Introduction
Circulating tumor cells (CTCs) are derived from the
primary tumor and circulate in the peripheral blood (1). They are considered to play a crucial
role in metastasis formation, which is the leading cause of
cancer-related death. Therefore, the detection and molecular
biological analysis of CTCs may enhance the diagnosis and treatment
of patients with cancer (2–4). Among the various CTC-detection devices
currently available, only the CellSearch system (Menarini Silicon
Biosystems Spa) has been approved by the Food and Drug
Administration for clinical use (5),
and has produced highly reproducible results and demonstrated
clinical relevance in breast, colorectal, prostate and lung cancer
(6–11). However, as CellSearch is an
epithelial cell adhesion molecule (EpCAM)-dependent cell-capture
system, it fails to identify CTCs in non-epithelial tumors with low
EpCAM expression, such as malignant pleural mesothelioma (MPM)
(12). To overcome this diagnostic
gap, in our previous studies, a new microfluidic device system,
named as the ‘Universal CTC-Chip’, was developed, which enables
EpCAM-independent cell capture by attaching various antibodies to a
large number of microposts on the chip surface (13,14). In
addition, the clinical significance of CTCs in MPM was found and
CTC capture was possible using an antibody against podoplanin, a
well-known diagnostic marker of MPM (14–16).
However, podoplanin expression is lower in non-epithelioid MPM
(30-75%) compared with that in epithelioid MPM (80-100%) (17–20),
which may reduce the efficiency of this diagnostic approach.
Furthermore, our previous study showed that CTCs were detected in
92.3% (12/13 patients) of epithelioid MPM cases compared with that
in 33.3% (3/9 patients) of non-epithelioid MPM cases (16), suggesting that some CTC populations
do not express podoplanin, particularly those of the
non-epithelioid subtype. Epidermal growth factor receptor (EGFR) is
a 170 kDa transmembrane protein with intrinsic tyrosine kinase
activity that regulates cell growth (21). EGFR is overexpressed in several
malignancies, including non-epithelioid MPM (22–25). In
recent years, cetuximab, a chimeric antibody targeting EGFR, was
found to be an effective cell-capture antibody compared with that
in other EGFR antibodies, due to its low dissociation constant and
strong cell adhesion ability (26).
In the present study, the following was investigated: i) EGFR
expression in the MPM cell lines; ii) the cell-capture efficiency
of a CTC-chip coated with cetuximab; and iii) whether an antibody
cocktail of podoplanin (clone NZ-1.2) and cetuximab could enhance
CTC capture in all histological subtypes of MPM.
Materials and methods
This research was conducted with the approval of the
Ethics Committee of the University of Occupational and
Environmental Health, Japan (approval no. H26-15).
Cell lines
A total of 5 human cell lines representing different
histological subtypes of MPM were used: Epithelioid MPM:
ACC-MESO-1, ACC-MESO-4, and NCI-H226; biphasic MPM: MSTO-211H; and
sarcomatoid MPM: NCI-H28. The ACC-MESO-1 and ACC-MESO-4 cell lines
were purchased from the Riken BioResource Research Center, while
the NCI-H226, MSTO-211H, and NCI-H28 cell lines were purchased from
the American Type Culture Collection. All the cell lines were
cultured in RPMI-1640 (FUJIFILM Wako Pure Chemical Corporation),
supplemented with 10% fetal bovine serum (Thermo Fisher Scientific,
Inc.) at 37°C in a humidified incubator with 5% CO2.
Flow cytometry analysis
To analyze EGFR expression, the cell lines were
incubated with an anti-EGFR antibody (1:100 dilution; clone 528;
cat. no. sc-120; Santa Cruz Biotechnology, Inc.) for 1 h at room
temperature. The cells were subsequently incubated with goat
anti-mouse IgG antibody conjugated with fluorescein isothiocyanate
for 30 min at room temperature (1:20 dilution; cat. no. 349031; BD
Biosciences). Flow cytometry analysis was performed using an EC800
Cell Analyzer (Sony Biotechnology, Inc.), and the data was analyzed
using FlowJo software version 10 (Becton-Dickinson and Company).
The mean fluorescence intensity was calculated as the ratio of the
positive control to that of the negative control (PBS with 1%
bovine serum albumin; Nacalai Tesque, Inc.).
CTC-chip preparation
The CTC-chip was first coated with antibodies in a
two-step process as previously described (14), then used for the experiments.
Briefly, the CTC-chip was incubated with the base antibody
overnight at 4°C followed by incubation with the capture antibody
at room temperature for 1 h.
In the present study, the CTC-chip was built based
on the following three combinations: i) previously established
NZ-1.2-chip (16) with goat anti-rat
IgG (200 µg/ml; cat. no. 3052-01; SouthernBiotech) as the base
antibody and rat anti-human podoplanin antibody (5,000 µg/ml; clone
NZ-1.2) (27) as the capture
antibody; i) Cetuximab-chip with goat anti-human IgG (200 µg/ml;
cat. no. 2043-01; SouthernBiotech) as the base antibody and
cetuximab (5,000 µg/ml; Bristol-Myers Squib Company) as the capture
antibody and iii) Cocktail-chip with goat anti-rat IgG (200 µg/ml)
+ goat anti-human IgG (200 µg/ml) as base antibodies and rat
anti-human podoplanin antibody (5,000 µg/ml) + cetuximab (2,500
µg/ml) as capture antibodies.
Sample preparation and evaluation of
cell-capture efficiency in the MPM cell lines
Sample preparation and evaluation of cell-capture
efficiency was performed as previously described (14). Tumor cells labeled using the
CellTrace CSFE cell proliferation kit (Thermo Fisher Scientific,
Inc.) were suspended in 1 ml blood collected from a healthy
volunteer (obtained from author MK; single venous blood collection
from the elbow; 100 cells/ml). This sample was added to each
CTC-chip system at a constant flow rate (1.0 ml/h) and monitored
using a fluorescence microscope (CKX41; Olympus Corporation). The
total number of cells added to the CTC-chip (N-total) was
determined by counting the number of cells that passed through the
inlet of the CTC-chip, whereas the number of captured cells
(N-captured) was determined by counting the number of cells that
remained on the CTC-chip. Cell-capture efficiency was calculated as
the N-captured/N-total ×100 (%), and the mean ± SE was calculated
for each sample. Each experiment was performed in triplicate.
