Introduction
Adamantinomatous craniopharyngioma (ACP) is a benign
tumor, but may exhibit invasive characteristics clinically
(1,2). ACP occurs in saddle areas and is
hypothesized to develop from the Rathke's pouch (1,2). For CP,
following the first successful removal via surgery ~100 years ago,
coupled with advances in surgical techniques and endocrine care,
the mortality rate of patients has decreased from ~100-5% (3–6). Despite
advancements in the treatment of ACP, there is still a high risk of
recurrence and surgical complications due to its unique anatomical
location (close proximity to the optic chiasma, optic nerve,
pituitary stalk, internal carotid artery, hypothalamus and middle
cerebral artery) and high risk of compression or perforation of
arteries and other important structures, including the
hypothalamus, pituitary stalk and optic chiasm (7,8). Thus,
it is important to identify alternative treatments. Recently,
considerable efforts have been made to analyze the genome,
transcriptome and proteome of these tumors to identify potential
therapeutic targets (9–11). However, studies on this region of the
CP are limited. Understanding the molecular mechanisms underlying
the development of ACP, as well identifying potentially relevant
biomarkers of metastasis may assist in the management of
patients.
Currently, high-throughput data analysis
technologies, including RNA sequencing and gene expression data
chip technology, are widely used to study the molecular mechanisms
underlying tumorigenesis. Abnormal mRNA expression and
differentially expressed genes (DEGs) can be detected by mRNA
expression chips (9–11). Microarray technology has been used to
identify DEGs, some of which have assisted in understanding the
development and progression of malignant tumors (12). However, studies using gene expression
microarray platforms to determine the gene expression profile in
ACP tissues are limited, despite being commonly used for other
types of tumors (12–14). The integrated gene expression
database, Gene Expression Omnibus (GEO) provides a method for
mining multiple tumor gene expression profiles for bioinformatics
analysis (15). Matrix
metalloproteinase (MMP)12, also termed macrophage metalloelastase,
is a type of metalloproteinase secreted as part of an inflammatory
response, and was initially identified in 1975 (16–18).
Previous studies have demonstrated that MMP12 gene knockout in
homozygous mice results in physiological abnormalities in
macrophages, such as a decrease in sensitivity to cigarette smoke,
and mice had smaller litter sizes (19,20).
Furthermore, a recent study reported that inhibition of MMP12 may
be a novel means of antiviral therapy (21).
MMP12 is expressed in several types of cancer,
including colorectal cancer, gastric cancer, lung cancer and liver
cancer, as well as ACP (22–26). MMP12 can degrade the extracellular
matrix and vascular components, thus promoting tumor invasion and
migration (26,27). Conversely, other study have reported
that MMP12 may also inhibit tumor growth, invasion and migration
(28,29). MMP12 is expressed in macrophages, as
well as tumor cells (29). Recent
studies have demonstrated that MMP12 expressed in the periphery of
the tumor suppresses tumor growth, whereas that expressed within
the tumor promotes its growth (28,29).
Thus, investigating MMP12 expression in ACP, and whether inhibition
of MMP12 in ACP can inhibit the proliferation of tumor cells, is
considered important.
MMP408 is an effective selective MMP12 inhibitor
(30,31). Previous studies have reported that
MMP408 can exert specific inhibitory effects on MMP12 at both mRNA
and protein levels, and MMP12 can affect the inflammatory response
due to its expression on macrophages (30,31).
Inflammation serves an important role in the occurrence and
development of ACP (32). Thus, it
was speculated that MMP12 may exhibit beneficial effects for
management of ACP.
In the present study, an integrated bioinformatics
approach was used to identify the DEGs between ACP and normal
tissues, based on datasets obtained from the GEO database (GSE94349
and GSE68015), to identify novel biomarkers associated with
progression and pathogenesis of ACP. The present study focused on
MMP12 based on the findings of a previous study (33). In addition, the effects of inhibiting
MMP12 in vitro were assessed.
Materials and methods
Microarray data
ACP gene expression databases were obtained from
NCBI GEO (ncbi.nlm.nih.gov/geo). For the present study, two GEO
datasets, GSE94349 (34) and
GSE68015 (35), which contained 48
tumor samples and 48 normal samples were used.
DEGs
The limma package (versions 3.46.0; Bioconductor) in
R was used to screen the differences between tumor samples and
normal samples in the respective datasets. A Log2
(foldchange) |>1| and FDR (False Discovery Rate) <0.05 were
used as the cut-off criteria for the DEGs. The common DEGs in the
GSE94349 and GSE68015 datasets were identified via the intersection
function in R. The difference in MMP12 expression between the two
groups was compared using the ggplot2 R package (versions 3.3.3;
Bioconductor).
Function and pathway analyses of
DEGs
The clusterProfiler software package (versions 3.
18.1; Bioconductor) was used to perform Gene Ontology (GO) and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analyses. GO includes biological processes (BP), cellular
components (CC) and molecular functions (MF) (36). Enrichment analysis was performed to
determine the biological significance of the DEGs.
Interaction and regulatory network
establishment
The Search Tool for the Retrieval of Interacting
Genes/Proteins database (https://www.researchgate.net/figure/STRING-Search-Tool-for-the-Retrieval-of-Interacting-Genes-Proteins-network-analysis-of_fig2_234098958)
was used to construct the protein-protein interaction (PPI) network
of the DEGs, and an interaction score of 0.4 was set as the
threshold. The MOCD plug-in version 3.6.1 (Cytoscape) was used to
select the aim module: Modules with MCODE scores >5, degree
cut-offof2, node score cut-off of 0.2, max depth=100 and k-score=2
were presented. Hub genes were screened out if they had a degree of
connectivity ≥42.
Patients and sample collection
Tumor samples were collected and preserved following
surgery at the Department of Neurosurgery, The First Affiliated
Hospital of Nanchang University (NanChang, China). Tumor samples
were collected after nasal endoscopic resection and stored at 4°C
for transporting. A total of six tumor samples, from three men and
three women were obtained. The mean age of the patients was 36
years (age range, 8–68 years), including three men and three women.
Patients pathologically diagnosed with ACP were included in the
present study, while patients who were associated with other
diagnoses were excluded. All specimens were pathologically and
clinically diagnosed as ACP by three pathologists. The present
study was approved by The Research Ethics Committee of Nanchang
University [NanChang, China; First Affiliated Hospital of Nanchang
University (2020) Medical Research Ethics Review (No. 160)] and
written informed consent was provided by all patients prior to the
study start.
