Introduction
According to data published in 2018 by the American
Cancer Society, colorectal cancer (CRC) is the second most common
type of cancer, with an incidence rate of 10.2%, and the fourth
most common cause of cancer-associated mortality worldwide
(1), accounting for ~900,000 deaths
per year in 2018 (2,3). In addition to sex, ageing and
geographical location, numerous other risk factors have been
associated with the increasing incidence of CRC (4). For example, both hereditary and
environmental factors are highly associated with the development of
CRC (5). According to the mutational
origin, CRC can be classified into sporadic, inherited and familial
forms (6). Angiogenesis is a
complex, tightly regulated process. Several molecules, including
VEGF, Notch, angiopoietins and integrins, are known to orchestrate
the process (7). Among these
factors, VEGF is essential for the induction of physiological and
pathological angiogenesis (8).
Upregulated expression levels of VEGF have been reported to promote
the uncontrolled formation of blood tumor vessels, which has been
revealed to be associated with CRC invasiveness, metastasis and
poor prognosis (9). Therefore, the
majority of current anti-angiogenic treatments for CRC target the
VEGF/VEGFR signaling pathway (10).
Mutant angiogenic VEGF may provide a genomic basis for the
diversity of the tumor-host response and suggests the importance of
targeting mutant VEGF with antisense oligonucleotides, since all
different VEGF isoforms would have to be neutralized to prevent
angiogenesis (11).
IL-8, through binding to its receptors, has been
revealed to promote migration, invasion, proliferation and
angiogenesis in numerous cancer cell types, including
HIF-1α-deficient colon cancer and estrogen receptor-negative breast
cancer cells (12). Notably, IL-8
signaling has been demonstrated to promote the malignant
progression of tumors and has been associated with inflammatory
signaling pathways and gastric cancer (13). SNPs in genes within the IL-8
signaling pathway [IL-8, C-X-C motif chemokine receptor (CXCR)1 and
CXCR2] have been reported to be associated with CRC risk (14). The upregulated expression levels of
variants of VEGF and IL-8 have been previously revealed to be
independently associated with decreased tumor recurrence in
patients with stage III CRC, and the analysis of the patients'
angiogenic potential based on VEGF and IL-8 genotypes has been
suggested to potentially further enhance the efficacy of
anti-angiogenic treatment (15). By
contrast, IL-27, a member of the IL-12 cytokine family, has
demonstrated antitumor activity via stimulation of cytotoxic T
cells and natural killer cell function, regulating the
differentiation of T cell subsets, and suppressing dendritic cell
function and angiogenesis (16). In
addition, IL-27 gene polymorphisms have been revealed to increase
the risk of CRC development (17).
The expression levels of hERG, which encodes a
potassium channel, have been demonstrated to be highly upregulated
in CRC cases with hyperplastic lesions in the colon (18) and in CRC cell lines (19). However, to the best of our knowledge,
the association between IL-8, IL-27 and VEGF, and the expression
levels of hERG remains unknown. Therefore, the present study aimed
to determine the changes in white blood cell (WBC) counts, serum
levels of CEA, and the concentrations and mutational status of
IL-8, IL-27 and VEGF genes in patients with CRC, and to investigate
their association with the expression levels of hERG.
Patients and methods
Patient samples
The present investigation was based on a
case-control study. Study samples were obtained from the Oncology
Department of the Hiwa Hospital (Sulaimanya, Kurdistan Region,
Iraq) and the Oncology Departments of the Rizgary, Nanakaly and PAR
Hospitals (Erbil, Kurdistan Region, Iraq). The present study was
approved by the Human Ethics Committee of the College of Science,
Salahaddin University-Erbil (N0:4/2/2002; date, 16/6/2019; Erbil,
Iraq) and the study was conducted according to the principles of
the Declaration of Helsinki. Written informed consent was obtained
from all participants for the use of their blood and tissues prior
to participation. Venous blood and tissue samples were obtained
from 80 patients with CRC (40 male and 40 female patients) and 80
healthy individuals (50 male and 30 female patients). For tissue
collection, control samples were obtained from the aforementioned
healthy individuals who had negative results following endoscopy
and CEA tests. Patients and healthy individuals were recruited
between August 2019 and February 2020, and the median age of
patients was 55 years (range, 20–70 years), while healthy
individuals had a median age of 52 years (range, 23–70 years).
Patients with CRC preferably detected by screening and colonoscopy
diagnosis were included. The following exclusion criteria were
used: i) Patients with underlying immunodeficiency disorders; and
ii) immunodeficient individuals who had other co-morbidities that
could introduce heterogeneity to the sample, such as additional
acquired brain injury, arthritis, chronic obstructive pulmonary
disease, asthma, diabetes mellitus, ankylosing spondylitis,
connective tissue diseases and other inflammatory diseases.
Tissue and blood collection
All tissue samples were obtained following endoscopy
or resection surgery at the PAR Hospital (Erbil, Iraq), while blood
samples were collected in a tube containing PBS at the Oncology
Department of the Hiwa Hospital (Sulaimanya, Kurdistan Region,
Iraq) and the Rizgary Hospital Oncology Department, Nanakaly
Hospital, Erbil and PAR Hospital (Erbil, Kurdistan, Iraq), then
directly transferred to the Advanced Cancer Biology Laboratory,
Department of Biology, College of Science, Salahaddin
University-Erbil, Erbil, Iraq for DNA and RNA extraction
procedures. Concurrently, blood samples were collected by
phlebotomy under the aseptic technique. Blood from both patients
and healthy individuals was aspirated into a 5-ml syringe,
transferred into a tube and then processed at the Advanced Cancer
Biology Laboratory within 1 h. Subsequently, the blood was placed
into a clot activator tube for serum preparation. Samples were
centrifuged at 1,300 × g for 5 min in a refrigerated centrifuge at
4°C and the separated sera were preserved in an Eppendorf tube,
which was stored at −80°C until required for further analysis.
WBC measurements
Total WBC and differential leukocyte counts were
determined according to the manufacturer's protocol using an
automated Coulter Ac.T diff Hematology analyzer (Beckman Coulter,
Inc.).
Estimation of CEA concentration
The concentration of CEA was determined using a
Cobas e411 analyzer (Roche Diagnostics) and a ready-to-use reagent
kit (Elecsys CEA; Roche Diagnostics) in individual cassettes
according to the manufacturer's protocol. This detection method
relies on using electrochemiluminescence to measure the
immunoreactivity (20).
Determination of IL-8, IL-27 and VEGF
concentrations
The serum concentrations of IL-8, IL-27 and VEGF
were determined using Human Interleukin 8 ELISA Kit (cat. no.
E-EL-H6008), Human Interleukin 27 ELISA Kit (cat. no. E-EL-H2338)
and Human Vascular Endothelial Growth Factor ELISA Kit (cat. no.
E-EL-H1600) (Elabscience, Inc.), respectively, using the
sandwich-ELISA method. All reagents and antibodies used for ELISA
were included in the kits. The micro-ELISA plates were pre-coated
with antibodies specific to IL-8, IL-27 and VEGF. Standards or
samples were added to the micro-ELISA plate wells and incubated
with the anti-IL-8, anti-IL-27 and anti-VEGF antibodies. A
biotinylated detection antibody specific for IL-8, IL-27 or VEGF,
avidin-HRP conjugate and substrate were then added to each well.
The micro-ELISA plate that contained IL-8, IL-27 and VEGF,
biotinylated detection antibodies and the avidin-HRP conjugate
appeared blue. The enzyme-substrate reaction was then stopped with
1 M H2SO4 solution and the color subsequently
turned yellow. The absorbance measured spectrophotometrically with
an ELISA reader (BioTek Instruments, Inc.) at a wavelength of 450
nm was proportional to the concentration of IL-8, IL-27 and VEGF.
