Introduction
Hepatocellular carcinoma (HCC) is the most common
subtype of liver cancer with a steady increase in incidence and
remains the fifth most common cancer worldwide (1). Every year, >780,000 novel HCC cases
are reported globally, causing ~745,000 mortalities (2,3).
Patients at different stages of HCC have tried different treatment
strategies, but the median overall survival of patients is only
6–20 months (3). Therefore, it is
urgent to find an effective treatment.
Long non-coding RNAs (lncRNAs, >200 nucleotides
in length) are RNA transcripts without protein-coding capacity but
with the ability to regulate gene expression and participate in
human diseases, including osteoarthritis (4), non-alcoholic fatty liver disease
(5) and rheumatoid arthritis
(6). lncRNAs play an important role
in a variety of physiological and pathological processes such as
cell proliferation and apoptosis, cell proliferation and
senescence, immune activation and inactivation (7,8).
Multiple lncRNAs have been confirmed by previous studies to play a
vital role in HCC, such as TMPO-AS1 (9), MYLK-AS1 (10) and DLX6-AS1 (11). However, there are relatively few
reports on the role of lncRNA MT1JP in HCC, and its ability to
predict the prognosis of this disease is still limited.
MicroRNA (miRNA/miR) is a type of endogenous
non-coding single-stranded small RNA, which is widely distributed
in eukaryotes. It is composed of 19–25 nucleotides (12). A growing body of research over the
past decades has demonstrated that miRNAs can act as tumor
suppressors or promoters during the development of HCC (13,14).
miR-32 is abnormally expressed in gastric cancer (15), glioma (16) and cervical cancer(17). However, the
function and mechanism of miR-32 in HCC needs further
exploration.
PTEN is a tumor suppressor gene with critical roles
in diverse biological processes, such as cell cycle regulation and
apoptosis (18). In cancer biology,
PTEN mainly inhibits the PI3K/Akt signaling pathway, which is the
main cell survival pathway, to induce cell death and suppress tumor
growth (19). It is known that PTEN
in HCC can be targeted by oncogenic miRNAs, such as miR-32
(20). The current study
investigated the expression levels of MT1JP and its prognostic
value for patients with HCC, and explored whether the PTEN/miR-32
axis was involved in HCC progression via interaction with lncRNA
MT1JP. The present study may identify a novel therapeutic target
for HCC.
Materials and methods
Study patients
This study included 64 patients with HCC (36 males,
28 females; age range, 33–67 years old; mean age, 51.8±6.0 years
old) selected from the 136 patients with HCC admitted to the Union
Hospital Affiliated to Tongji Medical College of Huazhong
University of Science and Technology (Wuhan, China) between April
2010 and April 2014. All patients were treated with surgical
resection of the primary tumors and/or targeted therapies,
radiotherapies and chemotherapies according to their conditions.
The study was approved by the review board of the Ethics Committee
of the aforementioned hospital. Inclusion criteria included: i)
patients had not received radiofrequency ablation preoperatively;
ii) newly diagnosed HCC case and iii) completed treatment and a
5-year follow-up in Union Hospital Affiliated to Tongji Medical
College of Huazhong University of Science and Technology. Exclusion
criteria included: i) Recurrent patients with HCC, ii) previous
history of cancer, iii) family history of cancer and iv) other
clinical disorders. All 64 patients with HCC provided written
informed consent.
Tissue samples, treatment and
follow-up
Before initiation of therapies, liver biopsy was
performed under the guidance of MRI to collect both HCC and
non-tumor (within 3 cm of the tumor) tissues. All tissues were
snap-frozen and then transferred into a −80°C freezer before use.
HCC tissue samples were sectioned at 5-µm. The clinicopathological
features of the patient specimens and the histological diagnosis of
all HCC and normal tissues were independently reviewed and approved
by two experienced pathologists. Pathologists performed a blinded
assessment of the specimens. The TNM stage was evaluated based on
the American Cancer Joint Commission Cancer Staging Manual
(21). From the day of admission,
all patients were followed-up for 5 years in a monthly manner
through telephone to record their survival. The overall survival
was defined as the day of diagnosis to the day of death or last
follow-up. Patients who died of other causes or were lost during
the follow-up were excluded. The 5-year overall survival rate was
calculated as the proportion of the survivors among all patients
with HCC at 5 years after hepatectomy.
