Introduction
Gastric cancer (GC) is the fifth most common type of
malignancy worldwide, with >1,000,000 new cases estimated to
occur each year (1). As GC is often
diagnosed at an advanced stage, the associated mortality rate
remains at a high level, and as such, the disease ranks as the
third leading cause of cancer-related mortality worldwide,
corresponding to 784,000 recorded deaths in 2018 (2). In China, GC is the second largest
health burden (3). The most
important risk factor for GC is Helicobacter pylori
infection, and although morbidity rates have decreased, the number
of Helicobacter pylori infections is increasing (4). GC incidence rates are currently
increasing in young individuals in high-income countries, and as
the population ages, more cases of GC are expected in the future
(5). Patients with early-stage GC do
not exhibit specific symptoms, and metastases are locally advanced
or distant by the time of diagnosis (6). GC treatment includes surgery,
chemotherapy, radiotherapy and gene therapy; however, the prognosis
of patients with GC remains poor, with a median overall survival
time of 10–12 months (7). Therefore,
it is imperative to identify biomarkers associated with the early
diagnosis and underlying mechanisms of GC progression.
Circular RNAs (circRNAs) are single-stranded,
covalently closed RNA molecules that are produced as a set of
transcripts from pre-mRNAs through a process known as reverse
splicing (8). Characterized by their
stable presence in tissues and cells, circRNAs are important
regulators of gene expression, while dysregulated circRNA
expression has been reported in almost all types of cancer
(9). circRNA_0074027 (circ_0074027)
is considered to exert a cancer-promoting effect; for example,
circ_0074027 has been reported to contribute to the progression of
non-small cell lung cancer through the microRNA
(miRNA/miR)-335-5p-mediated upregulation of cullin 4B expression
(10). circ_0074027 has also been
shown to be upregulated in non-small cell lung cancer, and
functions as a competing endogenous RNA to accelerate cellular
proliferation, apoptosis and invasiveness (11). Similarly, there is evidence that
circ_0074027 plays an oncogenic role in glioblastoma, promoting the
proliferation and invasiveness of glioblastoma cells (12). However, to the best of our knowledge,
the role and underlying mechanisms of circ_0074027 in GC are not
yet fully understood.
The aim of the present study was to determine the
association between the circ_0074027 expression level, and the
migration and proliferation capacities of GC cells, as well as the
mechanism of action underlying its functions in GC.
Materials and methods
Cell lines and culture
The AGS, MKN45, HGC-27 and SNU-5 human GC cell
lines, and the normal gastric mucosa cell line, GES-1, were
obtained from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. The cells were cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Invitrogen; Thermo Fisher Scientific, Inc.), 1% penicillin and 1%
streptomycin, in a 37°C humidified incubator with 5%
CO2.
Transfection
MKN45 cells were transfected with small interfering
(si)RNAs targeting circ_0074027 (si-circ-1,
5′-TAAGCACCTGGCGCAGGGACT−3′; si-circ-2,
5′-AAGCACCTGGCGCAGGGACTG−3′; 10 nM), si-negative control (si-NC,
5′-GCAGCTGACTACAGGCAGCGC−3′, 10 nM), the eukaryotic translation
initiation factor 4A3 (EIF4A3) overexpression vector (pc-EIF4A3; 10
nM) or the control vector (pcDNA3.1; 10 nM). All vectors were
purchased from Shanghai GenePharma Co., Ltd., and all transfections
were carried out using Lipofectamine® RNAiMAX
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Following incubation at 37°C (5%
CO2) for 6 h, the culture medium was replaced with fresh
medium containing 10% FBS. The culture was continued in the
original environment, and subsequent experimentation was conducted
within 24 h.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Total cellular RNA was isolated using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), and reverse transcribed into cDNA using the PrimeScript™ 1st
Strand cDNA Synthesis Kit (Takara Biotechnology Co., Ltd.)
according to the manufacturer's protocol. The reaction conditions
for reverse transcription were as follows: 95°C for 15 sec, 60°C
for 1 min and 72°C for 10 min, 35 cycles in total. Subsequently,
qPCR was using iTaq™ Universal SYBR® Green Supermix
(Bio-Rad Laboratories, Inc.) on an ABI 7500 system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using the following
thermocycling conditions: Initial denaturation at 95°C for 10 min;
followed by 40 cycles of 95°C for 10 sec, 60°C cycles for 20 sec
and 70°C cycles for 30 sec. The relative expression of genes was
calculated using the 2−ΔΔCq method (13). with GAPDH expression as the internal
control for normalization. The sequences of the primers are as
follows: circ_0074027 forward, 5′-GCGTCCCTGTGTATGTTGGA-3′ and
reverse, 5′-GTCTGTCTTAAAGCGACAGCG−3′; paired like homeodomain 1
(PITX1) forward, 5′-GCTACCCCGACATGAGCA−3′ and reverse,
5′-GTTACGCTCGCGCTTACG−3′); and GAPDH forward,
5′-GTGGGCATCAATGGATTTGG-3′ and reverse,
5′-ACACCATGTATTCCGGGTCAAT-3′.
RNase R digestion
A total of 10 U RNase R (Geneseed Biotech Co., Ltd.)
was added to 2.5 µg total RNA extracted from MKN45 cells, and
incubated at 37°C for 30 min, followed by RT-qPCR to determine the
expression levels of circ_0074027 and PITX1.