Clinical evaluation of CTCs in
patients with MPM
Peripheral blood samples were collected from 19
patients with MPM between January 2018 and August 2020, to assess
CTCs at diagnosis or prior to treatment. A total of 6 ml blood was
collected in a collection tube (BD Vacutainer EDTA-2K; BD
Biosciences), and after sufficient suspension, 1 ml blood was added
to both the NZ-1.2- and Cocktail-chips. The characteristics of the
patients with MPM are summarized in Table I. Clinical stage was determined
according to the guidelines of the International Mesothelioma
Interest Group, version 8 (28). All
patients provided written informed consent to participate in the
study. The cells captured on 2 of the CTC-chips were incubated with
a primary antibody, a rabbit anti-cytokeratin (CK) antibody (1:100
dilution; cat. no. ab9377; Abcam) and a mouse anti-CD45 antibody
(1:100 dilution; cat. no. 304002; clone HI30; BioLegend, Inc.) for
1 h at room temperature, followed by incubation with 30 min of
incubation at room temperature with a secondary antibody, an Alexa
Fluor 594 anti-rabbit IgG antibody (1:100 dilution; cat. no.
A-11037; Thermo Fisher Scientific, Inc.) and an Alexa Fluor 488
anti-mouse IgG antibody (1:100 dilution; cat. no. A-11029; Thermo
Fisher Scientific, Inc.) containing 1 µg/ml Hoechst 33342 (cat. no.
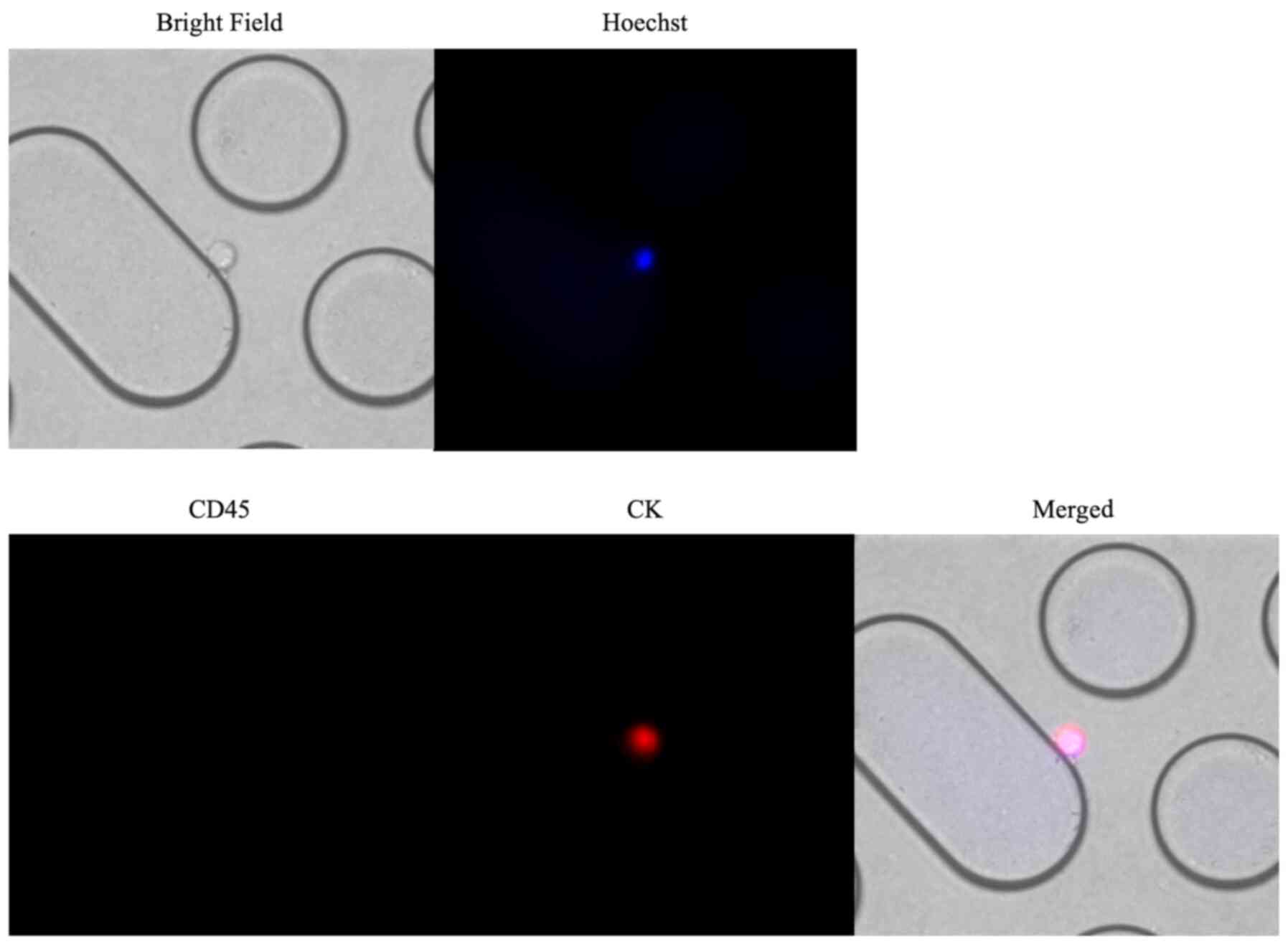
4082; Cell Signaling Technology, Inc.) Cells with round-to-oval
morphology, Hoechst 33342-positive nuclei, CK-positive staining in
the cytoplasm, and CD45-negative staining were identified as CTCs
using a fluorescence microscope at ×10 magnification (DMi8-S2G;
Leica Microsystems), and CTCs were independently identified by two
investigators who were blinded to the clinical data. Survival
analysis according to the CTC-counts in NZ-1.2-chip and
Cocktail-chip was also performed. The median follow-up was 175
(range, 28–1,067) days.
 | Table I.Patient characteristics (n=19). |
Table I.
Patient characteristics (n=19).
| Characteristic | Value |
|---|
| Mean age (range),
years | 69.0 (55–78) |
| Sex, n (%) |
|
| Male | 19 (100.0) |
|
Female | 0 (0.0) |
| Tumor laterality, n
(%) |
|
|
Right | 13 (68.4) |
| Left | 6 (31.6) |
| TNM stage, n (%) |
|
| IA | 2 (10.5) |
| IB | 7 (36.8) |
| II | 2 (10.5) |
| IIIA | 1 (5.3) |
| IIIB | 4 (21.1) |
| IV | 3 (15.8) |
| Histology, n
(%) |
|
|
Epithelioid | 10 (52.6) |
|
Non-epithelioid | 9 (47.4) |
|
Biphasic | 4 (44.4) |
|
Sarcomatous | 5 (55.6) |
In addition, to evaluate non-specific detection,
peripheral blood samples were collected from five healthy
individuals (6 ml; single venous blood collection from the elbow)
and used to detect CTCs in the same manner as aforementioned. The
healthy volunteers were recruited from our laboratory staff and
provided formal written informed consent to participate. Data
collection from the healthy subjects was also included in the
Ethics Committee approval (approval no. H26-15).