Subsequently, the concentrations in the samples were calculated by
comparing the absorbances of the samples to the standard curve.
DNA extraction and quantification
Genomic DNA was purified from CRC tissues using the
Tissue Genomic DNA Mini Kit (cat. no. GT050/100/300; Geneaid
Biotech Ltd.) according to the manufacturer's protocol. The eluted
DNA was assessed for concentration (A260) and purity (A260/A280
ratio) on a Nanodrop spectrophotometer (Thermo Fisher Scientific,
Inc.). The Nanodrop instrument was blanked with the elution buffer,
and the DNA purity was based on an optimal A260/A280 ratio of
1.8.
Genotype determination
The present study investigated three commonly
studied variants of IL-8, IL-27 and VEGF. Three different locations
were selected on the basis of published reports on reference gene
expression profiles and previous databases (21–23) as
follows: IL-8 [3′-untranslated region (UTR), rs4073], IL-27 (exon
region, rs17855750) and VEGF (5′-UTR, rs2010963).
First, the purified DNA was separately amplified
using ready to use master mix (containing Taq DNA
polymerase, dNTPs, MgCl2 and reaction buffer; Promega
Corporation) for each genetic polymorphism using PCR on a Techne
TC-512 gradient thermal cycler (Cole-Parmer, Ltd.) using the
following primers: IL-8 rs4073 forward,
5′-CTAAGAGCAGTAACAGTTCCTAG-3′ and reverse,
5′-CATTATGTCAGAGGAAATTCCACG-3′; IL-27 rs17855750 forward,
5′-GTTCCCTTCCTTCTAAGCT-3′ and reverse, 5′-GAAGGTCAGGGAAACATCAGG-3′;
and VEGF forward, 5′-CTCGGTGCTGGAATTTGATATTC-3′ and reverse,
5′-CAAAAGCAGGTCACTCACTTTGC-3′. The following thermocycling
conditions were used for PCR: Initial denaturation at 95°C for 5
min; followed by 35 cycles of 95°C for 30 sec, annealing at 55°C
for 30 sec and elongation at 72°C for 1 min; and a final extension
step at 72°C for 5 min. All PCR products were separated via 3%
agarose gel electrophoresis, compared with the 50 bp DNA marker
(MiniSizer 50 bp DNA Ladder; cat. no 11200; Norgen Biotek Corp.)
and stained with safe dye (Safe DNA Gel Stain dye; Add Bio, Inc.)
before casting into the tray. Gels were visualized using a gel
documentation system (UV Transilluminator UST-20M-8K; Biostep
GmbH).
Following the PCR procedure, the product was sent
for sequencing to Immunogen Center, Erbil, Iraq (using the same
forward primers for each specific region in the gene) on an
automatic 3130 genetic analyzer (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Sequence data has been included in a curated
data repository (GenBank database; http://www.ncbi.nlm.nih.gov/genbank/). Nucleotide
sequence data reported are available in the GenBank database under
the following accession numbers: MW672394 (www.ncbi.nlm.nih.gov/nuccore/MW672394.1/),
MW672395 (www.ncbi.nlm.nih.gov/nuccore/MW672395.1/) and
MW672396 (www.ncbi.nlm.nih.gov/nuccore/MW672396.1/).
Analysis of the Sanger sequencing data was performed using the
Mutation Surveyor software package 5.1 (SoftGenetics, LLC) by
comparing with reference genes (AF385628.2, EF064720.1 and
AH001553.2).
Reverse transcription-qPCR
Total RNA was extracted from blood and tissues using
TRIzol® reagent (cat. no. 15596026; Invitrogen; Thermo
Fisher Scientific, Inc.). Total RNA was reverse transcribed into
cDNA using a RevertAid First Strand cDNA Synthesis Kit (cat. no.
K1621; Thermo Fisher Scientific, Inc.). Reverse transcription was
performed at 25°C for 5 min, followed by incubation for 60 min at
42°C. Custom primer was designed to amplify specific regions in the
hERG gene and the sequences used were: Forward,
5′-CACCTCCTCGTTGGCATTG-3′ and reverse, 5′-GCTGGCTGTGGTGGACCT-3′.
For reverse transcription, in addition to patient's samples,
RT-negative, positive control (GAPDH) and no template negative
control were setup. qPCR was subsequently performed using a
SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories,
Inc.) on a Rotor-Gene® Q Real-Time PCR cycler (Qiagen
GmbH). GAPDH served as a reference gene (forward primer,
5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse primer,
5′-GAAGATGGTGATGGGATTTC-3′). The relative expression levels were
analyzed using the 2−ΔΔCq method (24). The qPCR cycling conditions were as
follows: 2 min at 95°C, followed by 40 cycles of denaturation at
95°C for 5 sec, annealing at 60°C for 30 sec and extension at 72°C
for 45 sec, and final extension at 72°C for 5 min.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 6.0 software (GraphPad Software, Inc.). Statistical
differences in the serological data between patients with CRC
(n=80) and healthy individuals (n=80) were determined using a
U-Mann Whitney test. Normality tests, namely D'Agostino and Pearson
omnibus, Shapiro-Wilk and KS normality tests, were performed for
all data. The area under the curve (AUC) for CEA concentration
(n=30) was calculated using a receiver operating characteristic
curve. The data are presented either as the median and
interquartile range or as mean ± SEM. The strength of the
correlation between the expression levels of hERG and the serum
concentration of IL-8 and IL-27 was calculated using a
nonparametric Spearman correlation test. P<0.05 was considered
to indicate a statistically significant difference.