HCC cells and transfections
Human HCC cell line SNU-398 (American Type Culture
Collection) was used. A mixture of 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and 90% RPMI 1640 medium was used to cultivate
cells. Cells were cultivated under the conditions of 37°C, 95%
humidity and 5% CO2. Cells were harvested at the
confluence of 70–80% to perform cell transfections. The
overexpression pcDNA3.1-MT1JP (MT1JP), pcDNA3.1-PTEN (PTEN) and
control pcDNA3.1 recombinant plasmids were constructed by Shanghai
GenePharma Co., Ltd. miRNA-NC and miR-32 mimic were obtained from
Guangzhou RiboBio Co., Ltd. Lipofectamine 2000® (Sangon
Biotech Co., Ltd.) was used to transfect 35 nM miRNA (miRNA-NC or
miR-32) or 2 µg plasmid for 24 h in six-well plates. The control
(C) cells for all transfections were untransfected cells. At 24 h
post-transfection at the room temperature, cells were harvested to
perform subsequent experiments. The sequences were as follows:
miR-32 Mimic, 5′-AGUAUACGUCCUGCAGUCAGCU-3′) and miRNA-NC,
5′-UGCUCCGGACGUGUGAGGUGT-3′.
Reverse transcription-quantitative
(RT-qPCR)
Total RNA from tissues and cells was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) following the manufacturer's protocol. Total RNA was reverse
transcribed into cDNA using oligo (dT)15, random primers
(GenScript), M-MLV reverse transcriptase, dNTPs, 5X buffer and
RNase inhibitor (all Tiangen Biotech Co., Ltd.) at 25°C for 10 min,
42°C for 50 min and 80°C for 10 min. QuantiTect SYBR®
Green PCR kit (Qiagen, Inc.) was used to prepare qPCR mixtures for
measuring the expression levels of MT1JP and PTEN. GAPDH was used
as the endogenous control of MT1JP and PTEN. To measure the
expression levels of miR-32, all steps were performed using
All-in-One™ miRNA qRT-PCR Detection kit (GeneCopoeia, Inc.). U6 was
used as the endogenous control of miR-32. The following
thermocycling conditions were used for the qPCR: Initial
denaturation at 94°C for 5 min; followed by 40 cycles at 94°C for
10 sec, 60°C for 20 sec and 72°C for 30 sec; and final extension at
72°C for 150 sec. The following primer pairs were used for the
qPCR: MT1JP forward: 5′-GCAAAGGGACGTCGGAGA-3′ and reverse:
5′-TCCAGGTTGCAGTTGTT-3′; PTEN forward: 5′-TGGCTTCGACTTAG-3′ and
reverse: 5′-GGTTGGTTATGGTCTTTAAAAGG-3′; miR-32 forward:
5′-GCGGCGTATTGCACATTACT-3′ and reverse: 5′-TCGTATCCAGTGCAGGGTC-3′;
GAPDH forward: 5′-AGATCATCAGCAATGCCTCCT-3′ and reverse:
5′-TGAGTCCTTCCACGATACCAA-3′ and U6 forward: 5′-CTCGCTTCGGCAGCACA-3′
and reverse: 5′-AACGCTTCACGAATTTGCGT-3′. The 2−ΔΔCq
method (22) was used for data
analysis and all experiments were performed in three
replicates.
Western blotting
SNU-398 cells were counted, and total proteins in
3×105 cells were extracted using RIPA solution (Sangon
Biotech Co., Ltd.). Protein samples were incubated in boiling water
for 10 min to denature proteins, followed by protein lysates (30
µg/lane) of each group were separated using 10% SDS-PAGE. Proteins
were then transferred to PVDF membranes and 5% non-fat milk was
used to block membranes at room temperature for 80 min. Rabbit
polyclonal PTEN (1:1,400; cat. no. ab31392; Abcam) and GAPDH
(1:1,400; cat. no. ab37168; Abcam) primary antibodies were
incubated with membranes at 4°C for 18 h, followed by incubation
with goat anti-rabbit IgG H+L (1:1,000; cat. no. ab6721; Abcam) at
room temperature for 2 h. After that, membranes were incubated with
RapidStep™ ECL detection reagent (EMD Millipore) at room
temperature for 10 min to develop signals. Data was processed using
ImageJ v1.46 software (National Institutes of Health).
Cell proliferation assay
SNU-398 cells were counted, and 3×104
cells per well were mixed with 1 ml mixture of 10% FBS and 90% RPMI
1640 medium to prepare single-cell suspensions. Cells were
cultivated in a 96-well plate (0.1 ml per well) under conditions of
37°C, 95% humidity and 5% CO2. Cell Counting Kit-8
solution (Sigma-Aldrich; Merck KGaA) was added into each well (10
µl per well) at 4 h before the end of cell culture. After cell
culture, 10 µl DMSO was added and optical density values were
measured at 450 nm wavelength using a microplate reader (Thermo
Fisher Scientific, Inc.).
Statistical analysis
Statistical analyses were performed using SPSS
version 23 (IBM Corp.). Data were expressed as mean values ±
standard deviation of three biological replicates. Associations
were analyzed by linear regression. Differences between two types
of tissue and among different cell groups were analyzed using
paired t-tests and ANOVA (one-way) in combination with the Tukey's
test, respectively. Survival analyses were performed by dividing
the 64 patients with HCC into high and low MT1JP level groups
(n=32) according to the median of MT1JP expression (2.34) in HCC
tissues. Kaplan-Meier plotter and log-rank tests were used to plot
and compare survival curves. P<0.05 was considered to indicate a
statistically significant difference.
