Introduction
The prognosis of locally advanced rectal cancer
(LARC) depends largely on the tumor stage at diagnosis. In the
preoperative setting, the post-treatment tumor, node and metastasis
(TNM) stage and pathological complete response to preoperative
radiochemotherapy (pRCT) have been reported to be associated with
disease-free survival (1). However,
these factors cannot be determined prior to surgery. Predictors of
tumor response and long-term outcome that are identifiable before
surgery are necessary for designing a customized management plan
for each patient to increase the survival rate. Therefore, it is
important to identify additional biomarkers to predict and select
patients who will respond favorably to treatment. In addition to
improving outcomes, studies on biomarkers may also provide novel
insights into the molecular mechanism of LARC.
Small heterodimer partner (SHP; also known as NR0B2)
is an orphan member of the nuclear receptor superfamily that
contains a putative ligand-binding domain (LBD), without the
classical DNA-binding domain (2,3). SHP has
been shown to bind to specific activating molecules through LBDs
and interact with other coactivators and corepressors to mediate
transcriptional regulation. Due to this ability, dysfunctional SHP
signaling leads to a wide variety of metabolic, reproductive and
proliferative disorders (4,5). The role of SHP has been extensively
studied in liver and breast cancer (6). In liver cancer, previous studies have
reported that SHP exhibits potent tumor suppressive activity by
inhibiting cellular growth and increasing the sensitivity of tumor
cells to apoptotic stimuli (7–9). In
addition, a study of SHP−/− mice suggested that SHP
plays a critical role in tumor suppression by repressing the
transcription of cell proliferation-associated genes (8). Furthermore, another mechanism of SHP
has been indicated to be the inhibition of estrogen-related
receptor γ, resulting in the interruption of Notch3 signaling
through the activation of miR-206 (10). In breast cancer, it has been
consistently reported that there are close associations between SHP
and estrogen-related signaling (11–13). SHP
blocks estrogen action by inhibiting estrogen receptor-mediated
transcriptional activation and inducing peroxisome
proliferator-activated receptor γ, which is an effective inhibitor
of aromatase expression (11,12,14,15).
Induction of the farnesoid X receptor-SHP-liver receptor homolog-1
pathway has been suggested as a potential new therapeutic approach
for the repression of tumor growth and induction of apoptosis in
breast cancer (16).
The role of inflammation in cancer is well
established (17). Cancer-associated
inflammation can increase the risk of cancer and impact the
progression and treatment response of patients with various types
of cancer, including colorectal, prostate and bladder cancer
(17). A meta-analysis demonstrated
that markers of the systemic inflammatory response, such as the
C-reactive protein (CRP) level, neutrophil-lymphocyte ratio (NLR),
lymphocyte-monocyte ratio (LMR) and platelet-lymphocyte ratio (PLR)
could be useful in predicting treatment response and monitoring
progression in patients with colorectal cancer (18–21). In
addition, local inflammatory markers, such as the tumor-to-stroma
ratio, Klintrup-Makinen score and Galon immunoscore can predict the
prognosis of patients with colorectal cancer (22,23).
Therefore, there is a growing interest in biochemical mediators
linking systemic and tumor inflammatory responses.
SHP has been shown to play an intricate role in the
prevention of excessive inflammation by regulating the innate
immune system (24–27). However, the role of SHP expression in
cancer-associated inflammation and the clinical outcome of rectal
cancer has not yet been investigated. In the present study, SHP
expression and its association with systemic inflammatory markers,
treatment response and survival in patients with rectal
adenocarcinoma (READ) were investigated using a combination of
bioinformatics and immunohistochemistry.
Materials and methods
Bioinformatics analysis
Gene Set Enrichment Analysis (GSEA) was performed as
previously described (28). Briefly
the mRNA-Seq profiles
(illuminahiseq_rnaseqv2-RSEM_genes_normalized) and clinical data of
patients with READ were obtained from Firehose (https://gdac.broadinstitute.org/). The Cancer
Genome Atlas (TCGA) RNA-Seq data were cross-referenced with the
clinical information recorded for the patients. Patients with
missing clinical data and/or expression values were excluded from
further analyses. Data from 95 samples were included in the study.
The mRNA-Seq data were normalized using the Rank Normalize module
in GenePattern (http://broadinstitute.org/cancer/software/genepattern).
To identify the best cutoff for inflammation by using GSEA, the
patients were then ranked based on SHP expression and divided into
high (30%, ranking 1–29) and low (70%, ranking 30–95) expression
groups according to rank. Phenotype labels were permuted 1,000
times, and a normalized P<0.05 and false discovery rate (FDR) of
<0.25 were selected as statistically significant
enrichments.
The OncoLnc database (http://www.oncolnc.org/) was used to determine whether
the expression of SHP was associated with the overall survival (OS)
of patients with READ. The SHP (NR0B2) gene was queried and a
Kaplan plot for READ was generated.
Analysis of LARC datasets was carried out
essentially as previously described (28). The SHP expression patterns were
downloaded from the GSE15781 dataset for the comparison of pre-pRCT
LARC and post-pRCT LARC patient groups (29). To compare the univariate differential
expression in each dataset, RNA data were analyzed using the
Wilcoxon-Mann-Whitney test.
Tumor Immune Estimation Resource (TIMER) is a
resource for the systematic analysis of immune infiltrates for
various types of cancer (https://cistrome.shinyapps.io/timer/) (30). TIMER applies a previously published
statistical method of deconvolution to infer the abundance of
tumor-infiltrating immune cells from gene expression profiles
(30). The association of SHP
expression with the abundance of various immune cells was analyzed
using the TIMER database. In addition, the association between SHP
expression and the gene expression of immune-suppressive molecules
was investigated.
Patients and pretreatment
evaluation
Between March 2003 and December 2011, 89 patients
with LARC who had been treated with pRCT at the Chungnam National
University Hospital (Daejeon, Republic of Korea) and for whom
pre-treatment tissue blocks were available were retrospectively
enrolled in the study. Eligibility for the study was determined
based on the following criteria: i) Histological evidence of READ;
ii) tumor extension through the bowel wall (T3-T4) or pelvic lymph
node involvement without evidence of distant metastasis; and iii)
presence of a resectable tumor. The exclusion criteria were as
follows: i) Patients who had previous history of other cancers; ii)
patients who had received previous curative resection for any
colorectal tumor lesion; and iii) patients who had distant
metastasis in the initial diagnosis. Patient and tumor
characteristics are listed in Table
I. Essential pre-treatment workups included a complete history,
physical examination, complete blood count, serum chemistry,
carcinoembryonic antigen (CEA) level analysis, chest radiography,
abdominal/pelvic computed tomography (CT) and colonoscopy with
biopsy. The TNM stage prior to clinical treatment was determined
mainly by CT imaging. An informed consent form was signed by all
patients. Biopsied tumor tissues were collected consecutively from
patients at Chungnam National University Hospital between March
2003 and December 2011, and were analyzed following study approval
from the institutional review board (IRB) of Chungnam National
University Hospital (IRB no. 2017-07-037).
 | Table I.Association of SHP with
clinicopathological features in patients with locally advanced
rectal cancer. |
Table I.
