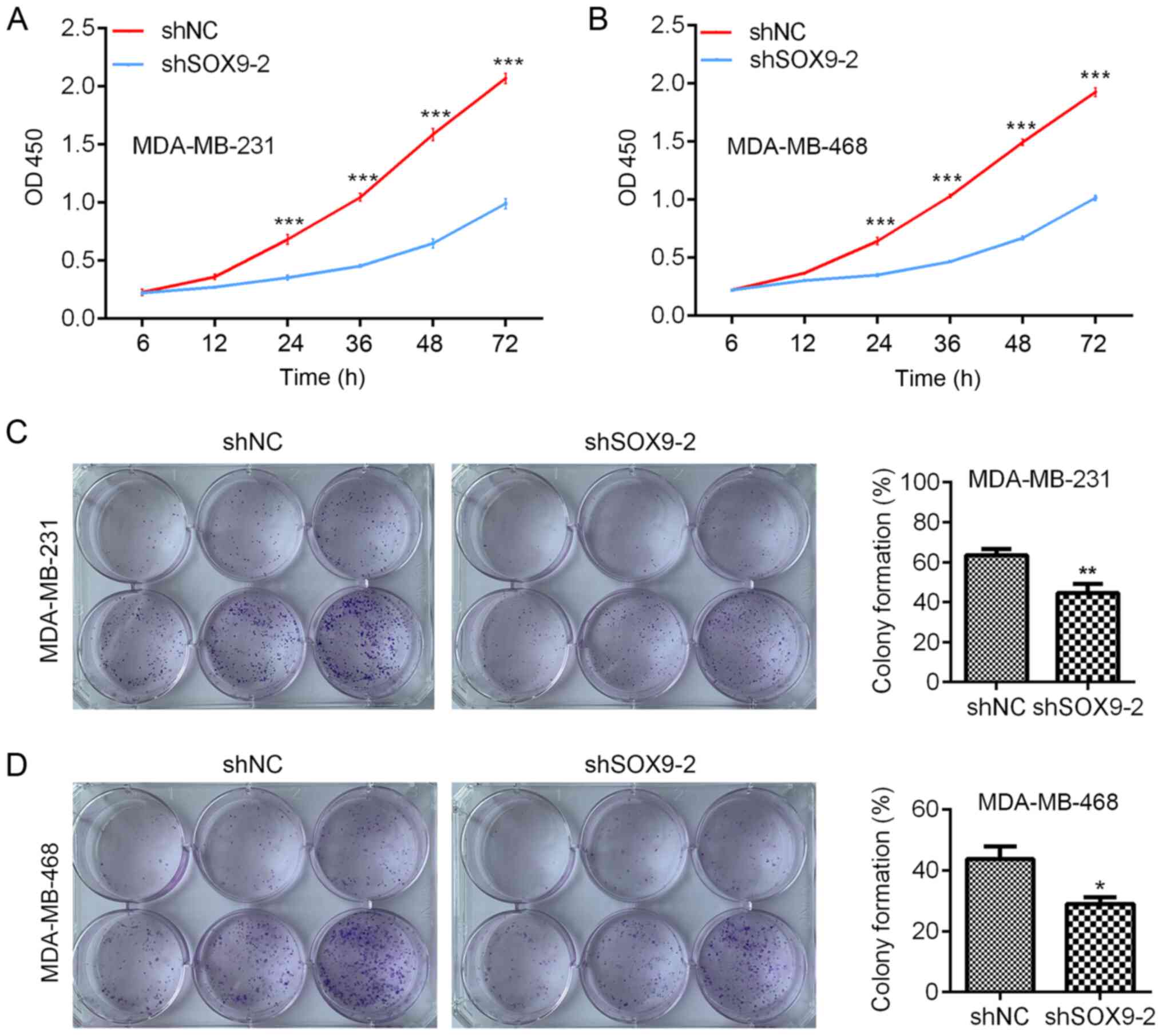
Since SOX9 was efficiently silenced by shSOX9-2,
shNC- and shSOX9-2-transfected MDA-MB-231 and MDA-MB-468 cells were
subjected to cell proliferation analysis. CCK-8 assay was performed
to analyze cell proliferation. The results indicated that, compared
with the shNC, SOX9-knockdown significantly decreased the
proliferation of MDA-MB-231 and MDA-MB-468 cells (Fig. 3A). To validate the results, colony
formation was evaluated in shNC- and shSOX9-2-transfected
MDA-MB-231 and MDA-MB-468 cells. Compared with shNC cells, shSOX9-2
cells exhibited significantly decreased colony formation (Fig. 3B). The current results suggested that
SOX9-knockdown suppressed the proliferation and colony formation of
TNBC cells.