Results
MT1JP is downregulated in HCC and
predicts poor survival
To illustrate the role of MT1JP in the progression
of HCC, the expression of MT1JP and the basic information, such as
clinical pathological features of patients with HCC, are displayed
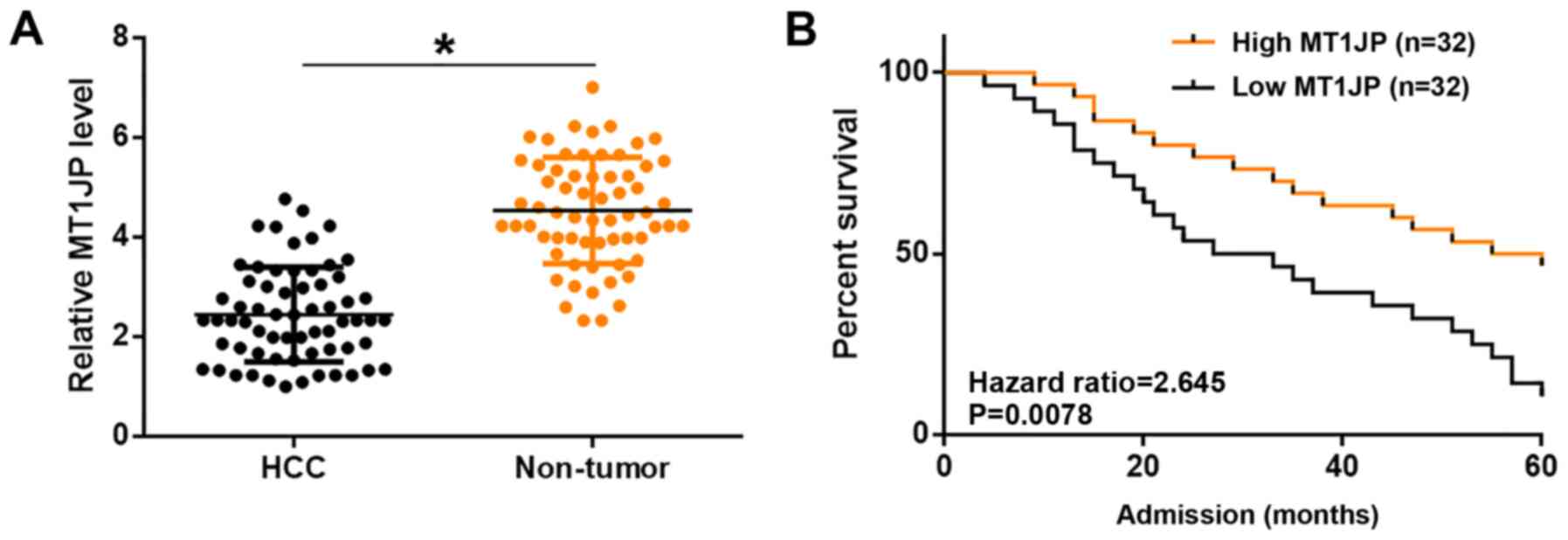
in Table I. The expression levels of
MT1JP in non-tumor and HCC tissues were measured by qPCR and
compared using a paired t-test. Compared with non-tumor tissues,
significantly lower expression levels of MT1JP were observed in HCC
tissues (P<0.05; Fig. 1A).
Compared with patients in the high MT1JP level group, a
significantly lower overall 5-year survival rate was observed in
the low MT1JP level group (P=0.0078; Fig. 1B). It is worth noting that the
expression levels of MT1JP were not significantly affected by HBC
and HCV infections as well as clinical stages (data not shown).
 | Table I.Association between MT1JP expression
and clinicopathological features of patients with hepatocellular
carcinoma. |
Table I.
Association between MT1JP expression
and clinicopathological features of patients with hepatocellular
carcinoma.