Association of SHP with
clinicopathological features in patients with locally advanced
rectal cancer.
|
|
| SHP expression, n
(%) |
|---|
|
|
|
|
|---|
|
Characteristics | Total, n (%) | Low (n=52) | High (n=37) | P-value |
|---|
| Age, years |
|
|
| 0.824 |
|
<60 | 57 (64.0) | 34 (65.4) | 23 (62.2) |
|
|
≥60 | 32 (36.0) | 18 (34.6) | 14 (37.8) |
|
| Sex |
|
|
| 0.809 |
|
Male | 65 (73.0) | 37 (71.2) | 28 (75.7) |
|
|
Female | 24 (27.0) | 15 (28.8) | 9 (24.3) |
|
| Tumor distance from
anal verge, cm |
|
|
| 0.822 |
|
<6 | 58 (65.2) | 33 (63.5) | 25 (67.6) |
|
| ≥6 | 31 (34.8) | 19 (36.5) | 12 (32.4) |
|
| CEA before RCT,
ng/ml |
|
|
| 0.822 |
| ≤5 | 58 (65.2) | 33 (63.5) | 25 (67.6) |
|
|
>5 | 31 (34.8) | 19 (36.5) | 12 (32.4) |
|
| cT stage |
|
|
| 0.224 |
|
T2-3 | 76 (85.4) | 42 (80.8) | 34 (91.9) |
|
| T4 | 13 (14.6) | 10 (19.2) | 3 (8.1) |
|
| cN stage |
|
|
| 0.004 |
| N
(−) | 6 (6.7) | 0 (0.0) | 6 (16.2) |
|
| N
(+) | 83 (93.3) | 52 (100.0) | 31 (83.8) |
|
Radiotherapy treatment and evaluation
of tumor response
Radiation was delivered via 6- and 10-MV photons
using a three-field technique (posterior and bilateral fields) in
most patients. Treatment was planned via computerized dosimetry,
and a dose of 1.8 Gy/fraction was prescribed to cover the planning
target volume. Radiotherapy was administered 5 days per week, once
a day, at 1.8 Gy/fraction. Pelvic radiotherapy was performed with
45 Gy in 25 fractions over a period of 5 weeks, and followed by a
booster dose of 5.4 Gy, administered to the primary tumor in 3
fractions using 2 lateral fields. The clinical target volume
contained the primary tumor, the mesorectum, the presacral space
and lymph nodes, including the perirectal, presacral, internal
iliac and/or external iliac nodes, as indicated. For the entire
pelvic field, the superior border was located at the L5-S1
interspace, and the inferior border was located 3–4 cm below the
primary tumor. The lateral border was located 1.5 cm outside the
true bony pelvis. For the lateral fields, the posterior margin was
1.5 cm behind the anterior bony sacral margin, and the anterior
border generally comprised the anterior acetabulum. Preoperative
chemotherapy was administered concurrently with radiation therapy.
Patients received oral chemotherapy consisting of 2 cycles of
capecitabine and leucovorin, according to the chemotherapy protocol
of Chungnam National University Hospital. At ~6 weeks after the
completion of pRCT, the patients underwent definitive surgery.
Surgical management included a sphincter-preservation approach,
whenever possible, via the total mesorectal excision technique.
Pathological evaluation of surgical specimens, including the
primary tumor and resected nodes, was performed by a specialist
pathologist. The pathological TNM staging, histological grading for
LARC and modified Ryan scheme for tumor regression score were
determined at the time of surgical resection and were based on the
8th edition of the American Joint Committee on Cancer staging
system (31). The complete absence
of residual tumor cells in the primary tumor was defined as a
pathologic complete response (pCR).
Immunohistochemistry (IHC)
Rectal adenocarcinoma tissues were fixed with 10%
buffered formalin for 24 h at room temperature. The expression of
SHP was analyzed by the IHC of paraffin-embedded tissue sections
from patients with LARC. Sections from paraffin blocks with a
thickness of 3 µm were used for IHC. Endogenous peroxidase blocking
(0.03% H2O2) was performed for 10 min at room
temperature. A rabbit polyclonal antibody against NR0B2 (cat. no.
ab186874; Abcam) was diluted at 1:400 with antibody diluent (DaKo
antibody diluent, cat. no. S3022; Dako; Agilent Technologies,
Inc.), and tissue sections were incubated in the mixture overnight
at 4°C in a humid chamber then washed 3 times with 0.05% TBS-T.
After washing, samples were incubated in 100 µl secondary antibody
[EnVision + Single Reagents horse-radish peroxidase (HRP); cat. no.
K4003; Dako; Agilent Technologies, Inc.] for an additional 20 min
at room temperature followed by additional washing. The reaction
products were visualized after 5 min in diaminobenzidine plus a
substrate-chromogen solution. The slides were counterstained with
Meyer's hematoxylin for 30 sec at room temperature and mounted.
Careful rinses with several changes of PBS were performed between
each stage of the procedure. Two experienced pathologists who had
no access to clinical information examined and scored the slides by
using a light microscope (BX51; Olympus Corporation). The nuclear
staining of the tumor was divided into 4 grades based on staining
intensity: Score 0, no staining; score 1, weak staining; score 2,
intermediate staining; score 3, strong staining. In cases of
heterogeneous staining within samples, the higher score was
selected if >50% of the cells had a higher staining intensity.
Cases with a score of 0 or 1 were categorized into a low expression
group, whereas those with a score of 2 and 3 were categorized into
a high expression group.
RNA extraction from formalin-fixed
paraffin-embedded (FFPE) samples and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from 10-µm slices of FFPE
samples from potentially eligible patients (n=12) using a Qiagen
RNeasy® FFPE kit (Qiagen, Inc.) following the
manufacturer's protocol. RNA quantitation and purity were measured
using a NanoDrop™ ND-8000 spectrophotometer (Thermo Fisher
Scientific, Inc.).
cDNA was synthesized by RT using SuperScript™ II
Reverse Transcriptase (cat. no. 18064; Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. qPCRs
were carried out using Rotor-Gene SYBR-Green PCR kits (cat. no.
204074; Qiagen, Inc.) in a Rotor-Gene Q 2plex system (cat. no.
9001620; Qiagen, Inc.). The samples were amplified for 40 cycles as
follows: 95°C for 10 sec and 60°C for 30 sec. To analyze the qPCR
data, relative quantification was performed using the
2−ΔΔCq method with human GAPDH as the internal control
gene; data are expressed as relative fold-changes (32). The sequences of human SHP and GAPDH
primers were as follows: SHP forward, 5′-TCCTCTTCAACCCCGATGTG-3′
and reverse, 5′-CAGGGTTCCAGGACTTCACAC-3′: GAPDH forward,
5′-TCGGAGTCAACGGATTTGGT-3′ and reverse,
5′-TTCCCGTTCTCAGCCTTGAC-3′.
Statistical analysis
The associations between clinicopathological or
hematological factors and SHP levels were analyzed using Pearson's
χ2 test and Fisher's exact test, and survival curves
were created by the Kaplan-Meier method. The prognostic value of
SHP expression was evaluated using the log-rank test for univariate
analysis and the Cox proportional hazards model for multivariate
analyses. A backward stepwise selection of covariates was used for
the Cox proportional hazards model, and P<0.1 was defined as the
threshold for covariate inclusion. P<0.05 was considered to
indicate a statistically significant difference. All statistical
analyses were conducted using PASW statistical software (version
17.0; SPSS, Inc.).