Primary cell culture
Fluorescence microscopy (magnification, ×200) was
performed to confirm the tumor parenchyma for the subsequent steps.
Tissue samples were repeatedly washed with PBS and subsequently cut
into 2–3 mm3 thick sections. Tissue section were
digested using trypsin containing streptomycin and penicillin
(0.25%) and collagenase II (1 mg/ml) for 45 min (all purchased from
Beijing Solarbio Science & Technology Co., Ltd.). The samples
were centrifuged at 1,000 × g for 10 min at 4°C, and the
supernatant was discarded. Cells were resuspended in high glucose
medium (Beijing Solarbio Science & Technology Co., Ltd.)
supplemented with 15% fetal bovine serum (FBS, Gibco; Thermo Fisher
Scientific, Inc.) and incubated at 37°C with 5% CO2.
Immunofluorescence
ACP cells were washed with PBS and fixed in 75%
ethanol for 20 min at room temperature. Cells were re-washed and
blocked with 5% BSA at 37°C for 1 h. Cells were subsequently
incubated with monoclonal anti-pan-cytokeratin (pan-CK, 1:100; cat.
no. ab215838; Abcam) and anti-vimentin (VIM; 1:100; cat. no. J144;
Invitrogen; Thermo Fisher Scientific, Inc.) antibodies overnight at
4°C. Following incubation, cells were counterstained with DAPI (10
µg/ml; Sigma-Aldrich; Merck KGaA) at 23°C for 10 min and observed
under a confocal laser scanning microscope (fluorescence
microscope, ×200). For the MMP408 and control groups,
anti-β-catenin (1:100; cat. no. C7207; Sigma-Aldrich; Merck KGaA)
was used.
Cell proliferation assay
ACP cells were seeded into 24-well plates at a
density of 3×103 cells/well, and in 9 wells, cells were
treated with 2 nmol/ml MMP408 (37°C, Sigma-Aldrich; Merck KGaA) for
24, 48 or 72 h (three wells/condition) as the treatment group, and
the remaining wells were treated for different periods of time (24,
48 or 72 h) at 37°C, as the control group. DMEM solution containing
10 µl Cell Counting Kit-8 (CCK-8, 10:1, cat. no. 40203ES60;
Shanghai Yeasen Biotechnology Co., Ltd.) was added to each well and
cells were incubated for an additional 2.5 h in the dark. Cell
proliferation was subsequently analyzed at a wavelength of 450 nm,
using a microplate reader (Enspire 23001489, PerkinElmer, Inc.).
The number of cells in each treatment condition was determined,
based on the absorbance.
Cell migration assay
The wound healing assay was performed to assess the
migratory ability of ACP cells. ACP cells were seeded into 6-well
plates at a density of 5×105 cells/well and incubated in
media supplemented with 15% FBS (37) (Gibco; Thermo Fisher Scientific,
Inc.), at 37°C for 4 h. Once the cells reached confluence (the
cells cover the plate), the monolayers were scratched with a 200 µl
pipette tip and washed three times with PBS. Cells were incubated
in DMEM supplemented with 15% FBS (37) at 37°C for 48 h (Gibco; Thermo Fisher
Scientific, Inc.) and the treatment group was treated with 2
nmol/ml MMP408 (Sigma-Aldrich; Merck KGaA) at 37°C for 48 g. Images
were taken at 0 and 48 h using an inverted fluorescence microscope
(magnification, ×200).
Colony formation assay
ACP cells were seeded into6-wells plates at a
density of 500 cells/well, with three wells as the control group
and the other three wells as the treatment group. All cells were
incubated in DMEM supplemented with 15% FBS (37) (Gibco; Thermo Fisher Scientific,
Inc.). at 37°C for 24 h. The treatment groups were treated with 2
nmol/ml MMP408 at 37°C for 2 weeks (Sigma-Aldrich; Merck KGaA).
Cells were cultured for 2 weeks, fixed with 4% paraformaldehyde for
30 min at 23°C stained with 50% crystal violet dye solution at 23°C
for 2 h and imaged. Cell colonies were observed under a
fluorescence microscope (magnification, ×200); a colony was counted
if it contained ≥50 cells.
Western blotting
Cells were collected on ice (4°C), lysed using lysis
buffer (Beyotime Institute of Biotechnology) and subsequently
centrifuged at 12,000 × g for 30 min at 23°C to retain the
supernatant. The BCA protein analysis kit (Beyotime Institute of
Biotechnology) was used to determine protein concentration. Protein
lysates (45 µg) were separated by SDS-PAGE (7.5%), transferred onto
PVDF (EMD Millipore) membranes and blocked with 5% skimmed milk at
4°C overnight. The membranes were incubated with primary antibodies
against c-Myc (1:1,000; cat. no. 10828-1-AP; ProteinTech Group,
Inc.), c-Jun (1:2,000; cat. no. 24909-1-AP; ProteinTech Group,
Inc.), Wisp1 (1:1,000; cat. no. 18166-1-AP; ProteinTech Group,
Inc.), β-catenin (1:5,000; cat. no. 51067-2-AP; ProteinTech Group,
Inc.) and GAPDH (1:5,000; cat. no. 10494-1-AP; ProteinTech Group,
Inc.) overnight at 4°C. The membranes were washed three times with
tris-buffered saline with Tween-20 (for 10 min each) and
subsequently incubated with HRP-conjugate goat anti-rabbit IgG
secondary antibody (1:10,000; cat. no. 31460; Thermo Fisher
Scientific, Inc.). Protein bands were visualized using enhanced
chemiluminescence reagent (EMD Millipore) and densitometry analysis
was performed using ImageJ software (Image-Pro Plus 6.0; National
Institutes of Health) and the data were normalized to expression of
the internal control GAPDH.