|
|
| MT1JP expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
variables | Value, n | Low, n=32 | High, n=32 | P-value |
|---|
| Sex |
|
|
| 0.325 |
| Male | 36 | 23 | 13 |
|
|
Female | 28 | 9 | 19 |
|
| Age, years |
|
|
| 0.269 |
|
<50 | 32 | 13 | 19 |
|
| ≥55 | 32 | 19 | 13 |
|
| HBsAg |
|
|
| 0.221 |
|
Negative | 28 | 15 | 13 |
|
|
Positive | 36 | 17 | 19 |
|
| Tumor size, cm |
|
|
| 0.026 |
|
<5 | 35 | 9 | 26 |
|
| ≥5 | 29 | 23 | 6 |
|
| TNM stage |
|
|
| 0.025 |
|
I/II | 37 | 15 | 22 |
|
|
III/IV | 27 | 17 | 10 |
|
Expression of MT1JP is associated with
the expression of PTEN and miR-32 in HCC
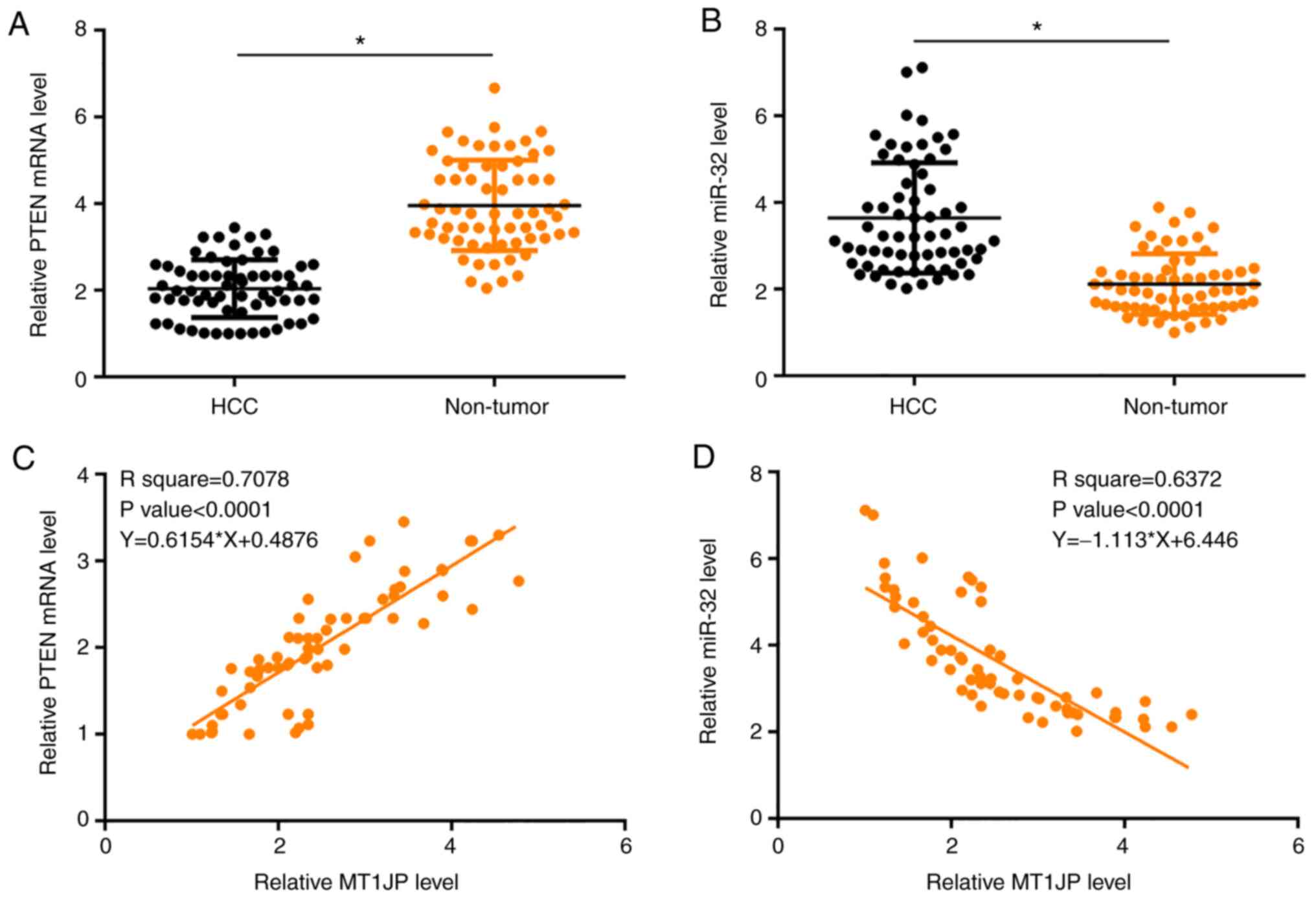
The expression levels of PTEN and miR-32 were also
measured by qPCR and compared using paired t-tests between
non-tumor and HCC tissues. Compared with non-tumor tissues,
significantly lower expression levels of PTEN (Fig. 2A) and significantly higher expression
levels of miR-32 (Fig. 2B) were
observed in HCC tissues (both P<0.05). Regression analysis
showed that the expression of MT1JP in HCC tissues was positively
associated with the expression of PTEN (Fig. 2C) and negatively associated with the
expression of miR-32 (Fig. 2D).
