Introduction
Long noncoding RNAs (lncRNAs) have been recognized
as important functional agents of the genome, containing intricate
structural and informational capacity (1–3).
Although these transcripts have little or no protein-coding
capability, lncRNAs regulate gene expression through multiple
mechanisms including chromatin modification and both
transcriptional and post-transcriptional regulation (1–3).
Chromosome 8q24, a large gene desert known for its paucity of
coding regions contains the lncRNA Colon Cancer Associated
Transcript 2 (CCAT2), located in close proximity to the MYC
oncogene (2,4–6). LncRNA
CCAT2 was initially recognized to have oncogenic effects in
colorectal cancer, and has since been linked to various cancers,
including ovarian cancer (2,4–12). CCAT2
harbors the rs6983267: NR_109834.1:n.662G>T SNP and has been
shown to interact near the MYC promoter and regulate gene
expression, mediating tumor metastasis and growth in colon, lung,
breast, prostate, endometrial and gastric cancers (2,4–9).
Ovarian cancer continues to be one of the leading
causes of death from gynecological cancers accounting for over
200,000 deaths per year worldwide (13). Thus, there have been multiple efforts
to discover novel biomarkers and therapeutic targets, including
lncRNAs, for this cancer. The rs6983267 SNP within CCAT2 has such
potential due to its allele-specific features which are
hypothesized to affect MYC expression (2,4–6,8–10,14,15).
Ghoussaini et al were the first to associate the 8q24.21.a
locus with increased risk of ovarian cancer (12). Further investigation by Huang et
al found that CCAT2 expression was significantly higher in
ovarian cancer tissue with high CCAT2 gene expression correlating
to poor prognostic parameters and shorter overall and disease-free
survival (7). Other studies revealed
similar results, showing that CCAT2 expression was upregulated in
ovarian cancer cells, and knock down of CCAT2 expression in
vitro significantly repressed proliferation and promoted
apoptosis in certain cell lines (11). Specifically, the G allele of
rs6983267 was shown to significantly increase a women's risk of
ovarian cancer (10).
Although there has been a growing body of evidence
linking ovarian cancer risk with CCAT2 expression, there is little
information about the association of the rs6983267 risk allele and
clinical outcomes in ovarian cancer patients. The aim of this study
is to assess the association between genotypes at the rs6983267
locus (within lncRNA CCAT2) and clinical outcomes in patients with
high-grade ovarian cancer (HGSOC), including survival and response
to chemotherapy.
Materials and methods
A retrospective genetic association study was
conducted using genomic DNA from ninety-eight patients with HGSOC
at the University of Iowa Hospitals and Clinics. The genomic DNA
originated from flash frozen tumor tissues stored in the Department
of Obstetrics and Gynecology Gynecologic Oncology Bank (IRB,
ID#200209010 and ID#201804817) which is part of the Women's Health
Tissue Repository (IRB, ID#201809807). All tissues archived in the
Women's Health Tissue Repository were originally obtained from
adult patients under written informed consent in accordance with
the University of Iowa IRB guidelines. Genomic DNAs were purified
from frozen tumor tissues using the DNeasy Blood and Tissue Kit
according to the manufacturer's (Qiagen) recommendations.
Clinical data
Clinical and pathological data were collected from
the electronic medical record. Clinical variables previously
observed to be associated with chemo-response were included in the
data collection (16). Only baseline
clinical and pathological characteristics which can be obtained
before starting initial chemotherapy were included.
Genotyping
The genomic region around rs6983267 was amplified
via standard PCR on a BioRad T-100 thermal cycler using primers
detailed in Fig. S1. The reagents
used in each PCR sample included: 3 µl 10X reaction buffer with 1.5
mM MgCl2, 1 µl 10 mM dNTPs, 1 µl (10 pmole) of each
primer, and 0.5 µl Taq polymerase (manufactured by New England
BioLabs) with a final reaction volume of 30 µl. The thermal cycler
was programmed for five minutes at 95°C for initial denaturation,
followed by 35 cycles of 30 sec at 95°C for denaturation, 30 sec at
55°C for annealing, 30 sec at 72°C for extension, and seven minutes
at 72°C for the final extension. The PCR amplicon was purified
using the QIAGEN QIAquick PCR amplification kit. The purified
amplicons were sequenced using conventional Sanger sequencing
carried out on an Applied Biosystems Model 3730×l capillary
sequencer in the Genome Facility at the University of Iowa
Institute of Human Genomics. The results provided the genotype (GG,
GT, TT) of each patient for the rs6983267 SNP.
Association with rs6983267
genotypes
Univariate logistic regression was used to explore
the association between the clinical outcomes and biological
variables and the rs6983267 genotypes (genotypes containing any G
allele and the TT genotype). This was performed to assess
advantageous characteristics in HGSOC survival. Clinical outcomes
analyzed included: Age, body mass index (BMI), Charlson
Co-morbidity Index, pre-operative CA-125, cancer stage, disease in
the upper abdomen by imaging (other than the omentum), disease in
chest by imaging, tumor grade, residual disease after surgery,
removal of pelvic lymph nodes, removal of para-aortic lymph nodes,
neoadjuvant chemotherapy, number of chemotherapy cycles delivered,
dose dense chemotherapy, and death by disease. Biological variables
included: CCAT2, MYC, MYCL, MYCN, MYCBP (MYC Binding Protein),
MYCBP2 (MYC Binding Protein 2), MYCBP2-AS1 (MYC Binding Protein 2
Antisense RNA 1), MYCBPAP (MYC Binding Protein Associated Protein),
MYCNUT (MYCN Upstream Transcript), MYCNOS (MYCN Opposite Strand),
and MYCT1 (MYC Target 1). Variables in the univariable analysis
with a p-value <0.10 were introduced in the multivariate
logistic regression model. This P-value was used to create a more
inclusive multivariate model (17).
The multivariable logistic regression model was used to assess
independent association of clinical and biological variables with
the rs6983267 genotypes (genotypes containing any G allele and the
TT genotype). Variables with a P-value <0.05 in the multivariate
analysis were considered significant.