Statistical analysis
Statistical analysis was performed using SPSS
version 19.0 software (IBM Corp.). All experiments were performed
in triplicate and data are presented as the mean ± standard
deviation. Paired Student's t-test was used to compare differences
between two groups, while one-way ANOVA followed by Tukey's post
hoc test were used to compare differences between multiple groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
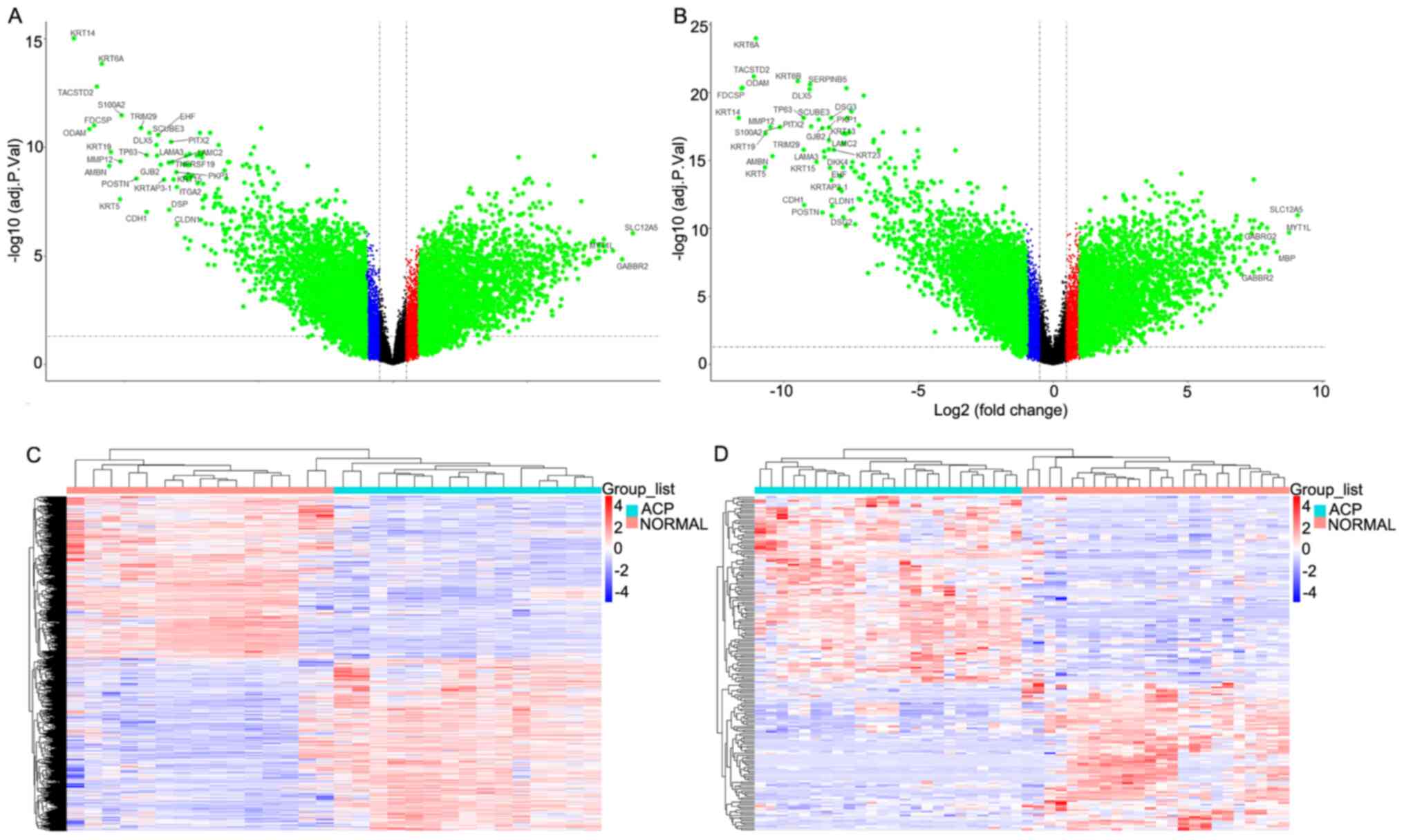
Identification of DEGs
The heatmaps and volcano plots were generated to
identify the downregulated and upregulated genes in the GEO
datasets (GSE94394 and GSE68015). As presented in Fig. 1A, MMP12 expression in the GSE94394
dataset was significantly higher in the normal group compared with
the ACP group (~1,000×; P<0.05). As presented in Fig. 1B, MMP12 expression in the GSE68015
dataset was significantly higher in the ACP group compared with the
normal group (~1,700×; P<0.05). The differences in gene
expression between the normal and ACP groups are presented in
Fig. 1C and D.
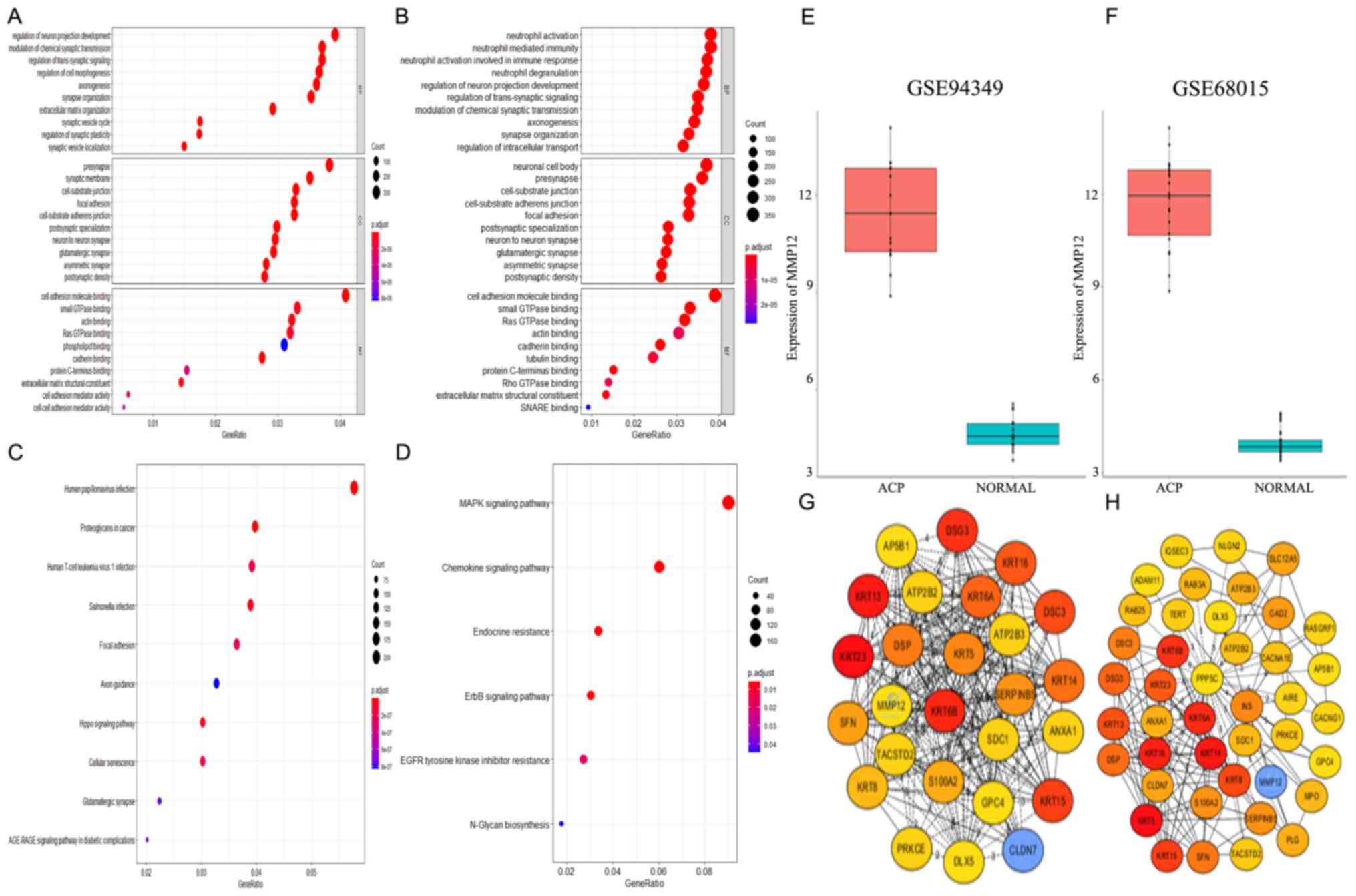
MMP12 expression in the ACP group
compared with the normal group and construction of the PPI
network
MMP12 expression levels in the two datasets are
presented in Fig. 2E and F. The
results demonstrated that MMP12 was significantly higher in the ACP
group compared with the normal group (P<0.05), which is
consistent with a previous study (9). The PPI networks are presented in
Fig. 2G and H. Although MMP12 was
not one of the top 10 dysregulated genes (top 26 and 42 in the
GSE94394 and GSE68015 datasets, respectively), the results
demonstrated that MMP12 has complex associations with several other
genes.
GO and KEGG pathway enrichment
analyses of the DEGs
GO and KEGG pathway enrichment analyses were
performed using the clusterProfiler package, with a threshold of
P<0.05 (Fig. 2A). Enriched BP, CC