MT1JP upregulates PTEN through
downregulation of miR-32 in HCC cells
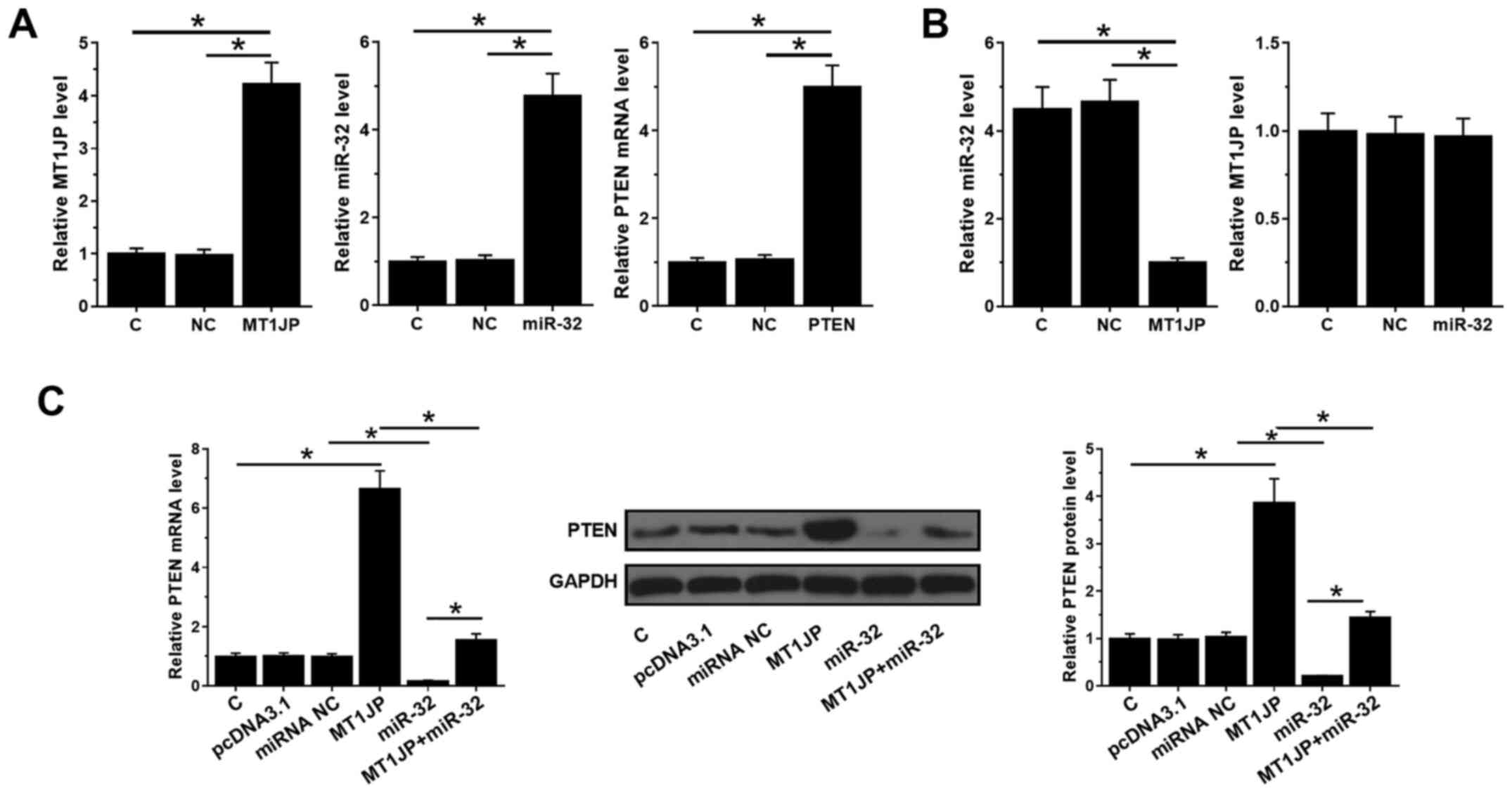
MT1JP and PTEN expression vectors, as well as miR-32
mimic, were transfected into SNU-398 cells and their expression
levels were measured by qPCR at 24 h post-transfection. Compared
with C and NC, the expression levels of MT1JP, miR-32 and PTEN were
significantly increased (all P<0.05; Fig. 3A). Moreover, overexpression of MT1JP
decreased the expression levels of miR-32 (P<0.05), whilst
overexpression of miR-32 did not affect the expression of MT1JP
(Fig. 3B). In addition,
overexpression of miR-32 inhibited the mRNA and protein levels of
PTEN (P<0.05), which could be reversed by simultaneous
upregulation of MT1JP (Fig. 3C).