Evaluation of podoplanin and EGFR
expression using immunohistochemical staining in the primary
lesions
Serial 4-µm-thick sections were cut from each 10%
formalin-fixed and paraffin-embedded primary tumor specimen
collected by pleural biopsy or radical surgery, then evaluated
using immunohistochemistry staining according to standard
protocols. Sections were heated in 0.01 M citrate buffer (pH 6.0;
cat. no. RM102-C; LSI Medience Corporation) at 98°C for 15 min for
antigen retrieval and incubated in 3% hydrogen peroxide (cat. no.
081-04215; FUJIFILM Wako Pure Chemical Corporation) for 10 min to
inactivate endogenous peroxidase. After blocking with Protein Block
Serum-Free (cat. no. X090930-2; Agilent Technologies, Inc.) for 15
min, sections were incubated with mouse anti-podoplanin monoclonal
antibody (clone D2-40; cat. no. 413451; pre-diluted antibody;
Nichirei Biosciences, Inc.) and rabbit anti-EGFR monoclonal
antibody (1:50 dilution; clone D38B1; cat. no. 4267S; Cell
Signaling Technology, Inc.) for 1 h. Sections were then washed and
incubated with Histofine Simple Stain MAX PO (MULTI) (cat. no.
424152; Nichirei Biosciences, Inc.) for 30 min. Thereafter,
sections were visualized with DAB+ Liquid (cat. no. K346811;
Agilent Technologies, Inc.) for 10 min and counterstained with
hematoxylin for 1 min (cat. no. 30002; Muto Pure Chemicals Co.,
Ltd.). All steps after the antigen retrieval step were performed at
room temperature.
Statistical analysis
Differences between continuous variables were
evaluated using a Wilcoxon signed rank test for paired data, that
was not normally distributed. The correlation between two variables
was analyzed using Spearman's rank correlation analysis. The
Kaplan-Meier method was used to estimate the probability of
survival, with survival differences being analyzed using the
log-rank test. P<0.05 was considered to indicate a statistically
significant difference. All statistical analyses were performed
using SPSS software (version 27.0; IBM Corp.).
Results
Cell-capture efficiency of the three
types of CTC-chip
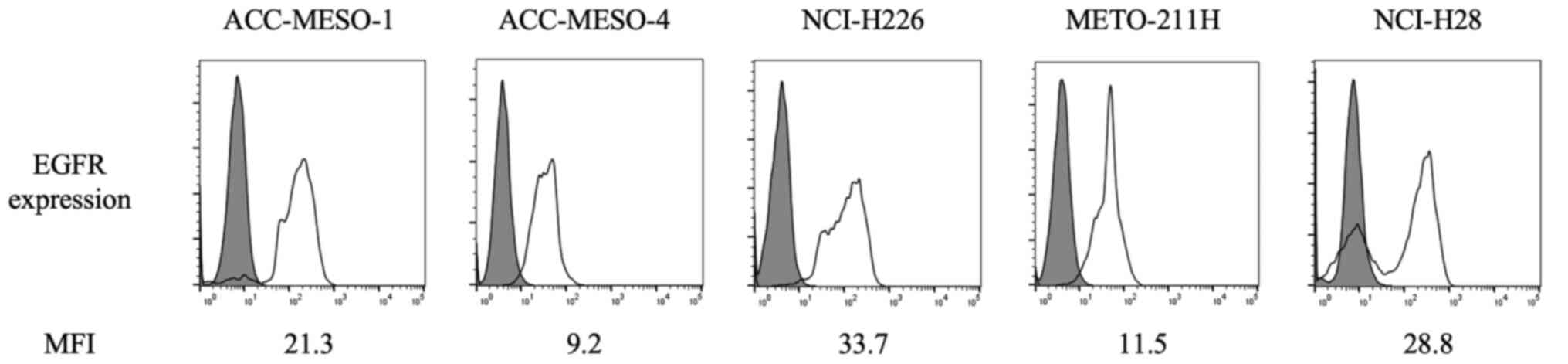
The flow cytometry results of EGFR expression are
shown in Fig. 1. EGFR was detected
in all the cell lines, including biphasic and sarcomatoid MPM
subtypes, which have low podoplanin expression.
The optimal concentration of cetuximab to be used on
the newly designed CTC-chip was determined using the NCI-H226 cell
line. The cell-capture efficiencies were 89.2, 89.1 and 102.5% at
cetuximab concentrations of 500, 1,000 and 5,000 µg/ml,
respectively (data not shown). Therefore, 5,000 µg/ml was used as
the optimal cetuximab concentration for the further experiments.
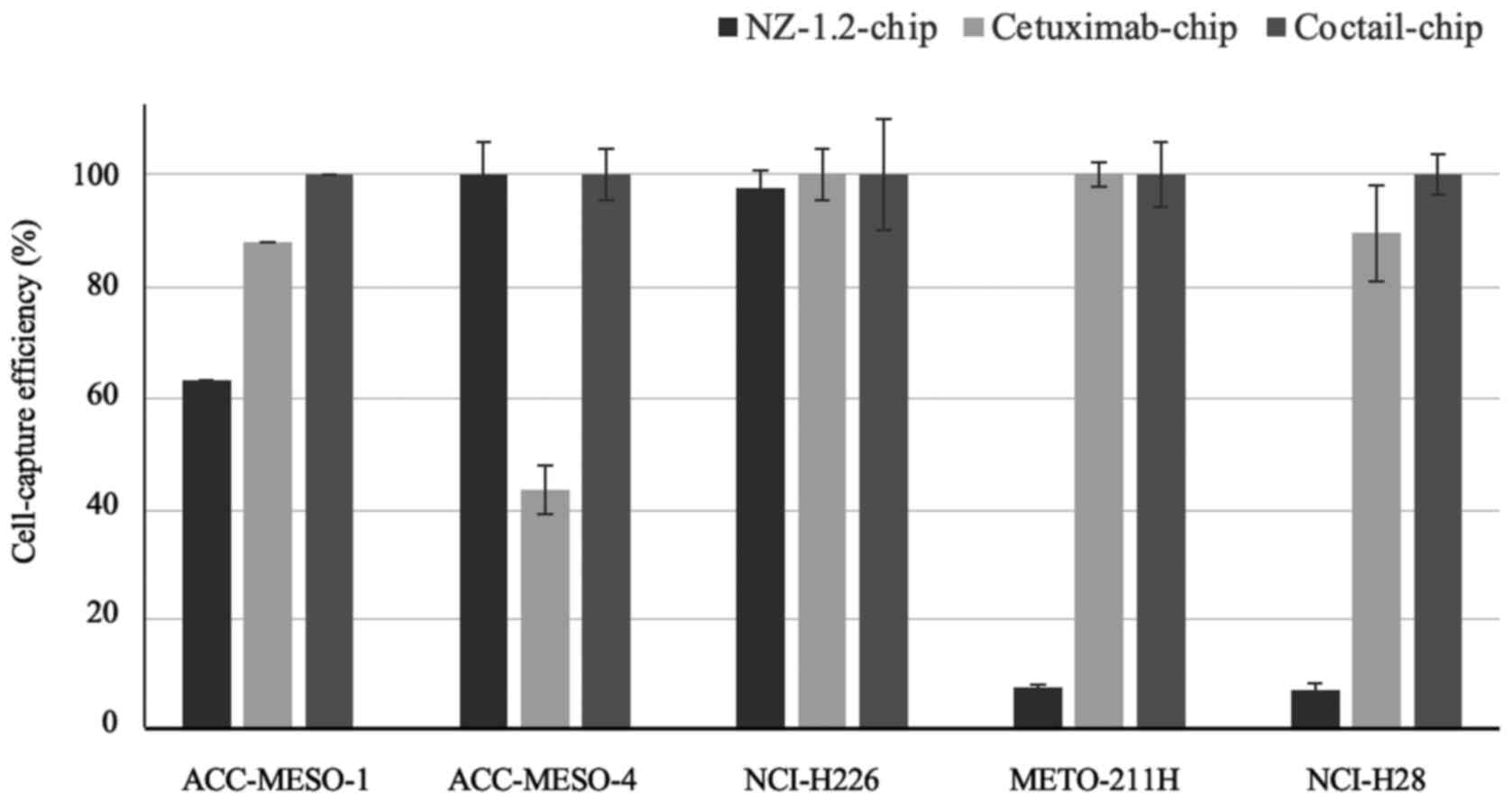
The cell-capture efficiencies of the three types of CTC-chips are
shown in Fig. 2. The epithelioid MPM
cell lines, with high podoplanin expression, were effectively
captured by the NZ-1,2-chip containing podoplanin, but the
cell-capture efficiency for the non-epithelioid cell lines was low.