Results
Total WBC count
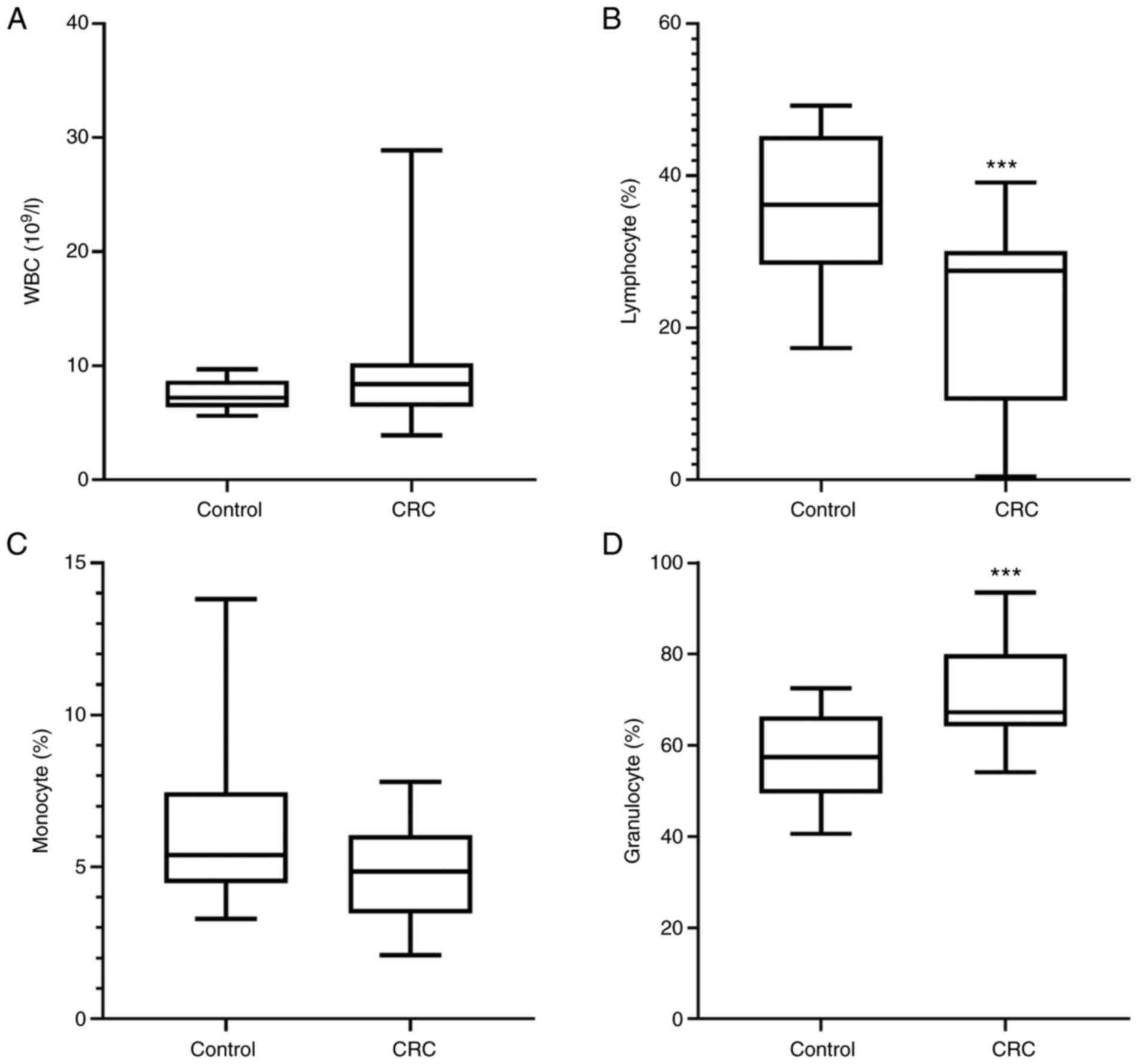
The percentage of granulocytes was significantly
increased (P<0.001), whereas the percentage of lymphocytes was
significantly decreased (P<0.001) in patients with CRC compared
with in healthy individuals (Fig. 1B and
D). However, there were no changes observed in the total WBC
count and percentage of monocytes between the patients with CRC and
healthy individuals (Fig. 1A and
C).
Tumor markers in patients with CRC and
healthy individuals
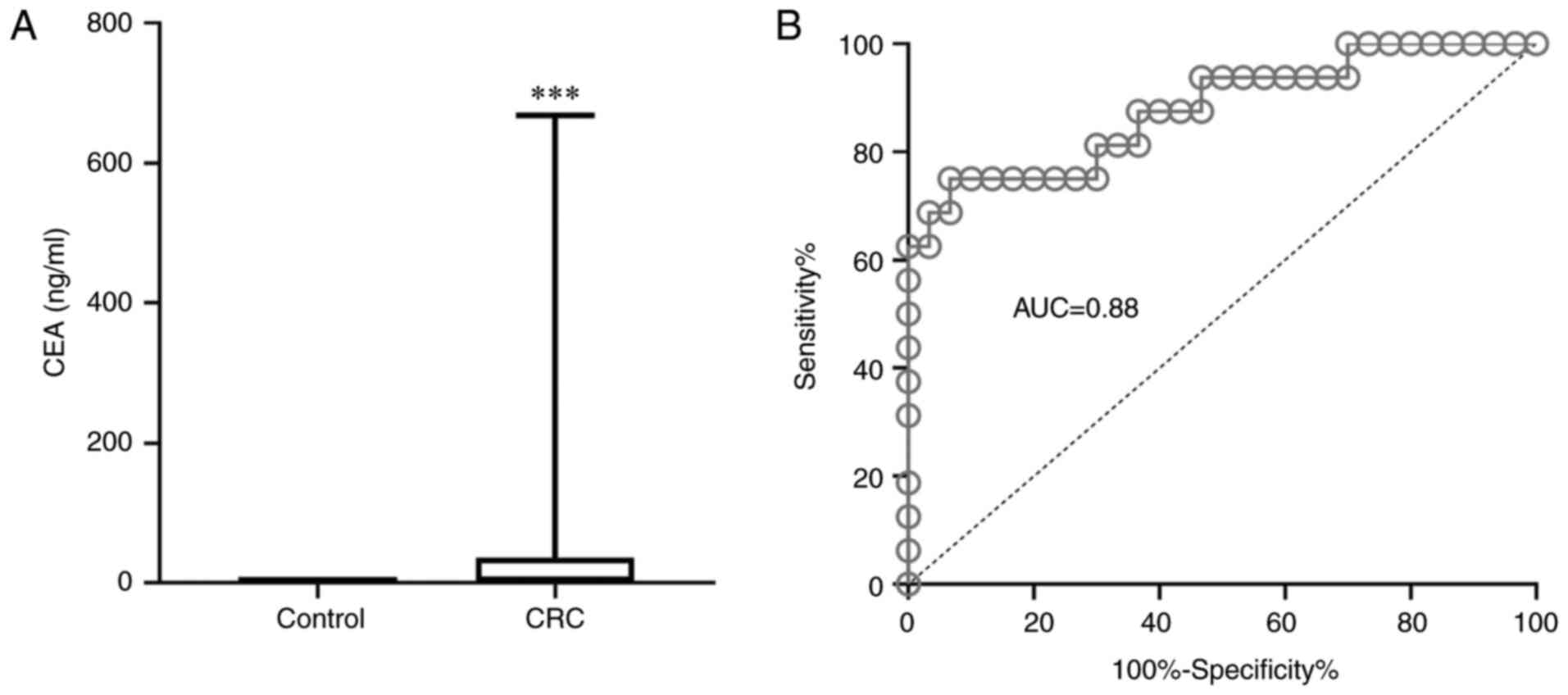
The serum levels of CEA in patients with CRC were
significantly increased compared with the levels in healthy
individuals (P<0.001). Furthermore, CEA was revealed to be an
efficient biomarker for CRC (AUC, 0.88; Fig. 2).