MT1JP inhibits HCC cell proliferation
through PTEN and miR-32
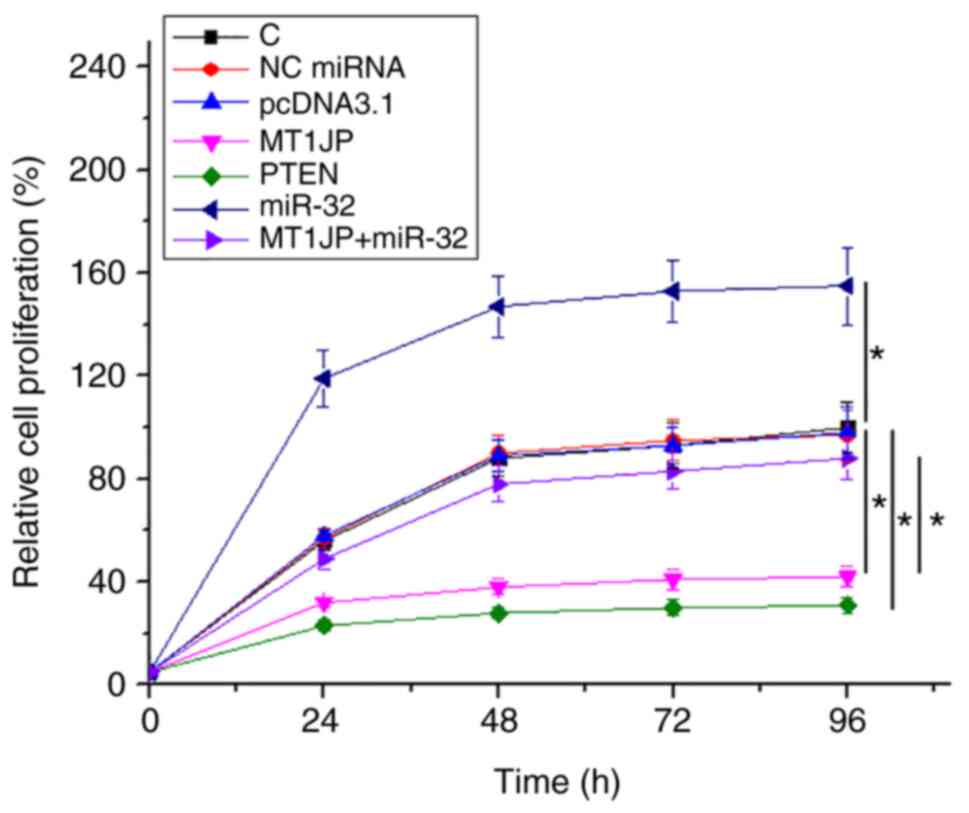
The effects of overexpression of MT1JP, miR-32 and
PTEN on the proliferation of SNU-398 cells were analyzed using a
cell proliferation assay. Compared with NC and C, overexpression of
MT1JP and PTEN resulted in significantly inhibited proliferation of
HCC cells (P<0.05). miR-32 promoted cell proliferation.
Moreover, the suppressive effect of MT1JP on cell proliferation was
reversed by miR-32 (P<0.05; Fig.
4).
Discussion
The present study investigated the function of MT1JP
in HCC. It was found that MT1JP was downregulated in HCC and was
associated with the survival of patients with HCC. In HCC, MT1JP
may have upregulated PTEN through miR-32 to inhibit the
proliferation of HCC cells.
MT1JP is shown to be downregulated in tissues of
different types of cancer (23),
such as gastric cancer (24), glioma
(25) and bladder cancer (26). A recent study has demonstrated the
inhibitory effect of MT1JP on the progression of HCC (27). This study found that MT1JP promotes
apoptosis and inhibits cell proliferation and migration by
inhibiting the expression of miR-24-3p. However, the study did not
thoroughly explore the function of miR-24-3p target genes and the
interaction of MT1JP, miR-24-3p and miR-24-3p target genes. Based
on this, the present study explored another miRNA (miR-32)
regulated by MT1JP and revealed that MT1JP promoted the expression
of the miR-32 target gene PTEN. This was investigated by
downregulating the expression of miR-32, thereby exerting an
antitumor effect, which which inhibited the proliferation of HCC
cells.
PTEN is a dual phosphatase with both protein and
lipid phosphatase activities (28).
Gao et al (29) revealed that
PTEN is concomitantly downregulated in breast cancer tissues and
cell lines, and overexpression of PTEN inhibits the progression of
breast cancer. Wang et al (30) reported that PTEN inhibits the
development of gastric cancer. The present study observed reduced
expression levels of PTEN after the overexpression of miR-32, which
further confirmed the targeting of PTEN by miR-32. The miR-32/PTEN
axis can be modulated by certain lncRNAs, such as GAS5, to regulate
cancer cell malignant behaviors (31). The present research showed that MT1JP
can upregulate PTEN by inhibiting the expression of miR-32, thereby
inhibiting cell proliferation. Moreover, previous studies have
found that the PTEN promoter contains a p53-binding element. Thus,
p53-mediated cell death requires coordinated repression of the
cellular survival machinery via direct activation of PTEN
transcription by p53 (32). Another
study has shown that MT1JP enhances p53 translation by binding to
the RNA binding protein TIAR, thereby regulating p53-related
pathways such as cell cycle, apoptosis and proliferation (23). Therefore, MTIJP may also inhibit
tumor growth through P53/PTEN signaling.