The Cetuximab-chip, containing the anti-EGFR antibody, had high
cell-capture efficiency in the majority of the cell lines, except
for the ACC-MESO-4 cell line, in which there was a 40% cell-capture
efficiency. The Cocktail-chip, containing antibodies targeting both
podoplanin and cetuximab, reached 100% of a cell-capture efficiency
in all the MPM cell lines; therefore, it was used for clinical
evaluation.
Clinical evaluation of the
Cocktail-chip in patients with MPM
Representative images of immunofluorescent staining
of CTCs captured using the Cocktail-chip in patients with malignant
pleural mesothelioma are shown in Fig.
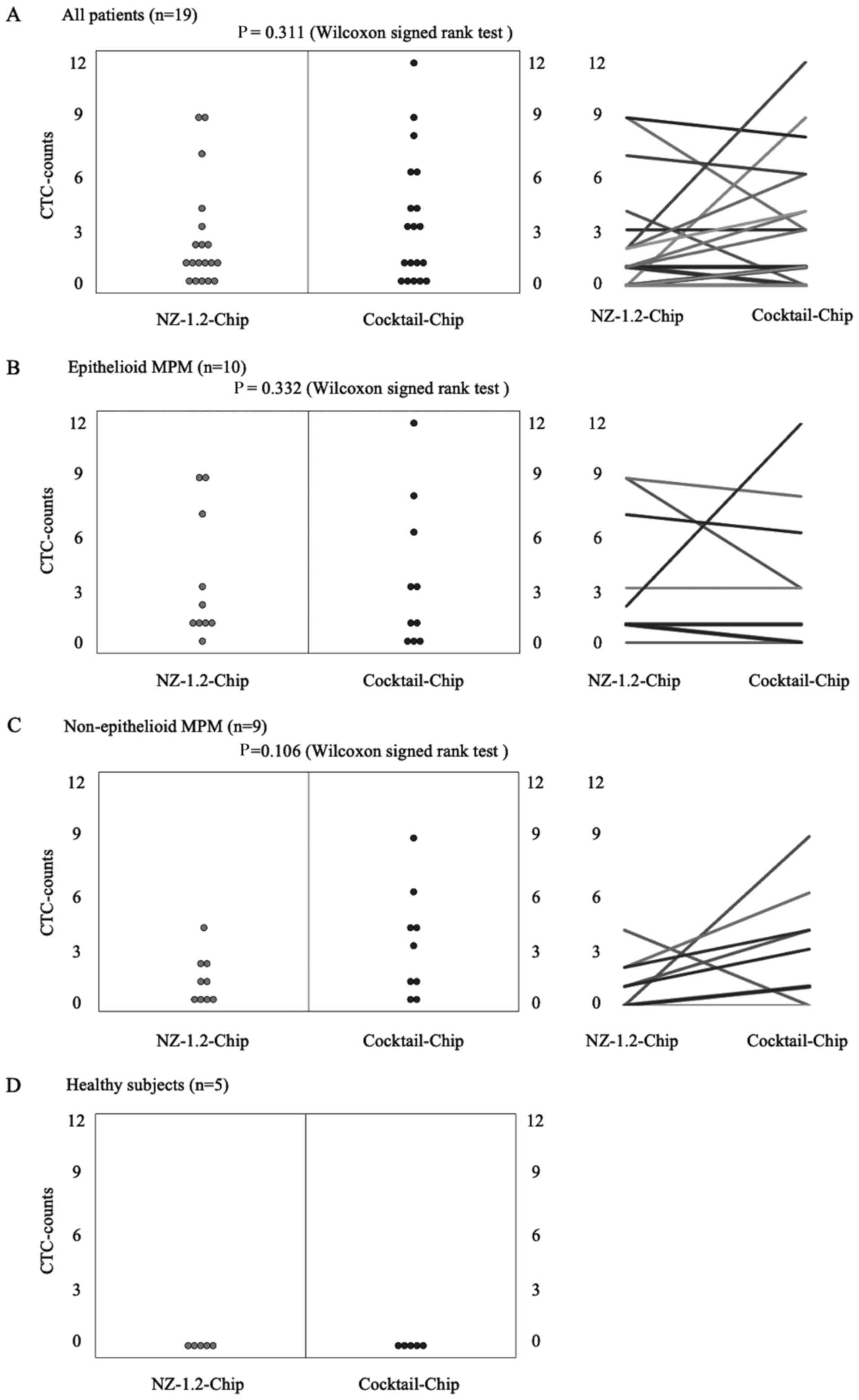
3. Fig. 4 shows the distribution
of CTC-counts using the NZ-1.2- and Cocktail-chips for each sample.
No cells were detected in the five healthy subjects, and no
non-specific detection was observed (Fig. 4D). CTCs were detected in 73.7% of the
samples (14/19 patients) with both the NZ-1.2- and Cocktail-chips.