Serum IL-8, IL-27 and VEGF
concentrations
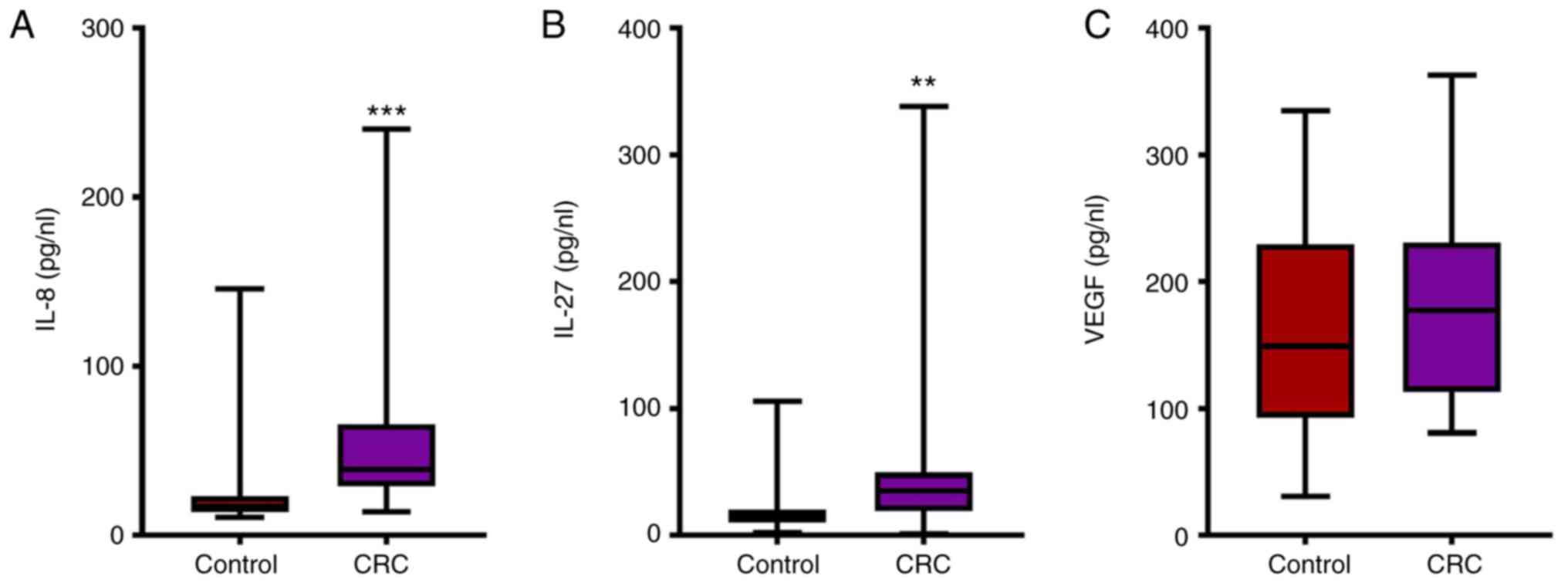
In patients with CRC, the serum concentrations of
IL-8 (median, 38.72; range, 28.95–38.72) and IL-27 (median, 35.18;
interquartile range, 18.96–49.46) were significantly increased
compared with healthy individuals (IL-8 median, 16.96;
interquartile range, 13.88–23.11; and IL-27 median, 14.65;
interquartile range, 10.17–20.37, respectively; Fig. 3A and B). However, no statistically
significant differences were observed in the VEGF concentration
between patients with CRC (median, 177.40; interquartile range,
113.00–230.90) and healthy individuals (median, 149.00;
interquartile range, 92.66–229.60; Fig.
3C).
Genetic polymorphisms of IL-8, IL-27
and VEGF
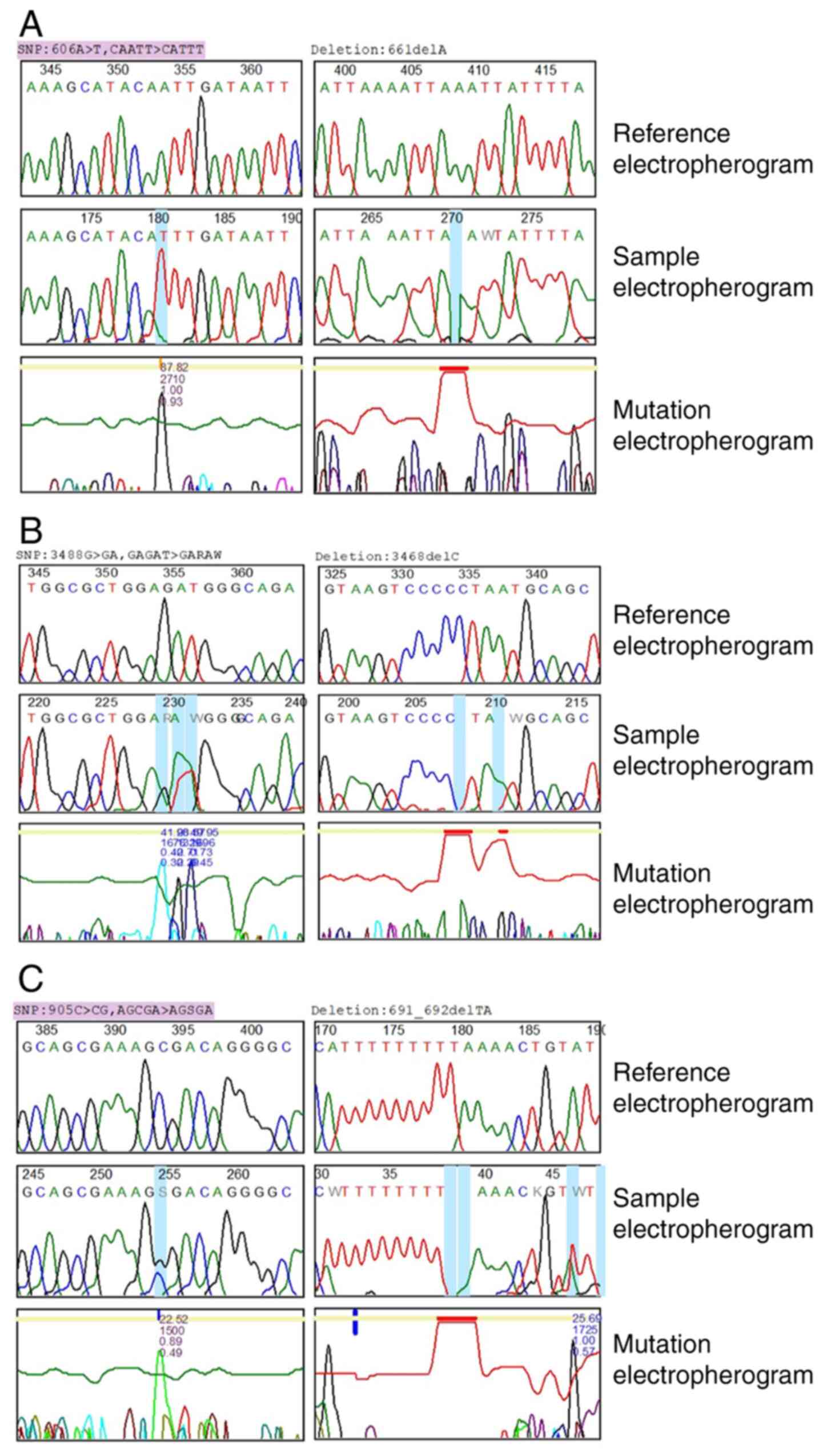
In total, 31 mutations were identified in the three
genes (Table I). In Fig. 4, the rows represent reference, sample
and mutation electropherograms, respectively. In VEGF, eight
mutations were recorded, 13 mutations were identified in IL-27 and
10 mutations were identified for IL-8. In detail, in VEGF, two
types of genomic mutations were recognized: Substitutions (C>CG,
A>AT, T>G and C>CT) and deletions (A and TA). The
homozygous variant mutation 905C>CG$23 on chromosome position
6:43738350 has been previously reported in an external public
database, dbSNP (https://www.ncbi.nlm.nih.gov/snp/). In the present
study, the variant percentage for this mutation was 83.3%. On the
other hand, to the best of our knowledge, the remaining mutational
variants of VEGF have not been reported in external databases.
 | Table I.Variants identified in patients with
colorectal cancer using Mutation Surveyor software. |
Table I.