Survival analysis
Survival analysis for patients with the different
genotypes at the rs6983267 SNP (GG, GT, TT) and MYC and CCAT2
expressions was performed using the Cox proportional hazard model.
Survival analysis was performed using a log-rank test for a model
with three genotypes (GG, GT and TT), and for a model comparing the
homozygous TT genotype with genotypes containing the most frequent
allele, G (GG or GT). Survival assessment of clinical variables
(age, BMI, tumor grade, FIGO stage, pre-operative CA-125,
neoadjuvant chemotherapy, residual disease after surgery, and
optimal surgery) also were performed using the Cox proportional
hazard ratio (HR). Clinical and biological variables associated in
the univariate analysis with a P≤0.10, were introduced in a Cox
Proportional hazard ratio multivariable model (17). Proportional hazards assumptions were
assessed for the final survival model. Variables with a P-value
<0.05 in the multivariate Cox model were considered
significant.
Gene expression of the biological variables listed
above was determined from previous RNA sequencing experiments using
the same patients (GEO accession number GSE156699) (18,19).
Power calculation
With 98 samples, and SNP (rs6983267) frequencies of
69% for GG/GT and 31% for TT, our study had a power of 79% to find
differences in survival of >30% at 5 years when comparing
between genotypes, with an α error of 0.05. Statistical analysis,
power calculations and graphics were performed with R statistical
package and computer environment (20). R packages survival, stats, and
survcomp were used for the statistical analyses (21).
Statistical analysis. A univariate logistic
regression was used to assess the association between the clinical
and biological variables and the rs6983267 genotypes. Variables
from this analysis (P≤0.1) were introduced into a multivariate
logistic regression to assess independent association of clinical
and biological variables with the rs6983267 genotypes. A log-rank
test was used to analyze survival for a model assessing the
rs6983267 genotypes (GG, GT, and TT) and for a model assessing the
homozygous TT genotype and genotypes containing the most frequent
allele, G (GG and GT). Survival assessment of clinical variables
was performed using the Cox Proportional hazard ratio. Significant
variables from this analysis were introduced into a multivariable
Cox Proportional hazard ratio model.
Results
Association with rs6983267
genotype
To assess which characteristics were associated with
the rs6983267 genotype that showed advantages in HGSOC survival
(TT), we performed univariate logistic regression analyses
(Table I). Significant clinical and
biological variables associated with the TT genotype were
introduced into the multivariate logistic regression model and
included: Age (OR=0.95, 95% CI 0.92–0.99, P=0.010), pre-operative
CA-125 (OR=1.00, 95% CI 1.001–1.002, P=0.042), cancer stage
(OR=2.49, 95% CI 1.08–6.15, P=0.040), and response to chemotherapy
(OR=2.91, 95% CI 1.03–9.17, P=0.052). In the multivariate logistic
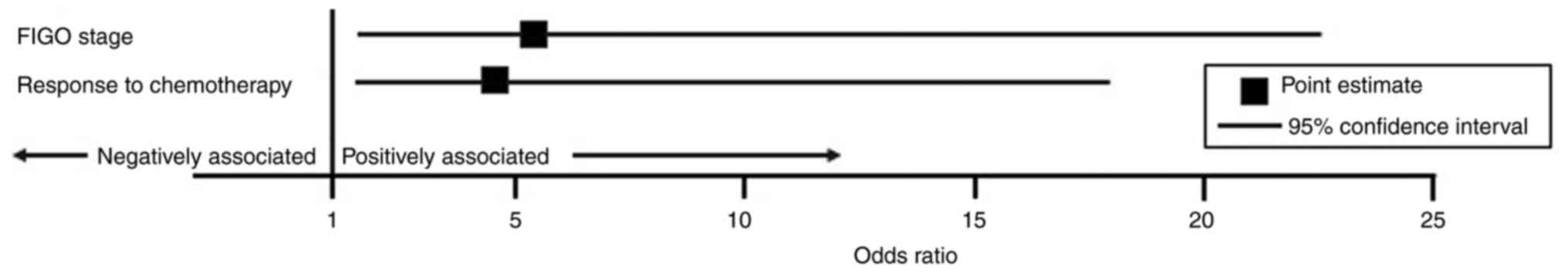
regression model (Fig. 1; Table II) TT genotype was independently
associated with FIGO stage and response to chemotherapy. HGSOC
patients with a TT genotype at the rs6983267 locus were over five
times more likely to have a higher FIGO stage (OR=5.34, 95% CI
1.50–22.62, P=0.014) and were four times more likely to respond to
chemotherapy (OR=4.51, 95% CI 1.40–18.00, P=0.018) compared to
individuals with a GG or GT genotype.
 | Table I.Table of patient characteristics
analyzed using univariate logistic regression to demonstrate
association with the TT genotype. |
Table I.
Table of patient characteristics
analyzed using univariate logistic regression to demonstrate
association with the TT genotype.