and MF terms were used to obtain a better understanding of the
biological functions of overlapping DEGs. The results demonstrated
that the significantly enriched GO terms for BP were ‘neutrophil
activation’, ‘neutrophil mediated immunity’, ‘neutrophil activation
involved in immune response’, ‘neutrophil degranulation’,
‘regulation of neuron projection development’, ‘regulation of
trans-synaptic signaling’, ‘modulation of chemical synaptic
transmission’, ‘axonogenesis’, ‘synapse organization’ and
‘regulation of intracellular transport’ (Fig. 2B). Furthermore, the significantly
enriched GO terms for CC were ‘neuronal cell body’, ‘presynapse’,
‘cell-substrate junction’, ‘cell-substrate adherens junction’,
‘focal adhesion’, ‘postsynaptic specialization’, ‘neuron to neuron
synapse’, ‘glutamatergic synapse’, ‘asymmetric synapse’ and
‘postsynaptic density’ (Fig. 2B).
The significantly enriched GO terms for MF were ‘cell adhesion
molecule binding’, ‘small GTPase binding’, ‘Ras GTPase binding’,
‘actin binding’, ‘cadherin binding’, ‘tubulin binding’, ‘protein
C-terminus binding’, ‘Rho GTPase binding’, ‘extracellular matrix
structural constituent’ and ‘SNARE binding’ (Fig. 2B). KEGG pathway enrichment analysis
demonstrated that the DEGs were primarily enriched in the ‘MAPK
signaling pathway’, ‘chemokine signaling pathway’, ‘endocrine
resistance’, ‘ErbB signaling pathway’, ‘EGFR tyrosine kinase
inhibitor resistance’, ‘N-glycan biosynthesis’, ‘human
papillomavirus infection’, ‘proteoglycans in cancer’, ‘human T-cell
leukemia virus 1 infection’, ‘salmonella infection’, ‘focal
adhesion’, ‘axon guidance’, ‘hippo signaling pathway’, ‘cellular
senescence’, ‘glutamatergic synapse’ and ‘AGE-RAGE signaling
pathway’ in diabetic complications (Fig.
2C and D). These significantly enriched terms and pathways may
provide further insight into potentially druggable targets for
management of DEGs.