Variants identified in patients with
colorectal cancer using Mutation Surveyor software.
| Gene | Chromosome
position | Mutation | Mutation
genotype | Heterozygous/homo
zygous | Variants | Variant, % | External
database |
|---|
| IL-8 | 4:74606024 | Substitution | A>T | Homozygous | 606A>T$88 | 100.0 | dbSNP:4073 |
|
| 4:74606126 | Substitution | T>TA | Heterozygous | 708T>TA$10 | 16.7 | Not found |
|
| 4:74605882 | Substitution | T>TA | Heterozygous | 464T>TA$10 | 50.0 | Not found |
|
|
4:74606163_4:74606164 | Insertion | T | Heterozygous |
745_746het_insT$6 | 16.7 | Not found |
|
| 4:74606091 | Duplication | G | Heterozygous | 673het_dupG$4 | 16.7 | Not found |
|
| 4:74606168 | Substitution | G>A | Homozygous | 750G>A$14 | 33.3 | Not found |
|
| 4:74605911 | Substitution | A>AT | Heterozygous | 493A>AT$24 | 14.3 | Not found |
|
| 4:74605919 | Substitution | A>AC | Heterozygous | 501A>AC$24 | 7.1 | Not found |
|
| 4:74606079 | Deletion | A | Homozygous | 661delA$10 | 8.3 | Not found |
|
| 4:74606078 | Substitution | A>T | Homozygous | 660A>T,$16 | 8.3 | Not found |
| IL-27 | 16:28515168 | Substitution | G>GA | Heterozygous | 3488G>GA$42 | 20.0 | Not found |
|
| 16:28515167 | Substitution | A>AT | Heterozygous | 3489A>AT$29 | 20.0 | Not found |
|
| 16:28515166 | Substitution | T>TA | Heterozygous | 3490T>TA$50 | 20.0 | Not found |
|
| 16:28515188 | Deletion | C | Homozygous | 3468delC$10 | 8.3 | Not found |
|
| 16:28515185 | Deletion | A | Homozygous | 3471delA$11 | 9.1 | Not found |
|
| 16:28515163 | Deletion | G | Heterozygous |
3493het_delG$10 | 16.7 | Not found |
|
| 16:28515270 | Substitution | A>AG | Heterozygous |
3386A>AG,45R>R/G$17 | 5.6 | Not found |
|
|
16:28515322_16:28515323 | Insertion | K | Homozygous |
3333_3334insK$11 | 5.6 | Not found |
|
| 16:28515320 | Substitution | G>C | Homozygous |
3336G>C,28G>G$8 | 5.6 | Not found |
|
| 16:28515319 | Substitution | A>AT | Heterozygous |
3337A>AT,28G>G/G$31 | 16.7 | Not found |
|
| 16:28515349 | Substitution | C>CT | Heterozygous |
3307C>CT,18P>P/P$30 | 5.6 | Not found |
|
|
16:28515214_16:28515215 | Deletion | GC | Heterozygous |
3441_3442delGC$10 | 6.2 | Not found |
|
| 16:28515195 | Deletion | A | Heterozygous |
3461het_delA,$5 | 7.7 | Not found |
| VEGF | 6:43738350 | Substitution | C>CG | Heterozygous | 905C>CG$23 | 83.3 | dbSNP:2010963 |
|
|
6:43738136_6:43738137 | Deletion | TA | Heterozygous |
691_692delTA$12 | 20.0 | Not found |
|
| 6:43738145 | Substitution | A>AT | Heterozygous | 700A>AT$26 | 20.0 | Not found |
|
| 6:43738147 | Substitution | T>G | Homozygous | 702T>G$9 | 16.7 | Not found |
|
| 6:43738165 | Substitution | A>AT | Heterozygous | 720A>AT$21 | 16.7 | Not found |
|
| 6:43738200 | Deletion | A | Heterozygous | 755het_delA$4 | 12.5 | Not found |
|
| 6:43738161 | Deletion | A | Homozygous | 716delA$11 | 16.7 | Not found |
|
| 6:43738204 | Substitution | C>CT | Heterozygous | 759C>CT$17 | 14.3 | Not found |
With regards to IL-27, the present data revealed two
distinctive features. First, IL-27 exhibited the highest rate of
mutations among the three genes, and second, to the best of our
knowledge, none of the mutations have been previously reported in
an external database. The analysis of the sequencing data of IL-27
revealed three types of mutation: Substitution (G>GA, A>AT,
T>TA, A>AG, G>C and C>CT), deletions (C, A, G, GC and
A) and insertions (K), and both homozygous and heterozygous variant
mutations were identified. The heterozygous variant deletions,
3493het_delG$10 and 3461het_delA, $5, were observed in chromosome
positions 16:28515163 and 16:28515195, respectively.
The analysis of the sequencing data of IL-8 revealed
four different types of genomic mutations: Substitutions (A>T,
T>TA, G>A, A>AT and A>AC), insertions (T), deletions
(A) and duplications (G). The homozygous variant mutation
606A>T$88 on chromosome position 4:74606024 has been previously
reported in external public databases, and its variant percentage
in the present study was 100%. The heterozygous insertion
745_746het_insT$6 and the heterozygous duplication 673het_dupG$4
were identified on chromosome positions 4:74606163_4:74606164 and
4:74606091, respectively.
Expression levels of hERG and
correlation with IL-8 and IL-27
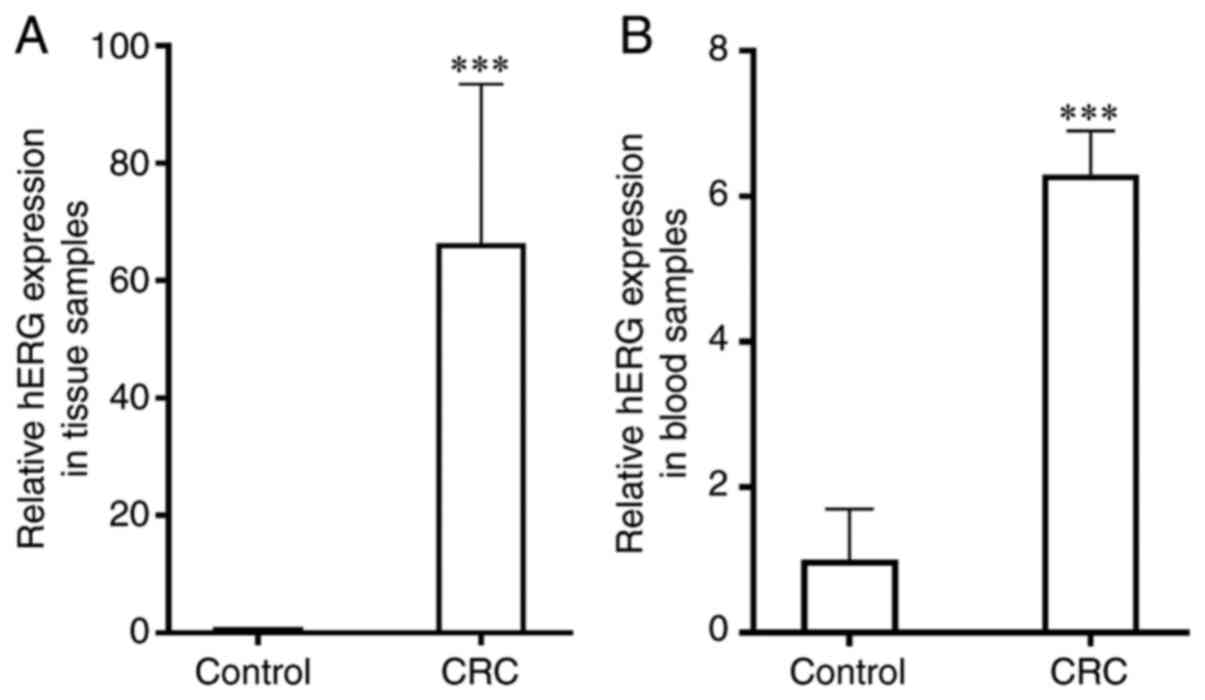
The relative mRNA expression levels of hERG were
significantly upregulated in the tissue (66.4±27.1) and blood
samples (6.3±0.6) from patients with CRC compared with healthy
individuals (1.0±0.0 and 1.0±0.7, respectively; P<0.001;
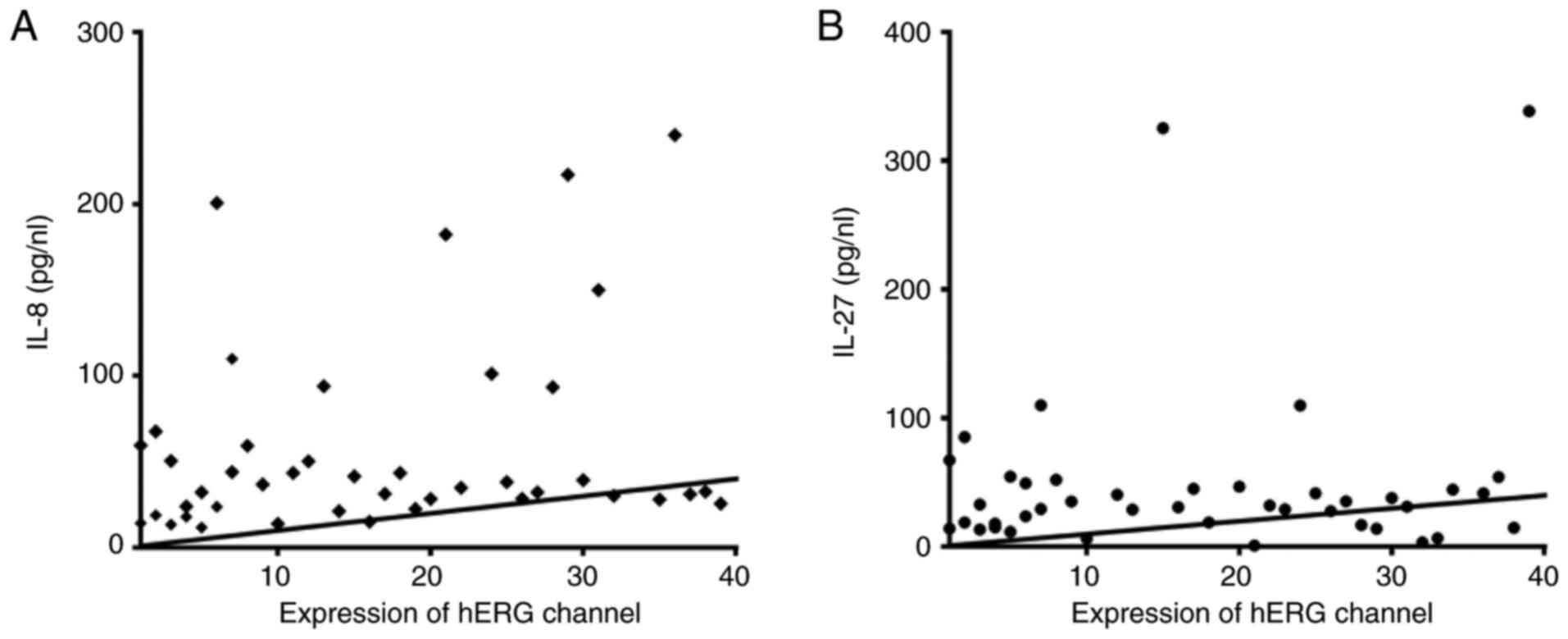
Fig. 5). A positive non-significant
correlation was identified between the expression levels of hERG
and IL-8 (r=0.43; P=0.42) and IL-27 (r=0.029; P=0.99; Fig. 6).
Discussion
To the best of our knowledge, a link has been