| Variable | G-allele (GG, TG)
N=68 | TT genotype,
N=30 | OR (95% CI) | P-value |
|---|
| Mean age,
years | 63 | 55 | 0.95
(0.92–0.99) | 0.010a |
| Mean body mass
index, kg/m2 | 27.1 | 25.7 | 0.97
(0.89–1.04) | 0.427 |
| Charlson
comorbidity index |
|
|
|
|
|
Low | 6 | 5 |
|
|
|
Medium | 40 | 15 | 0.68
(0.20–2.29) | 0.536 |
|
High | 6 | 2 | 0.60
(0.07–3.90) | 0.605 |
| Mean pre-operative
CA-125 | 1674.14 | 4719.79 | 1.00
(1.001–1.002) | 0.042a |
| FIGO stage |
|
|
|
|
| Stage
I–II | 4 | 0 | 2.49
(1.08–6.15) | 0.040a |
| Stage
III | 50 | 18 |
|
|
| Stage
IV | 14 | 11 |
|
|
| Disease in upper
abdomen by imaging (other than omentum) |
|
|
|
|
| Large
bowel | 2 | 1 | 1.00
(0.42–2.50) | 0.992 |
|
Spleen | 0 | 0 |
|
|
|
Portahepatis | 2 | 2 |
|
|
|
Mesenteric | 3 | 0 |
|
|
| Disease in chest by
imaging | 0 | 6 | 1.2×108
(8.3×10−6-NA) | 0.991 |
| Grade |
|
|
|
|
| 1 | 0 | 0 | 1.22
(0.44–3.79) | 0.712 |
| 2 | 16 | 6 |
|
|
| 3 | 48 | 22 |
|
|
| Residual disease
after surgery |
|
|
|
|
|
Microscopic | 13 | 3 | 0.47
(0.01-1,61) | 0.269 |
|
Macroscopic | 55 | 27 |
|
|
|
Optimal | 46 | 16 | 0.55
(0.23–1.32) | 0.178 |
|
Suboptimal | 22 | 14 |
|
|
| Removal of pelvic
lymph nodes | 13 | 2 | 0.30
(0.05–1.19) | 0.132 |
| Removal of
para-aortic lymph nodes | 8 | 1 | 0.26
(0.14–1.51) | 0.212 |
| Neoadjuvant
chemotherapy | 9 | 5 | 1.38 (0.39,
4.44) | 0.600 |
| Response to
chemotherapy |
|
|
|
|
|
Responders | 25 | 19 | 2.91
(1.03–9.17) | 0.052a |
|
Non-responders | 23 | 6 |
|
|
| Number of cycles
delivered |
|
|
|
|
| <6
cycles | 12 | 2 | 1.05
(0.80–1.37) | 0.686 |
| >6
cycles | 54 | 26 |
|
|
| Dose dense
chemotherapy | 1 | 2 | 4.79 | 0.209 |
| Death by
disease | 55 | 24 | 1.45
(0.44–105.5) | 0.594 |
| CCAT2b | 1.96 | 2.15 | 1.10
(0.81–1.49) | 0.552 |
| MYCBPb | 3.34 | 3.32 | 1.1×1014
(NA-4.2×10174) | 0.991 |
| MYCLb | 8.83 | 9.02 | 1.08
(0.82–1.49) | 0.575 |
| MYCBP2b | 12.12 | 12.03 | 0.79
(0.39–1.58) | 0.509 |
|
MYCBP2-AS1b | 3.40 | 3.44 | 2.08
(0.38–11.12) | 0.376 |
|
MYCBPAPb | 4.90 | 4.68 | 0.87
(0.61–1.23) | 0.441 |
| MYCNUTb | 3.32 | 3.34 | 2.96×108
(NA-2.3×10−179) | 0.991 |
| MYCNOSb | 3.51 | 3.47 | 0.84
(0.26–2.17) | 0.739 |
| MYCNb | 6.91 | 7.58 | 1.22
(0.96–1.56) | 0.109 |
| MYCT1b | 5.61 | 5.38 | 0.88
(0.62–1.22) | 0.451 |
| MYCb | 10.63 | 11.01 | 1.27
(0.90–1.84) | 0.189 |
 | Table II.Multivariate logistic regression of
patients with the TT genotype compared with patients with genotypes
containing any G allele. |
Table II.
Multivariate logistic regression of
patients with the TT genotype compared with patients with genotypes
containing any G allele.
| Variable | Odds ratio | 95% confidence
interval | P-value |
|---|
| Age, years | 0.99 | 0.95–1.05 | 0.853 |
| Pre-operative
CA-125 | 1.00 | 0.999–1.0002 | 0.145 |
| FIGO stage | 5.34 | 1.50–22.62 | 0.014a |
| Response to
chemotherapy | 4.51 | 1.40–18.00 | 0.018a |
Survival analysis
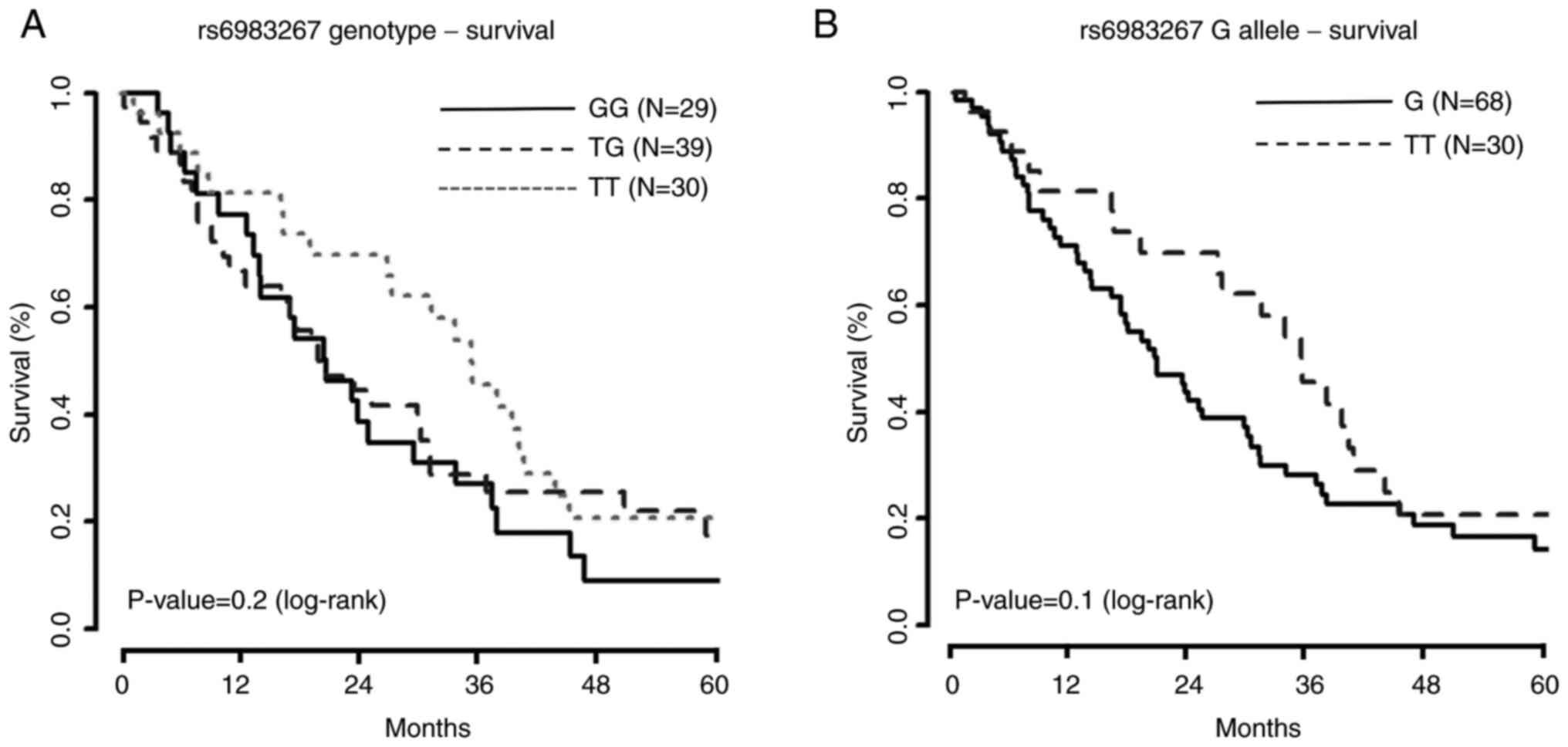
Analysis comparing survival among the three
genotypes (GG, GT, and TT) revealed no difference in survival
(P=0.20). When comparing survival curves between patients with G
allele presence (GG or GT) and patients with the TT genotype, there
was stronger evidence for differences in survival (P=0.10), even
after applying several weighted Kaplan-Meier tests due to
late-crossing survival curves (Fig.