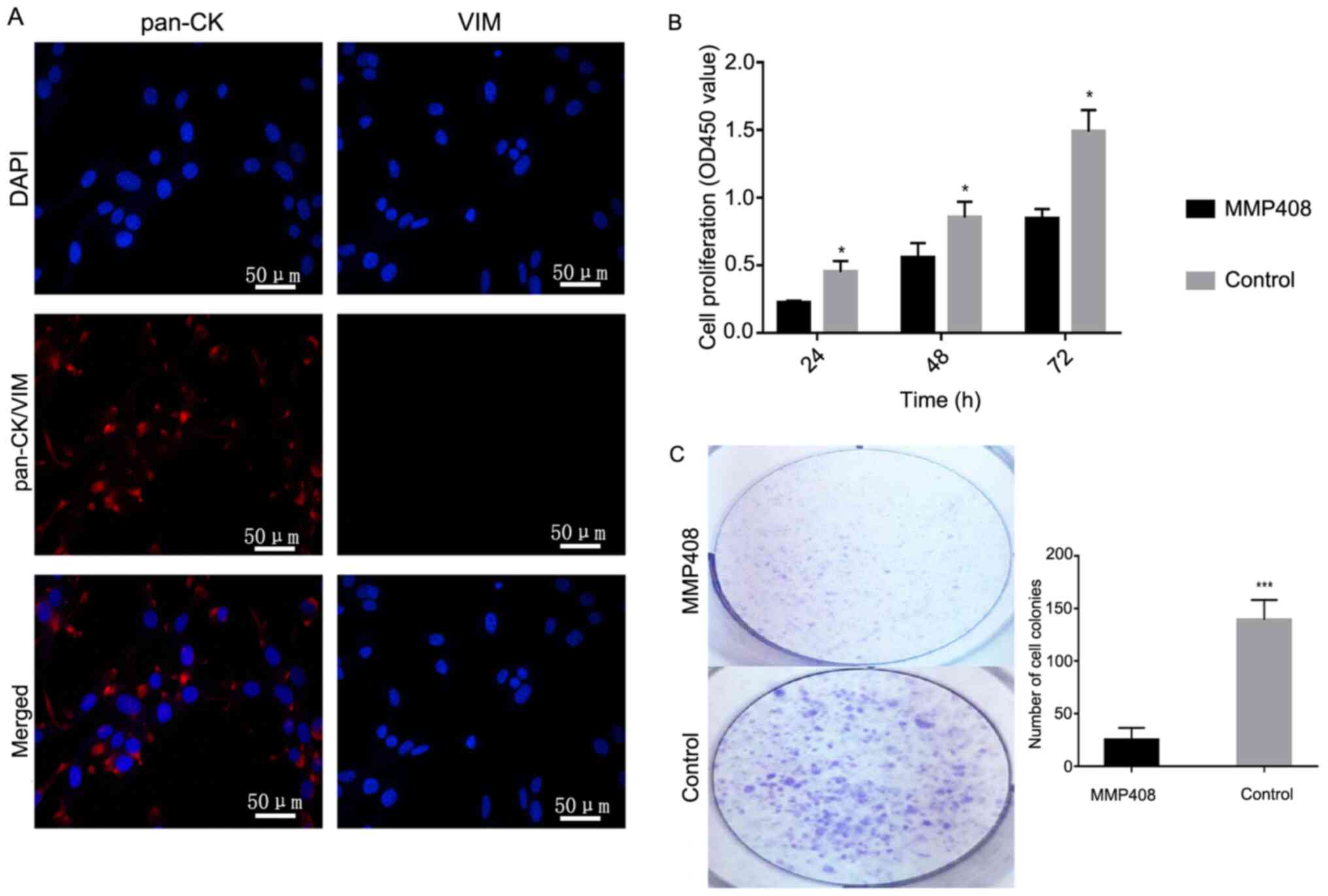
Establishment and identification of an
ACP cell line
Primary ACP cells which were obtained from tissues
pathologically confirmed to be ACP were successfully cultured for
subsequent functional assays in vitro. Keratins are
established molecular markers for tumors of an epithelial origin
(38–40) and Vim is an important marker to
identify epithelial tumor cells because epithelial tumors do not
express Vim (41). Thus, pan-CK and
VIM expression levels were used as positive and negative controls
of analysis of the primary ACP cell via immunofluorescence
analysis. The results demonstrated that pan-CK was primarily
expressed in the cytoplasm of ACP cells (Fig. 3A; left), while VIM expression was not
observed (Fig. 3A; right). These
results suggest that the primary cells were ACP cells, and they
were not contaminated by CP associated fibroblasts.
MMP12 promotes ACP proliferation and
migration in vitro
To investigate the effect of MMP408 on the
proliferation and migration of ACP cells, the CCK-8, colony
formation and wound healing assays were performed. The results of
the CCK-8 assay demonstrated that MMP408 significantly decreased
the proliferation of ACP cells compared with the control group
(P<0.05; Fig. 3B). The results of
the colony formation assay indicated that the number of cell
colonies in the treated group was significantly lower compared with
the control group (P<0.001; Fig.
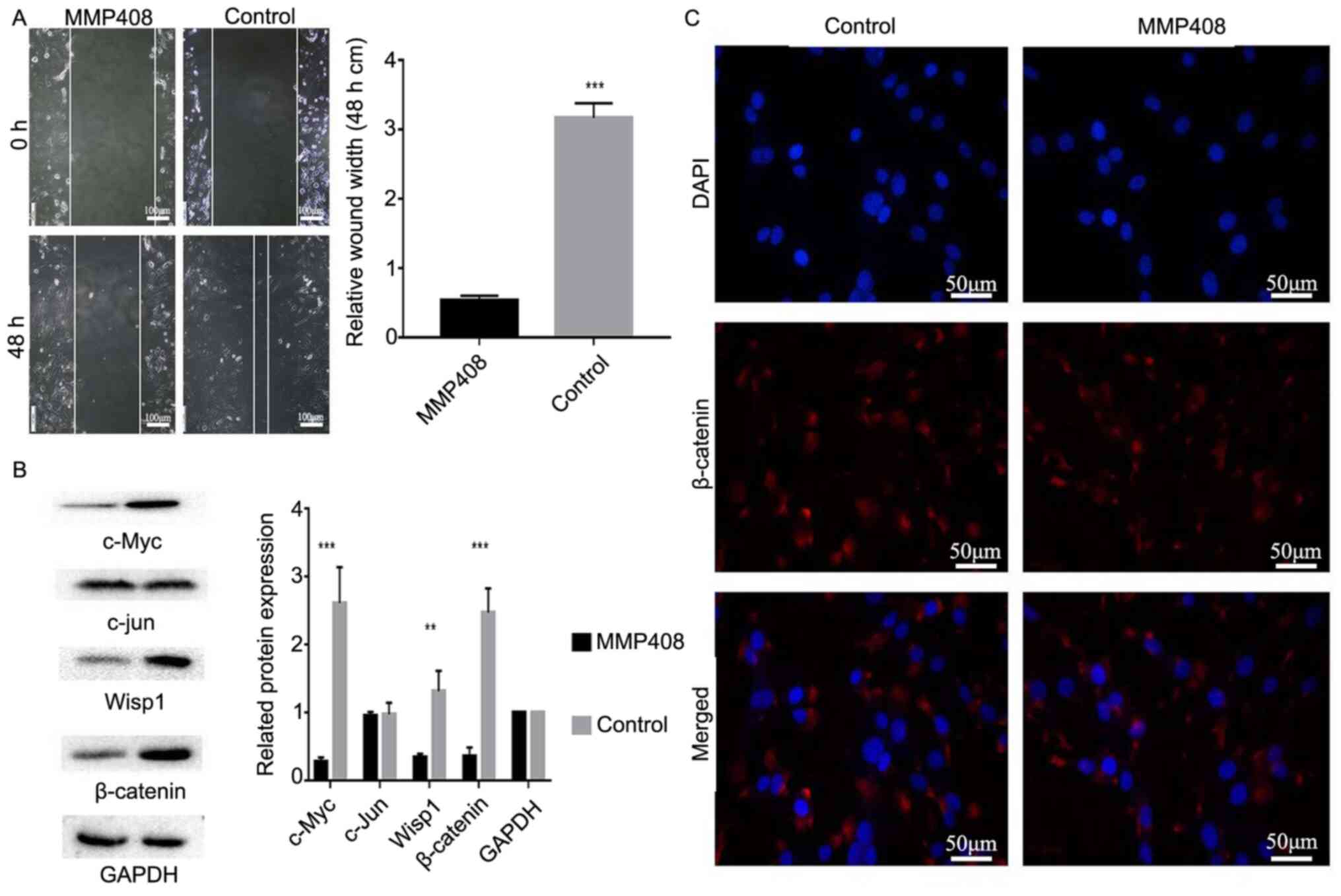
3C). The results of the wound healing assay demonstrated that
MMP408 significantly inhibited the migratory ability of ACP cells
(P<0.001; Fig. 4A).