observed between serum WBC count and CEA levels in patients with
CRC (25). This association could be
used as a predictive factor not only in CRC but also in other types
of cancer. It is crucial to identify predictive and prognostic
factors in tumor development because this could help in finding the
most effective treatments. The findings of this investigation
matched those of Tsai et al (26), who found that certain WBC cells,
notably the neutrophil-lymphocyte ratio (NLR) and CEA levels, could
be prognostic and predictive markers for advanced-stage CRC.
Increased expression of neutrophils and lymphocytes is associated
with systemic inflammation and a signal of impaired cell-mediated
immunity, respectively, according to the best explanation for their
relationship (27). NLR expression
is high, which inhibits lymphokine-activated killer cells (28). As a result, it may enhance the risk
of tumor metastasis in patients with CRC (29). The presence of more inflammatory
cells in the tumor microenvironment may explain why these cells,
particularly neutrophils, act as an angiogenic activator in
metastatic CRC (30). According to a
previous study, elevated CEA levels in the blood are linked to a
poor prognosis, a decreased survival rate, and advanced metastatic
stage in cancer cells (31). Lee
et al (32) demonstrated that
a high NLR was mainly observed in an advanced stage of tumor
progression, which suggested its potential use as a predictor of
the incidence and mortality of CRC. Increased levels of WBC are
known to enhance the levels of inflammation, which increases the
likelihood of tumor metastasis (33). The levels of CEA have been reported
to be elevated in metastatic tumors (34). Therefore, the association between CEA
levels and the NLR may be a valuable predictive factor to monitor
metastatic CRC, which could be extended for use in other types of
cancer.
The levels of IL-8, a proinflammatory cytokine,
which has angiogenic activity, are reportedly increased in numerous
types of tumor, including CRC (12).
In CRC, IL-8 has been revealed to act on cancer cells via their
receptors to promote migration, invasion and proliferation, in
addition to angiogenesis in vivo (12). In the present study, the
concentration of IL-8 was significantly increased in patients with
CRC compared with in healthy individuals. The increase in the
concentration of IL-8 has been previously reported in CRC tissues
and cell lines (35); the results
revealed that the concentrations of IL-8 were increased in CRC
tissues compared with healthy colon tissues. SNPs of driver genes
have been reported to serve a role in the development of multiple
neoplasms, including CRC (36). The
sequencing results in the present study identified several SNPs in
the IL-8 gene in the CRC tissues: Seven mutations, one insertion,
one deletion and one duplication mutation. Bondurant et al
(14) reported that the SNP, rs4073,
in IL-8 is associated with an increased CRC risk. Furthermore,
another previous study involving Malaysian patients with CRC
revealed that the homozygous variant of IL-8-251AA was associated
with a higher susceptibility of CRC compared with the homozygous
wild-type-251TT genotype (37). The
IL-8-251T>A polymorphism has been reported to be associated with
a higher promoter activity, which in turn increases the levels of
IL-8 and promotes the proangiogenic effects of IL-8, and ultimately
increases tumor cell proliferation (38).
In the present study, the levels of IL-27 were
increased in patients with CRC compared with in healthy
individuals. These findings were not consistent with previous
studies, which reported that IL-27 served as a tumor suppressor in
several types of cancer, including B16F10 melanoma and Lewis lung
carcinoma (39,40). Data collected from DNA sequencing in
the present study revealed multiple mutations in three different
exon regions in the IL-27 gene. These results are controversial;
the levels of IL-27 were increased in patients with CRC, whereas
several genetic mutations, including SNPs, insertions and deletions
were observed in several regions of the IL-27 gene. In normal cell
environments, IL-27 acts as a tumor suppressive and anti-angiogenic
factor (41). However, based on the
DNA sequencing data in the present study, IL-27 was revealed to be
mutated in the tumor microenvironment of patients with CRC. These
findings suggested that IL-27 may lose its tumor suppressive role,
which also provides an explanation for the high serum levels of
IL-27 in the patients with CRC. A previous study revealed that the
IL-27 polymorphism is associated with a high risk of CRC (17).
IL-27 exerts a tumor-suppressive role in several
types of cancer cells by inhibiting tumor cell proliferation,
angiogenesis, invasion and survival, in addition to activating
antitumor immune responses (42).
