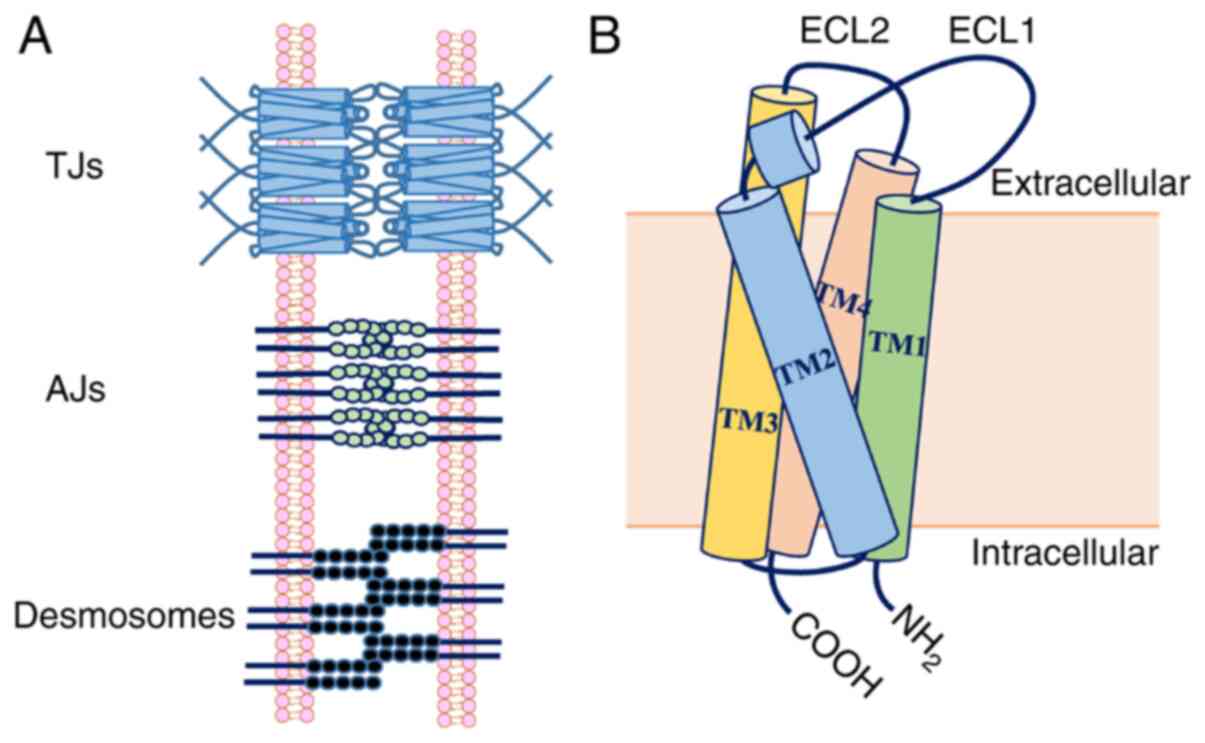
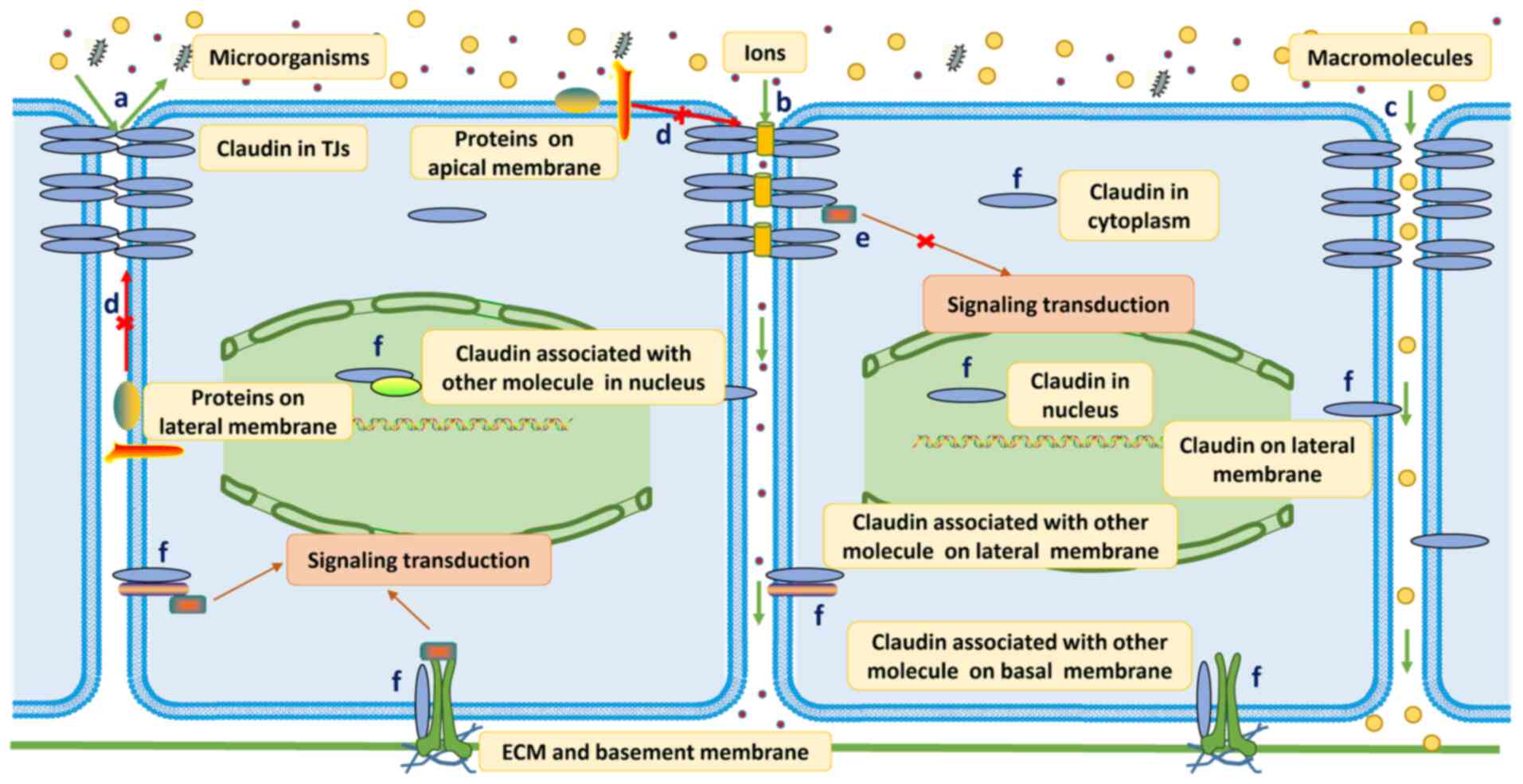
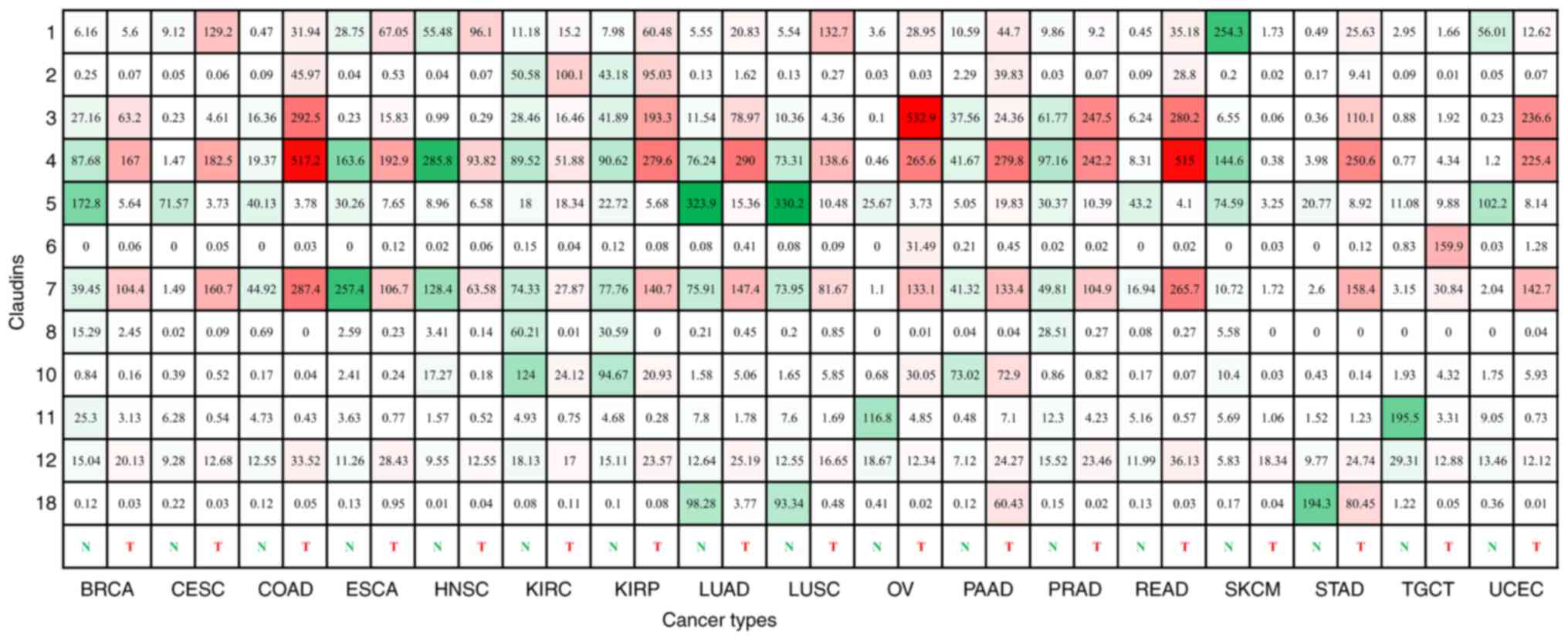
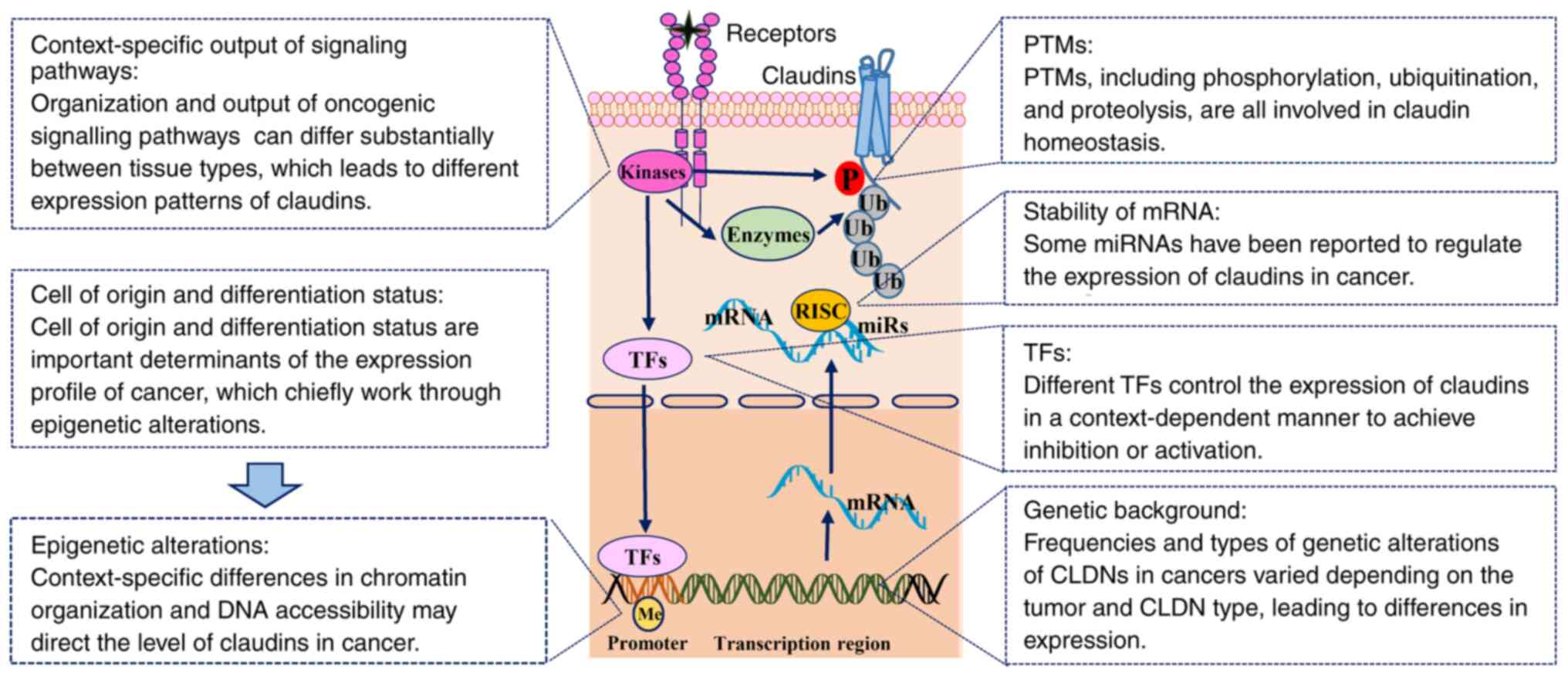
|
1
|
Furuse M, Sasaki H, Fujimoto K and Tsukita
S: A single gene product, claudin-1 or −2, reconstitutes tight
junction strands and recruits occludin in fibroblasts. J Cell Biol.
143:391–401. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Singh AB, Uppada SB and Dhawan P: Claudin
proteins, outside-in signaling, and carcinogenesis. Pflugers Arch.
469:69–75. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hashimoto Y, Tachibana K, Krug SM,
Kunisawa J, Fromm M and Kondoh M: Potential for tight junction
protein-Directed drug development using claudin binders and
angubindin-1. Int J Mol Sci. 20:40162019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schneeberger EE and Lynch RD: The tight
junction: A multifunctional complex. Am J Physiol Cell Physiol.