Cell counting kit-8 (CCK-8) assay
MKN45 cells were seeded into 96-well plates
(5×103 cells/well) and incubated at 37°C for 0, 24, 48
and 72 h, following the addition of si-circ_0074027 or
co-transfection with pc-EIF4A3. Next, 10 µl CCK-8 reagent (Beijing
Solarbio Science & Technology Co., Ltd.) was added to each well
and incubated for a further 2.5 h at 37°C. The absorbance was
measured at a wavelength of 450 nm using a microplate reader
(SPECTROstar Nano; BMG Labtech GmbH).
Colony formation assay
MKN45 cells were seeded into 6-well plates
(1×103 cells/well) and cultured in DMEM at 37°C for 10
days, replacing the medium every 3 days. Subsequently, the cells
were fixed with 4% paraformaldehyde for 30 min, stained with 0.5%
crystal violet (Beijing Solarbio Science & Technology Co.,
Ltd.) for 10 min (both at room temperature), and the number of
colonies with >50 cells was manually counted using a light
microscope (Olympus Corporation).
Wound-healing assay
MKN45 cells were seeded into 6-well culture plates
(2×105 cells/well) and cultured at 37°C until reaching
90% confluency. The monolayer was then scratched with a sterile
pipette tip, and the cells were cultured in serum-free medium
(Beijing Solarbio Science & Technology Co., Ltd.). Wound
distances were photographed after 0 and 48 h using a light
microscope (magnification, ×100; Olympus Corporation).
Transwell assay
Transfected cells (1×104 cells/well) were
seeded into the upper chamber of a Transwell insert pre-coated with
Matrigel® (Corning, Inc.) in serum-free medium. Complete
medium containing 10% FBS was added to the lower chamber. Following
a 12-h incubation at 37°C, residual cells on the surface were
removed, and the migratory cells were fixed with 4%
paraformaldehyde, followed by staining with 0.1% crystal violet for
15 min at 37°C. The migration level was observed by counting the
cells under a light microscope (magnification, ×100; Olympus
Corporation).
RNA binding immunoprecipitation (RIP)
assay
The Magna RIP RNA Binding Protein
Immunoprecipitation kit (Product ID: RIP; MilliporeSigma) was used
for RIP, according to the manufacturer's instructions. MKN45 cell
lysates were collected and 100 µl lysate per reaction was incubated
with NC IgG (1:500; cat. no. ab172730) or anti-EIF4A3 (1:1,000;
cat. no. ab180573) antibody (both Abcam) conjugated to protein A/G
magnetic beads (EMD Millipore) at 4°C overnight. Immunoprecipitated
RNA was then extracted and detected by RT-qPCR to confirm the
enrichment of binding targets.
Protein isolation and western blot
analysis
Total cellular proteins in MKN45 cells were
extracted using RIPA lysis buffer (Beyotime Institute of
Biotechnology) and the protein concentration was determined using a
BCA protein assay kit (Beyotime Institute of Biotechnology).
Proteins (30 ng per lane) were separated via 10% SDS-PAGE, after
which the samples were transferred to a polyvinylidene difluoride
membrane (Beyotime Institute of Biotechnology). The membranes were
blocked with 5% skimmed milk for 2 h at room temperature, followed
by an overnight incubation at 4°C with the primary antibodies.
Following the primary antibody incubation, the membranes were
incubated with a goat anti-mouse secondary antibody (1:5,000; cat.
no. S0001; Affinity Biosciences,) for 2 h at room temperature.
Signals were visualized using an ECL kit (Beyotime Institute of
Biotechnology, Inc.) and ImageJ software (v1.8.0; National
Institutes of Health) was used for densitometric analysis. The
primary antibodies were as follows: Anti-EIF4A3 (1:1,000; cat. no.
ab180573; Abcam) and anti-GAPDH (1:2,500; cat. no. ab9485;
Abcam).
Bioinformatics analysis
To further elucidate its potential mechanism in GC,
the Circular RNA Interactome (https://circinteractome.irp.nia.nih.gov/) was used to
predict the factors which bound to circ_0074027. In order to
analyze the overall level of differential expression of EIF4A3 in
normal subjects and patients with stomach adenocarcinoma, clinical
data were obtained from the Gene Expression Profile Interaction
Analysis (GEPIA) database (http://gepia.cancer-pku.cn/). The database has
definitive online genomic analysis technology based on large-scale
sequencing, while omitting information that may identify
patients.
Statistical analysis
Data are presented as the mean ± SD of at least
three independent experiments. Significant differences between
groups were determined using an independent unpaired t-test or
one-way ANOVA followed by Tukey's post hoc test. Statistical
analyses were performed using GraphPad Prism 6 software (GraphPad
Software, Inc.), and P<0.05 was considered to indicate a
statistically significant difference.