Furthermore, the NZ-1.2- and Cocktail-chips detected CTCs in 90
(9/10 patients) and 70% (7/10 patients) of samples with epithelioid
MPM, and in 55.6 (5/9 patients) and 77.7% (7/9 patients) of samples
with non-epithelioid MPM, respectively. No clusters, such as
clusters of tumor cells or clusters of tumor cells and white blood
cells, were observed. The median CTC-counts using the NZ-1.2- and
Cocktail-chips were 1 (range, 0–9) and 3 (range, 0–12) in the
overall population (P=0.311), 1.5 (range, 0–9) and 2 (range, 0–12)
in epithelioid MPM (P=0.332), and 1 (range, 0–4) and 3 (range, 0–9)
in non-epithelioid MPM (P=0.106), respectively. There was no
significant difference between the two chips; however, the
Cocktail-chip detected more CTCs in non-epithelioid MPM, suggesting
it was more effective at detecting CTCs. In addition, the
Cocktail-chip achieved a CTC-detection efficiency equivalent to
that of the conventional NZ-1.2-chip in epithelioid MPM, that is
there was no change in the number of epithelial cells (Fig. 4B).
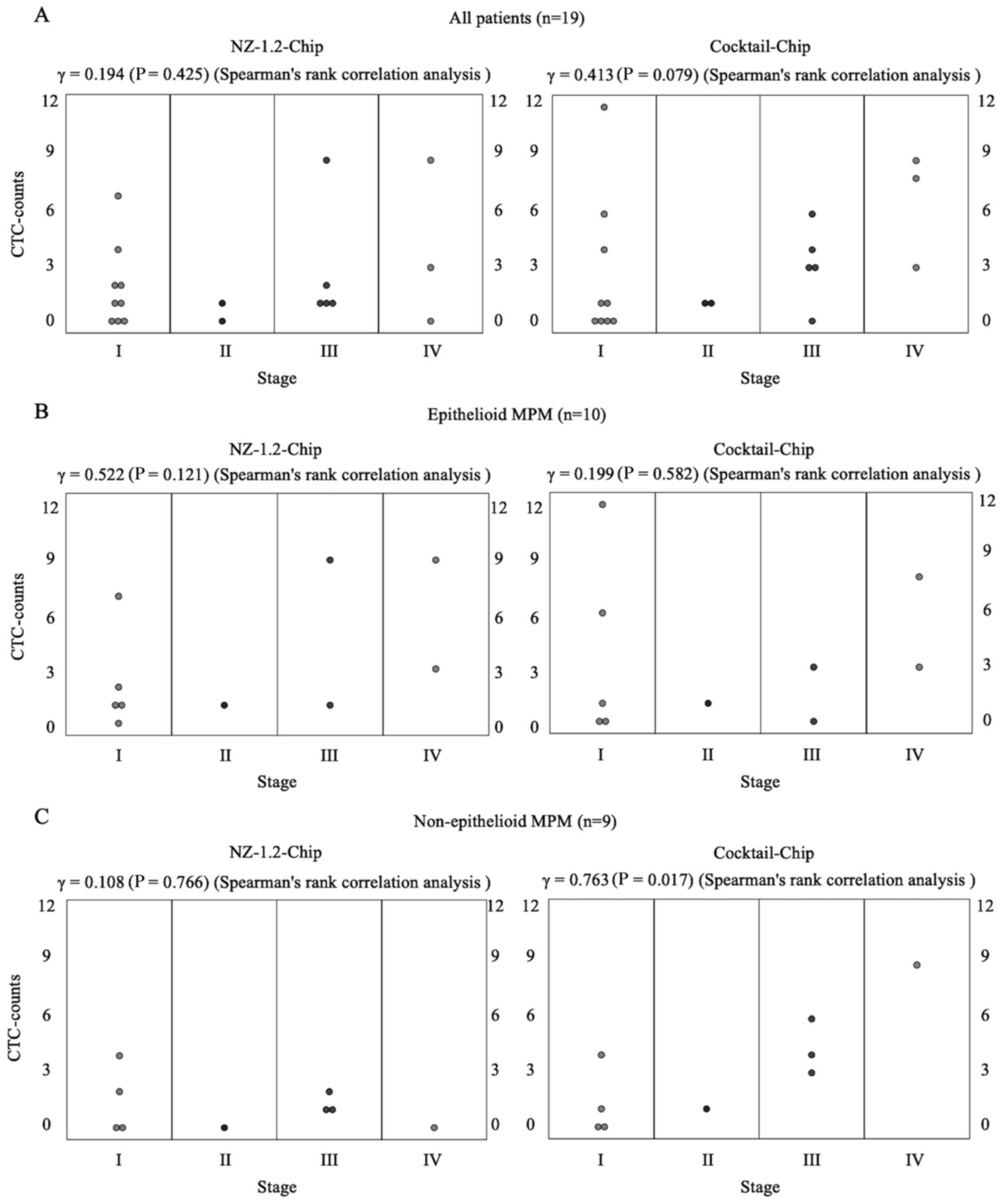
The correlation coefficient between CTC-counts and
clinical stage in all patients was 0.194 (P=0.425) with the
NZ-1.2-Chip and 0.413 (P=0.079) with the Cocktail-chip. Based on
the histology data, the correlation coefficients in epithelioid and
non-epithelioid MPM were 0.522 (P=0.121) and 0.199 (P=0.582) for
CTC-counts with the NZ-1.2-Chip and 0.108 (P=0.766) and 0.763
(P=0.017) with the Cocktail-chip, respectively (Fig. 5). These data indicated that the
CTC-counts using the Cocktail-chip were significantly correlated
with the clinical stage of non-epithelioid MPM.