2; Table SI). The results of
univariate survival analysis for clinical and biological variables
are summarized in Table III. The
majority of patient deaths were attributed to HGSOC (84%, 46 out of
55).
 | Table III.Univariate survival analysis of the
effect of variables on survival time in patients with high-grade
serous ovarian cancer. |
Table III.
Univariate survival analysis of the
effect of variables on survival time in patients with high-grade
serous ovarian cancer.
| Variable
(Ref.) | HR | 95% confidence
interval | P-value |
|---|
| rs6983267 (Ref: G
presence) | 0.665 | 0.40–1.10 | 0.100a |
| Age, years | 1.029 | 1.02–1.04 |
<0.001a |
| Body mass index,
kg/m2 | 0.997 | 0.98–1.02 | 0.698 |
| Grade | 0.821 | 0.61–1.10 | 0.191 |
| FIGO stage | 1.366 | 1.06–1.76 | 0.015a |
| Pre-operative
CA-125 | 1.000 | 1.00–1.00 | 0.680 |
| Neoadjuvant
chemotherapy (ref: no) | 2.249 | 1.50–3.38 |
<0.001a |
| Residual disease
(ref: micro) | 2.056 | 1.48–2.85 |
<0.001a |
| Optimal surgery
(ref: yes) | 1.586 | 1.26–2.00 |
<0.001a |
| Colon
cancer-associated transcript 2 | 1.000 | 0.84–1.19 | 0.997 |
| MYC | 1.091 | 0.89–1.33 | 0.391 |
Based on the univariate survival analysis, the
following variables were introduced into the multivariate Cox
proportional hazard ratio model: Genotype TT at the rs6983267 locus
(reference: G allele) (HR=0.665, 95% CI 0.40–1.10, P=0.100), age
(HR=1.029, 95% CI 1.02–1.04, P<0.001), FIGO stage (HR=1.366, 95%
CI 1.06–1.76, P=0.015), neoadjuvant chemotherapy (reference: No
neoadjuvant chemotherapy) (HR=2.249, 95% CI 1.50–3.38, P<0.001),
residual disease (reference: Microscopic disease) (HR=2.056, 95% CI
1.48–2.85, P<0.001), and optimal surgery (reference: Yes)
(HR=1.586, 95% CI 1.26–2.00, P<0.001). The multivariate survival
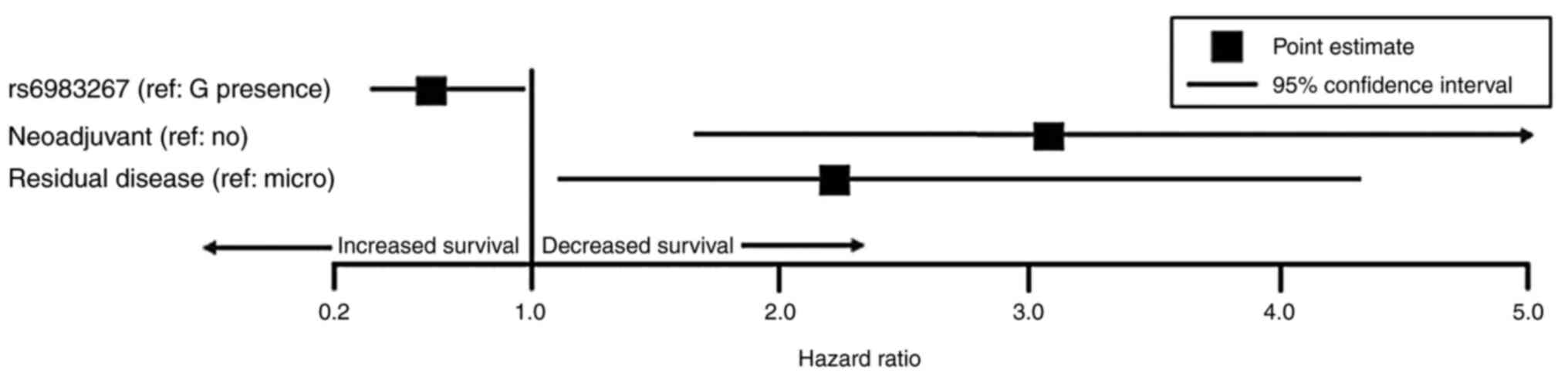
analysis (Fig. 3; Table IV) demonstrated that patients with
the TT genotype had improved survival time compared to patients
with genotypes containing any G allele (HR=0.59, 95% CI 0.36–0.97,
P=0.039;), even after accounting for other significant co-variates,
like neoadjuvant chemotherapy (HR=3.06, 95% CI 1.64–5.69, P=0.039)
and residual disease after surgery (HR=2.21, 95% CI 1.13–4.33,
P=0.021).
 | Table IV.Multivariable Cox proportional hazard
ratio analysis of the significant variables. |
Table IV.