Results
Bioinformatics analysis of SHP
expression in READ
Numerous studies have shown that SHP prevents or
controls acute inflammatory responses in innate immune cells
(24–27). These studies have led to the
hypothesis that SHP may play a role in the regulation of
cancer-associated inflammation. If this hypothesis is correct, a
low SHP expression should inhibit an adequate immune response in
READ. To test this, GSEA was performed by comparing high and low
SHP mRNA expression groups in hallmark gene sets using TCGA
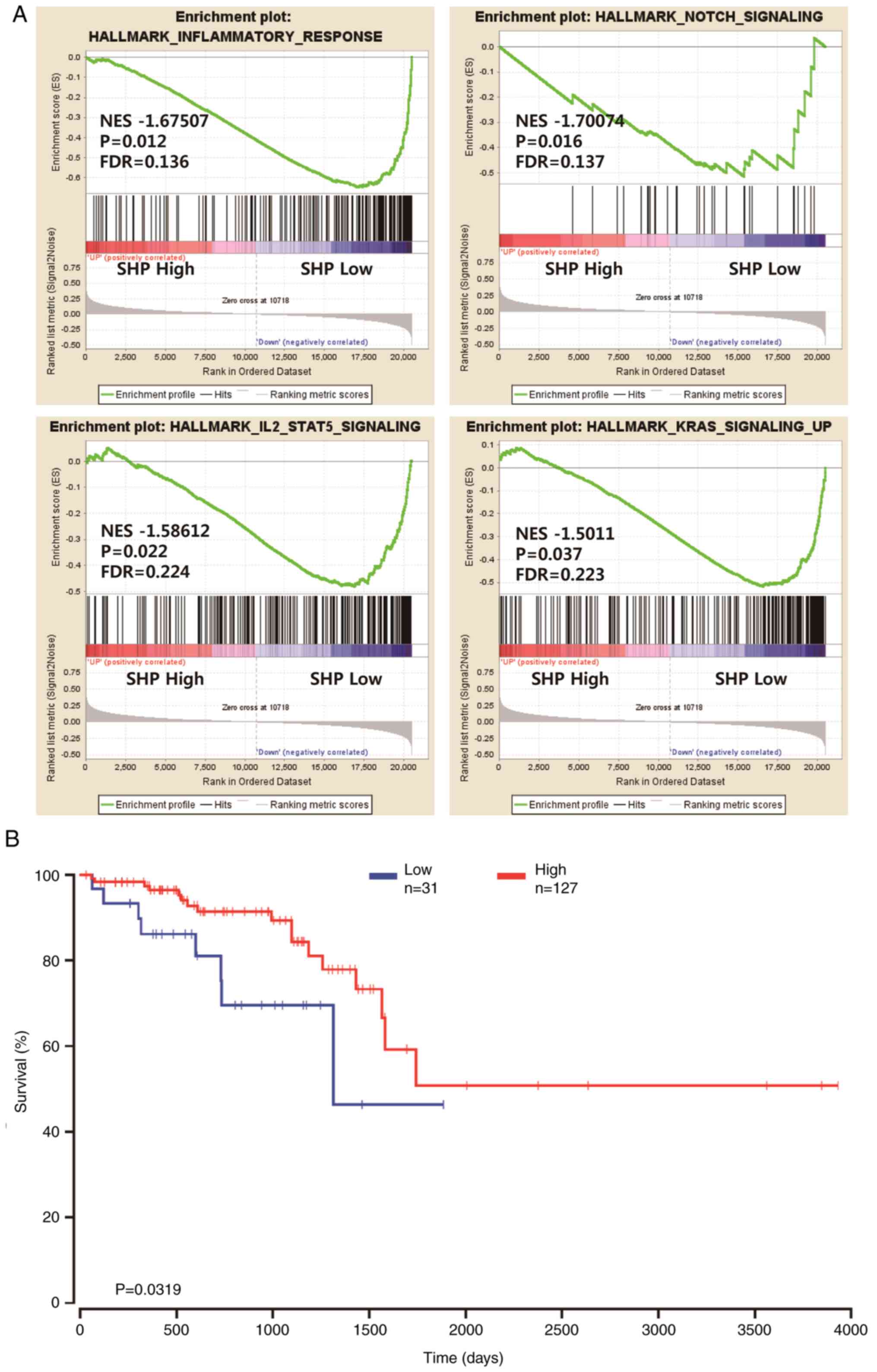
mRNA-Seq data (Fig. 1A). GSEA
revealed a significant difference (FDR, <0.25; nominal
P<0.05) between the two groups with regard to enrichment with
genes from the MSigDB Collection (h.all.v6.2.symbols.gmt), with
details shown in Fig. 1A. Moreover,
Fig. 1A shows that inflammatory
responses [normalized enrichment score (NES), −1.67507], NOTCH
signaling (NES, −1.70074), IL2-STAT5 signaling (NES, −1.58612) and
KRAS signaling (NES, −1.5011) were differentially enriched in the
SHP low expression group.
The GSEA results suggest that several pathways
associated with cancer progression were upregulated in patients
with READ who had low SHP expression. Therefore, the OncoLnc
database was used to investigate whether the mRNA expression level
of SHP was a prognostic factor for READ. The data from the OncoLnc
database, which stores gene expression data and clinical
information for READ, were analyzed. The results revealed that
patients with READ who had low SHP mRNA expression levels had a
worse prognosis than those with high SHP mRNA levels (P=0.0319;
Fig. 1B).
Association between SHP expression and
patient characteristics
Since GSEA and survival analysis rely on the
expression profile of mRNA, the SHP protein level was examined in
clinical samples derived from patients with LARC treated with pRCT.
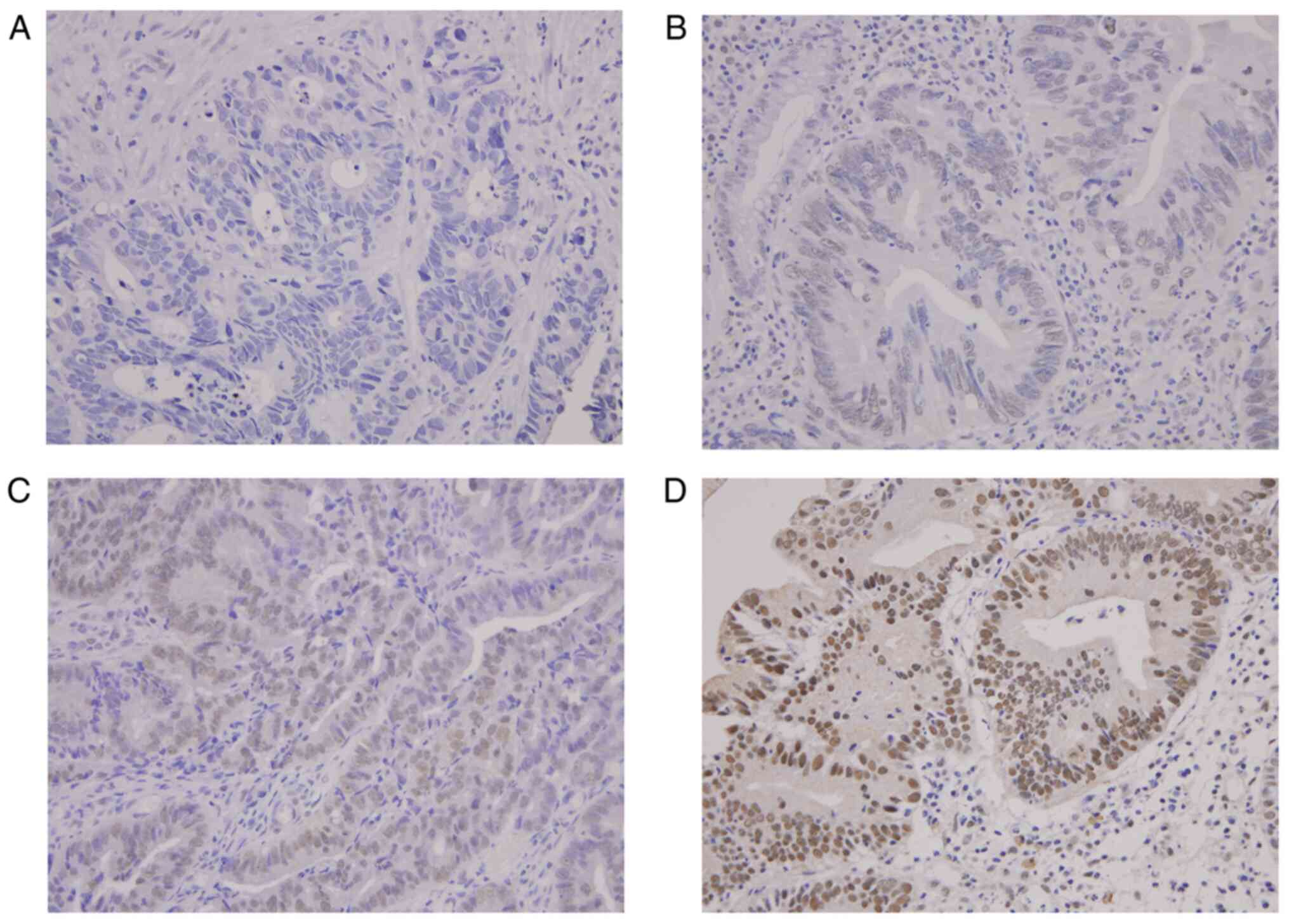
IHC showed that SHP was expressed mainly in the nuclei but also in
the cytoplasm. Representative SHP staining results are shown in
Fig. 2. A high level of SHP
expression was detected in the tumor tissues of 37/89 patients with
READ (41.6%), whereas 52 patients were categorized into the low
expression group (Table I).
The clinicopathological characteristics of the 89
patients with LARC according to SHP expression are presented in
Table I. The median age of the
patients was 62 years (range, 33–81 years). Low SHP expression was
positively associated with clinical N (cN) stage [N (−) vs. N (+);
P=0.004]. No significant association was identified between SHP
expression and other clinicopathological variables, including age,
sex, tumor distance from the anal verge and clinical T (cT)
stage.