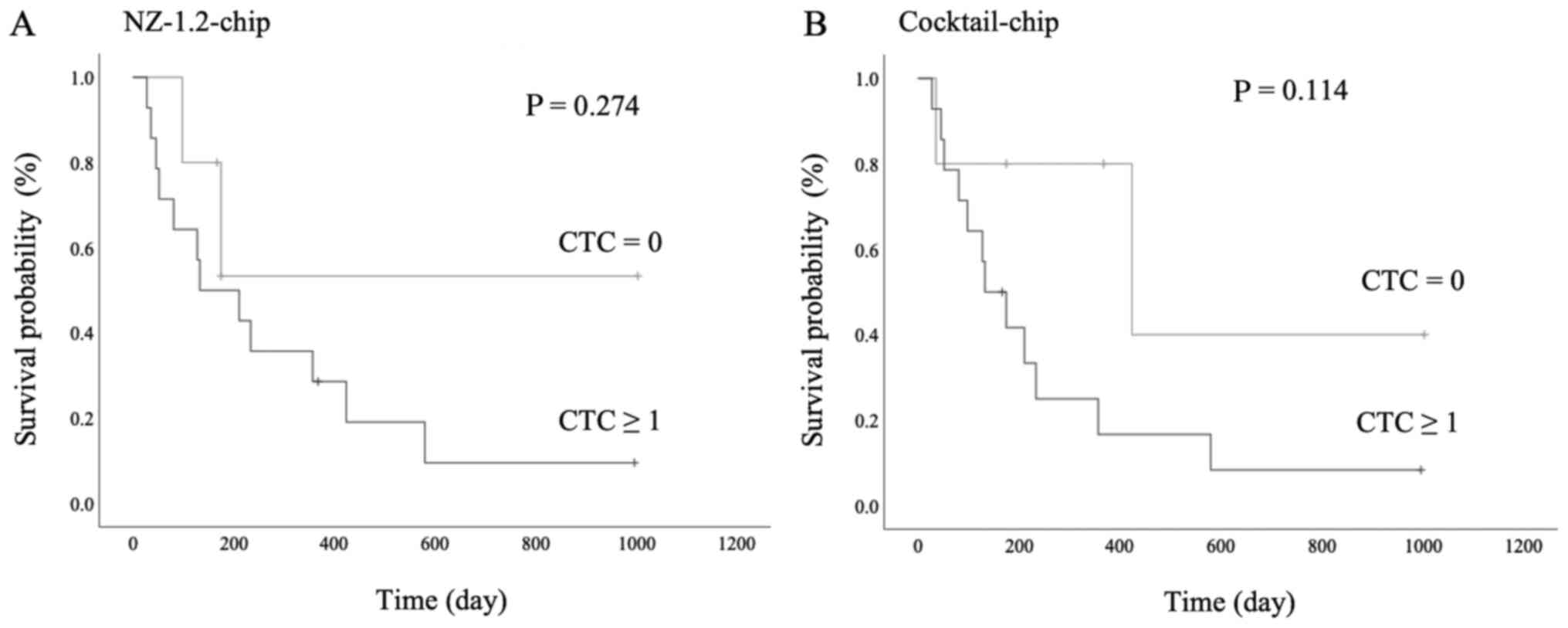
The survival analysis is shown in Fig. 6. There was no statistically
significant difference between the presence of CTCs and prognosis
in both the NZ-1.2- and Cocktail-chips; however, patients with CTCs
detected had a poorer prognosis (P=0.274 and P=0.114,
respectively). Furthermore, in patients with stage I MPM, one
patient without CTCs detected survived without progression for
>3 years following surgery, whereas 2 patients with CTCs
detected showed early progression and died following treatment.
More specifically, case 1 (NZ-1.2-chip:2, Cocktail-chip:12) died 4
months following chemotherapy due to tumor progression, including
distant metastasis, and case 2 (NZ-1.2-chip:2, Cocktail chip:4)
died 4 months following surgery due to locally advanced tumor
progression (data not shown).
Evaluation of podoplanin and EGFR
expression using immunohistochemical staining in primary
lesions
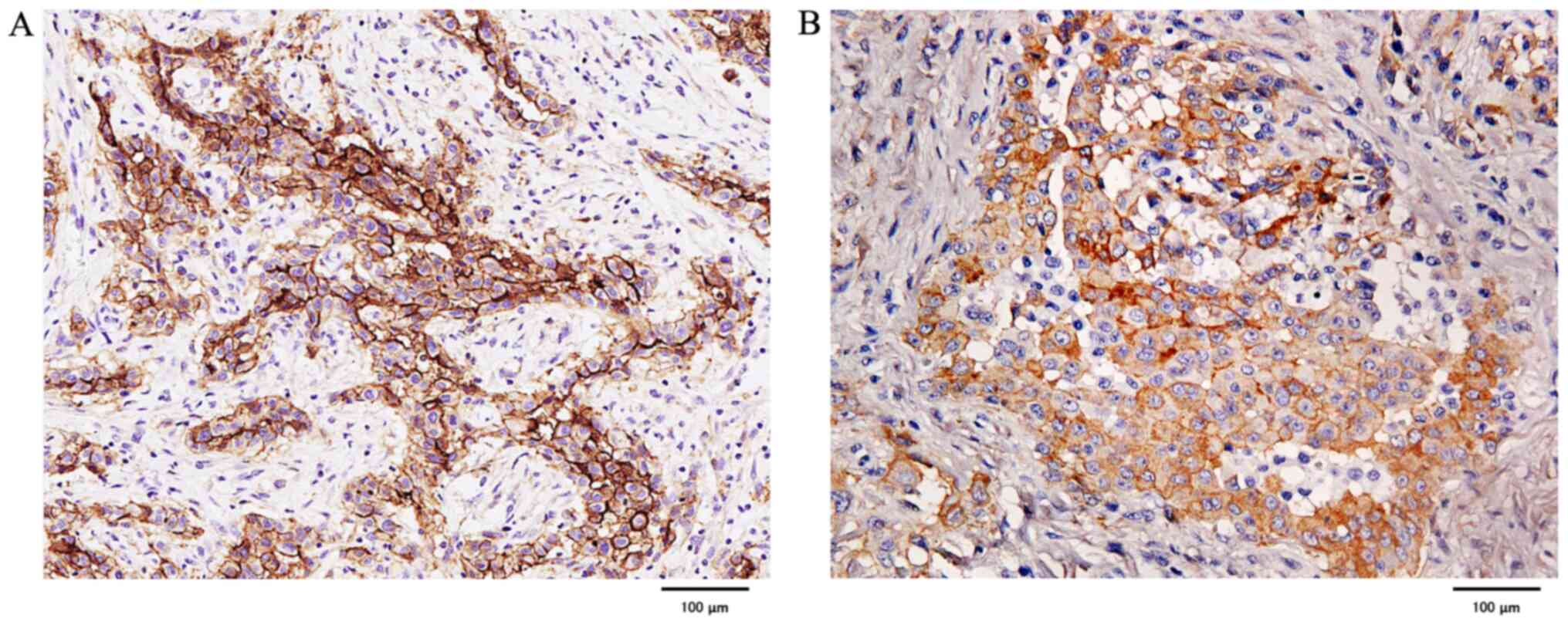
Podoplanin staining was strongly positive in all
cases of epithelioid MPM, whereas it was negative or weakly
positive in 3 out of 5 cases of sarcomatoid MPM. In biphasic MPM,
the epithelial component was strongly positive in all four cases,
whereas the sarcoma component was negative or weakly positive in
some cases. In contrast, there were no negative cases of EGFR, and
all were strongly positive except for 2 epithelioid and 1 biphasic
MPM, which were weakly positive (Fig.
7).