Multivariable Cox proportional hazard
ratio analysis of the significant variables.
| Variable
(Ref.) | Hazard ratio | 95% confidence
interval | P-value |
|---|
| rs6983267 (ref: G
presence) | 0.59 | 0.36, 0.97 | 0.039a |
| Age, years | 1.02 | 0.999, 1.04 | 0.057 |
| FIGO stage | 1.09 | 0.66, 1.80 | 0.747 |
| Neoadjuvant
chemotherapy (ref: no) | 3.06 | 1.64, 5.69 |
<0.001a |
| Residual disease
(ref: micro) | 2.21 | 1.13, 4.33 | 0.021a |
| Optimal surgery
(ref: yes) | 0.73 | 0.44, 1.20 | 0.215 |
Discussion
In this study we showed that HGSOC patients with the
TT genotype at the rs6983267 locus had improved survival time when
compared to HGSOC patients with any G allele (GG and GT genotypes).
This is despite patients with the TT genotype having a higher FIGO
stage. A possible explanation for this outcome is patients with the
TT genotype responded better to initial standard chemotherapy. The
mechanism by which this occurs is unclear; however, an example of
this phenomenon can be seen in BRCA and homologous repair deficient
(HRD) ovarian cancer patients. In epithelial ovarian cancer, the
strongest known genetic risk factor is BRCA1 and BRCA2 germline
mutations, which account for ~10% of cases (22,23). Yet
BRCA-associated epithelial ovarian cancer patients have been shown
to have greater 5-year survival when compared to sporadic mutations
(22,23). This has been attributed to improved
response to chemotherapy due to a process coined ‘synthetic
lethality’ in which simultaneous impairment of two DNA repair
pathways leads to cytotoxicity and cell death (23–25).
PARP inhibitors, which block the repair of DNA single-strand
breaks, have been shown to be 100–1,000 times more effective in
cells deficient in BRCA1 or BRCA2 (25). This same response has been
demonstrated in HGSOC patients who exhibit aberrations in other
homologous recombination repair genes (24–26).
Further studies to assess the mechanisms and pathophysiology
specific to the role of lncRNA CCAT2 in ovarian cancer may be able
to characterize other treatments or maintenance therapies that have
greater efficacy based on the rs6983267 genotype.
Ghoussaini et al associated two other SNPs
within 8q24 with ovarian cancer, rs10505477 and rs10808556, that
may also warrant further investigation (12). SNP rs10505477 is located within long
non-coding RNA Cancer Susceptibility Candidate 8 (CASC8), which is
in the chromosome 8q24 locus (27).
Several studies have looked at clinical outcomes in patients with
gastric and lung cancer in relation to SNP rs10505477. In patients
with gastric cancer undergoing cisplatin chemotherapy, GA and AA
genotypes of rs10505477 were correlated with poorer overall
survival, compared with the GG genotype (28). Similarly, the rs10505477 and
rs6983267 polymorphisms have been shown to respond to
platinum-based chemotherapy in lung cancer (29,30). In
addition, several studies have suggested a strong linkage
disequilibrium between the SNP rs10505477 and the SNP rs6983267
(27,31). Wu et al analyzed the
association between CCAT2 and CASC8 polymorphisms, suggesting
effects from both variants play a role in hepatocellular carcinoma
risk (32). It may be the case that
several polymorphisms contribute to the development of ovarian
cancer, treatment response, and overall survival. The collective
effects of several polymorphisms within 8q24 have yet to be
addressed in ovarian cancer and may provide interesting
insight.
There are limited studies regarding SNP rs10808556.
Tong et al conducted a meta-analysis evaluating twenty-eight
variants in 8q24 and their association with cancer risk. They found
that rs10808556 was significantly associated with colorectal cancer
risk (33). Another study assessed
the relationship between rs10808556 and thyroid carcinoma risk;
however, findings did not suggest an association (34). Similar to rs6983267, numerous studies
involving SNPs within 8q24 focus on cancer susceptibility. Our
study highlights the need to not only look at the role that SNPs
play in the onset of disease but also the response to therapeutic
interventions and therefore prognosis.
Studies involving 8q24 often revolve around MYC
activity in relation to cancer risk-associated SNPs. While these
SNPs have been associated with increased cancer risk, there have
been uncertainties regarding MYC's role behind this observation.
Goode et al analyzed common variants at 8q24 in relation to
ovarian cancer and noted significant SNPs for ovarian cancer were
located in a gene desert relatively far from the 3′ end of MYC
(35). SNP rs6983267, for instance,
is located 335 kb from MYC, its closest gene (5,9,36–39). It
was suggested that MYC may not be the target gene for ovarian
cancer or that these polymorphisms were capable of influencing MYC
from a distance (35). However, in
colorectal and prostate cancer tissues, evidence revealing
long-range physical interaction between rs6983267 and MYC was
discovered (36,40). Furthermore, risk loci within 8q24
appeared to act in a tissue-dependent manner such that the risk
loci associated with prostate cancer, for instance, interacted with
MYC in prostate cancer cells but not breast or colon cancer cells
(39). It was concluded that
rs6983267, along with other risk loci within 8q24, likely acts as a
tissue-specific cis-regulatory enhancer element, leading to
increased expression of MYC (36,39). It
has been proposed that the mechanism behind this is similar to what
is seen with the Colon Cancer Associated Transcript 1 locus (CCAT1)
located 515 kb upstream from MYC within a colorectal cancer
super-enhancer. CCAT1 encodes two lncRNAs: Colon Cancer Associated
Transcript 1 long isoform (CCAT1-L) and Colon Cancer Associated
Transcript 1 short isoform (CCAT1-S) (6). LncRNAs CCAT1-L and CCAT1-S facilitate
the formation of chromatin looping, which allow for MYC interaction
with its enhancers (6,37). In fact, interaction between various
cancer risk variants within 8q24 and the MYC oncogene via chromatin
looping has been observed in multiple studies (5,37,39). In
addition, chromatin looping allows the lncRNAs to accumulate around
the MYC locus and carry out their role in MYC regulation (6).