286:C1213–C1228. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Niessen CM: Tight junctions/adherens
junctions: Basic structure and function. J Invest Dermatol.
127:2525–2532. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Meng W and Takeichi M: Adherens junction:
Molecular architecture and regulation. Cold Spring Harb Perspect
Biol. 1:a0028992009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kottke MD, Delva E and Kowalczyk AP: The
desmosome: Cell science lessons from human diseases. J Cell Sci.
119((Pt 5)): 797–806. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Farquhar MG and Palade GE: Junctional
complexes in various epithelia. J Cell Biol. 17:375–412. 1963.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zihni C, Mills C, Matter K and Balda MS:
Tight junctions: From simple barriers to multifunctional molecular
gates. Nat Rev Mol Cell Biol. 17:564–580. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lal-Nag M and Morin PJ: The claudins.
Genome Biol. 10:2352009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tsukita S, Tanaka H and Tamura A: The
Claudins: From tight junctions to biological systems. Trends
Biochem Sci. 44:141–152. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Suzuki H, Nishizawa T, Tani K, Yamazaki Y,
Tamura A, Ishitani R, Dohmae N, Tsukita S, Nureki O and Fujiyoshi
Y: Crystal structure of a claudin provides insight into the
architecture of tight junctions. Science. 344:304–307. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Suzuki H, Tani K, Tamura A, Tsukita S and
Fujiyoshi Y: Model for the architecture of claudin-based
paracellular ion channels through tight junctions. J Mol Biol.
427:291–297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Saitoh Y, Suzuki H, Tani K, Nishikawa K,
Irie K, Ogura Y, Tamura A, Tsukita S and Fujiyoshi Y: Tight
junctions. Structural insight into tight junction disassembly by
Clostridium perfringens enterotoxin. Science. 347:775–778.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shinoda T, Shinya N, Ito K, Ohsawa N,
Terada T, Hirata K, Kawano Y, Yamamoto M, Kimura-Someya T, Yokoyama
S and Shirouzu M: Structural basis for disruption of claudin
assembly in tight junctions by an enterotoxin. Sci Rep.
6:336322016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakamura S, Irie K, Tanaka H, Nishikawa K,
Suzuki H, Saitoh Y, Tamura A, Tsukita S and Fujiyoshi Y:
Morphologic determinant of tight junctions revealed by claudin-3
structures. Nat Commun. 10:8162019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shen L, Weber CR, Raleigh DR, Yu D and
Turner JR: Tight junction pore and leak pathways: A dynamic duo.
Annu Rev Physiol. 73:283–309. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Otani T and Furuse M: Tight junction
structure and function revisited. Trends Cell Biol. 30:805–817.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ahmad R, Kumar B, Chen Z, Chen X, Müller
D, Lele SM, Washington MK, Batra SK, Dhawan P and Singh AB: Loss of
claudin-3 expression induces IL6/gp130/Stat3 signaling to promote
colon cancer malignancy by hyperactivating Wnt/β-catenin signaling.
Oncogene. 36:6592–6604. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou B, Flodby P, Luo J, Castillo DR, Liu
Y, Yu FX, McConnell A, Varghese B, Li G, Chimge NO, et al:
Claudin-18-mediated YAP activity regulates lung stem and progenitor
cell homeostasis and tumorigenesis. J Clin Invest. 128:970–984.
2018. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hagen SJ: Non-canonical functions of
claudin proteins: Beyond the regulation of cell-cell adhesions.
Tissue Barriers. 5:e13278392017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu Z, Kim DH, Fan J, Lu Q, Verbanac K,
Ding L, Renegar R and Chen YH: A non-tight junction function of
claudin-7-Interaction with integrin signaling in suppressing lung
cancer cell proliferation and detachment. Mol Cancer. 14:1202015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nübel T, Preobraschenski J, Tuncay H,
Weiss T, Kuhn S, Ladwein M, Langbein L and Zöller M: Claudin-7
regulates EpCAM-mediated functions in tumor progression. Mol Cancer
Res. 7:285–299. 2009. View Article : Google Scholar
|
|
24
|
Pope JL, Bhat AA, Sharma A, Ahmad R,
Krishnan M, Washington MK, Beauchamp RD, Singh AB and Dhawan P:
Claudin-1 regulates intestinal epithelial homeostasis through the
modulation of Notch-signalling. Gut. 63:622–634. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu F, Koval M, Ranganathan S, Fanayan S,
Hancock WS, Lundberg EK, Beavis RC, Lane L, Duek P, McQuade L, et
al: Systems proteomics view of the endogenous human claudin protein
family. J Proteome Res. 15:339–359. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Milatz S and Breiderhoff T: One gene, two
paracellular ion channels-claudin-10 in the kidney. Pflugers Arch.