Since the aforementioned GSEA results indicate that
SHP expression is negatively associated with inflammatory
responses, the associations between SHP protein expression and
systemic inflammatory markers were determined. The associations
between SHP expression and hematological characteristics of
inflammation in the patients are presented in Table II. Among the various inflammatory
markers, low SHP expression was associated with the following
hematological parameters: Neutrophil counts (P=0.023), lymphocyte
counts (P=0.024) and NLR values (P=0.023). Similarly, patients with
low SHP expression tended to present with more highly elevated
erythrocyte sedimentation rates (ESRs) than those with high SHP
expression (Fig. S1). However, no
association was detected between SHP and either platelet counts or
PLR values.
 | Table II.Association of SHP with hematologic
parameters in patients with locally advanced rectal cancer. |
Table II.
Association of SHP with hematologic
parameters in patients with locally advanced rectal cancer.
|
|
| SHP expression, n
(%) |
|---|
|
|
|
|
|---|
|
Characteristics | Total, n (%) | Low (n=52) | High (n=37) | P-value |
|---|
| Neutrophil
count |
|
|
| 0.023 |
| Below
the median | 45 (50.6) | 21 (40.4) | 24 (64.9) |
|
| Above
the median | 44 (49.4) | 31 (59.6) | 13 (35.1) |
|
| Lymphocyte
count |
|
|
| 0.024 |
| Below
the median | 44 (49.4) | 23 (44.2) | 21 (56.8) |
|
| Above
the median | 45 (50.6) | 29 (55.8) | 16 (43.2) |
|
| Platelet count |
|
|
| 0.761 |
| Below
the median | 45 (50.6) | 27 (51.9) | 18 (48.6) |
|
| Above
the median | 44 (49.4) | 25 (48.1) | 19 (51.4) |
|
| NLR |
|
|
| 0.023 |
| Below
the median | 45 (50.6) | 21 (40.4) | 24 (64.9) |
|
| Above
the median | 44 (49.4) | 31 (59.6) | 13 (35.1) |
|
| PLR |
|
|
| 0.900 |
| Below
the median | 45 (50.6) | 26 (50.0) | 19 (51.4) |
|
| Above
the median | 44 (49.4) | 26 (50.0) | 18 (48.6) |
|
Association of SHP expression with pCR
to RCT
Next, the relationship between treatment response
and patient characteristics was determined using Fisher's exact
tests. Following pRCT, a pCR was observed in 19 patients (21.3%).
The associations between pCR and patient characteristics, including
SHP expression and clinical/hematological factors, are presented in
Table III. The nuclear expression
of SHP was significantly higher in patients with pCR, as compared
with those without (67.6 vs. 32.4%; P=0.038). In addition, a
significant association was detected between pathologic tumor
response and other factors, including the low NLR (66.7 vs. 33.3%;
P=0.009) and PLR (66.7 vs. 33.3%; P=0.009). No significant
association was identified between pCR and age, sex, tumor distance
from the anal verge, cT or cN stage, and neutrophil, lymphocyte and
platelet counts.
 | Table III.Analysis of predictive factors
associated with pathologic complete response. |
Table III.
Analysis of predictive factors
associated with pathologic complete response.
|
| Pathologic complete
response, n (%) |
|---|
|
|
|
|---|
|
| No | Yes |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | 70 (78.7%) | 19 (21.3) | P-value |
|---|
| Age, years |
|
| 0.790 |
|
<60 | 44 (77.2) | 13 (22.8) |
|
|
≥60 | 26 (81.3) | 6 (18.7) |
|
| Sex |
|
| 0.576 |
|
Male | 50 (76.9) | 15 (23.1) |
|
|
Female | 20 (83.3) | 4 (16.7) |
|
| Tumor distance from
anal verge, cm |
|
| 0.588 |
|
<6 | 47 (81.0) | 11 (19.0) |
|
| ≥6 | 23 (83.9) | 8 (16.1) |
|
| CEA before RCT,
ng/ml |
|
| 0.429 |
| ≤5 | 44 (75.9) | 14 (24.1) |
|
|
>5 | 26 (83.9) | 5 (16.1) |
|
| cT stage |
|
| 0.727 |
|
T2-3 | 59 (77.6) | 17 (22.4) |
|
| T4 | 11 (84.7) | 2 (15.4) |
|
| cN stage |
|
| 1.000 |
| N
(−) | 5 (83.3) | 1 (16.7) |
|
| N
(+) | 65 (78.3) | 18 (21.7) |
|
| SHP |
|
| 0.038 |
|
Low | 45 (86.5) | 7 (13.5) |
|
|
High | 25 (67.6) | 12 (32.4) |
|
| Neutrophil
count |
|
| 0.606 |
| Below
the median | 34 (75.6) | 11 (24.4) |
|
| Above
the median | 36 (81.8) | 8 (18.2) |
|
| Lymphocyte
count |
|
| 0.302 |
| Below
the median | 37 (84.1) | 7 (15.9) |
|
| Above
the median | 33 (73.3) | 12 (26.7) |
|
| Platelet count |
|
| 0.606 |
| Below
the median | 34 (75.6) | 11 (24.4) |
|
| Above
the median | 36 (81.8) | 8 (18.2) |
|
| NLR |
|
| 0.009 |
| Below
the median | 30 (66.7) | 15 (33.3) |
|
| Above
the median | 40 (90.9) | 4 (9.1) |
|
| PLR |
|
| 0.009 |
| Below
the median | 30 (66.7) | 15 (33.3) |
|
| Above
the median | 40 (90.9) | 4 (9.1) |
|
To validate the relationship between the mRNA
expression of SHP and pCR in patients with LARC, RT-qPCR analysis
of LARC tissues was conducted. As shown in Fig. S2, the mRNA expression of SHP was
significantly higher (P=0.0283) in patients that achieved a pCR to
pRCT, as compared with those in the non-responder group.
Additionally, whether the expression level of SHP is affected by
pRCT was investigated. To evaluate the change in SHP expression
between pretreatment and post-pRCT surgical specimens, a publicly
available transcriptome dataset (GSE15781) was analyzed (Fig. S3). A non-significant reduction in
the mRNA expression of SHP was observed in residual cancer tissues
following pRCT treatment (Fig.
S3).
Association between SHP expression and
survival
The median follow-up time was 54.0 months (range,
16–88 months) for all patients and 58.5 months (range, 16–88
months) for the surviving patients. Local and distant failure were
observed in 13 (14.6%) and 24 (27.0%) cases, respectively. The
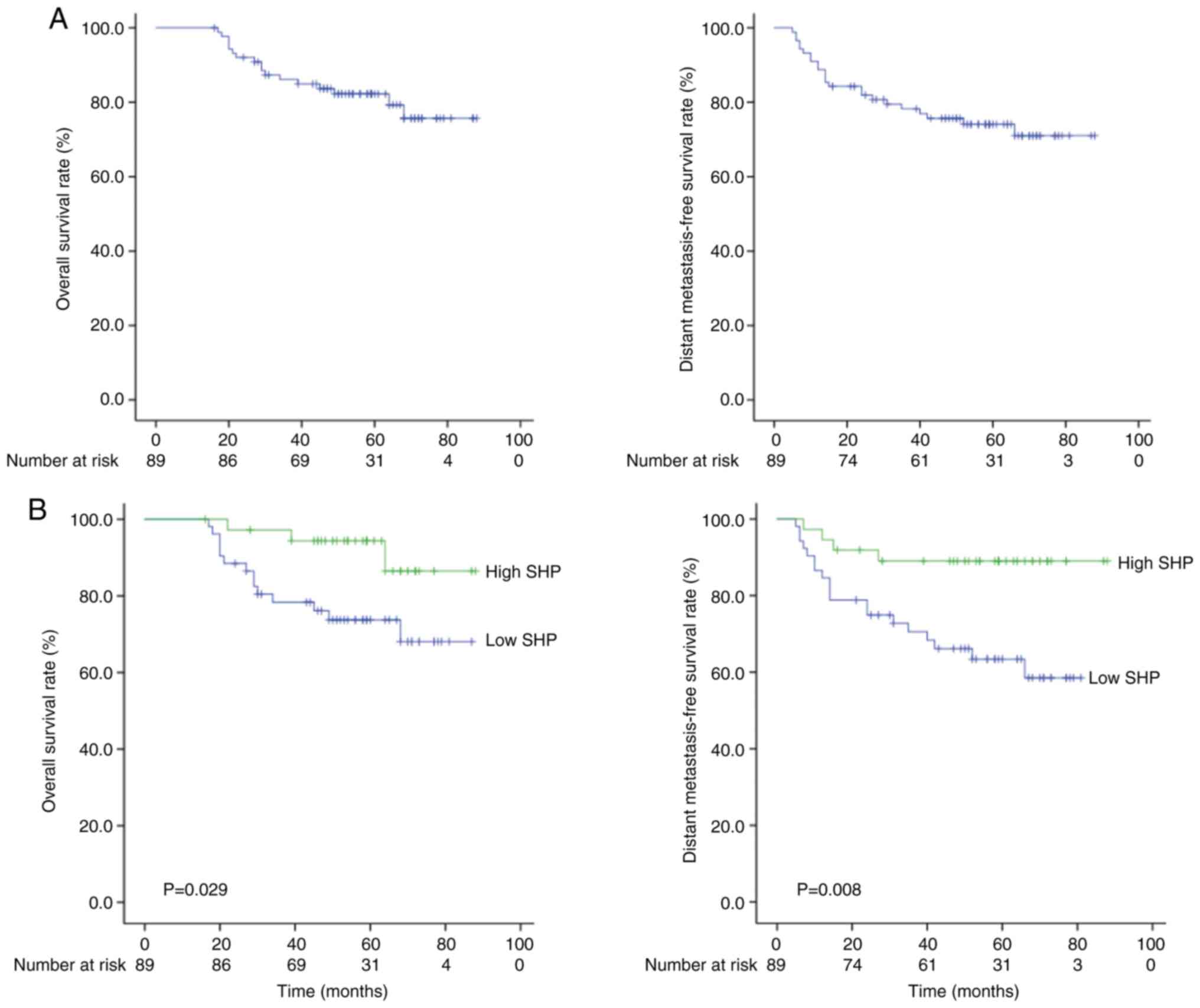
5-year OS and distant metastasis-free survival (DMFS) rates were
81.3 and 73.3%, respectively (Fig.