SOX9 regulates apoptosis and the cell
cycle in TNBC cells
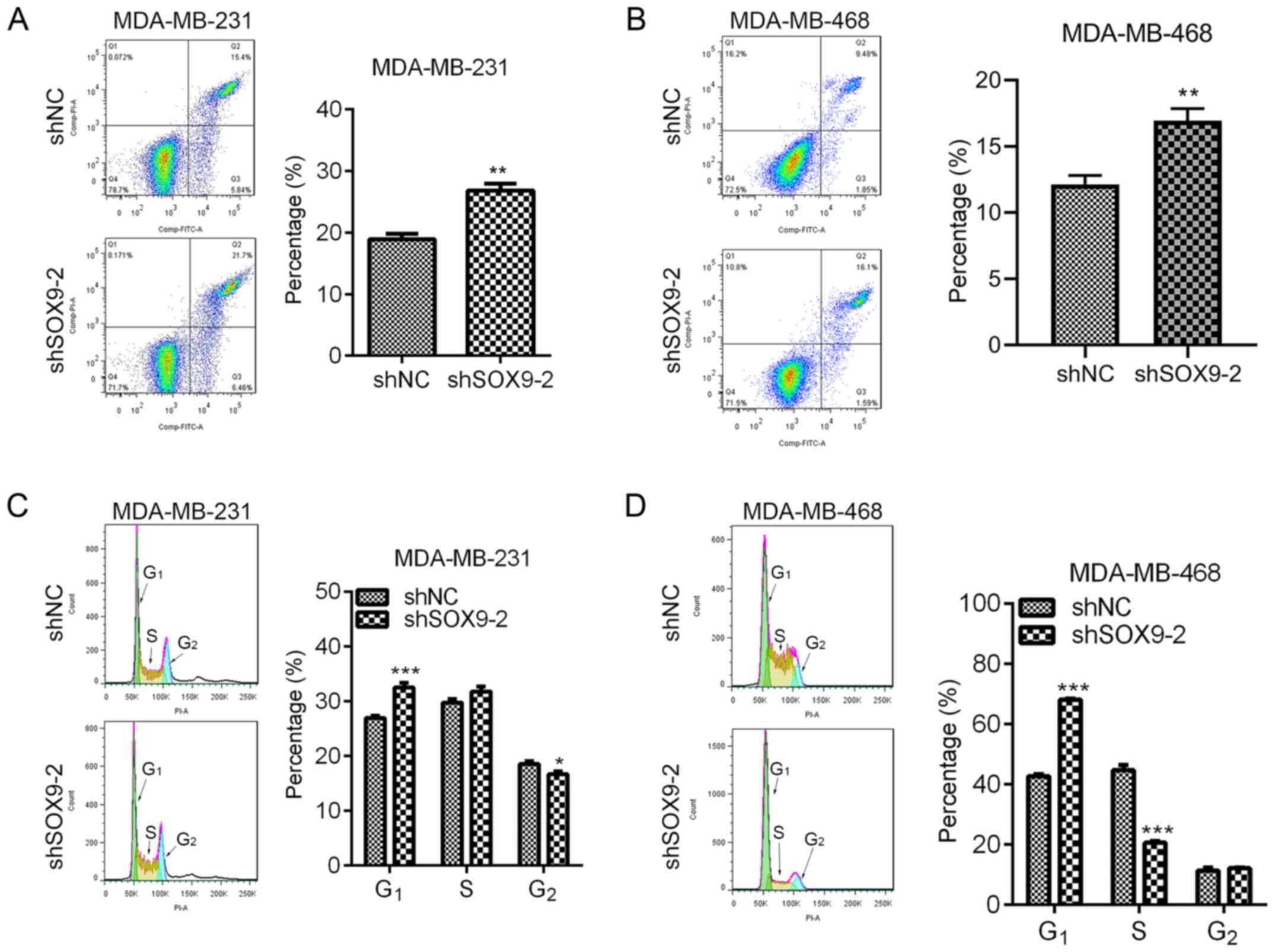
Decreased apoptosis and accelerated cell cycle
progression are common features of cancer cells (18). Whether SOX9 regulated apoptosis and
the cell cycle was explored by staining the cells with PI/Annexin
V-APC and PI, respectively. In MDA-MB-231 and MDA-MB-468 cells,
SOX9-knockdown significantly increased apoptosis (Fig. 4A and B). Additionally, SOX9-knockdown
resulted in significantly increased G1 cell cycle phase
in both cell lines, compared with the shNC (Fig. 4C and D). However, the S phase in
MDA-MB-231 cells remained unchanged, while it was significantly
decreased in MDA-MB-468 cells after SOX9-knockdown, compared with
that in the shNC-transfected cells (Fig.
4C and D). The G2 phase was significantly decreased
in shSOX9-2-transfected MDA-MB-231 cells compared with that in the
shNC-transfected cells, but unchanged in shSOX9-2-transfected
MDA-MB-468 cells (Fig. 4C and D).