469:115–121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Zhang Y, Hu D, Gong T, Xu R and Gao
J: Analysis of the expression and genetic alteration of CLDN18 in
gastric cancer. Aging (Albany NY). 12:14271–14284. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Milatz S: A novel claudinopathy based on
claudin-10 mutations. Int J Mol Sci. 20:53962019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Blanchard AA, Zelinski T, Xie J, Cooper S,
Penner C, Leygue E and Myal Y: Identification of claudin 1
transcript variants in human invasive breast cancer. PLoS One.
11:e01633872016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ben-David U, Nudel N and Benvenisty N:
Immunologic and chemical targeting of the tight-junction protein
Claudin-6 eliminates tumorigenic human pluripotent stem cells. Nat
Commun. 4:19922013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Reinhard K, Rengstl B, Oehm P, Michel K,
Billmeier A, Hayduk N, Klein O, Kuna K, Ouchan Y, Wöll S, et al: An
RNA vaccine drives expansion and efficacy of claudin-CAR-T cells
against solid tumors. Science. 367:446–453. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Erdélyi-Belle B, Török G, Apáti Á, Sarkadi
B, Schaff Z, Kiss A and Homolya L: Expression of tight junction
components in hepatocyte-like cells differentiated from human
embryonic stem cells. Pathol Oncol Res. 21:1059–1070. 2015.
View Article : Google Scholar
|
|
33
|
Fujita H, Chiba H, Yokozaki H, Sakai N,
Sugimoto K, Wada T, Kojima T, Yamashita T and Sawada N:
Differential expression and subcellular localization of claudin-7,
−8, −12, −13, and −15 along the mouse intestine. J Histochem
Cytochem. 54:933–944. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
D'Souza T, Sherman-Baust CA, Poosala S,
Mullin JM and Morin PJ: Age-related changes of claudin expression
in mouse liver, kidney, and pancreas. J Gerontol A Biol Sci Med
Sci. 64:1146–1153. 2009. View Article : Google Scholar
|
|
35
|
Perdomo-Ramirez A, Aguirre M, Davitaia T,
Ariceta G, Ramos-Trujillo E; RenalTube Group, ; Claverie-Martin F:
Characterization of two novel mutations in the claudin-16 and
claudin-19 genes that cause familial hypomagnesemia with
hypercalciuria and nephrocalcinosis. Gene. 689:227–234. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ouban A: Claudin-1 role in colon cancer:
An update and a review. Histol Histopathol. 33:1013–1019.
2018.PubMed/NCBI
|
|
37
|
Wang K, Xu C, Li W and Ding L: Emerging
clinical significance of claudin-7 in colorectal cancer: A review.
Cancer Manag Res. 10:3741–3752. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dhawan P, Singh AB, Deane NG, No Y, Shiou
SR, Schmidt C, Neff J, Washington MK and Beauchamp RD: Claudin-1
regulates cellular transformation and metastatic behavior in colon
cancer. J Clin Invest. 115:1765–1776. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Blanchard AA, Skliris GP, Watson PH,
Murphy LC, Penner C, Tomes L, Young TL, Leygue E and Myal Y:
Claudins 1, 3, and 4 protein expression in ER negative breast
cancer correlates with markers of the basal phenotype. Virchows
Arch. 454:647–656. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cherradi S, Ayrolles-Torro A, Vezzo-Vié N,
Gueguinou N, Denis V, Combes E, Boissière F, Busson M,
Canterel-Thouennon L, Mollevi C, et al: Antibody targeting of
claudin-1 as a potential colorectal cancer therapy. J Exp Clin
Cancer Res. 36:892017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Matsuoka T, Mitomi H, Fukui N, Kanazawa H,
Saito T, Hayashi T and Yao T: Cluster analysis of claudin-1 and −4,
E-cadherin, and β-catenin expression in colorectal cancers. J Surg
Oncol. 103:674–686. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bhat AA, Ahmad R, Uppada SB, Singh AB and
Dhawan P: Claudin-1 promotes TNF-α-induced epithelial-mesenchymal
transition and migration in colorectal adenocarcinoma cells. Exp
Cell Res. 349:119–127. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bornholdt J, Friis S, Godiksen S, Poulsen
SS, Santoni-Rugiu E, Bisgaard HC, Lothe IM, Ikdahl T, Tveit KM,
Johnson E, et al: The level of claudin-7 is reduced as an early
event in colorectal carcinogenesis. BMC Cancer. 11:652011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tanaka M, Shibahara J, Fukushima N,
Shinozaki A, Umeda M, Ishikawa S, Kokudo N and Fukayama M:
Claudin-18 is an early-stage marker of pancreatic carcinogenesis. J
Histochem Cytochem. 59:942–952. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Stratton MR: Exploring the genomes of
cancer cells: Progress and promise. Science. 331:1553–1558. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gonzalez-Perez A, Perez-Llamas C, Deu-Pons
J, Tamborero D, Schroeder MP, Jene-Sanz A, Santos A and Lopez-Bigas
N: IntOGen-mutations identifies cancer drivers across tumor types.