However, several previous studies have reported that IL-27 serves a
protumorigenic role (41–43). Diakowska et al (44) reported that the serum levels of IL-27
are increased in gastrointestinal tract cancer, which is associated
with a higher lymph node status. Similarly, Lu et al
(45) revealed that in patients with
breast cancer, elevated serum IL-27 levels are associated with VEGF
expression levels and clinical stage. A similar inhibitory role of
IL-27 has been reported in prostate cancer cells, in which high
levels of IL-27 are associated with the inhibition of prostate
cancer cell survival and proliferation (46). These findings, combined with the DNA
sequencing findings of the present study, suggested that the
protumorigenic role of IL-27 may be another possible explanation
for the high serum levels of IL-27 in patients with CRC. Therefore,
CRC may be used as an example to demonstrate the dual role of IL-27
in normal and tumor cells.
Among several angiogenic factors, VEGF is an
essential factor in mCRC (47). In
the present study, DNA sequencing data revealed several DNA
abnormalities, including SNPs, and insertion and deletion mutations
in three different exon regions of the VEGF gene. The results of
ELISA revealed that the levels of VEGF in patients with CRC were
not statistically significantly different compared with the
controls. These results were not consistent with the findings of a
number of previous studies, which reported that VEGF may be a
crucial factor for inducing the proliferation of endothelial cells
and its expression levels are upregulated in the majority of
patients with mCRC (48,49). On the other hand, Jubb et al
(50) reported that the levels of
VEGF in patients with CRC following bevacizumab treatment were
downregulated compared with the control individuals. The
explanation for the changes in the expression levels of VEGF not
being statistically significant in patients with mCRC in the
present study may be due to the fact that the majority of the
patients with mCRC were undergoing chemotherapy. Bevacizumab can
directly interact with VEGF and block its biological activity,
which may eventually inhibit its crucial role as a proangiogenic
factor in CRC development (47).
In the present study, upregulation of hERG channel
expression could be a suitable biomarker for detecting patients
with CRC, since hERG channels are frequently upregulated in solid
cancer types and such expression is associated with
clinicopathological features (18).
An important role of the hERG channel in CRC carcinogenesis in
vivo is that early upregulation of the expression of the hERG
gene marks precancerous lesions that undergo malignant progression
(51). Since there has been no
previous study linking the expression levels of the hERG channel to
IL-8, IL-27 or VEGF in patients with CRC, the present study was the
first to link the expression levels of the hERG channel to IL-8,
IL-27 or VEGF in patients with CRC. This holds great promise for
future pharmacological targeted CRC therapy. A limitation of the
present study was that only the serum concentrations of IL-8 and
IL-27 was measured. Therefore, future studies should focus on
assessing the molecular signaling pathways in CRC tissues and cell
lines to provide a more comprehensive association between hERG
expression and interleukin signaling.
In conclusion, the findings of the present study
indicated that the genetic variations of IL-8, IL-27 and VEGF might
serve important roles in the development and angiogenesis of CRC.
These changes were suggested to be associated with the upregulated
expression levels of the hERG channel gene. These data may enhance
the current understanding of the molecular mechanisms of CRC
angiogenesis and may help determine a novel target for the
treatment of CRC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MNA and FAQ performed experiments. MNA and AS
designed the experiments, analyzed data and co-wrote the
manuscript. MNA, FAQ and AS confirm the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was authorized and approved by the
Human Ethics Committee of Salahaddin University-Erbil, Erbil, Iraq.
Patients provided informed written informed consent.
Patient consent for publication
All patients provided written informed consent for
the publication of data in the present study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wen J, Min X, Shen M, Hua Q, Han Y, Zhao
L, Liu L, Huang G, Liu J and Zhao X: ACLY facilitates colon cancer
cell metastasis by CTNNB1. J Exp Clin Cancer Res. 38:4012019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hirata A, Hatano Y, Niwa M, Hara A and
Tomita H: Heterogeneity of colon cancer stem cells. Adv Exp Med
Biol. 1139:115–126. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rawla P, Sunkara T and Barsouk A:
Epidemiology of colorectal cancer: Incidence, mortality, survival,
and risk factors. Prz Gastroenterol. 14:89–103. 2019.PubMed/NCBI
|
|
4
|
Wong MC, Ding H, Wang J, Chan PS and Huang
J: Prevalence and risk factors of colorectal cancer in Asia. Intest
Res. 17:317–329. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thanikachalam K and Khan G: Colorectal
cancer and nutrition. Nutrients. 11:1642019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fearon ER and Vogelstein B: A genetic
model for colorectal tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun W: Angiogenesis in metastatic
colorectal cancer and the benefits of targeted therapy. J Hematol
Oncol. 5:632012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lan J, Li H, Luo X, Hu J and Wang G: BRG1
promotes VEGF-A expression and angiogenesis in human colorectal
cancer cells. Exp Cell Res. 360:236–242. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ceci C, Atzori MG, Lacal PM and Graziani
G: Role of VEGFs/VEGFR-1 signaling and its inhibition in modulating
tumor invasion: Experimental evidence in different metastatic
cancer models. Int J Mol Sci. 21:13882020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Deng F, Zhou R, Lin C, Yang S, Wang H, Li
W, Zheng K, Lin W, Li X, Yao X, et al: Tumor-secreted dickkopf2
accelerates aerobic glycolysis and promotes angiogenesis in
colorectal cancer. Theranostics. 9:1001–1014. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Uthoff SM, Duchrow M, Schmidt MHH, Broll
R, Bruch HP, Strik MW and Galandiuk S: VEGF isoforms and mutations
in human colorectal cancer. Int J Cancer. 101:32–36. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ning Y and Lenz HJ: Targeting IL-8 in
colorectal cancer. Expert Opin Ther Targets. 16:491–497. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Waugh DJ and Wilson C: The interleukin-8
pathway in cancer. Clin Cancer Res. 14:6735–6741. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bondurant KL, Lundgreen A, Herrick JS,
Kadlubar S, Wolff RK and Slattery ML: Interleukin genes and
associations with colon and rectal cancer risk and overall
survival. Int J Cancer. 132:905–915. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lurje G, Zhang W, Schultheis AM, Yang D,
Groshen S, Hendifar AE, Husain H, Gordon MA, Nagashima F, Chang HM
and Lenz HJ: Polymorphisms in VEGF and IL-8 predict tumor
recurrence in stage III colon cancer. Ann Oncol. 19:1734–1741.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liang Y, Chen Q, Du W, Chen C, Li F, Yang
J, Peng J, Kang D, Lin B, Chai X, et al: Epstein-barr virus-induced
gene 3 (EBI3) blocking leads to induce antitumor cytotoxic T
lymphocyte response and suppress tumor growth in colorectal cancer
by bidirectional reciprocal-regulation STAT3 signaling pathway.
Mediators Inflamm. 2016:32141052016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lyu S, Ye L, Wang O, Huang G, Yang F, Liu
Y and Dong S: IL-27 rs153109 polymorphism increases the risk of
colorectal cancer in Chinese Han population. Onco Targets Ther.
8:1493–1497. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lastraioli E, Lottini T, Bencini L,
Bernini M and Arcangeli A: hERG1 potassium channels: Novel
biomarkers in human solid cancers. Biomed Res Int. 2015:8964322015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Crociani O, Zanieri F, Pillozzi S,
Lastraioli E, Stefanini M, Fiore A, Fortunato A, D'Amico M,
Masselli M, De Lorenzo E, et al: hERG1 channels modulate integrin
signaling to trigger angiogenesis and tumor progression in
colorectal cancer. Sci Rep. 3:33082013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wei PL, Lee LT, Tseng LM and Huang KW:
Validation of assaying carcinoembryonic antigen in human serum by
using immunomagnetic reduction. Sci Rep. 8:100022018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vislovukh A, Vargas TR, Polesskaya A and
Groisman I: Role of 3′-untranslated region translational control in
cancer development, diagnostics and treatment. World J Biol Chem.