MMP12 promotes the expression levels
of β-catenin, c-Myc and Wisp1
The Wnt/β-catenin signaling pathway is involved in
the progression and development of several types of tumors
(42). β-catenin serves an important
role in tumor invasion and development. MMP12 may interact with the
Wnt/β-catenin signaling pathway to regulate the development of the
central nervous system (43). c-Myc
expression was also assessed in the present study. The results
demonstrated that MMP408 significantly decreased the protein
expression levels of c-Myc, Wisp1 and β-catenin (P<0.01 and
P<0.001), but not c-Jun (Fig.
4B).
Immunofluorescence analysis indicated that treatment
with MMP408 decreased β-catenin expression in the nucleus (Fig. 4C), where it normally interacts with
target genes involved in tumor progression (30–32).
Taken together, these results suggest that MMP12 may promote the
translocation of β-catenin into the nucleus by regulating β-catenin
and c-Myc, where it drives transcription of its target genes, and
induces proliferation and migration of ACP cells.
Discussion
Although ACP is a benign tumor, it accounts for 3%
of all and 4% of childhood cranial tumors, with an annual incidence
ranging from 5–20 cases per 100,000,000 (44–46).
However, due to its unique anatomical structure and the risk of
serious postoperative complications (7,8), it is
urgent to improve surgical removal and non-surgical treatment
methods. However, the number of studies on drugs targeting genes
uniquely upregulated in ACP are limited. It has been demonstrated
that MAPK/ERK inhibitors can inhibit progression of ACP tumors;
however, related research is still in its infancy (45). Thus, further studies are required to
understand the unique genome of ACP. In addition, due to various
factors, bioinformatics-based analyses of ACP is limited.
In the present study, the gene expression profiles
of the GSE94349 and GSE68015 datasets were integrated to identify
the DEGs between ACP tissues and normal tissues using
bioinformatics analysis. GO and KEGG pathway enrichment analyses
were performed using the clusterProfiler R package. The enriched GO
terms for BP were ‘neutrophil activation’, ‘neutrophil mediated
immunity’ and ‘neutrophil activation involved in immune response’,
and KEGG pathway analysis demonstrated that these DEGs were
enriched in the ‘chemokine signaling pathway’, ‘endocrine
resistance’ and ‘human T-cell leukemia virus 1 infection’ in both
groups, which are all associated with inflammation in the tumor
(32). MMP12 can affect tumor
inflammatory response by affecting the secretion and expression of
macrophages (46). A previous study
reported that inflammation plays a key role in the occurrence and
progression of ACP (32). In
addition, MMP12 plays an important role in the occurrence and
progression of ACP (32). Notably,
in the KEGG pathway enrichment analysis, it was confirmed that EGFR
tyrosine kinase inhibitor resistance exists in ACP. However, the
specific mechanism and situation under which this resistance is
exhibited requires further investigation, which may also be
relevant for further research and development of targeted drugs for
ACP.
The progression of cancer cells primarily manifests
as increased cell proliferation and migration (19,29,37,43,47). In
the present study, the wound healing, cell proliferation and colony
formation assays demonstrated that MMP408 inhibited the
proliferative and migratory abilities of ACP cells. Previous
studies have demonstrated that MMP408 can inhibit MMP12 expression
at both mRNA and protein levels (26–29).
Thus, it was speculated that MMP12 can inhibit the proliferation
and migration of ACP cells. However, the lack of experimental
verification of the inhibition of MMP12 expression/enzymatic
activity by MMP408 is a limitation of the present study.
Currently, the predominant hypothesis by which ACP
develops/progresses is overactivation of the Wnt/β-Catenin
signaling pathway; thus, the expression of β-catenin is considered
an important indicator of ACP (48).
β-catenin is a cytoplasmic protein that mediates gene transcription
and cell-cell adhesion (48). In the
absence of the Wnt ligand, β-catenin undergoes proteasomal
degradation as a result of interactions between Axin, glycogen
synthase kinase 3β and anaphase-promoting complex. β-catenin is
considered an important regulator of ACP, and in most tumors, c-Myc
promotes the proliferation of cancer cells by acting on SLC1A5. Jun
can regulate tumor proliferation and other functions via AP-1. It
has been reported that high Wisp1 expression is positively
associated with the proliferation and invasion of several types of
tumors, and is negatively associated with prognosis. β-catenin is
prevented from activating the transcription factors T-cell factor
(TCF) and lymphoid enhancer factor (LEF), and c-Myc, c-Jun and
Wisp1 are downstream proteins of TCF/LEF (32,47–49). The
expression of these genes has important effects on ACP cell
functions, such as proliferation and invasion (32,47–49). In
the present study, immunofluorescence and western blot analyses
demonstrated that the expression levels of β-catenin, c-Myc and
Wisp1 significantly decreased in ACP cells when MMP12 expression
was inhibited. Thus, inhibition of MMP12 can inhibit the
transcription and expression of related proteins in ACP, and thus
inhibit the occurrence and progression of ACP.
Recent studies have reported that MMP12 expression
in ACP is >820× of that in the surrounding normal tissues, and
thus may be an important potential target for ACP therapy in the
future (9,38,48). In
the present study, MMP12 expression significantly varied between
the ACP and normal groups. Although MMP12 was not amongst the top
10 genes that were differentially expressed between the ACP and
normal groups, it did not affect the role of MMP12 in ACP, as there
were several associations between MMP12 and the top 10 dysregulated
genes.
MMP12 is predominantly expressed in the cell nuclei
and the extracellular matrix (28,29).