Nat Methods. 10:1081–1082. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Martínez-Jiménez F, Muiños F, Sentís I,
Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, Mularoni L, Pich O,
Bonet J, Kranas H, et al: A compendium of mutational cancer driver
genes. Nat Rev Cancer. 20:555–572. 2020. View Article : Google Scholar
|
|
48
|
Paschoud S, Bongiovanni M, Pache JC and
Citi S: Claudin-1 and claudin-5 expression patterns differentiate
lung squamous cell carcinomas from adenocarcinomas. Mod Pathol.
20:947–954. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Schaefer MH and Serrano L: Cell
type-specific properties and environment shape tissue specificity
of cancer genes. Sci Rep. 6:207072016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Honda H, Pazin MJ, D'Souza T, Ji H and
Morin PJ: Regulation of the CLDN3 gene in ovarian cancer cells.
Cancer Biol Ther. 6:1733–1742. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Di Cello F, Cope L, Li H, Jeschke J, Wang
W, Baylin SB and Zahnow CA: Methylation of the claudin 1 promoter
is associated with loss of expression in estrogen receptor positive
breast cancer. PLoS One. 8:e686302013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chiang SK, Chang WC, Chen SE and Chang LC:
DOCK1 regulates growth and motility through the RRP1B-claudin-1
pathway in claudin-low breast cancer cells. Cancers (Basel).
11:17622019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li CP, Cai MY, Jiang LJ, Mai SJ, Chen JW,
Wang FW, Liao YJ, Chen WH, Jin XH, Pei XQ, et al: CLDN14 is
epigenetically silenced by EZH2-mediated H3K27ME3 and is a novel
prognostic biomarker in hepatocellular carcinoma. Carcinogenesis.
37:557–566. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Martínez-Estrada OM, Cullerés A, Soriano
FX, Peinado H, Bolós V, Martínez FO, Reina M, Cano A, Fabre M and
Vilaró S: The transcription factors Slug and Snail act as
repressors of Claudin-1 expression in epithelial cells. Biochem J.
(394(Pt 2)): 449–457. 2006. View Article : Google Scholar
|
|
55
|
Bhat AA, Sharma A, Pope J, Krishnan M,
Washington MK, Singh AB and Dhawan P: Caudal homeobox protein Cdx-2
cooperates with Wnt pathway to regulate claudin-1 expression in
colon cancer cells. PLoS One. 7:e371742012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Feng J, Cen J, Li J, Zhao R, Zhu C, Wang
Z, Xie J and Tang W: Histone deacetylase inhibitor valproic acid
(VPA) promotes the epithelial mesenchymal transition of colorectal
cancer cells via up regulation of Snail. Cell Adh Migr. 9:495–501.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kohno Y, Okamoto T, Ishibe T, Nagayama S,
Shima Y, Nishijo K, Shibata KR, Fukiage K, Otsuka S, Uejima D, et
al: Expression of claudin7 is tightly associated with epithelial
structures in synovial sarcomas and regulated by an Ets family
transcription factor, ELF3. J Biol Chem. 281:38941–38950. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Farkas AE, Hilgarth RS, Capaldo CT,
Gerner-Smidt C, Powell DR, Vertino PM, Koval M, Parkos CA and
Nusrat A: HNF4alpha regulates claudin-7 protein expression during
intestinal epithelial differentiation. Am J Pathol. 185:2206–2218.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Baltzegar DA, Reading BJ, Brune ES and
Borski RJ: Phylogenetic revision of the claudin gene family. Mar
Genomics. 11:17–26. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Honda H, Pazin MJ, Ji H, Wernyj RP and
Morin PJ: Crucial roles of Sp1 and epigenetic modifications in the
regulation of the CLDN4 promoter in ovarian cancer cells. J Biol
Chem. 281:21433–21444. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li CF, Chen JY, Ho YH, Hsu WH, Wu LC, Lan
HY, Hsu DS, Tai SK, Chang YC and Yang MH: Snail-induced claudin-11
prompts collective migration for tumour progression. Nat Cell Biol.
21:251–262. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liu M, Yang J, Zhang Y, Zhou Z, Cui X,
Zhang L, Fung KM, Zheng W, Allard FD, Yee EU, et al: ZIP4 promotes
pancreatic cancer progression by repressing ZO-1 and claudin-1
through a ZEB1-dependent transcriptional mechanism. Clin Cancer
Res. 24:3186–3196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Sun X, Cui S, Fu X, Liu C, Wang Z and Liu
Y: MicroRNA-146-5p promotes proliferation, migration and invasion
in lung cancer cells by targeting claudin-12. Cancer Biomark.