Overall, SOX9 silencing caused cell cycle arrest at the
G1 phase.
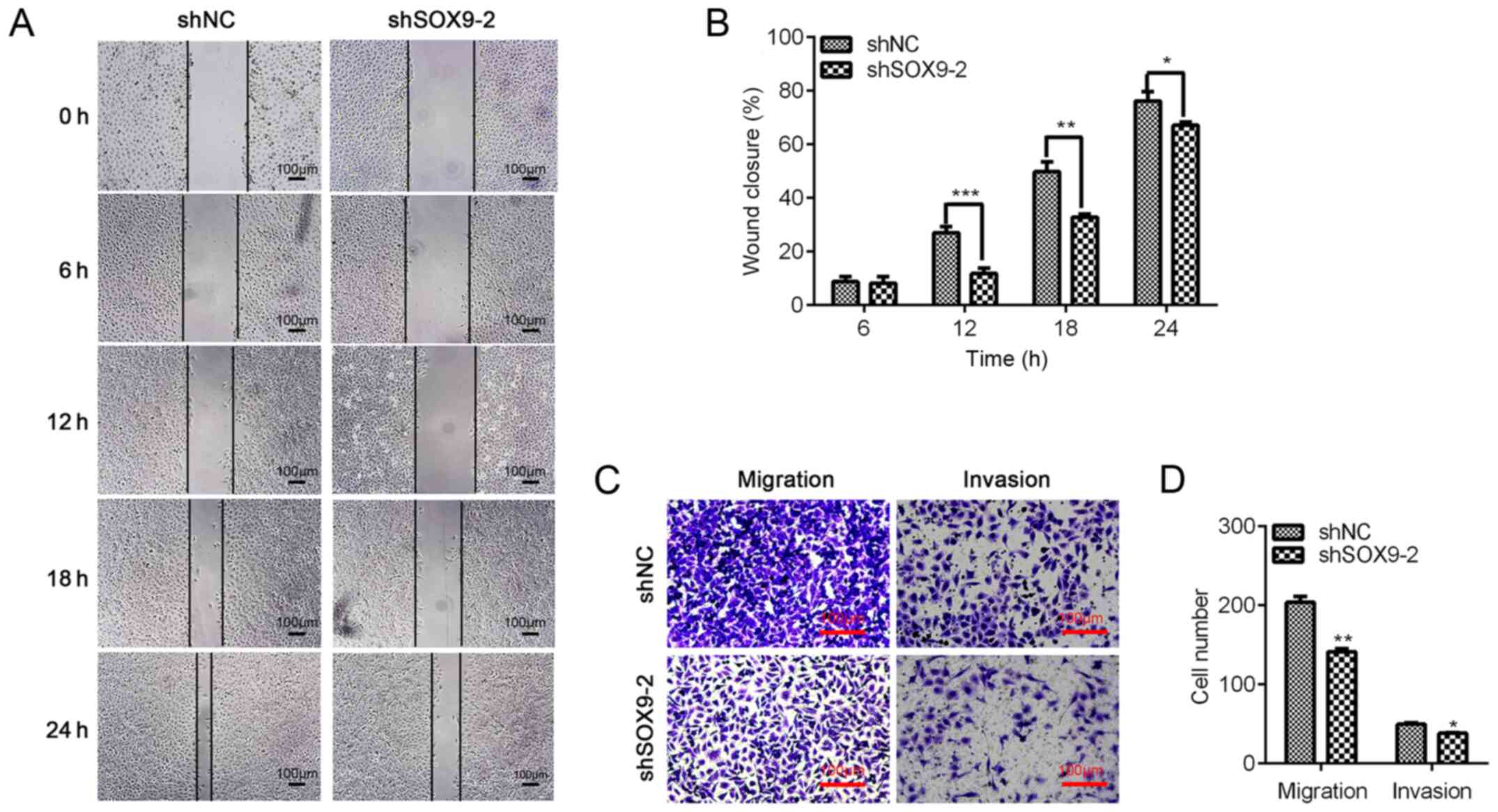
Downregulation of SOX9 inhibits
migration and invasion of TNBC cells
TNBC is a malignant tumor with a high metastasis
rate (19). It has been reported
that SOX9 contributes to the migration and invasion of various
cancer cells (17,20,21).
Therefore, the present study explored whether SOX9-knockdown
regulated the migration of TNBC cells using wound-healing assays.
The results revealed that cell migration was significantly
repressed by SOX9-knockdown at 12, 18 and 24 h (Fig. 5A and B). To validate the effect of
SOX9 on TNBC cell migration and invasion, Transwell assays without
or with Matrigel-coated chambers, respectively, were performed. The
results indicated that, compared with the shNC, SOX9-knockdown
significantly decreased the migration and invasion of TNBC cells
(Fig. 5C and D). The results
suggested that SOX9 may be essential for TNBC cell migration and
invasion.
RNA sequencing of dysregulated genes
after SOX9-knockdown in TNBC cells
To elucidate the downstream factors regulated by
SOX9, RNA sequencing was performed in control and SOX9-knockdown
cells. The results revealed that SOX9-knockdown led to the
upregulation and downregulation of numerous genes (Table SI). The top three upregulated genes
included nucleophosmin (NPM1), thioredoxin reductase 1 (TXNRD1) and
succinate dehydrogenase complex subunit D (SDHD), while the most
downregulated genes were nuclear receptor binding SET domain
protein 2 (NSD2), eukaryotic translation initiation factor 4γ1
(EIF4G1) and glycogen phosphorylase L (PYGL) (Table SII). The expression levels of these
genes in breast invasive carcinoma (BRCA) tissues were analyzed
based on data from TCGA database. The results indicated that the
expression levels of NPM1, TXNRD1, NSD2 and EIF4G1 were upregulated
in BRCA tissues, while PYGL and SDHD was downregulated in BRCA
tissues compared with in normal tissues (Fig. 6).