Results
Circ_0074027 is highly expressed in GC
cell lines
To investigate the expression level of circ_0074027,
GC cell lines (AGS, MKN45, HGC-27 and SNU-5) and GES-1 cells were
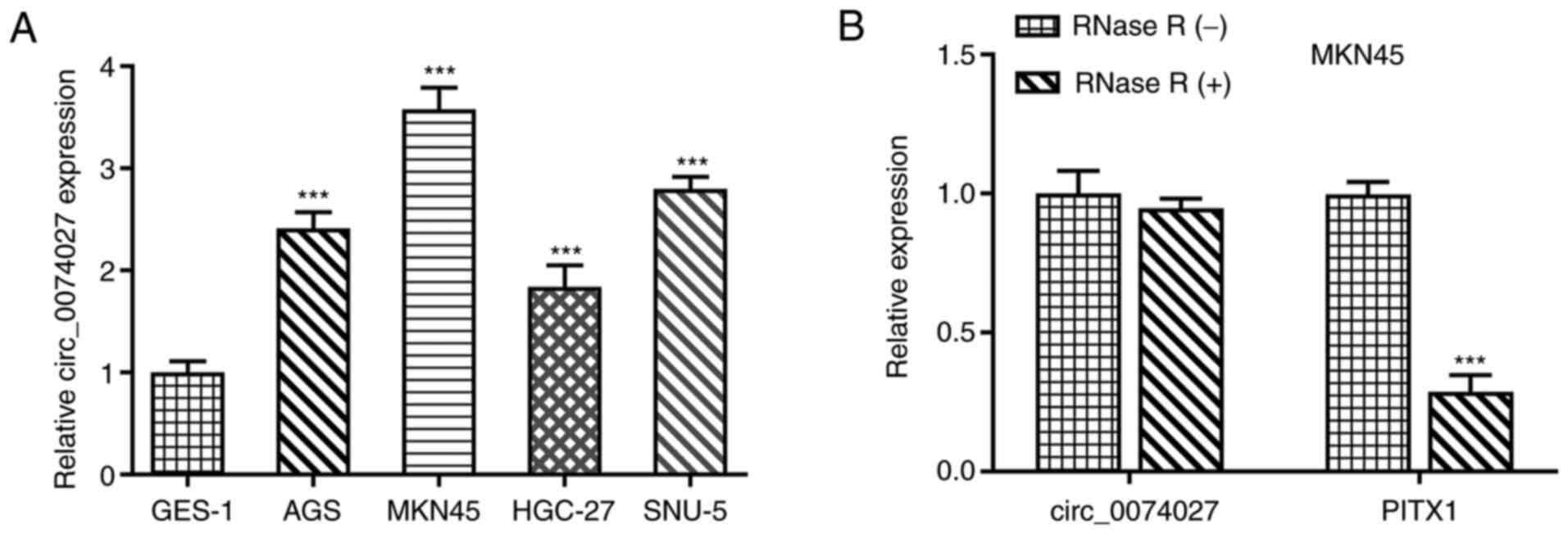
subjected to RT-qPCR. As shown in Fig.
1A, compared with GES-1 cells, circ_0074027 was highly
expressed in the GC cell lines, particularly in MKN45 cells. Thus,
MKN45 cells were selected for use in subsequent experiments. RNase
R digestion assays subsequently revealed that linear PITX1 (as the
paired linear form of circ_0074027), could be digested by RNase R.
However, circ_0074027 was resistant to RNase R digestion in MKN45
cells (Fig. 1B), suggesting that
circ_0074027 features a closed-loop structure. These results
indicated that circ_0074027 expression was upregulated in GC cell
lines.
Circ_0074027-knockdown inhibits the
proliferation and motility of GC cells
In order to investigate the biological function of
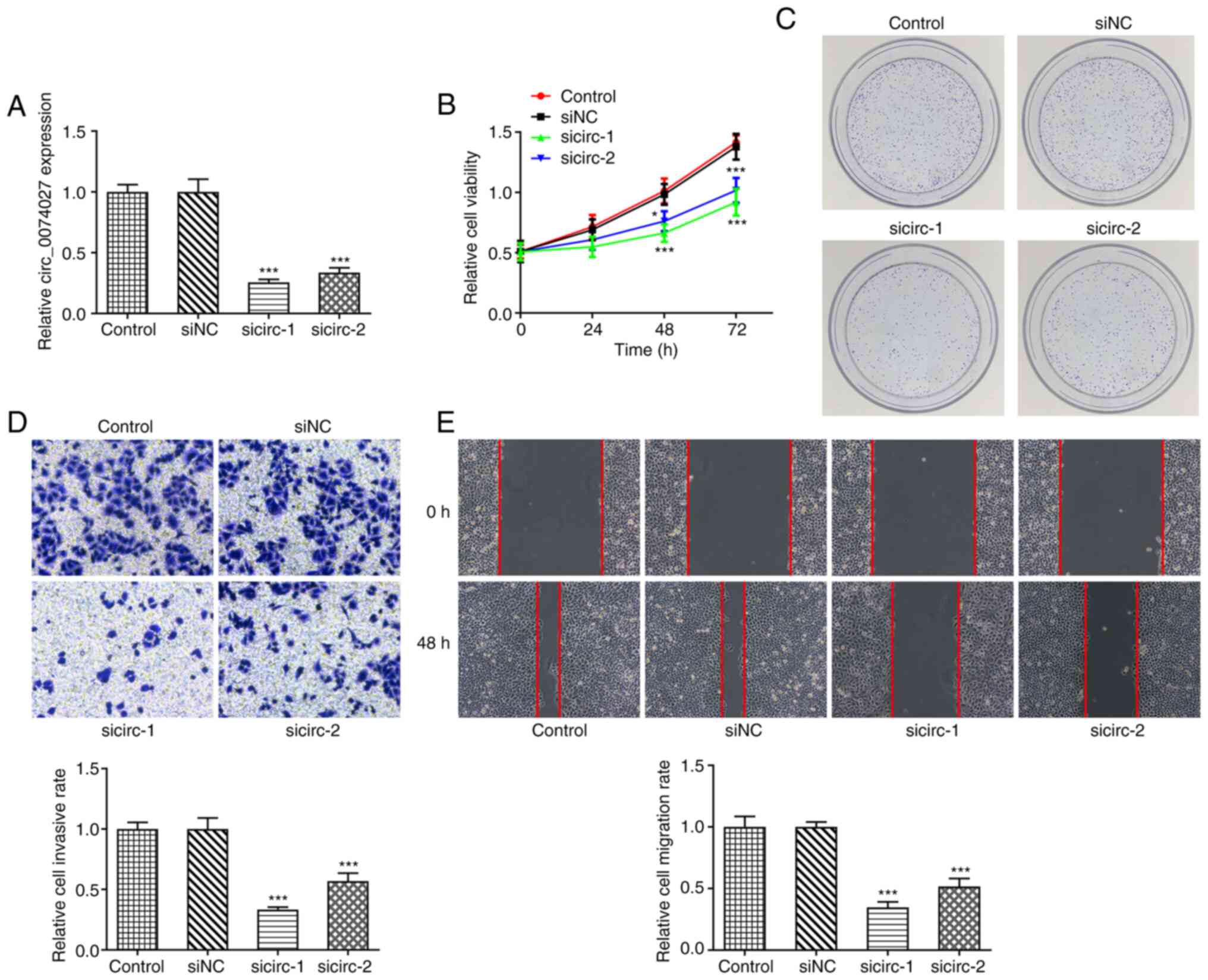
circ_0074027 in GC, MKN45 cells were transfected with
si-circ_0074027. The results of RT-qPCR analysis revealed that the
knockdown efficiency of si-circ-1 was greater than that of
si-circ-2 (Fig. 2A). As revealed by
CCK-8 analysis, the proliferative capacity of MKN45 cells was
significantly inhibited following circ_0074027 silencing, compared
with the si-NC group, with a more pronounced effect observed with
si-circ-1 (Fig. 2B). Similarly, the
results of the colony formation assay indicated that
circ_0074027-knockdown markedly decreased the colony formation
ability of MKN45 cells, compared with the si-NC group, with fewer
colonies observed following transfection with si-circ-1 than with
si-circ-2 (Fig. 2C). The results of