Previous studies (28,29) have reported that MMP12 possesses dual
functions. Its expression in the cell nucleus can promote tumor
proliferation, while its expression in the extracellular matrix can
inhibit tumor growth (9). Thus,
determining the mode of action of MMP12 in ACP is important in any
study of its role in cancer. In the present study, inhibiting MMP12
expression decreased the proliferative and migratory abilities of
ACP cells. In addition, the expression levels of the related
proteins in vitro decreased, suggesting that MMP12 promotes
ACP proliferation and migration in vitro. The effects of
MMP12 inhibition should be determined in vivo (9,50).
In conclusion, the present study performed
bioinformatics analysis of ACP datasets to determine the important
characteristics of ACP. Taken together, these results support the
notion that MMP12 serves an important role in ACP and may be
involved in drug resistance. The in vitro experiments
demonstrated that MMP408 decreased the proliferation and migration
of ACP cells, suggesting the therapeutic potential of targeting
MMP12 in management of ACP. However, further studies are required
to validate the results presented here.
Acknowledgements
Not applicable.
Funding
The present study was funded by National Natural
Science Foundation of China (grant no. 82060246).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
ML and TH conceived and designed the experiments,
analyzed the data and prepared the manuscript. ML, LZ and SL
performed the experiments. LF, LY, XW, CY, YB, SL, ZT, SZ, BT, EZ,
SX and CC contributed to data collection and analysis. TH and ML
and LZ confirmed the authenticity of all the raw data. All authors
have read and approved the final version.
Ethics approval and consent to
participate
The present study was approved by The Research
Ethics Committee of Nanchang University [First Affiliated Hospital
of Nanchang University (2020) Medical Research Ethics Review (No.
160)] and written informed consent was provided by all patients
prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Martinez-Barbera JP and Buslei R:
Adamantinomatous craniopharyngioma: Pathology, molecular genetics
and mouse models. J Pediatr Endocrinol Metab. 28:7–17. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kasai H, Hirano A, Llena JF and Kawamoto
K: A histopathological study of craniopharyngioma with special
reference to its stroma and surrounding tissue. Brain Tumor Pathol.
14:41–45. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Barkhoudarian G and Laws ER:
Craniopharyngioma: History. Pituitary. 16:1–8. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Laws ER Jr: Transsphenoidal microsurgery
in the management of craniopharyngioma. J Neurosurg. 52:661–666.
1980. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Laws ER Jr: Transsphenoidal removal of
craniopharyngioma. Pediatr Neurosurg. 21 (Suppl 1):S57–S63. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yaşargil MG, Curcic M, Kis M, Siegenthaler
G, Teddy PJ and Roth P: Total removal of craniopharyngiomas.
Approaches and long-term results in 144 patients. J Neurosurg.
73:3–11. 1990. View Article : Google Scholar
|
|
7
|
Alvarez M: Craniopharyngiomas. J Neurosci
Nurs. 38:362–368. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Karavitaki N, Cudlip S, Adams CB and Wass
JA: Craniopharyngiomas. Endocr Rev. 27:371–397. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gump JM, Donson AM, Birks DK, Amani VM,
Rao KK, Griesinger AM, Kleinschmidt-DeMasters BK, Johnston JM,
Anderson RC, Rosenfeld A, et al: Identification of targets for
rational pharmacological therapy in childhood craniopharyngioma.
Acta Neuropathol Commun. 3(30)2015.PubMed/NCBI
|
|
10
|
Xia Z, Liu W, Li S, Jia G, Zhang Y, Li C,
Ma Z, Tian J and Gong J: Expression of matrix metalloproteinase-9,
type IV collagen and vascular endothelial growth factor in
adamantinous craniopharyngioma. Neurochem Res. 36:2346–2351. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Baker JC, Beddington RS and Harland RM:
Wnt signaling in Xenopus embryos inhibits bmp4 expression and
activates neural development. Genes Dev. 13:3149–3159. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li T, Gao X, Han L, Yu J and Li H:
Identification of hub genes with prognostic values in gastric
cancer by bioinformatics analysis. World J Surg Oncol. 16:1142018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi
ST, Siu HC, Deng S, Chu KM, Law S, et al: Whole-genome sequencing
and comprehensive molecular profiling identify new driver mutations
in gastric cancer. Nat Genet. 46:573–582. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiang P and Liu XS: Big data mining yields
novel insights on cancer. Nat Genet. 47:103–104. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang F, Qian X, Lu Z, Lai Y, Li Z, He C
and He Z: Identification of differentially expressed genes,
biological pathways and prognostic signature in bladder cancer. BMC
Urology. 10.21203/rs.3.rs-362542/v1.
|
|
16
|
Werb Z and Gordon S: Elastase secretion by
stimulated macrophages. Characterization and regulation. J Exp Med.
42:361–377. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Banda MJ and Werb Z: Mouse macrophage
elastase. Purifcation and characterization as a metalloproteinase.
Biochem J. 193:589–605. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
White RR, Norby D, Janoff A and Dearing R:
Partial purifcation and characterization of mouse peritoneal
exudative macrophage elastase. Biochim Biophys Acta. 612:233–244.
1980. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hautamaki RD, Kobayashi DK, Senior RM and
Shapiro SD: Requirement for macrophage elastase for cigarette
smoke-induced emphysema in mice. Science. 277:2002–2004. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shipley JM, Wesselschmidt RL, Kobayashi
DK, Ley TJ and Shapiro SD: Metalloelastase is required for
macrophage-mediated proteolysis and matrix invasion in mice. Proc
Natl Acad Sci USA. 93:3942–3946. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Marchant DJ, Bellac CL, Moraes TJ,
Wadsworth SJ, Dufour A, Butler GS, Bilawchuk LM, Hendry RG,
Robertson AG, Cheung CT, et al: A new transcriptional role for
matrix metalloproteinase-12 in antiviral immunity. Nat Med.
20:493–502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kerkelä E, Ala-Aho R, Jeskanen L, Rechardt
O, Grénman R, Shapiro SD, Kähäri VM and Saarialho-Kere U:
Expression of human macrophage metalloelastase (MMP-12) by tumor
cells in skin cancer. J Invest Dermatol. 114:1113–1119. 2000.