3A).
Since SHP expression was found to be significantly
associated with OS and DMFS, the effects of prognostic factors on
the two types of survival were analyzed. Table IV shows the association between
potential prognostic factors and OS, as determined by univariate
and multivariate analyses. In the univariate analysis, pCR and SHP
expression were significantly associated with OS. In addition, the
52 patients with low nuclear SHP expression exhibited poorer OS
rates than the 37 patients with high nuclear SHP expression (72.3
vs. 94.4%; Fig. 3A). Furthermore,
the pre-pRCT CEA level and the NLR were found to be marginally
significant prognostic factors for OS. Variables with P<0.1,
based on univariate analysis, were entered into a Cox proportional
hazards model for the multivariate analysis of OS. Multivariate
analysis confirmed that pre-pRCT SHP and CEA levels were
independent prognostic factors for OS at a marginal level of
significance. Several parameters were found to be associated with
DMFS (Table V). In addition, 52
patients with low nuclear SHP expression exhibited poorer DMFS
rates compared with the 37 patients with high nuclear SHP
expression (58.5 vs. 89.0%; Fig.
3B). In the univariate analysis, pCR and SHP expression were
significant prognostic factors for DMFS. In the multivariate
analysis, SHP expression remained significant (hazard ratio, 0.315;
95% confidence interval, 0.107–0.932; P=0.037).
 | Table IV.Prognostic factor analysis for
overall survival. |
Table IV.
Prognostic factor analysis for
overall survival.
|
|
| P-value |
|---|
|
|
|
|
|---|
| Prognostic
factor | 5-year overall
survival rate (%) | Univariate
analysis | Multivariate
analysis |
|---|
| Age, years |
| 0.266 |
|
|
<60 | 77.8 |
|
|
|
≥60 | 90.4 |
|
|
| Sex |
| 0.480 |
|
|
Male | 82.3 |
|
|
|
Female | 81.5 |
|
|
| Tumor distance from
anal verge, cm |
| 0.510 |
|
|
<6 | 83.3 |
|
|
| ≥6 | 80.3 |
|
|
| CEA before CRT,
ng/ml |
| 0.053 | 0.093 |
| ≤5 | 87.3 |
|
|
|
>5 | 72.8 |
|
|
| cT stage |
| 0.673 |
|
|
T2-3 | 82.0 |
|
|
| T4 | 80.9 |
|
|
| cN stage |
| 0.217 |
|
| N
(−) | 100.0 |
|
|
| N
(+) | 79.9 |
|
|
| Pathologic complete
response |
| 0.025 | 0.957 |
| No | 77.4 |
|
|
|
Yes | 100 |
|
|
| SHP |
| 0.029 | 0.087 |
|
Low | 73.7 |
|
|
|
High | 94.4 |
|
|
| Neutrophil
count |
| 0.686 |
|
| Below
the median | 83.3 |
|
|
| Above
the median | 81.2 |
|
|
| Lymphocyte
count |
| 0.543 |
|
| Below
the median | 81.6 |
|
|
| Above
the median | 82.6 |
|
|
| NLR |
| 0.059 | 0.357 |
| Below
the median | 87.4 |
|
|
| Above
the median | 77.1 |
|
|
| Platelet count |
| 0.247 |
|
| Below
the median | 79.0 |
|
|
| Above
the median | 85.6 |
|
|
| PLR |
| 0.838 |
|
| Below
the median | 80.8 |
|
|
| Above
the median | 83.5 |
|
|
 | Table V.Prognostic factor analysis for
distant metastasis-free survival. |
Table V.
Prognostic factor analysis for
distant metastasis-free survival.
|
|
| P-value |
|---|
|
|
|
|
|---|
| Prognostic
factor | 5-year distant
metastasis-free survival rate (%) | Univariate