MYC expression is regulated through the binding of
Wnt proteins to their receptors on the cell surface (39). This results in a signaling pathway
that stabilizes β-catenin and allows it to enter the nucleus to
bind to the TCF4 transcription factor (5,15,39).
Wright et el assessed the rs6983267 risk allele's effect on
chromatin loop formation in order to better elucidate whether
increased MYC expression was a result of alterations in loop
formation versus interactions with the TCF4 transcription factor.
It was shown that the loop does not alter in frequency of formation
or interactions in response to the rs6983267 SNP, and that the loop
exists regardless of which genotype is present. This suggested that
increased MYC activity is a result of increased TCF4 recruitment
and not by altered loop formation (37). Additionally, affinity for the TCF4
transcription factor was found to be higher for the G allele of
rs6983267 than the T allele (9,38).
Several studies have aimed to elucidate the
pathophysiology of the effect of lncRNA CCAT2 on MYC expression. It
has been proposed that lncRNA CCAT2, transcribed from the MYC-335
enhancer region involving the rs6983267 site, associates with TCF4
to augment its transcription activity, though the specific
mechanism is unknown. Binding of CCAT2 may alter protein structure
or modify the association of TCF4 and its partners within the
transcription complex in an allosteric manner. This in turn leads
to increased Wnt and MYC activity (6,15).
Alternatively, given that the G allele appears to increase
transcription of CCAT2 compared to the T allele in colorectal
cancer, it has been suggested that G, the risk allele of rs6983267,
changes the property of the final CCAT2 transcript, ultimately
influencing its binding capacity to TCF4 (4,6).
Our study supports the notion that increased MYC
expression is responsible for unregulated growth in ovarian cancer;
however, our results did not show a significant genotype-dependent
increase in MYC expression. In fact, direct evidence consistently
linking rs6983267 alleles to level of MYC expression have not been
demonstrated (5,9,36,37,39,40).
Several reasons have been proposed, including differential
expression of MYC among the different cell types that comprise an
organ. For example, MYC may be expressed at different levels in
epithelial cells, germ cells, and stromal cells within the ovary
(39). Another reason for this
discordance may be the inadequacies of our current technology to
pick up subtle differences in MYC transcription (5). Further, this association may not be
detected due to timing of risk elevation in relation to
presentation of clinical disease, which occurs earlier in the
disease course (5,36,40).
This was also described by Wasserman et al who analyzed the
cancer risk allele in prostate cancer (40). This study demonstrated that
allele-specific enhancer activity may be more active early in
development before tumorigenesis occurs (40). Capturing a protein level at a single
time point, in other words, may not correctly reflect the gene's
role in tumorigenesis. Similarly, obtaining tissue samples later in
the disease course, as was the case in our study, may not
accurately reflect the differences in MYC activity among different
genotypes, especially since our population consisted of patients
who had known ovarian cancer. In addition, the prior mentioned
studies focus on allele-specific MYC expression in relation to
cancer risk. Future studies assessing MYC expression after a
patient has been diagnosed with cancer in regard to clinical course
and response to treatment may prove to be beneficial.
Although differences in allele-specific MYC
transcription remain unclear, previous studies have suggested that
the T allele had a 2-fold increase in MYC transcription despite
extensive evidence associating the G allele with increased cancer
risk (4,6,7,10,11,15).
These seemingly contradictory findings were addressed by Sotelo
et al who connected MYC to a phenomenon called ‘intrinsic
tumor suppression’ (15). This
phenomenon, first proposed by Lowe et al, describes the
tight coupling of cell proliferation and cell death. In normally
functioning cells, mutations that drive cell proliferation also
possess the ability to activate senescence and apoptosis (41). Oncogenic MYC, for instance, has been
shown to trigger the ARF/p53 tumor suppressor pathway to induce
apoptosis when levels of MYC reach a certain threshold (41,42).
However, Murphy et al showed low levels of MYC failed to
activate the apoptotic transcriptional pathway, allowing for
tumorigenesis (42). Thus, it has
been theorized that low-level uninhibited MYC is more likely to
initiate oncogenesis than MYC that is overexpressed (42). Extending this theory to our findings,
it may be the case that the TT genotype activates the tumor
suppressor pathway to a greater extent than the GG or GT
genotype.
In this study variables with a P-value of <0.10
in the univariate analysis were included in the multivariate
analysis. A higher P-value than the traditional level of 0.05 was
chosen in order to decrease the risk of excluding a potentially
important variable (17). Several
other studies have utilized similar methods. Hoshimoto et al
assessed pre-operative factors associated with survival of
cholangiocarcinoma, and incorporated variables with P<0.10 in
the univariate analysis into the multivariate model (43). This was also utilized in a
publication by Chao et al, which assessed characteristics
associated with hepatocellular carcinoma survival after liver
transplant. Variables that had a P-value <0.10 in the univariate
analysis were included in the multivariate analysis (44). While this model allows for increased
inclusivity, it also increases the risk of introducing variables
that have a confounding effect on each other, resulting in high
intercorrelations among non-significant variables (17).
A limitation of this study was the relatively small
sample size and the retrospective nature of the design. Variables
such as the Charlson Comorbidity Index, cancer stage, and location
of metastatic disease contained subcategories that often had 0–2
cases for a particular genotype, limiting the data analysis. In
addition, it is unknown whether the rs6983267 SNP investigated in
this study resulted from a germline or somatic DNA mutation.
However, given the aim of this study was to analyze the association
of survival, rather than cancer risk, with the tumor genotype, our
conclusions should not change based on type of DNA mutation. This
study was strengthened by the data collection occurring at a single
tertiary medical center, which ensured consistency with study
protocol in all sample collection and analysis procedures. In
addition, due to the diversity of patients treated at a large
tertiary medical center, the 98 samples in this study likely
represent a broad array of clinical phenotypes in ovarian cancer,
although all of them shared a common ancestry (45).
To our knowledge, this is the first study to assess
the association between clinical outcomes in patients with HGSOC
and genotypes of the rs6983267 SNP within the lncRNA CCAT2. HGSOC
patients with the TT genotype at this locus had improved survival
time compared to patients with genotypes containing any G allele,
despite patients with the TT genotype being diagnosed at a more
advanced disease stage. Increased survival may be due to better
response to initial chemotherapy by patients with the TT genotype.