View Article : Google Scholar
|
|
23
|
Zhao X, Xu M, Cai Z, Yuan W, Cui W and Li
MD: Identifcation of LIFR, PIK3R1, and MMP12 as novel prognostic
signatures in gallbladder cancer using network-based module
analysis. Front Oncol. 9:3252019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Roman J: On the ‘TRAIL’ of a killer: MMP12
in lung cancer. Am J Respir Crit Care Med. 196:262–264. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Klupp F, Neumann L, Kahlert C, Diers J,
Halama N, Franz C, Schmidt T, Koch M, Weitz J, Schneider M and
Ulrich A: Serum MMP7, MMP10 and MMP12 level as negative prognostic
markers in colon cancer patients. BMC Cancer. 16:4942016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ng KT, Qi X, Kong KL, Cheung BY, Lo CM,
Poon RT, Fan ST and Man K: Overexpression of matrix
metalloproteinase-12 (MMP-12) correlates with poor prognosis of
hepatocellular carcinoma. Eur J Cancer. 47:2299–2305. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ella E, Harel Y, Abraham M, Wald H, Benny
O, Karsch-Bluman A, Vincent D, Laurent D, Amir G, Izhar U, et al:
Matrix metalloproteinase 12 promotes tumor propagation in the lung.
J Thorac Cardiovasc Surg. 155:2164–2175. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang W, Arii S, Gorrin-Rivas MJ, Mori A,
Onodera H and Imamura M: Human macrophage metalloelastase gene
expression in colorectal carcinoma and its clinicopathologic
signifcance. Cancer. 91:1277–1283. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kerkelä E, Ala-aho R, Klemi P, Grénman S,
Shapiro SD, Kähäri VM and Saarialho-Kere U: Metalloelastase
(MMP-12) expression by tumour cells in squamous cell carcinoma of
the vulva correlates with invasiveness, while that by macrophages
predicts better outcome. J Pathol. 198:258–269. 2002. View Article : Google Scholar
|
|
30
|
Chen S, Xie J, Zhao K, Ren L, Deng Y, Xie
X, Chen S, Xu H, Long X and Liu E: LPS aggravates lung inflammation
induced by RSV by promoting the ERK-MMP-12 signaling pathway in
mice. Respir Res. 21:1932020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li J, Wang JJ, Peng Q, Chen C, Humphrey
MB, Heinecke J and Zhang SX: Macrophage metalloelastase (MMP-12)
deficiency mitigates retinal inflammation and pathological
angiogenesis in ischemic retinopathy. PLoS One. 7:e526992012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Apps JR, Carreno G, Gonzalez-Meljem JM,
Haston S, Guiho R, Cooper JE, Manshaei S, Jani N, Hölsken A,
Pettorini B, et al: Tumour compartment transcriptomics demonstrates
the activation of inflammatory and odontogenic programmes in human
adamantinomatous craniopharyngioma and identifies the MAPK/ERK
pathway as a novel therapeutic target. Acta Neuropathol.
135:757–777. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gupta S, Bi WL, Giantini Larsen A,
Al-Abdulmohsen S, Abedalthagafi M and Dunn IF: Craniopharyngioma: A
roadmap for scientific translation. Neurosurg Focus. 44:E122018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gene Expression Omnibus: Series GSE94349,
. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE94349February
1–2017
|
|
35
|
Gene Expression Omnibus: Series GSE68015,
. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE68015April
18–2015
|
|
36
|
Wang F, Xue Q, Xu D, Jiang Y, Tang C and
Liu X: Identifying the hub gene in gastric cancer by bioinformatics
analysis and in vitro experiments. Cell Cycle. 19:1326–1337. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yin X, Liu Z, Zhu P, Wang Y, Ren Q, Chen H
and Xu J: CXCL12/CXCR4 promotes proliferation, migration, and
invasion of adamantinomatous craniopharyngiomas via PI3K/AKT signal
pathway. J Cell Biochem. 120:9724–9736. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Adamson TE, Wiestler OD, Kleihues P and
Yasargil MG: Correlation of clinical and pathological features in
surgically treated craniopharyngiomas. J Neurosurg. 73:12–17. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nelson WG, Battifora H, Santana H and Sun
TT: Specific keratins as molecular markers for neoplasms with a
stratified epithelial origin. Cancer Res. 44:1600–1603.
1984.PubMed/NCBI
|
|
40
|
Quentmeier H, Osborn M, Reinhardt J,
Zaborski M and Drexler HG: Immunocytochemical analysis of cell
lines derived from solid tumors. J Histochem Cytochem.
49:1369–1378. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang N: Vimentin and tumor diagnosis.
Zhonghua Bing Li Xue Za Zhi. 19:122–124. 1990.(In Chinese).
PubMed/NCBI
|
|
42
|
Zimmerli D, Hausmann G, Cantù C and Basler
K: Pharmacological interventions in the Wnt pathway: Inhibition of
Wnt secretion versus disrupting the protein-protein interfaces of
nuclear factors. Br J Pharmacol. 174:4600–4610. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chelluboina B, Nalamolu KR, Klopfenstein
JD, Pinson DM, Wang DZ, Vemuganti R and Veeravalli KK: MMP-12, a
promising therapeutic target for neurological diseases. Mol
Neurobiol. 55:1405–1409. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bunin GR, Surawicz TS, Witman PA,
Preston-Martin S, Davis F and Bruner JM: The descriptive
epidemiology of craniopharyngioma. Neurosurg Focus. 3:e11997.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Erfurth EM, Holmer H and Fjalldal SB:
Mortality and morbidity in adult craniopharyngioma. Pituitary.
16:46–55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ostrom QT, Gittleman H, Farah P, Ondracek
A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 15
(Suppl 2):ii1–ii56. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Brennan A, Leech JT, Kad NM and Mason JM:
Selective antagonism of cJun for cancer therapy. J Exp Clin Cancer
Res. 39:1842020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gurbuz I and Chiquet-Ehrismann R:
CCN4/WISP1 (WNT1 inducible signaling pathway protein 1): A focus on
its role in cancer. Int J Biochem Cell Biol. 62:142–146. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Panda S, Banerjee N and Chatterjee S:
Solute carrier proteins and c-Myc: A strong connection in cancer
progression. Drug Discovery Today. 25:891–900. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yang M, Zhang X, Liu Q, Niu T, Jiang L, Li
H, Kuang J, Qi C, Zhang Q, He X, et al: Knocking out matrix
metalloproteinase 12 causes the accumulation of M2 macrophages in
intestinal tumor microenvironment of mice. Cancer Immunol
Immunother. 69:1409–1421. 2020. View Article : Google Scholar : PubMed/NCBI
|