analysis | Multivariate
analysis |
|---|
| Age, years |
| 0.257 |
|
|
<60 | 70.6 |
|
|
|
≥60 | 80.4 |
|
|
| Sex |
| 0.950 |
|
|
Male | 74.8 |
|
|
|
Female | 70.8 |
|
|
| Tumor distance from
anal verge, cm |
| 0.355 |
|
|
<6 | 76.9 |
|
|
| ≥6 | 69.2 |
|
|
| CEA before CRT,
ng/ml |
| 0.438 |
|
| ≤5 | 76.5 |
|
|
|
>5 | 69.9 |
|
|
| cT stage |
| 0.821 |
|
|
T2-3 | 72.3 |
|
|
| T4 | 70.5 |
|
|
| cN stage |
| 0.542 |
|
| N
(−) | 83.3 |
|
|
| N
(+) | 73.3 |
|
|
| Pathologic complete
response |
| 0.024 | 0.099 |
| No | 68.3 |
|
|
|
Yes | 94.7 |
|
|
| SHP |
| 0.008 | 0.037 |
|
Low | 63.4 |
|
|
|
High | 89.0 |
|
|
| Neutrophil
count |
| 0.661 |
|
| Below
the median | 71.9 |
|
|
| Above
the median | 75.8 |
|
|
| Lymphocyte
count |
| 0.821 |
|
| Below
the median | 74.1 |
|
|
| Above
the median | 74.1 |
|
|
| NLR |
| 0.348 |
|
| Below
the median | 78.5 |
|
|
| Above
the median | 69.3 |
|
|
| Platelet count |
| 0.735 |
|
| Below
the median | 75.6 |
|
|
| Above
the median | 72.3 |
|
|
| PLR |
| 0.794 |
|
| Below
the median | 76.8 |
|
|
| Above
the median | 71.7 |
|
|
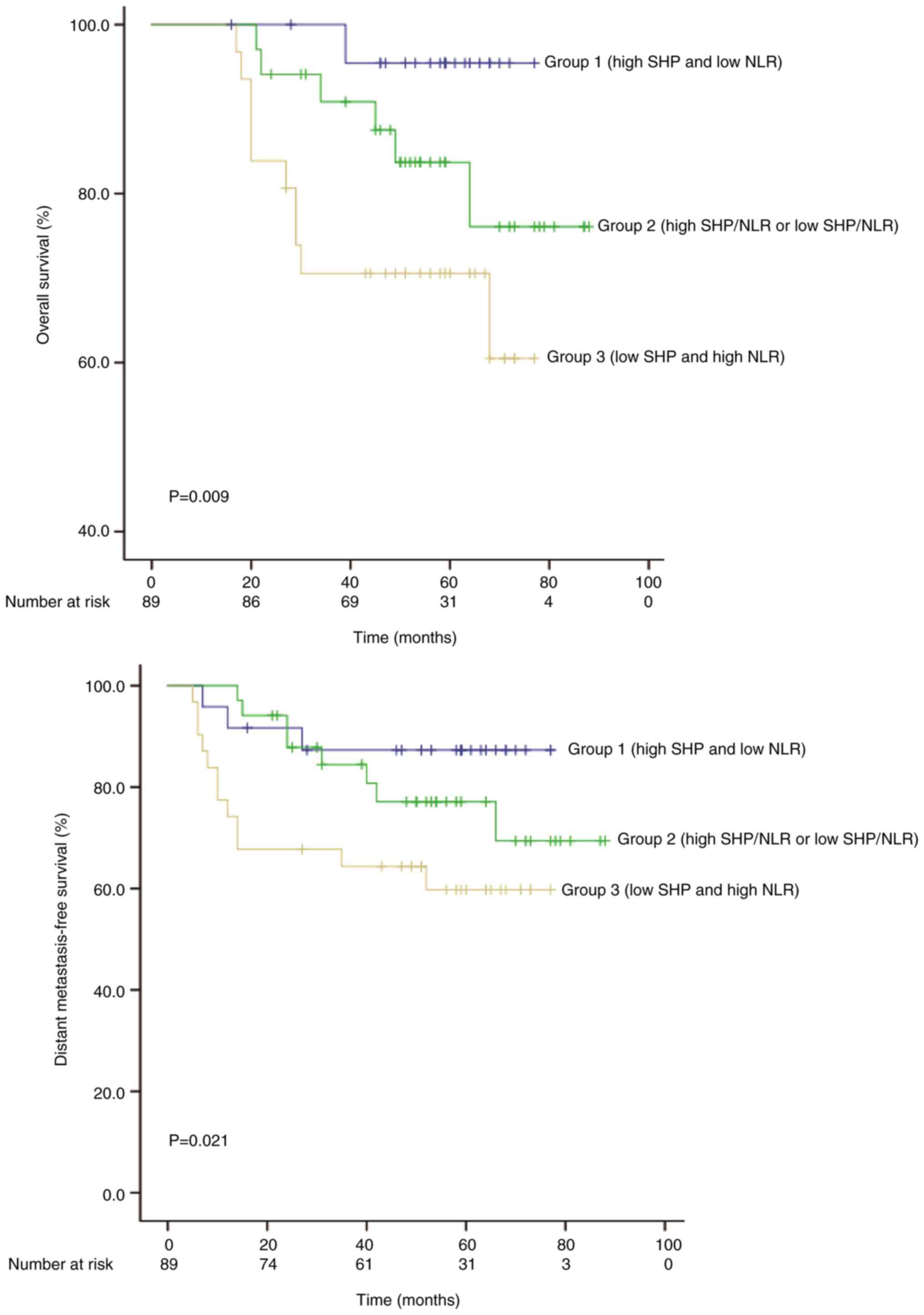
As Table II shows,
significant associations were identified between SHP protein
expression and various hematological parameters. Among the
hematological parameters associated with SHP, the NLR was a
predictor of pCR and OS. Therefore, the 89 cases of READ were
classified into three groups, according to the SHP expression and
NLR: Group 1, high SHP expression and low NLR (n=24); group 2, high
SHP expression and high NLR or low SHP expression and low NLR
(n=34); group 3, low SHP and high NLR (n=31). Kaplan-Meier analysis
(Fig. 4) indicated that patients
with a high NLR and low SHP expression had the shortest OS and
DMSF, and patients with a low NLR and a high SHP expression had the
longest OS and DMFS (P=0.009 and P=0.021, respectively; Fig. 4).
Correlation of SHP expression with
immune cell infiltration and programmed death-ligand 1 (PD-L1)
To investigate the relationship between SHP and
diverse immune infiltrating cells, the correlations between SHP and
immune marker sets of various READ immune cells in the TIMER
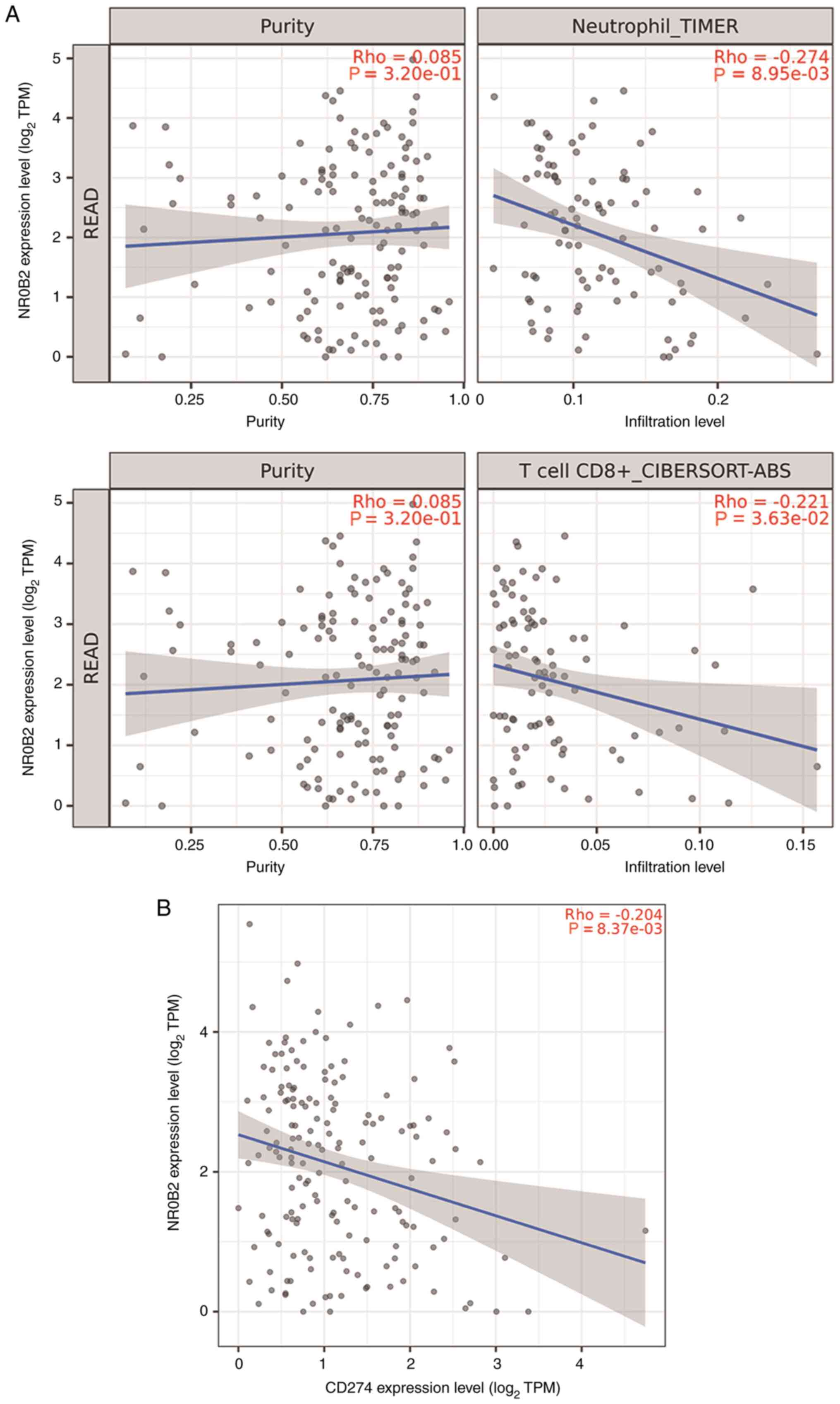
databases were determined. Notably, neutrophil (r=−0.274;
P=0.00895) and CD8+ T cell (r=−0.221; P=0.0363)
infiltration were negatively correlated with SHP in patients with
READ (Fig. 5A). Generally,
CD8+ T-cell infiltration is a favorable prognostic
factor in patients with READ (33).
However, patients with high PD-L1 expression levels and
CD8+ T cell densities have been reported to exhibit
impaired progression-free survival and OS (34). Therefore, the relationship between
SHP expression and the gene expression of immune-suppressive
molecules, including PD-L1 (also known as CD274), indoleamine
2,3-dioxygenase 1 and cytotoxic T-lymphocyte associated protein 4,
was examined in patients with READ. Of the various molecules
tested, a negative correlation was observed between SHP and PD-L1
(r=−0.204, P=0.00837; Fig. 5B).