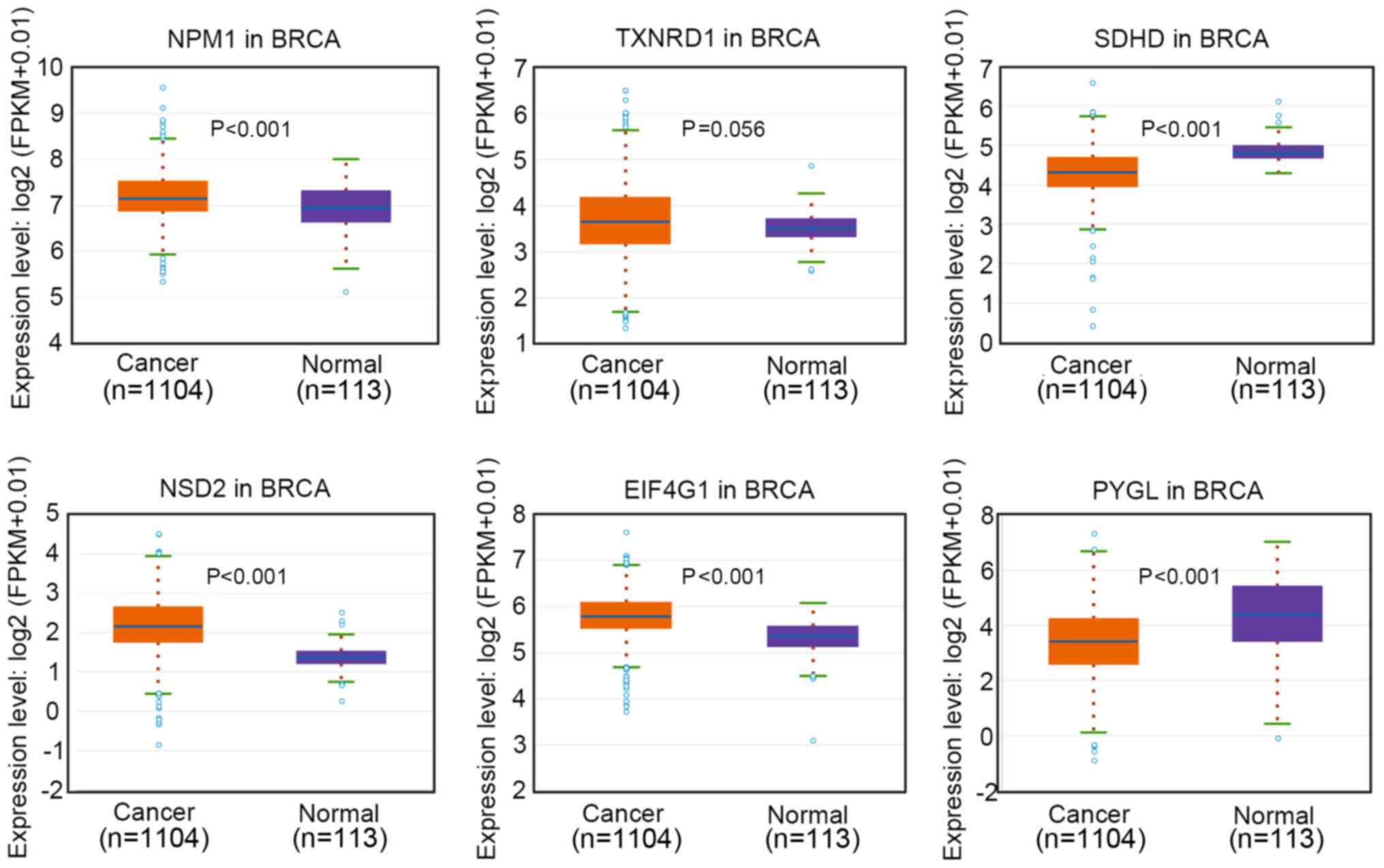 | Figure 6.Relative expression levels of NPM1,
TXNRD1, SDHD, NSD2, EIF4G1 and PYGL in BRCA based on The Cancer
Genome Atlas database. NPM1, nucleophosmin; TXNRD1, thioredoxin
reductase 1; SDHD, succinate dehydrogenase complex subunit D; NSD2,
nuclear receptor binding SET domain protein 2; EIF4G1, eukaryotic
translation initiation factor 4γ1; PYGL, glycogen phosphorylase L;
FPKM, fragments per kilobase of sequence per million mapped reads;
BRCA, breast invasive carcinoma. |
Discussion
Breast cancer poses the greatest threat to the
health of women worldwide. Breast cancer has become the second
leading cause of death in the United States, with 1,150,000 new
cases and 370,000 deaths each year (1). Although TNBC is not the most common
type of breast cancer, it is highly malignant and no effective
drugs are available against this deadly disease (19). Understanding the molecular events
driving this malignancy may help to gain insight into the
pathological process of the disease. In the present study, it was
revealed that SOX9 expression was higher in TNBC MDA-MB-231 and
MDA-MB-468 cells compared with in TNBC BT-549 and MDA-MB-436 cells.
SOX9 expression was critical for the proliferation and migration of
TNBC cells, including MDA-MB-231 and MDA-MB-436 cells.
SOX9-knockdown suppressed the proliferation, colony formation, cell
cycle, migration and invasion, and induced apoptosis of TNBC
cells.
SOX9 is a member of the SOX family, which controls
the expression of numerous genes, such as VGF nerve growth factor
inducible (22,23). Since it is a transcriptional factor,
drugs to inhibit SOX may have highly toxic side effects (22). Based on the current study, SOX9
functions as an oncogene in TNBC. The proliferation, colony
formation, migration and invasion of MDA-MB-231 and MDA-MB-436
cells were significantly suppressed by SOX9-knockdown. However,
knowing the function may not help in our understanding of the