Transwell and wound-healing assays revealed the role of
circ_0074027 in the migration of GC cells. Both the si-circ-1 and
si-circ-2 groups exhibited fewer positively-stained cells (Fig. 2D), suggesting that the silencing of
circ_0074027 suppressed the invasive and migration abilities of
MKN45 cells. After 48 h, circ_0074027-knockdown markedly delayed
wound closure compared with the si-NC group (Fig. 2E). These data demonstrated that the
circ_0074027 expression level was closely associated with the
proliferation and migration of GC cells, and that the reduction in
circ_0074027 expression exerted an anti-oncogenic effect.
Silencing of circ_0074027 leads to the
downregulation of EIF4A3 expression
To further elucidate the potential mechanisms of
circ_0074027 in GC, the Circular RNA Interactome was used to
predict factors that bound to EIF4A3 (Fig. 3A). An RIP assay was also performed to
confirm the prediction; circ_0074027 was found to be abundantly
enriched with EIF4A3 in MKN45 cells, relative to the NC IgG
(Fig. 3B). Furthermore, the GEPIA
results revealed higher expression of EIF4A3 in patients with
stomach adenocarcinoma, compared with healthy individuals (Fig. 3C). Therefore, it was hypothesized
that circ_0074027 may play a role in GC by binding to EIF4A3.
Subsequently, the expression of EIF4A3 in GC cell lines was
analyzed by RT-qPCR. The results revealed that EIF4A3 was more
highly expressed in GC cell lines compared with GES-1 cells, with
the highest expression observed in MKN45 cells (Fig. 3D). Following transfection with
si-circ_0074027, the expression of EIF4A3 in MKN45 cells was
decreased, and si-circ-1 was found to exert a more prominent effect