Discussion
In the present study, EGFR expression was confirmed
in all types of the MPM cancer cell lines. By combining with
cetuximab (Cocktail-chip), the cell-capture efficiency was higher
in non-epithelioid MPM-derived CTCs compared with that in the
conventional NZ-1.2-chip, which was designed to only detect
podoplanin.
Effective capture of rare CTCs presents technical
challenges. A common strategy is EpCAM-dependent isolation, as is
used in CellSearch, because epithelial tumor cells express EpCAM.
However, EpCAM-dependent methods may not be applicable to the
isolation of non-epithelial tumor cells (15), and CTCs in MPM have not been fully
investigated so far. In our previous studies, a CTC capture system
for MPM using anti-podoplanin antibodies (clone E1) (14,15) and
NZ-1.2 (16) was developed, and
demonstrated its clinical significance. However, that system did
not detect CTCs in non-epithelioid MPM, such as sarcomatoid and
biphasic subtypes, which are particularly aggressive (29). Therefore, the present study focused
on EGFR as it is widely expressed in several types of tumor, such
as gastric, liver, colorectal and lung cancer (21). EGFR expression was observed in
non-epithelioid MPM cell lines with poor podoplanin expression,
indicating that EGFR represents a suitable target for CTC
detection. The Cetuximab-chip showed high cell-capture efficiency;
however, this chip exhibited a low capture efficiency for one
epithelioid subtype cell line (ACC-MESO-4). Thus, to overcome this
possible detection limitation of cetuximab, a new chip was designed
by combining antibodies targeting podoplanin and cetuximab. This
dual detection approach had a high capture efficiency in all
histological MPM cell lines. In a clinical setting, the
Cocktail-chip also achieved a high cell-capture efficiency, as well
as correlation with clinical stage in non-epithelioid subtype MPM
cases. In addition, the immunostaining results showed that there
were several cases of poor expression of podoplanin in
non-epithelioid MPM. These results suggested that cetuximab could
be used in combination with podoplanin to support CTC detection in
the non-epithelioid subtype. Furthermore, the Cocktail-chip
achieved a CTC-detection efficiency equivalent to that of the
conventional NZ-1.2-chip in epithelioid MPM. Therefore, the results
from the present study suggested that the Cocktail-chip could be
used for CTC detection in all histological MPM cases.
In the present study, the prognostic analysis was
not sufficient, due to the low number of cases and the observation
period was short in some cases. There was no association between
CTC detection and prognosis; however, patients in which CTCs were
detected there was a poorer prognosis, which may indicate that CTC
detection could be of benefit in treatment selection, such as
prioritizing chemotherapy over surgery in patients in which CTCs
were detected in the early stage. In addition, distant metastasis
is a rare progression event in MPM and mainly occurs due to local
metastasis; however, based on the results for case 2 in the present
study and a reported case in which an increased number of CTCs
during CTC monitoring contributed to detection of local progression
(16), CTC detection in MPM may
indicate local progression as well as distant metastasis. These
clinical findings should be verified in further studies with larger
numbers of cases.
The present study has several limitations. First,
only 19 patients were included in this study; therefore, the cohort
is too small to draw a definitive conclusion on the efficiency of
the Cocktail-chip. Second, CTC detection and counting was performed
only once for each patient. Therefore, collecting peripheral blood
samples from newly diagnosed and treated patients with MPM is
currently ongoing, which will verify the prognosis and predictive
value of the general CTC-detection system. Third, there may have
been variability in the experiment as 1 ml blood from each patient
was used for each chip, and the experimental protocol was not fully
automated. Finally, in the current study, it was not validated
whether the cells captured with the CTC-chip were true MPM cells.
To resolve these system-related limitations, the application of a
fully automated system using the automated pipetting system
‘EDR-24LX’ (BIOTEC Co., Ltd.) is being evaluated and developing a
protocol for genetically analyzing cells captured by the CTC-chip
using the micromanipulator ‘M-152’ (NARISHIGE Group).
In conclusion, the novel Cocktail-chip was more
effective at capturing CTCs from all histological MPMs, including
the non-epithelioid subtype. Further studies may reveal the
clinical value of MPM-derived CTCs captured using the
Cocktail-chip. Based on the results from the present study, this
Cocktail-chip system may assist in the development of novel
diagnostic, therapeutic and prognostic options for monitoring MPM
progression.
Acknowledgements
Not applicable.
Funding
This study was supported by the University of
Occupational and Environmental Health Research Grant for Promotion
of Occupational Health (grant no. 902) and Japan Agency for Medical
Research and Development (AMED) under grant nos. JP20am0401013,
JP20am0101078, JP20ae0101028 and JP20bm1004001.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MK, KY, TO, YK and FT contributed to the conception
and design of the study. AT, SS, TK, MT and KK provided the samples
and analyzed the data. MK, RO and MM performed the cell
experiments. TO performed the CTC-Chip and provided technical
support. YK provided the capture antibodies. MK drafted the
manuscript. TO and YK revised manuscript for important intellectual
content. KY and FT supervised the study and revised the manuscript.
All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
This research was conducted with the approval of the
Ethics Committee of the University of Occupational and
Environmental Health, Japan (approval no. H26-15). All patients
provided written informed consent to participate in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CTCs
|
circulating tumor cells
|
|
EGFR
|
epidermal growth factor receptor
|
|
EpCAM
|
epithelial cell adhesion molecule
|
|
MPM
|
malignant pleural mesothelioma
|
References
|
1
|
Tanaka F, Yoneda K and Hasegawa S:
Circulating tumor cells (CTCs) in lung cancer: Current status and
future perspectives. Lung Cancer (Auckl). 1:77–84. 2010.PubMed/NCBI
|
|
2
|
Alix-Panabières C, Riethdorf S and Pantel
K: Circulating tumor cells and bone marrow micrometastasis. Clin
Cancer Res. 14:5013–5021. 2008. View Article : Google Scholar
|
|
3
|
Siravegna G, Marsoni S, Siena S and
Bardelli A: Integrating liquid biopsies into the management of
cancer. Nat Rev Clin Oncol. 14:531–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cabel L, Proudhon C, Gortais H, Loirat D,
Coussy F, Pierga JY and Bidard FC: Circulating tumor cells:
Clinical validity and utility. Int J Clin Oncol. 22:421–430. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Allard WJ, Matera J, Miller MC, Repollet
M, Connelly MC, Rao C, Tibbe AG, Uhr JW and Terstappen LW: Tumor
cells circulate in the peripheral blood of all major carcinomas but
not in healthy subjects or patients with nonmalignant diseases.
Clin Cancer Res. 10:6897–6904. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cristofanilli M, Budd GT, Ellis MJ,
Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ,
Terstappen LW, et al: Circulating tumor cells, disease progression,
and survival in metastatic breast cancer. N Engl J Med.