This study suggests individualized cancer outcomes are influenced
by patient genomic variation. We know that certain patients will
respond better to chemotherapy, but we are starting to untangle
some of the reasons why this may happen. Further studies are needed
to discern the intrinsic biological mechanisms of this observation
and its potential use as target therapy.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present work was supported in part by the
National Institutes of Health (grant nos. R01 CA99908 and R01
CA184101), The Basic Research Fund from the Department of
Obstetrics and Gynecology at the University of Iowa and by The
American Association of Obstetricians and Gynecologists Foundation
Bridge Funding Award.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DI, NC, JGB and ED conceived the study. ED designed
the methodology. ED and JGB validated the study. JGB performed the
formal analysis. DI and ED performed the investigation. JGB and ED
procured the resources. ED was responsible for data curation. DI
wrote the original draft manuscript. DI, NC, JGB and ED reviewed
and edited the draft manuscript. JBG visualized the study. JGB and
ED supervised the study, were project administrators and acquired
funding. JGB and ED confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The study was conducted according to the guidelines
of the Declaration of Helsinki, and was approved by The
Institutional Review Board (or Ethics Committee) of the University
of Iowa (approval nos. #200209010, #201804817, #201809807).
Informed consent was obtained from all subjects involved in the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Redis RS, Vela LE, Lu W, Ferreira de
Oliveira J, Ivan C, Rodriguez-Aguayo C, Adamoski D, Pasculli B,
Taguchi A, Chen Y, et al: Allele-specific reprogramming of cancer
metabolism by the long non-coding RNA CCAT2. Mol Cell. 61:6402016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mercer TR and Mattick JS: Structure and
function of long noncoding RNAs in epigenetic regulation. Nat
Struct Mol Biol. 20:300–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huppi K, Pitt JJ, Wahlberg BM and Caplen
NJ: The 8q24 gene desert: An Oasis of non-coding transcriptional
activity. Front Genet. 3:692012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Grisanzio C and Feedman ML: Chromosome
8q24-associated cancers and MYC. Genes Cancer. 1:555–559. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xiang JF, Yang L and Chen LL: The long
noncoding RNA regulation at the MYC locus. Curr Opin Genet Dev.
33:41–48. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang S, Qing C, Huang Z and Zhu Y: The
long non-coding RNA CCAT2 is up-regulated in ovarian cancer and
associated with poor prognosis. Diagn Pathol. 11:492016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao X, Wei X, Zhao L, Shi L, Cheng J,
Kang S, Zhang H, Zhang J, Li L, Zhang H and Zhao W: The rs6983267
SNP and long non-coding RNA CARLo-5 are associated with endometrial
carcinoma. Environ Mol Mutagen. 57:508–515. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim T, Cui R, Jeon YJ, Lee JH, Lee JH, Sim
H, Park JK, Fadda P, Tili E, Nakanishi H, et al: Long-range
interaction and correlation between MYC enhancer and oncogenic long
noncoding RNA CARLo-5. Proc Natl Acad Sci USA. 111:4173–4178. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han J, Zhou J, Yuan H, Zhu L, Ma H, Hang D
and Li D: Genetic variants within the cancer susceptibility region
8q24 and ovarian cancer risk in Han Chinese women. Oncotarget.
8:36462–36468. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hua F, Li CH, Chen XG and Liu XP: Long
noncoding RNA CCAT2 knockdown suppresses tumorous progression by
sponging miR-424 in epithelial ovarian cancer. Oncol Res.
26:241–247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ghoussaini M, Song H, Koessler T, Al Olama
AA, Kote-Jarai Z, Driver KE, Pooley KA, Ramus SJ, Kjaer SK, Hogdall
E, et al: Multiple loci with different cancer specificities within
the 8q24 gene desert. J Natl Cancer Inst. 100:962–966. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Barry KH, Moore LE, Sampson J, Yan L,
Meyer A, Oler AJ, Chung CC, Wang Z, Yeager M, Amundadottir L and
Berndt SI: DNA methylation levels at chromosome 8q24 in peripheral
blood are associated with 8q24 cancer susceptibility loci. Cancer
Prev Res (Phila). 7:1282–1292. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sotelo J, Esposito D, Duhagon MA, Banfield
K, Mehalko J, Liao H, Stephens RM, Harris TJ, Munroe DJ and Wu X:
Long-range enhancers on 8q24 regulate c-Myc. Proc Natl Acad Sci
USA. 107:3001–3005. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Newtson AM, Devor EJ and Gonzalez Bosquet
J: Prediction of epithelial ovarian cancer outcomes with
integration of genomic data. Clin Obstet Gynecol. 63:92–108. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bursac Z, Gauss CH, Williams DK and Hosmer
DW: Purposeful selection of variables in logistic regression.
Source Code Biol Med. 3:172008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Reyes HD, Devor EJ, Warrier A, Newtson AM,
Mattson J, Wagner V, Duncan GN, Leslie KK and Gonzalez-Bosquet J:
Differential DNA methylation in high-grade serous ovarian cancer
(HGSOC) is associated with tumor behavior. Sci Rep. 9:179962019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gonzalez Bosquet J, Devor EJ, Newtson AM,
Smith BJ, Bender DP, Goodheart MJ, McDonald ME, Braun TA, Thiel KW
and Leslie KK: Creation and validation of models to predict
response to primary treatment in serous ovarian cancer. Sci Rep.
11:59572021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
R Core Team R, . A Language and
environment for statistical computing. 2016.http://www.R-project.org/
|
|
21
|
Schroder MS, Culhane AC, Quackenbush J and
Haibe-Kains B: Survcomp: An R/Bioconductor package for performance
assessment and comparison of survival models. Bioinformatics.