molecular events downstream of SOX9 in TNBC. Therefore, RNA
sequencing was performed in the present study to illustrate the
downstream targets of SOX9. Several newly reported genes regulated
by SOX9 were identified, including NPM1, TXNRD1 and SDHD, which
were upregulated, and NSD2, EIF4G1 and PYGL, which were
downregulated by SOX9-knockdown. Mutations in NPM1 serve an
important role in acute myeloid leukemia (24,25).
NPM1 expression is upregulated in patients with TNBC, and
NPM1-knockdown suppresses TNBC cell proliferation (26), suggesting that NPM1 acts as an
oncogene in TNBC. This may imply that inhibition of TNBC cell
proliferation by SOX9-knockdown may not act via the regulation of
NPM1 expression. TXNRD1 and EIF4G1 expression is upregulated in
distinct malignancies, such hepatocellular carcinoma and lung
cancer (27–29). NSD2 upregulation promotes renal
cancer growth by activating Akt/Erk signaling (30). However, the function of SDHD and PYGL
remains unclear in cancer. Based on TCGA database, the present
study revealed that the expression levels of EIF4G1, NPM1, NSD2 and
TXNRD1 were upregulated, whereas those of PYGL and SDHD were
downregulated in breast cancer tissues compared with those in
normal tissues. Nevertheless, the expression levels of these genes
were analyzed in all breast cancer types from TCGA; therefore, the
expression profile among TNBC tissues and other types of breast
cancer requires further investigation. Therefore, SOX9 may promote
TNBC cell proliferation and migration via regulation of one of the
aforementioned genes. Nevertheless, future studies should be
performed to elucidate the downstream effectors of SOX9 in TNBC.
Future studies may be important for the treatment of patients with
TNBC with SOX9 upregulation.
In summary, the present study revealed that SOX9 was
critical for the proliferation, colony formation, migration and
invasion of TNBC cells. Mechanistically, NPM1, TXNRD1, SDHD, NSD2,
EIF4G1 or PYGL may be downstream effectors of SOX9 in TNBC.