351:781–791. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cohen SJ, Punt CJ, Iannotti N, Saidman BH,
Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, et
al: Relationship of circulating tumor cells to tumor response,
progression-free survival, and overall survival in patients with
metastatic colorectal cancer. J Clin Oncol. 26:3213–3221. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
de Bono JS, Scher HI, Montgomery RB,
Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ
and Raghavan D: Circulating tumor cells predict survival benefit
from treatment in metastatic castration-resistant prostate cancer.
Clin Cancer Res. 14:6302–6309. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Krebs MG, Sloane R, Priest L, Lancashire
L, Hou JM, Greystoke A, Ward TH, Ferraldeschi R, Hughes A, Clack G,
et al: Evaluation and prognostic significance of circulating tumor
cells in patients with non-small-cell lung cancer. J Clin Oncol.
29:1556–1563. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Miller MC, Doyle GV and Terstappen LW:
Significance of Circulating Tumor Cells Detected by the CellSearch
System in Patients with Metastatic Breast Colorectal and Prostate
Cancer. J Oncol. 2010:6174212010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tanaka F, Yoneda K, Kondo N, Hashimoto M,
Takuwa T, Matsumoto S, Okumura Y, Rahman S, Tsubota N, Tsujimura T,
et al: Circulating tumor cell as a diagnostic marker in primary
lung cancer. Clin Cancer Res. 15:6980–6986. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yoneda K, Tanaka F, Kondo N, Hashimoto M,
Takuwa T, Matsumoto S, Okumura Y, Tsubota N, Sato A, Tsujimura T,
et al: Circulating tumor cells (CTCs) in malignant pleural
mesothelioma (MPM). Ann Surg Oncol. 21 (Suppl 4):S472–S480. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ohnaga T, Shimada Y, Moriyama M, Kishi H,
Obata T, Takata K, Okumura T, Nagata T, Muraguchi A and Tsukada K:
Polymeric microfluidic devices exhibiting sufficient capture of
cancer cell line for isolation of circulating tumor cells. Biomed
Microdevices. 15:611–616. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chikaishi Y, Yoneda K, Ohnaga T and Tanaka
F: EpCAM-independent capture of circulating tumor cells with a
‘universal CTC-chip’. Oncol Rep. 37:77–82. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yoneda K, Kuwata T, Chikaishi Y, Mori M,
Kanayama M, Takenaka M, Oka S, Hirai A, Imanishi N, Kuroda K, et
al: Detection of circulating tumor cells with a novel microfluidic
system in malignant pleural mesothelioma. Cancer Sci. 110:726–733.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kuwata T, Yoneda K, Mori M, Kanayama M,
Kuroda K, Kaneko MK, Kato Y and Tanaka F: Detection of circulating
tumor cells (CTCs) in malignant pleural mesothelioma (MPM) with the
‘Universal’ CTC-Chip and an anti-podoplanin antibody NZ-1.2. Cells.
9:8882020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Husain AN, Colby TV, Ordóñez NG, Allen TC,
Attanoos RL, Beasley MB, Butnor KJ, Chirieac LR, Churg AM, Dacic S,
et al: Guidelines for Pathologic Diagnosis of Malignant
Mesothelioma 2017 Update of the Consensus Statement From the
International Mesothelioma Interest Group. Arch Pathol Lab Med.
142:89–108. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ordóñez NG: D2-40 and podoplanin are
highly specific and sensitive immunohistochemical markers of
epithelioid malignant mesothelioma. Hum Pathol. 36:372–380. 2005.
View Article : Google Scholar
|
|
19
|
Hinterberger M, Reineke T, Storz M, Weder
W, Vogt P and Moch H: D2-40 and calretinin - a tissue microarray
analysis of 341 malignant mesotheliomas with emphasis on
sarcomatoid differentiation. Mod Pathol. 20:248–255. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Padgett DM, Cathro HP, Wick MR and Mills
SE: Podoplanin is a better immunohistochemical marker for
sarcomatoid mesothelioma than calretinin. Am J Surg Pathol.
32:123–127. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Salomon DS, Brandt R, Ciardiello F and
Normanno N: Epidermal growth factor-related peptides and their
receptors in human malignancies. Crit Rev Oncol Hematol.
19:183–232. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Destro A, Ceresoli GL, Falleni M, Zucali
PA, Morenghi E, Bianchi P, Pellegrini C, Cordani N, Vaira V,
Alloisio M, et al: EGFR overexpression in malignant pleural
mesothelioma. An immunohistochemical and molecular study with
clinico-pathological correlations. Lung Cancer. 51:207–215. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dazzi H, Hasleton PS, Thatcher N, Wilkes
S, Swindell R and Chatterjee AK: Malignant pleural mesothelioma and
epidermal growth factor receptor (EGF-R). Relationship of EGF-R
with histology and survival using fixed paraffin embedded tissue
and the F4, monoclonal antibody. Br J Cancer. 61:924–926. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Govindan R, Ritter J and Suppiah R: EGFR
and HER-2 overexpression in malignant mesothelioma. Proc Am Soc
Clin Oncol. 20:31062001.
|
|
25
|
Govindan R, Kratzke RA, Herndon JE II,
Niehans GA, Vollmer R, Watson D, Green MR and Kindler HL: Cancer
and Leukemia Group B (CALGB 30101): Gefitinb in patients with
malignant mesothelioma: a phase II study by the Cancer and Leukemia
Group B. Clin Cancer Res. 11:2300–2304. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ohnaga T, Takei Y, Nagata T and Shimada Y:
Highly efficient capture of cancer cells expressing EGFR by
microfluidic methods based on antigen-antibody association. Sci
Rep. 8:120052018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kaji C, Tsujimoto Y, Kato Kaneko M, Kato Y
and Sawa Y: Immunohistochemical Examination of Novel Rat Monoclonal
Antibodies against Mouse and Human Podoplanin. Acta Histochem
Cytochem. 45:227–237. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rice D, Rusch V, Pass H, Asamura H, Nakano
T, Edwards J, Giroux DJ, Hasegawa S, Kernstine KH, Waller D, et al
International Association for the Study of Lung Cancer
International Staging Committee and the International Mesothelioma
Interest Group, : Recommendations for uniform definitions of
surgical techniques for malignant pleural mesothelioma: A consensus
report of the international association for the study of lung
cancer international staging committee and the international
mesothelioma interest group. J Thorac Oncol. 6:1304–1312. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Habougit C, Trombert-Paviot B, Karpathiou
G, Casteillo F, Bayle-Bleuez S, Fournel P, Vergnon JM, Tiffet O,
Péoc'h M and Forest F: Histopathologic features predict survival in
diffuse pleural malignant mesothelioma on pleural biopsies.
Virchows Arch. 470:639–646. 2017. View Article : Google Scholar : PubMed/NCBI
|