(Fig. 3E). These results indicated
that circ_0074027 was positively associated with EIF4A3 in GC.
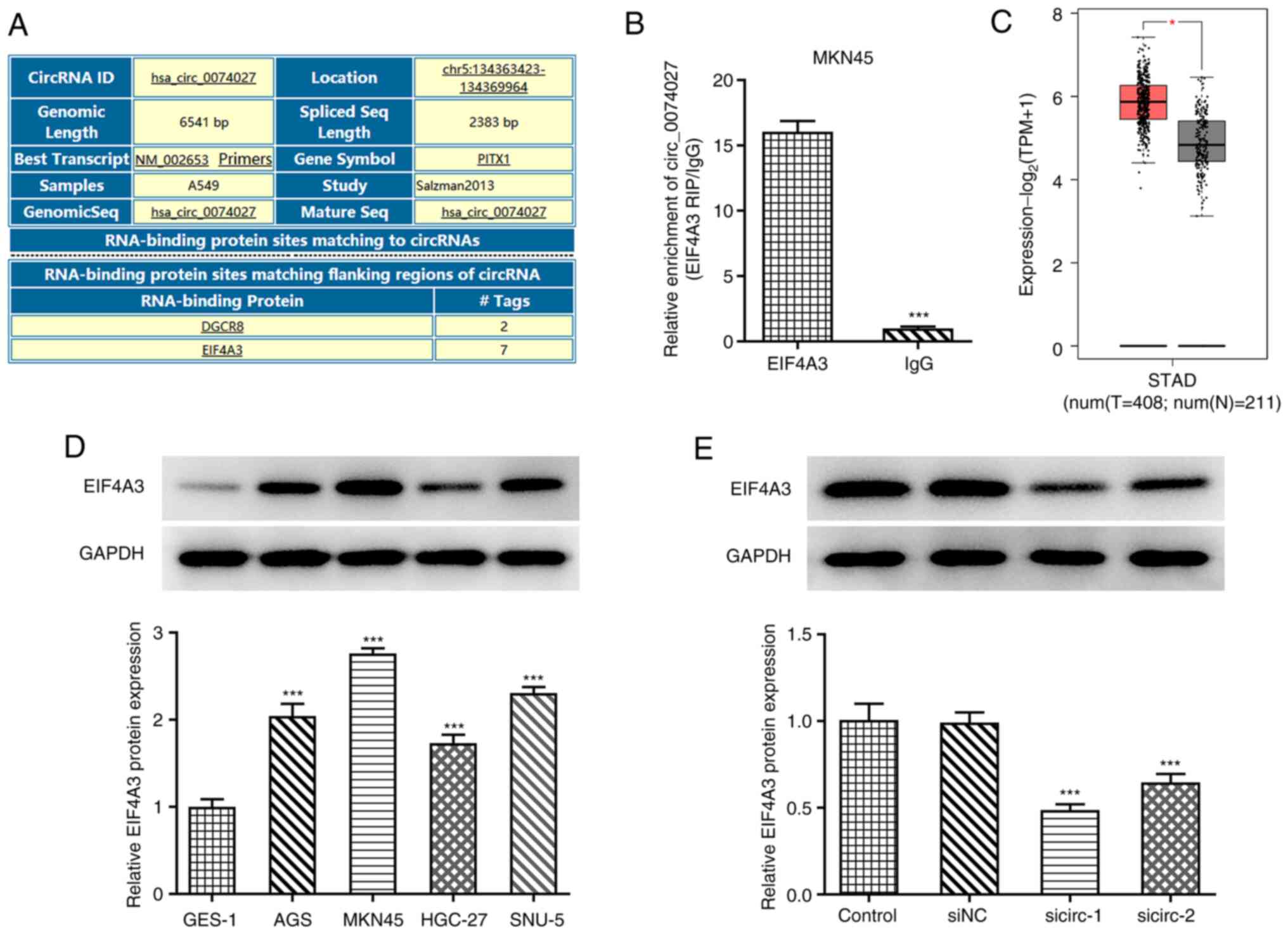 | Figure 3.circ_0074027 is positively associated
with EIF4A3. (A) Circular RNA Interactome analysis predicted genes
that bound to circ_0074027. (B) circ_0074027 with EIF4A3
enrichment, which was measured using an RNA immunoprecipitation
assay. (C) Gene expression of EIF4A3 profiled across tumor samples
and tissues from normal patients, acquired from the Gene Expression
Profiling Interactive Analysis database. (D) Expression of EIF4A3
was detected using RT-qPCR in gastric cancer cells following
transfection. (E) Expression of EIF4A3 was detected using RT-qPCR
in MKN45 cells following transfection with si-NC, si-circ-1 or
si-circ-2. *P<0.05 and ***P<0.001 vs. GES-1 or siNC. RT-qPCR,
reverse transcription-quantitative PCR; circ_0074027, circular
RNA_0074927; sicirc, small interfering RNA of circ_0074027; si,
small interfering RNA; NC, negative control; EIF4A3, eukaryotic
translation initiation factor 4A3; STAD, stomach
adenocarcinoma. |
Overexpression of EIF4A3 attenuates
the suppressive effects of circ_0074027 on GC cell
proliferation
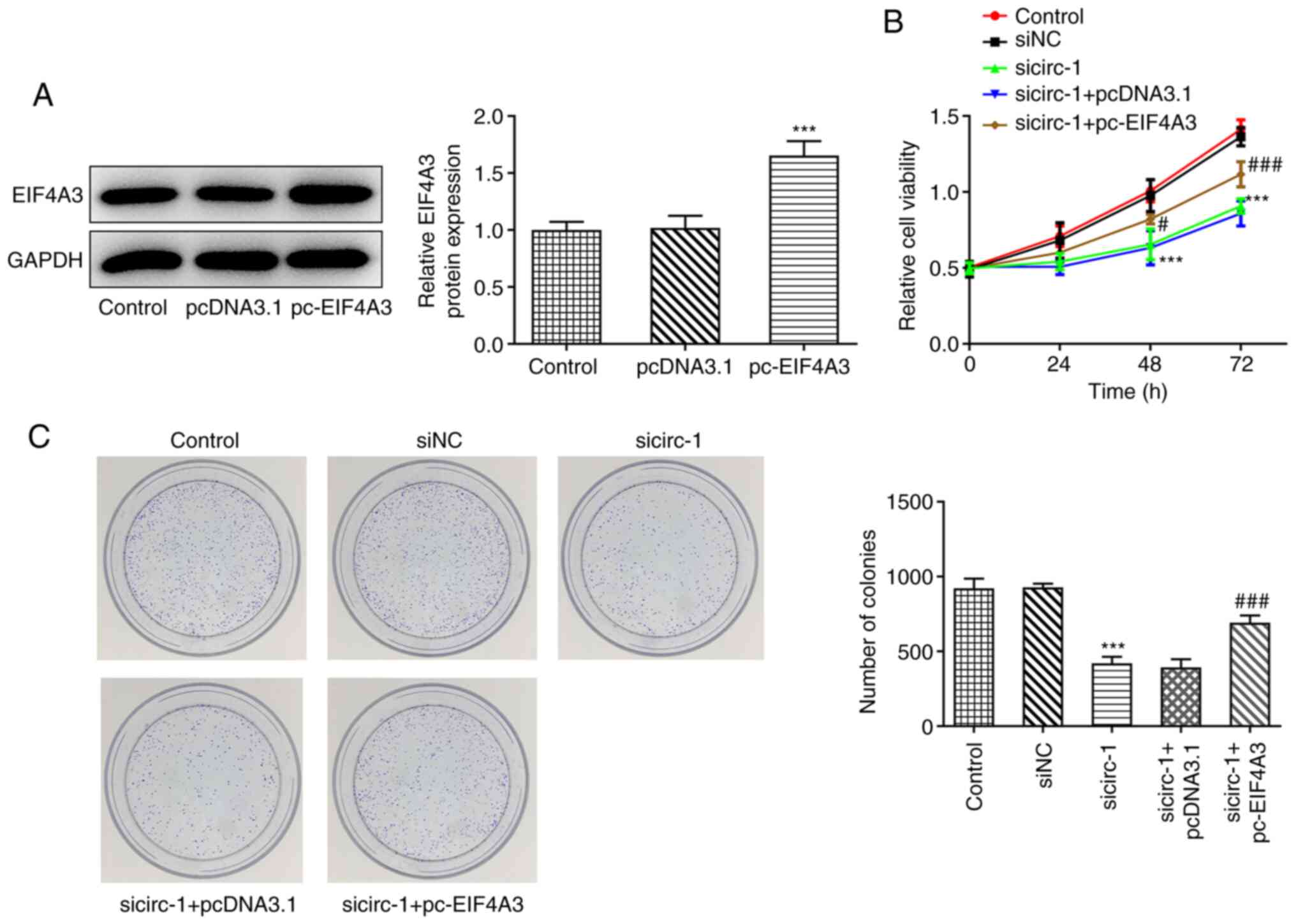
To further determine the role of circ_0074027 and
EIF4A3 in GC, the EIF4A3 overexpression plasmid (pc-EIF4A3) was
used in the following experiments. The results of western blot
analysis revealed that pc-EIF4A3 significantly elevated the
expression of EIF4A3, compared with pcDNA3.1 (Fig. 4A). As revealed by CCK-8 analysis,
MKN45 cells co-transfected with si-circ-1 and pc-EIF4A3 exhibited
greater proliferative ability than those co-transfected with
si-circ-1 and pcDNA3.1 (Fig. 4B).