27:3206–3208. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bolton KL, Chenevix-Trench G, Goh C,
Sadetzki S, Ramus SJ, Karlan BY, Lambrechts D, Despierre E,
Barrowdale D, McGuffog L, et al: Association between BRCA1 and
BRCA2 mutations and survival in women with invasive epithelial
ovarian cancer. JAMA. 307:382–390. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vencken PMLH, Kriege M, Hoogwerf D,
Beugelink S, van der Burg MEL, Hooning MJ, Berns EM, Jager A,
Collée M, Burger CW and Seynaeve C: Chemosensitivity and outcome of
BRCA1- and BRCA2-associated ovarian cancer patients after
first-line chemotherapy compared with sporadic ovarian cancer
patients. Ann Oncol. 22:1346–1352. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang X, Li X, Li W, Bai H and Zhang Z:
PARP inhibitors in ovarian cancer: Sensitivity prediction and
resistance mechanisms. J Cell Mol Med. 23:2303–2313. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ledermann JA, Drew Y and Kristeleit RS:
Homologous recombination deficiency and ovarian cancer. Eur J
Cancer. 60:49–58. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Frey MK and Pothuri B: Homologous
recombination deficiency (HRD) testing in ovarian cancer clinical
practice: A review of the literature. Gynecol Oncol Res Pract.
4:42017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma G, Gu D, Lv C, Chu H, Xu Z, Tong N,
Wang M, Tang C, Xu Y, Zhang Z, et al: Genetic variant in 8q24 is
associated with prognosis for gastric cancer in a Chinese
population. J Gastroenterol Hepatol. 30:689–695. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shen L, Du M, Wang C, Gu D, Wang M, Zhang
Q, Zhao T, Zhang X, Tan Y, Huo X, et al: Clinical significance of
POU5F1P1 rs10505477 polymorphism in Chinese gastric cancer patients
receving cisplatin-based chemotherapy after surgical resection. Int
J Mol Sci. 15:12764–12777. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu L, Chen SH, Lv QL, Sun B, Qu Q, Qin CZ,
Fan L, Guo Y, Cheng L and Zhou HH: Clinical significance of long
non-coding RNA CASC8 rs10505477 polymorphism in lung cancer
susceptibility, platinum-based chemotherapy response, and toxicity.
Int J Environ Res Public Health. 13:5452016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gong WJ, Yin JY, Li XP, Fang C, Xiao D,
Zhang W, Zhou HH, Li X and Liu ZQ: Association of
well-characterized lung cancer lncRNA polymorphisms with lung
cancer susceptibility and platinum-based chemotherapy response.
Tumour Biol. 37:8349–8358. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Haerian MS, Baum L and Haerian BS:
Association of 8q24.21 loci with the risk of colorectal cancer: A
systematic review and meta-analysis. J Gastroenterol Hepatol.
26:1475–1484. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu ER, Hsieh MJ, Chiang WL, Hsueh KC, Yang
SF and Su SC: Association of lncRNA CCAT2 and CASC8 gene
polymorphisms with hepatocellular carcinoma. Int J Environ Res
Public Health. 16:28332019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tong Y, Tang Y, Li S, Zhao F, Ying J, Qu
Y, Niu X and Mu D: Cumulative evidence of relationships between
multiple variants in 8q24 region and cancer incidence. Medicine
(Baltimore). 99:e207162020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cipollini M, Figlioli G, Garritano S,
Bramante S, Maiorano L, Gnudi F, Cecchini A, De Paola F, Damicis L,
Frixa T, et al: Risk of differentiated thyroid carcinoma and
polymorphisms within the susceptibility cancer region 8q24. Cancer
Epidemiol Biomarkers Prev. 22:2121–2125. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Goode EL, Chenevix-Trench G, Song H, Ramus
SJ, Notaridou M, Lawrenson K, Widschwendter M, Vierkant RA, Larson
MC, Kjaer SK, et al: A genome-wide association study identifies
susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet.
42:874–879. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pomerantz MM, Ahmadiyeh N, Jia L, Herman
P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M,
et al: The 8q24 cancer risk variant rs6983267 shows long-range
interaction with MYC in colorectal cancer. Nat Genet. 41:882–884.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wright JB, Brown SJ and Cole MD:
Upregulation of c-MYC in cis through a large chromatin loop linked
to a cancer risk-associated single-nucleotide polymorphism in
colorectal cancer cells. Mol Cell Biol. 30:1411–1420. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tuupanen S, Turunen M, Lehtonen R,
Hallikas O, Vanharanta S, Kivioja T, Björklund M, Wei G, Yan J,
Niittymäki I, et al: The common colorectal cancer predisposition
SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt
signaling. Nat Genet. 41:885–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ahmadiyeh N, Pomerantz MM, Grisanzio C,
Herman P, Jia L, Almendro V, He HH, Brown M, Liu XS, Davis M, et
al: 8q24 prostate, breast, and colon cancer risk loci show
tissue-specific long-range interaction with MYC. Proc Natl Acad Sci
USA. 107:9742–9746. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wasserman NF, Aneas I and Nobrega MA: An
8q24 gene desert variant associated with prostate cancer risk
confers differential in vivo activity to a MYC enhancer. Genome
Res. 20:1191–1197. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lowe SW, Cepero E and Evan G: Intrinsic
tumour suppression. Nature. 432:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Murphy DJ, Junttila MR, Pouyet L, Karnezis
A, Shchors K, Bui DA, Brown-Swigart L, Johnson L and Evan GI:
Distinct thresholds govern Myc's biological output in vivo. Cancer
Cell. 14:447–457. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hoshimoto S, Hishinuma S, Shirakawa H,
Tomikawa M, Ozawa I and Ogata Y: Association of preoperative
Platelet-to-Lymphocyte ratio with poor outcome in patients with
distal cholangiocarcinoma. Oncology. 96:290–298. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chao JS, Zhao SL, Ou-Yang SW, Qian YB, Liu
AQ, Tang HM, Zhong L, Peng ZH, Xu JM and Sun HC: Post-transplant
infection improves outcome of hepatocellular carcinoma patients
after orthotopic liver transplantation. World J Gastroenterol.
25:5630–5640. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Miller MD, Devor EJ, Salinas EA, Newtson
AM, Goodheart MJ, Leslie KK and Gonzalez-Bosquet J: Population
substructure has implications in validating next-generation cancer
genomics studies with TCGA. Int J Mol Sci. 20:11922019. View Article : Google Scholar : PubMed/NCBI
|