Discussion
In the present study, a data-driven approach was
used to investigate the role of SHP in READ using GSEA. The results
revealed the following information about the potential underlying
mechanism: A low SHP mRNA expression in READ was associated with
inflammatory responses, as well as NOTCH, IL2-STAT5 and KRAS
signalling. These signaling pathways are associated with treatment
resistance and poor prognosis (35–38). The
clinical significance of SHP expression in READ was also examined
using the OncoLnc database. Kaplan-Meier survival curves
demonstrated that a low SHP mRNA expression is a poor prognostic
factor in READ. Next, the prognostic significance of SHP expression
was investigated in LARC using IHC data from Chungnam National
University Hospital. Analysis of the IHC and clinical data from
this hospital confirmed the results obtained from public databases,
suggesting that SHP protein expression is a favorable prognostic
factor in LARC. Furthermore, the present results showed that the
mRNA and protein expression of SHP is associated with pCR and DMFS
in pRCT-treated LARC patients. These findings suggest that the
nuclear expression of SHP may be used as an indicator of favorable
prognosis for LARC patients who receive pRCT. To date, no clinical
studies have evaluated the prognostic role of SHP in LARC. To the
best of our knowledge, this study is the first to show the
predictive and prognostic significance of SHP expression in
LARC.
Cancer-related local and systemic inflammation has
been identified as a key player in tumor invasion and metastasis
(23,35,39). It
has been demonstrated that several biomarkers and hematological
indices are representative of the local immune response, including
tumor necrosis factor, inflammasomes, cytokines, chemokines and
transcription factors, as well as systemic inflammatory markers,
such as acute-phase proteins and circulating immune cells (23,40).
Among these, the NLR, PLR, LMR, albumin, ESR and CRP levels in
patients with cancer are common prognostic factors, due to their
ease of use in clinical practice (23,40). In
colorectal cancer, the NLR and PLR are associated with pCR or
primary tumor downstaging following pRCT (41–44).
Similarly, the results of the present study revealed that
hematological parameters, including the NLR and PLR, are
significant predictors of pCR following pRCT for rectal cancer. Due
to the detrimental effect of cancer-related inflammation on the
response to radiotherapy, there is substantial interest in
therapeutic strategies for manipulating the inflammatory response.
Thus, there is a growing interest in novel approaches for targeting
cancer-related inflammatory pathways in combination with radiation
therapy. A large variety of natural and synthetic compounds have
been reported to interfere with cancer-related inflammation through
the regulation of various molecular pathways, including NF-κB,
STAT3, hypoxia-inducible factor 1 and prostaglandin-endoperoxide
synthase 2 pathways, and are regarded as putative radiosensitizing
agents (45).
Our previous studies have demonstrated the
importance of SHP in the regulation of innate immune and
inflammatory responses against pathogen invasion (24–27).
However, no study has yet evaluated the role of SHP in cancer
immunology. In the present study, GSEA revealed that inflammatory
responses are differentially enriched in patients with READ and low
SHP expression. Furthermore, a negative association was identified
between SHP expression and the NLR. A previous study reported a
close association of immune suppression with an increased NLR
(46). These results suggest that
low SHP expression may generate an immunosuppressive tumor
microenvironment by promoting cancer-associated inflammation.
Another previous study reported that patients with gastric or
gastro-esophageal junction adenocarcinomas who have a high PD-L1
expression and high CD8+ T cell density have poor
survival, suggesting a potentially adaptive immune resistance
mechanism (34). In this situation,
cancer cells frequently escape immune cell recognition via the
upregulation of immune checkpoint molecules, such as PD-1/PD-L1 and
cytotoxic T-lymphocyte antigen 4 (47,48).
Similarly, despite the negative association between SHP expression
and CD8+ T cell infiltration, the present study observed
that patients with low SHP expression had a poor prognosis. Among
the various molecules studied, a negative association was
identified between SHP and PD-L1. Collectively, these results
suggest that low SHP expression is strongly associated with
immune-suppressive functions via the upregulation of PD-L1,
resulting in poor prognosis in LARC.
It is important to acknowledge the methodological
limitations of the present retrospective study. First, it included
a small number of patients from a single center. Furthermore, 46
LARC cases from the study period were excluded due to the
unavailability of paraffin blocks and only 89 LARC cases were
evaluated for SHP IHC status. Thus, the analysis may have been
subject to potential selection bias. Despite its limitations, the
study is of value because the primary data from the patients were
validated by various bioinformatics approaches.
In conclusion, the present pilot study assessed the
role of SHP expression in rectal cancer. The IHC results suggested
that the expression level of SHP is associated with systemic
inflammation, treatment outcomes and prognosis in LARC. The
bioinformatics analysis results support the observation that low
SHP expression is associated with cancer-related inflammation,
immunosuppression and poor prognosis. Therefore, these findings
provide valuable insights for the identification of potential
therapeutic targets and promising prognostic markers in rectal
cancer.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Professor Hueng-Sik
Choi (School of Biological Sciences and Technology, Chonnam
National University) for critical reading of the manuscript.
Funding
This study was supported by National Research
Foundation of Korea grants funded by the Korean Government
(Ministry of Science, ICT and Future Planning; grant nos.
2017R1A5A2015385 and NRF-2019M3E5D1A02068546) and by a grant from
2019 from Chungnam National University.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SK took part in conception and design of the study
and the acquisition, analysis and interpretation of data, and
drafted the manuscript. EKJ and JMK took part in conception and
design of the study, critical revision of the manuscript for
important intellectual content and supervision of the study. SK,
MJ, MKY, MJC and JSK took part in the acquisition of data and
supervision. SK, EKJ and JMK confirmed the authenticity of all the
raw data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The current study was a retrospective study with
approval from the institutional review board of Chungnam National
University Hospital (IRB no. 2017-07-037) and the contents of this
study are open to the public at the hospital. All methods were
performed in accordance with the 1975 Declaration of Helsinki. All
patients signed an informed consent form.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kuo LJ, Liu MC, Jian JJ, Horng CF, Cheng
TI, Chen CM, Fang WT and Chung YL: Is final TNM staging a predictor
for survival in locally advanced rectal cancer after preoperative
chemoradiation therapy? Ann Surg Oncol. 14:2766–2772. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Seol W, Choi HS and Moore DD: An orphan
nuclear hormone receptor that lacks a DNA binding domain and
heterodimerizes with other receptors. Science. 272:1336–1339. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Seol W, Chung M and Moore DD: Novel
receptor interaction and repression domains in the orphan receptor
SHP. Mol Cell Biol. 17:7126–7131. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Burris TP, Solt LA, Wang Y, Crumbley C,
Banerjee S, Griffett K, Lundasen T, Hughes T and Kojetin DJ:
Nuclear receptors and their selective pharmacologic modulators.