However, further experiments should be performed to clarify the
molecular mechanisms by which SOX9 promotes TNBC cell proliferation
and migration.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by Gansu Health
Industry Planning Project (grant no. GSWSKY2018-06).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Gene Expression Omnibus
repository, under the accession number PRJNA721654.
Authors' contributions
YFW and YXT designed the study, performed the
experiments, and drafted and revised the manuscript. HFD, XL, QQW
and CG conducted some experiments and revised the manuscript. YFW
and YXT confirm the authenticity of all the raw data. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sheikh A, Hussain SA, Ghori Q, Naeem N,
Fazil A, Giri S, Sathian B, Mainali P and Al Tamimi DM: The
spectrum of genetic mutations in breast cancer. Asian Pac J Cancer
Prev. 16:2177–2185. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Harbeck N, Penault-Llorca F, Cortes J,
Gnant M, Houssami N, Poortmans P, Ruddy K, Tsang J and Cardoso F:
Breast cancer. Nat Rev Dis Primers. 5:662019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Early Breast Cancer Trialists'
Collaborative Group (EBCTCG), ; Davies C, Godwin J, Gray R, Clarke
M, Cutter D, Darby S, McGale P, Pan HC, Taylor C, et al: Relevance
of breast cancer hormone receptors and other factors to the
efficacy of adjuvant tamoxifen: Patient-level meta-analysis of
randomised trials. Lancet. 378:771–784. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Piccart-Gebhart MJ, Procter M,
Leyland-Jones B, Goldhirsch A, Untch M, Smith I, Gianni L, Baselga
J, Bell R, Jackisch C, et al: Trastuzumab after adjuvant
chemotherapy in HER2-positive breast cancer. N Engl J Med.
353:1659–1672. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Garrido-Castro AC, Lin NU and Polyak K:
Insights into molecular classifications of triple-negative breast
cancer: Improving patient selection for treatment. Cancer Discov.
9:176–198. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lefebvre V, Dumitriu B, Penzo-Méndez A,
Han Y and Pallavi B: Control of cell fate and differentiation by
Sry-related high-mobility-group box (Sox) transcription factors.
Int J Biochem Cell Biol. 39:2195–2214. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gnerlich JL, Ding X, Joyce C, Turner K,
Johnson CD, Chen H, Abood GJ, Pappas SG and Aranha GV: Increased
SOX9 expression in premalignant and malignant pancreatic neoplasms.
Ann Surg Oncol. 26:628–634. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang JQ, Wei FK, Xu XL, Ye SX, Song JW,
Ding PK, Zhu J, Li HF, Luo XP, Gong H, et al: SOX9 drives the
epithelial-mesenchymal transition in non-small-cell lung cancer
through the Wnt/β-catenin pathway. J Transl Med. 17:1432019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang L, Zhang Z, Yu X, Li Q, Wang Q, Chang
A, Huang X, Han X, Song Y, Hu J, et al: SOX9/miR-203a axis drives
PI3K/AKT signaling to promote esophageal cancer progression. Cancer
Lett. 468:14–26. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mesquita P, Freire AF, Lopes N, Gomes R,
Azevedo D, Barros R, Pereira B, Cavadas B, Pópulo H, Boaventura P,
et al: Expression and clinical relevance of SOX9 in gastric cancer.
Dis Markers. 2019:82670212019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang H, Zhou Y, Mo J, Xiang Q, Qin M, Liu
W, Shang J, Yang Q, Xu W, Yang G, et al: SOX9 represses hepatitis B
virus replication through binding to HBV EnhII/Cp and inhibiting
the promoter activity. Antiviral Res. 177:1047612020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xue Y, Lai L, Lian W, Tu X, Zhou J, Dong
P, Su D, Wang X, Cao X, Chen Y and Wang Q: SOX9/FXYD3/Src axis is
critical for ER+ breast cancer stem cell function. Mol
Cancer Res. 17:238–249. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Trapnell C, Pachter L and Salzberg SL:
TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics.