There are certain limitations to the current study.
First, the effects of MT1JP/miR-32/PTEN on several signaling
pathways remains to be elucidate. MT1JP could target multiple
miRNAs to regulate its function in HCC, therefore other potential
target miRNAs should be investigated. This study lacks in
vivo trials, and our conclusions need to be validated in
vivo in the future.
In conclusion, MT1JP played a tumor-suppressive role
in HCC by upregulating PTEN through downregulation of miR-32 to
suppress HCC cell proliferation.
Acknowledgements
Not applicable.
Funding
The study was funded by The 2020 General Program
(Major Project) of Wuhan Municipal Health Commission (grant nos.
WX20B04) and The General Project of Natural Science Foundation of
Hubei Province in 2020 (grant no. 2020CFB796).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SZ and JX developed the project, carried out the
experiments and clinical studies, collected the data and wrote the
manuscript. QC and FZ collected the data, carried out the
experiments and clinical studies, analyzed the data and edited the
manuscript. HW and HG analyzed the data and carried out the
literature research. SZ and JX confirm the authenticity of all raw
data. All contributing authors have read and agreed to the final
version of the manuscript.
Ethics approval and consent to
participate
The Ethics Committee of Union Hospital Affiliated to
Tongji Medical College of Huazhong University of Science and
Technology approved the study (Wuhan, China). Written informed
consent was obtained from all individual participants included in
the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen Z, He M, Chen J, Li C and Zhang Q:
Long non-coding RNA SNHG7 inhibits NLRP3-dependent pyroptosis by
targeting the miR-34a/SIRT1 axis in liver cancer. Oncol Lett.
20:893–901. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Parkin DM and Steliarova-Foucher
E: Estimates of cancer incidence and mortality in Europe in 2008.
Eur J Cancer. 46:765–781. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu J and Xu Y: The lncRNA MEG3
downregulation leads to osteoarthritis progression via miR-16/SMAD7
axis. Cell Biosci. 7:692017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu J, Tang T, Wang GD and Liu B:
lncRNA-H19 promotes hepatic lipogenesis by directly regulating
miR-130a/PPARgamma axis in non-alcoholic fatty liver disease.
Biosci Rep. 39:BSR201817222019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang CA, Li JP, Yen JC, Lai IL, Ho YC,
Chen YC, Lan JL and Chang JG: lncRNA NTT/PBOV1 axis promotes
monocyte differentiation and is elevated in rheumatoid arthritis.
Int J Mol Sci. 19:28062018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Harries LW: Long non-coding RNAs and human
disease. Biochem Soc Trans. 40:902–906. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jiang MC, Ni JJ, Cui WY, Wang BY and Zhuo
W: Emerging roles of lncRNA in cancer and therapeutic
opportunities. Am J Cancer Res. 9:1354–1366. 2019.PubMed/NCBI
|
|
9
|
Wang Z, Huang D, Huang J, Nie K, Li X and
Yang X: lncRNA TMPO-AS1 exerts oncogenic roles in HCC through
regulating miR-320a/SERBP1 Axis. Onco Targets Ther. 13:6539–6551.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Teng F, Zhang JX, Chang QM, Wu XB, Tang
WG, Wang JF, Feng JF, Zhang ZP and Hu ZQ: lncRNA MYLK-AS1
facilitates tumor progression and angiogenesis by targeting
miR-424-5p/E2F7 axis and activating VEGFR-2 signaling pathway in
hepatocellular carcinoma. J Exp Clin Cancer Res. 39:2352020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu X, Peng D, Cao Y, Zhu Y, Yin J, Zhang
G, Peng X and Meng Y: Upregulated lncRNA DLX6-AS1 underpins
hepatocellular carcinoma progression via the miR-513c/Cul4A/ANXA10
axis. Cancer Gene Ther. 28:486–501. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wei F, Yang L, Jiang D, Pan M, Tang G,
Huang M and Zhang J: Long noncoding RNA DUXAP8 contributes to the
progression of hepatocellular carcinoma via regulating
miR-422a/PDK2 axis. Cancer Med. 9:2480–2490. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Komoll RM, Hu Q, Olarewaju O, von Döhlen
L, Yuan Q, Xie Y, Tsay HC, Daon J, Qin R, Manns MP, et al:
MicroRNA-342-3p is a potent tumour suppressor in hepatocellular