The results of the colony formation assay indicated that the
colony-forming ability of MKN45 cells was decreased following
circ_0074027-knockdown, compared with the control, while it was
significantly restored following co-transfection with pc-EIF4A3
(Fig. 4C). Collectively, these
findings indicated that elevated EIF4A3 expression reversed the
inhibitory effects of circ_0074027 on the proliferation of GC
cells.
Overexpression of EIF4A3 counteracts
the inhibitory effects on motility generated by
circ_0074027-knockdown
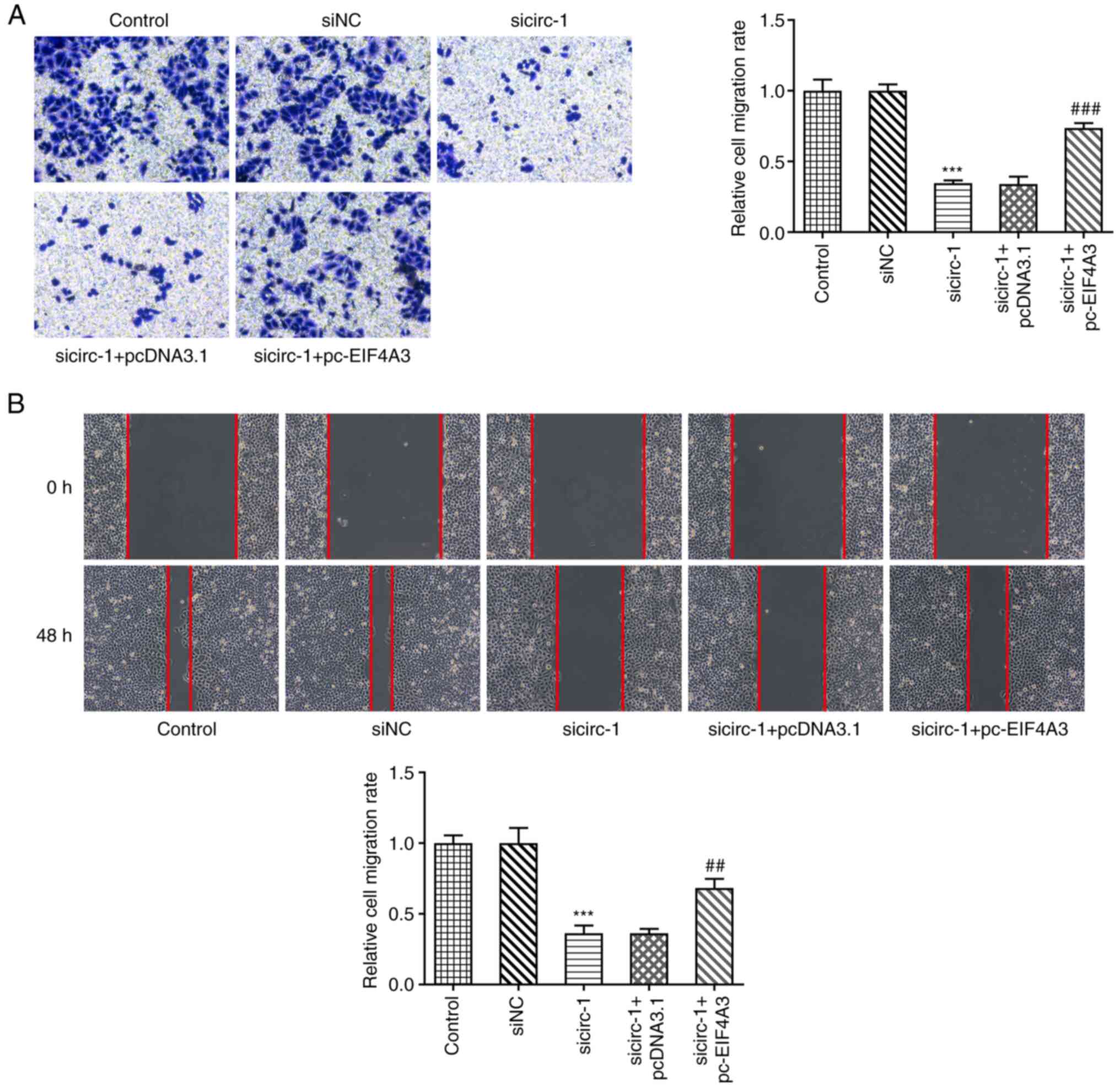
Further experiments were conducted to examine the
effects of circ_0074027 and EIF4A3 on the motility of GC cells.