25:1105–1111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jana S, Madhu Krishna B, Singhal J, Horne
D, Awasthi S, Salgia R and Singhal SS: SOX9: The master regulator
of cell fate in breast cancer. Biochem Pharmacol. 174:1137892020.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Carneiro BA and El-Deiry WS: Targeting
apoptosis in cancer therapy. Nat Rev Clin Oncol. 17:395–417. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Al-Mahmood S, Sapiezynski J, Garbuzenko OB
and Minko T: Metastatic and triple-negative breast cancer:
Challenges and treatment options. Drug Deliv Transl Res.
8:1483–1507. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Siu MKY, Jiang YX, Wang JJ, Leung THY, Han
CY, Tsang BK, Cheung ANY, Ngan HYS and Chan KKL: Hexokinase 2
regulates ovarian cancer cell migration, invasion and stemness via
FAK/ERK1/2/MMP9/NANOG/SOX9 signaling cascades. Cancers (Basel).
11:8132019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang W, Wu Y, Hou B, Wang Y, Deng D, Fu Z
and Xu Z: A SOX9-AS1/miR-5590-3p/SOX9 positive feedback loop drives
tumor growth and metastasis in hepatocellular carcinoma through the
Wnt/β-catenin pathway. Mol Oncol. 13:2194–2210. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Grimm D, Bauer J, Wise P, Krüger M,
Simonsen U, Wehland M, Infanger M and Corydon TJ: The role of SOX
family members in solid tumours and metastasis. Semin Cancer Biol.
67:122–153. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim JY, Bai Y, Jayne LA, Abdulkader F,
Gandhi M, Perreau T, Parikh SV, Gardner DS, Davidson AJ, Sander V,
et al: SOX9 promotes stress-responsive transcription of VGF nerve
growth factor inducible gene in renal tubular epithelial cells. J
Biol Chem. 295:16328–16341. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Prieto-Conde MI, Jiménez C, García-Álvarez
M, Ramos F, Medina A, Cuello R, Balanzategui A, Alonso JM,
Sarasquete ME, Queizán JA, et al: Identification of
relapse-associated gene mutations by next-generation sequencing in
low-risk acute myeloid leukaemia patients. Br J Haematol.
189:718–730. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen Y and Hu J: Nucleophosmin1 (NPM1)
abnormality in hematologic malignancies, and therapeutic targeting
of mutant NPM1 in acute myeloid leukemia. Ther Adv Hematol.
11:20406207198998182020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zeng D, Xiao Y, Zhu J, Peng C, Liang W and
Lin H: Knockdown of nucleophosmin 1 suppresses proliferation of
triple-negative breast cancer cells through activating
CDH1/Skp2/p27kip1 pathway. Cancer Manag Res. 11:143–156. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fu B, Meng W, Zeng X, Zhao H, Liu W and
Zhang T: TXNRD1 is an unfavorable prognostic factor for patients
with hepatocellular carcinoma. Biomed Res Int. 2017:46981672017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Poerschke RL and Moos PJ: Thioredoxin
reductase 1 knockdown enhances selenazolidine cytotoxicity in human
lung cancer cells via mitochondrial dysfunction. Biochem Pharmacol.
81:211–221. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jaiswal PK, Koul S, Palanisamy N and Koul
HK: Eukaryotic translation initiation factor 4 gamma 1 (EIF4G1): A
target for cancer therapeutic intervention? Cancer Cell Int.
19:2242019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Han X, Piao L, Xu X, Luo F, Liu Z and He
X: NSD2 promotes renal cancer progression through stimulating
Akt/Erk signaling. Cancer Manag Res. 12:375–383. 2020. View Article : Google Scholar : PubMed/NCBI
|