25:89–99. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang YB, Shi Q, Li G, Zheng JH, Lin J and
Qiu W: MicroRNA-488 inhibits progression of colorectal cancer via
inhibition of the mitogen-activated protein kinase pathway by
targeting claudin-2. Am J Physiol Cell Physiol. 316:C33–C47. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sun H, Cui C, Xiao F, Wang H, Xu J, Shi X,
Yang Y, Zhang Q, Zheng X, Yang X, et al: MiR-486 regulates
metastasis and chemosensitivity in hepatocellular carcinoma by
targeting CLDN10 and CITRON. Hepatol Res. 45:1312–1322. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhang SJ, Feng JF, Wang L, Guo W, Du YW,
Ming L and Zhao GQ: MiR-1303 targets claudin-18 gene to modulate
proliferation and invasion of gastric cancer cells. Dig Dis Sci.
59:1754–1763. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Cheng B, Rong A, Zhou Q and Li W: LncRNA
LINC00662 promotes colon cancer tumor growth and metastasis by
competitively binding with miR-340-5p to regulate CLDN8/IL22
co-expression and activating ERK signaling pathway. J Exp Clin
Cancer Res. 39:52020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhang XZ, Mao HL, Zhang SJ, Sun L, Zhang
WJ, Chen QZ, Wang L and Liu HC: lncRNA PCAT18 inhibits
proliferation, migration and invasion of gastric cancer cells
through miR-135b suppression to promote CLDN11 expression. Life
Sci. 249:1174782020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Krishnan M, Singh AB, Smith JJ, Sharma A,
Chen X, Eschrich S, Yeatman TJ, Beauchamp RD and Dhawan P: HDAC
inhibitors regulate claudin-1 expression in colon cancer cells
through modulation of mRNA stability. Oncogene. 29:305–312. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhou B, Moodie A, Blanchard AA, Leygue E
and Myal Y: Claudin 1 in breast cancer: New insights. J Clin Med.
4:1960–1976. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Tanaka M, Kamata R and Sakai R: EphA2
phosphorylates the cytoplasmic tail of Claudin-4 and mediates
paracellular permeability. J Biol Chem. 280:42375–42382. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Owari T, Sasaki T, Fujii K, Fujiwara-Tani
R, Kishi S, Mori S, Mori T, Goto K, Kawahara I, Nakai Y, et al:
Role of nuclear claudin-4 in renal cell carcinoma. Int J Mol Sci.
21:83402020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Heiler S, Mu W, Zöller M and Thuma F: The
importance of claudin-7 palmitoylation on membrane subdomain
localization and metastasis-promoting activities. Cell Commun
Signal. 13:292015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yuan M, Chen X, Sun Y, Jiang L, Xia Z, Ye
K, Jiang H, Yang B, Ying M, Cao J and He Q: ZDHHC12-mediated
claudin-3 S-palmitoylation determines ovarian cancer progression.
Acta Pharm Sin B. 10:1426–1439. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Mandel I, Paperna T, Volkowich A, Merhav
M, Glass-Marmor L and Miller A: The ubiquitin-proteasome pathway
regulates claudin 5 degradation. J Cell Biochem. 113:2415–2423.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Gong Y, Wang J, Yang J, Gonzales E, Perez
R and Hou J: KLHL3 regulates paracellular chloride transport in the
kidney by ubiquitination of claudin-8. Proc Natl Acad Sci USA.
112:4340–4345. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Willemsen LE, Hoetjes JP, van Deventer SJ
and van Tol EA: Abrogation of IFN-gamma mediated epithelial barrier
disruption by serine protease inhibition. Clin Exp Immunol.
142:275–284. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Lytle NK, Barber AG and Reya T: Stem cell
fate in cancer growth, progression and therapy resistance. Nat Rev
Cancer. 18:669–680. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Rahner C, Mitic LL and Anderson JM:
Heterogeneity in expression and subcellular localization of
claudins 2, 3, 4, and 5 in the rat liver, pancreas, and gut.
Gastroenterology. 120:411–422. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Paquet-Fifield S, Koh SL, Cheng L, Beyit
LM, Shembrey C, Mølck C, Behrenbruch C, Papin M, Gironella M,
Guelfi S, et al: Tight junction protein claudin-2 promotes
self-renewal of human colorectal cancer stem-like cells. Cancer
Res. 78:2925–2938. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Sahin U, Koslowski M, Dhaene K, Usener D,
Brandenburg G, Seitz G, Huber C and Türeci O: Claudin-18 splice
variant 2 is a pan-cancer target suitable for therapeutic antibody
development. Clin Cancer Res. 14:7624–7634. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Akizuki R, Maruhashi R, Eguchi H,
Kitabatake K, Tsukimoto M, Furuta T, Matsunaga T, Endo S and Ikari
A: Decrease in paracellular permeability and chemosensitivity to
doxorubicin by claudin-1 in spheroid culture models of human lung
adenocarcinoma A549 cells. Biochim Biophys Acta Mol Cell Res.