Pharmacol Rev. 65:710–778. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim MK, Chanda D, Lee IK, Choi HS and Park
KG: Targeting orphan nuclear receptor SHP in the treatment of
metabolic diseases. Expert Opin Ther Targets. 14:453–466. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Hagedorn CH and Wang L: Role of
nuclear receptor SHP in metabolism and cancer. Biochim Biophys
Acta. 1812:893–908. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y, Soto J, Park K, Viswanath G,
Kuwada S, Abel ED and Wang L: Nuclear receptor SHP, a death
receptor that targets mitochondria, induces apoptosis and inhibits
tumor growth. Mol Cell Biol. 30:1341–1356. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Y, Xu P, Park K, Choi Y, Moore DD
and Wang L: Orphan receptor small heterodimer partner suppresses
tumorigenesis by modulating cyclin D1 expression and cellular
proliferation. Hepatology. 48:289–298. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He N, Park K, Zhang Y, Huang J, Lu S and
Wang L: Epigenetic inhibition of nuclear receptor small heterodimer
partner is associated with and regulates hepatocellular carcinoma
growth. Gastroenterology. 134:793–802. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song G, Zhang Y and Wang L: MicroRNA-206
targets notch3, activates apoptosis, and inhibits tumor cell
migration and focus formation. J Biol Chem. 284:31921–31927. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Seol W, Hanstein B, Brown M and Moore DD:
Inhibition of estrogen receptor action by the orphan receptor SHP
(short heterodimer partner). Mol Endocrinol. 12:1551–1557. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Johansson L, Thomsen JS, Damdimopoulos AE,
Spyrou G, Gustafsson JA and Treuter E: The orphan nuclear receptor
SHP inhibits agonist-dependent transcriptional activity of estrogen
receptors ERalpha and ERbeta. J Biol Chem. 274:345–353. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lai K, Harnish DC and Evans MJ: Estrogen
receptor alpha regulates expression of the orphan receptor small
heterodimer partner. J Biol Chem. 278:36418–36429. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nishizawa H, Yamagata K, Shimomura I,
Takahashi M, Kuriyama H, Kishida K, Hotta K, Nagaretani H, Maeda N,
Matsuda M, et al: Small heterodimer partner, an orphan nuclear
receptor, augments peroxisome proliferator-activated receptor gamma
transactivation. J Biol Chem. 277:1586–1592. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rubin GL, Duong JH, Clyne CD, Speed CJ,
Murata Y, Gong C and Simpson ER: Ligands for the peroxisomal
proliferator-activated receptor gamma and the retinoid X receptor
inhibit aromatase cytochrome P450 (CYP19) expression mediated by
promoter II in human breast adipose. Endocrinology. 143:2863–2871.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Swales KE, Korbonits M, Carpenter R, Walsh
DT, Warner TD and Bishop-Bailey D: The farnesoid X receptor is
expressed in breast cancer and regulates apoptosis and aromatase
expression. Cancer Res. 66:10120–10126. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Woo HD, Kim K and Kim J: Association
between preoperative C-reactive protein level and colorectal cancer
survival: A meta-analysis. Cancer Causes Control. 26:1661–1670.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Choi N, Kim JH, Chie EK, Gim J and Kang
HC: A meta-analysis of the impact of neutrophil-to-lymphocyte ratio
on treatment outcomes after radiotherapy for solid tumors. Medicine
(Baltimore). 98:e153692019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song W, Wang K, Zhang RJ and Zou SB:
Prognostic value of the lymphocyte monocyte ratio in patients with
colorectal cancer: A meta-analysis. Medicine (Baltimore).
95:e55402016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tan D, Fu Y, Su Q and Wang H: Prognostic
role of platelet-lymphocyte ratio in colorectal cancer: A
systematic review and meta-analysis. Medicine (Baltimore).
95:e38372016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Park JH, Richards CH, McMillan DC, Horgan
PG and Roxburgh CSD: The relationship between tumour stroma
percentage, the tumour microenvironment and survival in patients
with primary operable colorectal cancer. Ann Oncol. 25:644–651.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Diakos CI, Charles KA, McMillan DC and
Clarke SJ: Cancer-related inflammation and treatment effectiveness.
Lancet Oncol. 15:e493–e503. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuk JM, Jin HS and Jo EK: Small
heterodimer partner and innate immune regulation. Endocrinol Metab
(Seoul). 31:17–24. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yuk JM, Shin DM, Lee HM, Kim JJ, Kim SW,
Jin HS, Yang CS, Park KA, Chanda D, Kim DK, et al: The orphan
nuclear receptor SHP acts as a negative regulator in inflammatory
signaling triggered by Toll-like receptors. Nat Immunol.
12:742–751. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang CS, Yuk JM, Kim JJ, Hwang JH, Lee CH,
Kim JM, Oh GT, Choi HS and Jo EK: Small heterodimer
partner-targeting therapy inhibits systemic inflammatory responses
through mitochondrial uncoupling protein 2. PLoS One. 8:e634352013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang CS, Kim JJ, Kim TS, Lee PY, Kim SY,
Lee HM, Shin DM, Nguyen LT, Lee MS, Jin HS, et al: Small
heterodimer partner interacts with NLRP3 and negatively regulates
activation of the NLRP3 inflammasome. Nat Commun. 6:61152015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Eun HS, Cho SY, Lee BS, Kim S, Song IS,
Chun K, Oh CH, Yeo MK, Kim SH and Kim KH: Cytochrome P450 4A11
expression in tumor cells: A favorable prognostic factor for
hepatocellular carcinoma patients. J Gastroenterol Hepatol.
34:224–233. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Snipstad K, Fenton CG, Kjaeve J, Cui G,
Anderssen E and Paulssen RH: New specific molecular targets for
radio-chemotherapy of rectal cancer. Mol Oncol. 4:52–64. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Weiser MR: AJCC 8th edition: Colorectal
cancer. Ann Surg Oncol. 25:1454–1455. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jochems C and Schlom J: Tumor-infiltrating
immune cells and prognosis: The potential link between conventional
cancer therapy and immunity. Exp Biol Med (Maywood). 236:567–579.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Thompson ED, Zahurak M, Murphy A, Cornish
T, Cuka N, Abdelfatah E, Yang S, Duncan M, Ahuja N, Taube JM, et
al: Patterns of PD-L1 expression and CD8 T cell infiltration in
gastric adenocarcinomas and associated immune stroma. Gut.
66:794–801. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Roxburgh CS and McMillan DC: Cancer and
systemic inflammation: Treat the tumour and treat the host. Br J
Cancer. 110:1409–1412. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tyagi A, Sharma AK and Damodaran C: A
review on notch signaling and colorectal cancer. Cells. 9:15492020.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen S-Y, Chen S, Feng W, Li Z, Luo Y and
Zhu X: A STING-related prognostic score predicts high-risk patients
of colorectal cancer and provides insights into immunotherapy. Ann
Transl Med. 9:142021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Arrington AK, Heinrich EL, Lee W, Duldulao
M, Patel S, Sanchez J, Garcia-Aguilar J and Kim J: Prognostic and
predictive roles of KRAS mutation in colorectal cancer. Int J Mol
Sci. 13:12153–12168. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Grivennikov SI, Greten FR and Karin M:
Immunity, inflammation, and cancer. Cell. 140:883–899. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hannisdal E, Tveit KM, Theodorsen L and
Høst H: Host markers and prognosis in recurrent rectal carcinomas
treated with radiotherapy. Acta Oncol. 33:415–421. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xiao B, Peng J, Zhang R, Xu J, Wang Y,
Fang Y, Lin J, Pan Z and Wu X: Density of CD8+ lymphocytes in
biopsy samples combined with the circulating lymphocyte ratio
predicts pathologic complete response to chemoradiotherapy for
rectal cancer. Cancer Manag Res. 9:701–708. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hodek M, Sirák I, Ferko A, Örhalmi J,
Hovorková E, Hadži Nikolov D, Paluska P, Kopecký J, Petera J and
Vošmik M: Neoadjuvant chemoradiotherapy of rectal carcinoma:
Baseline hematologic parameters influencing outcomes. Strahlenther
Onkol. 192:632–640. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kim IY, You SH and Kim YW:
Neutrophil-lymphocyte ratio predicts pathologic tumor response and
survival after preoperative chemoradiation for rectal cancer. BMC
Surg. 14:942014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lee JH, Song C, Kang SB, Lee HS, Lee KW
and Kim JS: Predicting pathological complete regression with
haematological markers during neoadjuvant chemoradiotherapy for
locally advanced rectal cancer. Anticancer Res. 38:6905–6910. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Multhoff G and Radons J: Radiation,
inflammation, and immune responses in cancer. Front Oncol.
2:582012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gonda K, Shibata M, Sato Y, Washio M,
Takeshita H, Shigeta H, Ogura M, Oka S and Sakuramoto S: Elevated
neutrophil-to-lymphocyte ratio is associated with nutritional
impairment, immune suppression, resistance to S-1 plus cisplatin,
and poor prognosis in patients with stage IV gastric cancer. Mol
Clin Oncol. 7:1073–1078. 2017.PubMed/NCBI
|
|
47
|
Topalian SL, Drake CG and Pardoll DM:
Immune checkpoint blockade: A common denominator approach to cancer
therapy. Cancer Cell. 27:450–461. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
McGranahan N, Furness AJ, Rosenthal R,
Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak
NJ, Hiley CT, et al: Clonal neoantigens elicit T cell
immunoreactivity and sensitivity to immune checkpoint blockade.
Science. 351:1463–1469. 2016. View Article : Google Scholar : PubMed/NCBI
|