5:40–57. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang YF and Zhao AD: Common polymorphisms
in IL-27 genes may contribute to risk of various human diseases in
Asian populations: A meta-analysis. Med Sci Monit. 22:766–775.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jain L, Vargo CA, Danesi R, Sissung TM,
Price DK, Venzon D, Venitz J and Figg WD: The role of vascular
endothelial growth factor SNPs as predictive and prognostic markers
for major solid tumors. Mol Cancer Ther. 8:2496–2508. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kang HY, Choe EK, Park KJ and Lee Y:
Factors requiring adjustment in the interpretation of serum
carcinoembryonic antigen: A cross-sectional study of 18,131 healthy
nonsmokers. Gastroenterol Res Pract. 2017:98589312017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tsai PL, Su WJ, Leung WH, Lai CT and Liu
CK: Neutrophil-lymphocyte ratio and CEA level as prognostic and
predictive factors in colorectal cancer: A systematic review and
meta-analysis. J Cancer Res Ther. 12:582–589. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen L, Deng H, Cui H, Fang J, Zuo Z, Deng
J, Li Y, Wang X and Zhao L: Inflammatory responses and
inflammation-associated diseases in organs. Oncotarget.
9:7204–7218. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mogensen TH: Pathogen recognition and
inflammatory signaling in innate immune defenses. Clin Microbiol
Rev. 22:240–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu W, He Y, Wang Y, Li X, Young J,
Ioannidis JPA, Dunlop MG and Theodoratou E: Risk factors and risk
prediction models for colorectal cancer metastasis and recurrence:
An umbrella review of systematic reviews and meta-analyses of
observational studies. BMC Med. 18:1722020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Singel KL and Segal BH: Neutrophils in the
tumor microenvironment: Trying to heal the wound that cannot heal.
Immunol Rev. 273:329–343. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yan C, Hu Y, Zhang B, Huang K, Zhao H, Ma
C, Li X, Tao D, Gong J and Qin J: The CEA-/lo colorectal cancer
cell population harbors cancer stem cells and metastatic cells.
Oncotarget. 7:80700–80715. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee YJ, Lee HR, Nam CM, Hwang UK and Jee
SH: White blood cell count and the risk of colon cancer. Yonsei Med
J. 47:646–656. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fares J, Fares MY, Khachfe HH, Salhab HA
and Fares Y: Molecular principles of metastasis: A hallmark of
cancer revisited. Signal Transduct Target Ther. 5:282020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Saito G, Sadahiro S, Kamata H, Miyakita H,
Okada K, Tanaka A and Suzuki T: Monitoring of serum
carcinoembryonic antigen levels after curative resection of colon
cancer: Cutoff values determined according to preoperative levels
enhance the diagnostic accuracy for recurrence. Oncology.
92:276–282. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Doll D, Keller L, Maak M, Boulesteix AL,
Siewert JR, Holzmann B and Janssen KP: Differential expression of
the chemokines GRO-2, GRO-3, and interleukin-8 in colon cancer and
their impact on metastatic disease and survival. Int J Colorectal
Dis. 25:573–581. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nguyen HT and Duong HQ: The molecular
characteristics of colorectal cancer: Implications for diagnosis
and therapy. Oncol Lett. 16:9–18. 2018.PubMed/NCBI
|
|
37
|
Ankathil R, Mustapha MA, Abdul Aziz AA,
Mohd Shahpudin SN, Zakaria AD, Abu Hassan MR and Musa KI:
Contribution of genetic polymorphisms of inflammation response
genes on sporadic colorectal cancer predisposition risk in
Malaysian patients-a case control study. Asian Pac J Cancer Prev.
20:1621–1632. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huang S, Mills L, Mian B, Tellez C,
McCarty M, Yang XD, Gudas JM and Bar-Eli M: Fully humanized
neutralizing antibodies to interleukin-8 (ABX–IL8) inhibit
angiogenesis, tumor growth, and metastasis of human melanoma. Am J
Pathol. 161:125–134. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yoshimoto T, Chiba Y, Furusawa J, Xu M,
Tsunoda R, Higuchi K and Mizoguchi I: Potential clinical
application of interleukin-27 as an antitumor agent. Cancer Sci.
106:1103–1110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Oniki S, Nagai H, Horikawa T, Furukawa J,
Belladonna ML, Yoshimoto T, Hara I and Nishigori C: Interleukin-23
and interleukin-27 exert quite different antitumor and vaccine
effects on poorly immunogenic melanoma. Cancer Res. 66:6395–6404.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kourko O, Seaver K, Odoardi N, Basta S and
Gee K: IL-27, IL-30, and IL-35: A cytokine triumvirate in cancer.
Front Oncol. 9:9692019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fabbi M, Carbotti G and Ferrini S: Dual
roles of IL-27 in cancer biology and immunotherapy. Mediators
Inflamm. 2017:39580692017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Park YJ, Ryu H, Choi G, Kim BS, Hwang ES,
Kim HS and Chung Y: IL-27 confers a protumorigenic activity of
regulatory T cells via CD39. Proc Natl Acad Sci USA. 116:3106–3111.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Diakowska D, Lewandowski A,
Markocka-Mączka K and Grabowski K: Concentration of serum
interleukin-27 increase in patients with lymph node metastatic
gastroesophageal cancer. Adv Clin Exp Med. 22:683–691.
2013.PubMed/NCBI
|
|
45
|
Lu D, Zhou X, Yao L, Liu C, Jin F and Wu
Y: Clinical implications of the interleukin 27 serum level in
breast cancer. J Investig Med. 62:627–631. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Di Carlo E, Sorrentino C, Zorzoli A, Di
Meo S, Tupone MG, Ognio E, Mincione G and Airoldi I: The antitumor
potential of Interleukin-27 in prostate cancer. Oncotarget.
5:10332–10341. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Izawa N, Shitara K, Masuishi T, Denda T,
Yamazaki K, Moriwaki T, Okuda H, Kondoh C, Nishina T, Makiyama A,
et al: Vascular endothelial growth factor (VEGF)-D and clinical
outcomes in metastatic colorectal cancer (mCRC) patients (pts)
treated with second-line FOLFIRI plus bevacizumab (Bev): A
biomarker study of the WJOG 6210G trial. J Clin Oncol. 38 (Suppl
4):S2262020. View Article : Google Scholar
|
|
48
|
Cao D, Hou M, Guan YS, Jiang M, Yang Y and
Gou HF: Expression of HIF-1alpha and VEGF in colorectal cancer:
Association with clinical outcomes and prognostic implications. BMC
Cancer. 9:4322009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Raluca BA, Cimpean AM, Cioca A, Cretu O,
Mederle O, Ciolofan A, Gaje P and Raica M: Endothelial cell
proliferation and vascular endothelial growth factor expression in
primary colorectal cancer and corresponding liver metastases. Asian
Pac J Cancer Prev. 16:4549–4553. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jubb AM, Hurwitz HI, Bai W, Holmgren EB,
Tobin P, Guerrero AS, Kabbinavar F, Holden SN, Novotny WF, Frantz
GD, et al: Impact of vascular endothelial growth factor-a
expression, thrombospondin-2 expression, and microvessel density on
the treatment effect of bevacizumab in metastatic colorectal
cancer. J Clin Oncol. 24:217–227. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fiore A, Carraresi L, Morabito A, Polvani
S, Fortunato A, Lastraioli E, Femia AP, De Lorenzo E, Caderni G and
Arcangeli A: Characterization of hERG1 channel role in mouse
colorectal carcinogenesis. Cancer Med. 2:583–594. 2013. View Article : Google Scholar : PubMed/NCBI
|