1865:769–780. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Kyuno D, Kojima T, Yamaguchi H, Ito T,
Kimura Y, Imamura M, Takasawa A, Murata M, Tanaka S, Hirata K and
Sawada N: Protein kinase Cα inhibitor protects against
downregulation of claudin-1 during epithelial-mesenchymal
transition of pancreatic cancer. Carcinogenesis. 34:1232–1243.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Li M, Zhang Y, Liu Z, Bharadwaj U, Wang H,
Wang X, Zhang S, Liuzzi JP, Chang SM, Cousins RJ, et al: Aberrant
expression of zinc transporter ZIP4 (SLC39A4) significantly
contributes to human pancreatic cancer pathogenesis and
progression. Proc Natl Acad Sci USA. 104:18636–18641. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Akimoto T, Takasawa A, Takasawa K, Aoyama
T, Murata M, Osanai M, Saito T and Sawada N: Estrogen/GPR30
signaling contributes to the malignant potentials of ER-negative
cervical adenocarcinoma via regulation of claudin-1 expression.
Neoplasia. 20:1083–1093. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Okada T, Konno T, Kohno T, Shimada H,
Saito K, Satohisa S, Saito T and Kojima T: Possibility of targeting
claudin-2 in therapy for human endometrioid endometrial carcinoma.
Reprod Sci. 27:2092–2103. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Domazetovic V, Iantomasi T, Bonanomi AG
and Stio M: Vitamin D regulates claudin-2 and claudin-4 expression
in active ulcerative colitis by p-Stat-6 and Smad-7 signaling. Int
J Colorectal Dis. 35:1231–1242. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Mattern J, Roghi CS, Hurtz M, Knäuper V,
Edwards DR and Poghosyan Z: ADAM15 mediates upregulation of
claudin-1 expression in breast cancer cells. Sci Rep. 9:125402019.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Kim WK, Kwon Y, Jang M, Park M, Kim J, Cho
S, Jang DG, Lee WB, Jung SH, Choi HJ, et al: β-catenin activation
down-regulates cell-cell junction-related genes and induces
epithelial-to-mesenchymal transition in colorectal cancers. Sci
Rep. 9:184402019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Rachakonda G, Vu T, Jin L, Samanta D and
Datta PK: Role of TGF-β-induced Claudin-4 expression through c-Jun
signaling in non-small cell lung cancer. Cell Signal. 28:1537–1544.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Chen YJ, You ML, Chong QY, Pandey V,
Zhuang QS, Liu DX, Ma L, Zhu T and Lobie PE: Autocrine human growth
hormone promotes invasive and cancer stem cell-like behavior of
hepatocellular carcinoma cells by STAT3 dependent inhibition of
CLAUDIN-1 expression. Int J Mol Sci. 18:12742017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Chang TL, Ito K, Ko TK, Liu Q,
Salto-Tellez M, Yeoh KG, Fukamachi H and Ito Y: Claudin-1 has tumor
suppressive activity and is a direct target of RUNX3 in gastric
epithelial cells. Gastroenterology. 138:255–265.e1-3. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Ashikari D, Takayama KI, Obinata D,
Takahashi S and Inoue S: CLDN8, an androgen-regulated gene,
promotes prostate cancer cell proliferation and migration. Cancer
Sci. 108:1386–1393. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Mima S, Tsutsumi S, Ushijima H, Takeda M,
Fukuda I, Yokomizo K, Suzuki K, Sano K, Nakanishi T, Tomisato W, et
al: Induction of claudin-4 by nonsteroidal anti-inflammatory drugs
and its contribution to their chemopreventive effect. Cancer Res.
65:1868–1876. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Iitaka D, Moodley S, Shimizu H, Bai XH and
Liu M: PKCδ-iPLA2-PGE2-PPARγ signaling cascade mediates TNF-α
induced Claudin 1 expression in human lung carcinoma cells. Cell
Signal. 27:568–577. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Morin PJ: Claudin proteins in human
cancer: Promising new targets for diagnosis and therapy. Cancer
Res. 65:9603–9606. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Singh P, Toom S and Huang Y: Anti-claudin
18.2 antibody as new targeted therapy for advanced gastric cancer.
J Hematol Oncol. 10:1052017. View Article : Google Scholar : PubMed/NCBI
|