Both Transwell and wound-healing assays (Fig. 5A and B) revealed that
circ_0074027-knockdown inhibited MKN45 cell migration, and that the
overexpression of EIF4A3 attenuated this effect. The si-circ-1 +
pcDNA3.1 group did not exhibit any marked differences compared with
the si-circ-1 group, whereas the si-circ-1 + pc-EIF4A3 group
exhibited a markedly enhanced migration rate. These results
demonstrated that the elevated expression of EIF4A3 counteracted
the inhibitory effects of circ_0074027-knockdown on the motility of
GC cells.
Discussion
CircRNAs, a newly identified group of non-coding
RNAs, have gained increasing attention in the field of cancer
research (14). Emerging evidence
has indicated the existence of thousands of endogenous circRNAs in
mammalian cells, which can modulate gene expression by binding to
miRNAs or other molecules, and suppressing their function at the
transcriptional or post-transcriptional level (15). In the present study, silencing
circ_0074027 was demonstrated to inhibit the proliferation and
migration of GC cells, and this effect was reversed by the
overexpression of EIF4A3.
Firstly, circRNAs were found to be present in GC
cells in the form of a closed loop, and to be highly expressed
therein. Based on previous studies, this result is in accordance
with the functions of circRNAs in tumors. For example, circ_0067934
was found to be expressed at higher levels in esophageal cancer
compared with normal tissues (16).
In addition, the expression level of circ_002059 in the plasma of
patients with post-operative GC has been shown to be higher than
that of patients with pre-operative GC (17). Moreover, circ_0005075 has been shown
to be aberrantly expressed in hepatocellular carcinoma tissues
(18). Previously, Qian et al
(12) revealed that circ_0074027
upregulation promoted glioblastoma cell proliferation and
invasiveness. Subsequently, circ_0074027 has been reported to
promote the progression of non-small cell lung cancer (10). For example, Duan et al
(19) revealed that circ_0074027
contributed to non-small cell lung cancer progression by positively
modulating RHOA via miR-2467-3p sequestration. Furthermore, Jiang
et al (20) demonstrated that
circ_0074027 promoted the progression of non-small cell lung cancer
via the miR-362-3p/clathrin heavy chain axis. Based on the
aforementioned research conclusions, the present study confirmed
that the overexpression of circ_0074027 had a potential promoting
effect on cancer, such as enhancing cancer cell proliferation and
migration. Then, the Circular RNA Interactome was used to predict
that EIF4A3 was able to bind circ_0074027. Furthermore, the
expression of EIF4A3 was found to be higher in GC. Thus, the
present study aimed to determine the biological function of
circ_0074027 in GC. The proliferation and migration of tumor cells
affect the malignant progression of tumors (21); thus, the effect of f
circ_0074027-knockdown on GC cells was analyzed using CCK-8, colony
formation, Transwell and wound-healing assays. Silencing of
circ_0074027 was found to inhibit the progression of GC cells. This
is consistent with the findings of previous studies, which
identified that circ_0074027 functioned as a tumor promoter in GC
cells (22). And circ_0074027 has
also been shown to contribute to the progression of GC (22).
In order to elucidate the potential mechanisms of
circ_0074027 in GC, the Circular RNA Interactome database was used
to screen for the gene with the most tags for RNA binding, EIF4A3.
A previous study implicated EIF4A3 as an important component of RNA
splicing (23). Moreover, as an RNA
binding protein, EIF4A3 regulates the expression of non-coding RNAs
in tumors (24), and bioinformatics
analysis revealed that EIF4A3 expression was upregulated at the
transcriptional level in common malignant tumors, such as breast,
lung and urinary cancers (25). In
the present study, the binding of circ_0074027 to EIF4A3 was
confirmed using an RIP assay. The expression of EIF4A3 in GC was
then verified through GEPIA, and GC cell lines expressing high
levels of EIF4A3. Notably, EIF4A3 was found to be positively
associated with circ_0074027. EIF4A3 has been shown to trigger the
proliferation, invasion and metastasis of glioblastoma multiforme
cells, and thus contributes to tumorigenesis (26). It has also been demonstrated that E2F
transcription factor 1 and EIF4A3 coordinate with circSEPT9 to
facilitate the development and progression of triple-negative
breast cancer through the circSEPT9/miR-637/LIF IL6 family cytokine
axis (27). Similarly, the results
of the present study confirmed the carcinogenic effect of EIF4A3.
The increased expression of EIF4A3 counteracted the inhibitory
effects of circ_0074027-knockdown on the proliferation and motility
of GC cells. Collectively, the present study provides a novel
insight into the functions of circ_0074027 in GC, which are
hypothesized to occur via the regulation of EIF4A3. However, the
study had certain limitations. For example, only in vitro
experiments were performed, and tissue samples were not used for
validation. In addition, the molecular mechanism of EIF4A3 in tumor
progression remains unclear. Thus, future studies are required to
further elucidate the mechanisms involved.
In conclusion, the findings of the present study
indicated that the expression of circ_0074027 and EIF4A3 were
upregulated in GC cells. circ_0074027 modulated GC cell
proliferation and migration through targeted binding of EIF4A3.