carcinoma. J Hepatol. 74:122–134. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Pan Q and Shao Z:
Tumor-Suppressive role of microRNA-202-3p in hepatocellular
carcinoma through the KDM3A/HOXA1/MEIS3 pathway. Front Cell Dev
Biol. 8:5560042021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao L, Han T, Li Y, Sun J, Zhang S, Liu
Y, Shan B, Zheng D and Shi J: The lncRNA SNHG5/miR-32 axis
regulates gastric cancer cell proliferation and migration by
targeting KLF4. FASEB J. 31:893–903. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Y, Wang J, An W, Chen C, Wang W, Zhu
C, Chen F, Chen H, Zheng W and Gong J: MiR-32 inhibits
proliferation and metastasis by targeting EZH2 in glioma. Technol
Cancer Res Treat. 18:15330338198541322019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu YJ, Zhou HG, Chen LH, Qu DC, Wang CJ,
Xia ZY and Zheng JH: MiR-32-5p regulates the proliferation and
metastasis of cervical cancer cells by targeting HOXB8. Eur Rev Med
Pharmacol Sci. 23:87–95. 2019.PubMed/NCBI
|
|
18
|
Carnero A, Blanco-Aparicio C, Renner O,
Link W and Leal JF: The PTEN/PI3K/AKT signalling pathway in cancer,
therapeutic implications. Curr Cancer Drug Targets. 8:187–198.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chalhoub N and Baker SJ: PTEN and the
PI3-kinase pathway in cancer. Annu Rev Pathol. 4:127–150. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yan SY, Chen MM, Li GM, Wang YQ and Fan
JG: MiR-32 induces cell proliferation, migration, and invasion in
hepatocellular carcinoma by targeting PTEN. Tumour Biol.
36:4747–4755. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Zhu Z, Huang S, Zhao Q, Huang C,
Tang Y, Sun C, Zhang Z, Wang L, Chen H, et al: lncRNA XIST
regulates proliferation and migration of hepatocellular carcinoma
cells by acting as miR-497-5p molecular sponge and targeting PDCD4.
Cancer Cell Int. 19:1982019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu L, Yue H, Liu Q, Yuan J, Li J, Wei G,
Chen X, Lu Y, Guo M, Luo J and Chen R: lncRNA MT1JP functions as a
tumor suppressor by interacting with TIAR to modulate the p53
pathway. Oncotarget. 7:15787–15800. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu Y, Zhang G, Zou C, Zhang H, Gong Z,
Wang W, Ma G, Jiang P and Zhang W: lncRNA MT1JP suppresses gastric
cancer cell proliferation and migration through
MT1JP/miR-214-3p/RUNX3 axis. Cell Physiol Biochem. 46:2445–2459.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen J, Lou J, Yang S, Lou J, Liao W, Zhou
R, Qiu C and Ding G: MT1JP inhibits glioma progression via negative
regulation of miR-24. Oncol Lett. 19:334–342. 2020.PubMed/NCBI
|
|
26
|
Yu H, Wang S, Zhu H and Rao D: lncRNA
MT1JP functions as a tumor suppressor via regulating miR-214-3p
expression in bladder cancer. J Cell Physiol. Feb 20–2019.(Epub
ahead of print). doi: 10.1002/jcp.28274. View Article : Google Scholar
|
|
27
|
Wu JH, Xu K, Liu JH, Du LL, Li XS, Su YM
and Liu JC: lncRNA MT1JP inhibits the malignant progression of
hepatocellular carcinoma through regulating AKT. Eur Rev Med
Pharmacol Sci. 24:6647–6656. 2020.PubMed/NCBI
|
|
28
|
Chen CY, Chen J, He L and Stiles BL: PTEN:
Tumor suppressor and metabolic regulator. Front Endocrinol
(Lausanne). 9:3382018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao X, Qin T, Mao J, Zhang J, Fan S, Lu Y,
Sun Z, Zhang Q, Song B and Li L: PTENP1/miR-20a/PTEN axis
contributes to breast cancer progression by regulating PTEN via
PI3K/AKT pathway. J Exp Clin Cancer Res. 38:2562019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang YN, Xu F, Zhang P, Wang P, Wei YN, Wu
C and Cheng SJ: MicroRNA-575 regulates development of gastric
cancer by targeting PTEN. Biomed Pharmacother. 113:1087162019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gao ZQ, Wang JF, Chen DH, Ma XS, Wu Y,
Tang Z and Dang XW: Long non-coding RNA GAS5 suppresses pancreatic
cancer metastasis through modulating miR-32-5p/PTEN axis. Cell
Biosci. 7:662017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Stambolic V, MacPherson D, Sas D, Lin Y,
Snow B, Jang Y, Benchimol S and Mak TW: Regulation of PTEN
transcription by p53. Mol Cell. 8:317–325. 2001. View Article : Google Scholar : PubMed/NCBI
|