This finding may help to elucidate the molecular mechanisms of GC
progression. In addition, circ_0074027 may prove to be a promising
prognostic biomarker for GC and a novel candidate for targeted GC
therapy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YW and HZ conceived and designed the study, acquired
and interpreted the data. YW was a major contributor in writing the
manuscript. YW and HZ confirm the authenticity of all the raw data.
Both authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Johnston FM and Beckman M: Updates on
management of gastric cancer. Curr Oncol Rep. 21:672019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee YC, Chiang TH, Chou CK, Tu YK, Liao
WC, Wu MS and Graham DY: Association between Helicobacter
pylori eradication and gastric cancer incidence: A systematic
review and meta-analysis. Gastroenterology. 150:1113–1124.e5. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Venerito M, Link A, Rokkas T and
Malfertheiner P: Gastric cancer-clinical and epidemiological
aspects. Helicobacter. 21 (Suppl 1):S39–S44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu J, Huang C, Sun Y, Su X, Cao H, Hu J,
Wang K, Suo J, Tao K, He X, et al: Effect of laparoscopic vs open
distal gastrectomy on 3-year disease-free survival in patients with
locally advanced gastric cancer: The CLASS-01 randomized clinical
trial. JAMA. 321:1983–1992. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Oliveira C, Pinheiro H, Figueiredo J,
Seruca R and Carneiro F: Familial gastric cancer: Genetic
susceptibility, pathology, and implications for management. Lancet
Oncol. 16:e60–e70. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Capel B, Swain A, Nicolis S, Hacker A,
Walter M, Koopman P, Goodfellow P and Lovell-Badge R: Circular
transcripts of the testis-determining gene Sry in adult mouse
testis. Cell. 73:1019–1030. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao
L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al: The
landscape of circular RNA in cancer. Cell. 176:869–881.e13. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu C, Ying J, Yu K, Shen W and Jiang M:
Circ_0074027 contributes to nonsmall cell lung cancer progression
by upregulating CUL4B expression through miR-335-5p. Cancer Biother
Radiopharm. Jun 22–2020.(Epub ahead of print). View Article : Google Scholar
|
|
11
|
Gao P, Wang Z, Hu Z, Jiao X and Yao Y:
Circular RNA circ_0074027 indicates a poor prognosis for NSCLC
patients and modulates cell proliferation, apoptosis, and invasion
via miR-185-3p mediated BRD4/MADD activation. J Cell Biochem.
121:2632–2642. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qian L, Guan J, Wu Y and Wang Q:
Upregulated circular RNA circ_0074027 promotes glioblastoma cell
growth and invasion by regulating miR-518a-5p/IL17RD signaling
pathway. Biochem Biophys Res Commun. 510:515–519. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T
and Shu Y: CircRNAs in cancer metabolism: A review. J Hematol
Oncol. 12:902019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang HD, Jiang LH, Sun DW, Hou JC and Ji
ZL: CircRNA: A novel type of biomarker for cancer. Breast Cancer.
25:1–7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xia W, Qiu M, Chen R, Wang S, Leng X, Wang
J, Xu Y, Hu J, Dong G, Xu PL and Yin R: Circular RNA
has_circ_0067934 is upregulated in esophageal squamous cell
carcinoma and promoted proliferation. Sci Rep. 6:355762016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang X, Song H, Zi Z, Kou J, Chen S, Dai
Y, Wang J, Yuan L and Gao K: Circ_0005075 promotes hepatocellular
carcinoma progression by suppression of microRNA-335. J Cell
Physiol. 234:21937–21946. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Duan Z, Wei S and Liu Y: Circ_0074027
contributes to non-small cell lung cancer progression through
positively modulating RHOA via sequestering miR-2467-3p. J Bioenerg
Biomembr. 53:223–233. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang Z, Yin J, Peng G and Long X:
Circ_0074027 contributes to the progression of non-small cell lung
cancer via microRNA-362-3p/clathrin heavy chain axis. Anticancer
Drugs. 32:1–10. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xue YF, Li M, Li W, Lin Q, Yu BX, Zhu QB
and Chen HJ: Roles of circ-CSPP1 on the proliferation and
metastasis of glioma cancer. Eur Rev Med Pharmacol Sci.
24:5519–5525. 2020.PubMed/NCBI
|
|
22
|
Klein CA: Cancer progression and the
invisible phase of metastatic colonization. Nat Rev Cancer.
20:681–694. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chan CC, Dostie J, Diem MD, Feng W, Mann
M, Rappsilber J and Dreyfuss G: eIF4A3 is a novel component of the
exon junction complex. RNA. 10:200–209. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu Y, Ren C and Yang L: Effect of
eukaryotic translation initiation factor 4A3 in malignant tumors.
Oncol Lett. 21:3582021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin Y, Zhang J, Cai J, Liang R, Chen G,
Qin G, Han X, Yuan C, Liu Z, Li Y, et al: Systematic analysis of
gene expression alteration and co-expression network of eukaryotic
initiation factor 4A-3 in cancer. J Cancer. 9:4568–4577. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang R, Zhang S, Chen X, Li N, Li J, Jia
R, Pan Y and Liang H: EIF4A3-induced circular RNA MMP9 (circMMP9)
acts as a sponge of miR-124 and promotes glioblastoma multiforme
cell tumorigenesis. Mol Cancer. 17:1662018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zheng X, Huang M, Xing L, Yang R, Wang X,
Jiang R, Zhang L and Chen J: The circRNA circSEPT9 mediated by E2F1
and EIF4A3 facilitates the carcinogenesis and development of
triple-negative breast cancer. Mol Cancer. 19:732020. View Article : Google Scholar : PubMed/NCBI
|