Introduction
Liver cancer has been predicted to be the sixth most
commonly diagnosed cancer and the fourth leading cause of
cancer-related deaths worldwide. Among different types of primary
liver cancer, hepatocellular carcinoma (HCC) is the most common,
comprising 75–85% of cases in adults (1). Ultrasonography and α-fetoprotein (AFP)
detection are the most widely employed techniques for the screening
and early diagnosis of HCC. However, the sensitivity of
ultrasonography for detecting early HCC is only 63%. The clinical
diagnostic accuracy of AFP is also inadequate due to its low
sensitivity and specificity, since 30–40% of patients with HCC are
serum-AFP-negative (2,3). Moreover, biomarkers for the accurate
diagnosis of HCC have not yet been reported. Therefore, it is
crucial to establish effective biomarkers expressed in both the
tissue and blood samples of patients with HCC. Furthermore, it is
important to understand the characteristics of HCC through gene
expression profiling in biomarker studies. Schulze et al
(4) identified 161 putative genetic
alterations in HCC using exome sequencing analysis. Using a series
of bioinformatics methods, Zhang et al (5) and Gao et al (6) investigated key genes and pathways known
to be closely associated with HCC. Moreover, the number of studies
evaluating the global gene expression profiles of HCC has markedly
increased in recent years. Therefore, identifying stably expressed
optimal internal controls is necessary for the accurate gene
expression profiling of HCC.
Recent studies have suggested that the measurement
of exosome markers is emerging as a novel and efficient method of
biomarker quantification as the various molecular constituents of
exosomes are closely connected with the original cells from which
the exosomes are derived (7–9). Exosomes are membrane-bound
nanometer-sized vesicles widely derived from cancer cells, and have
been highlighted as notable constituents of intercellular
communication (10,11). Therefore, exosomes can be considered
as a type of predictive biomarker. The study of gene expression
profiles, including those of exosomes, is commonly performed using
modalities such as cDNA microarrays, though it is difficult to
detect a small number of mRNA copies. As such, due lower economic
burden and increased accuracy, reverse transcription-quantitative
(RT-q) PCR is often used as an alternative, especially since it is
the only technology that can detect mRNA copies at low expression
levels (12).
RT-qPCR is a rapid, sensitive and accurate method
used to detect gene expression. The technique is based on the
normalization of target gene expression within a biological
material with any stably-expressed internal reference gene in the
same material. Therefore, selection of appropriate reference genes
is one of the most important factors for ensuring the accuracy of
RT-qPCR analysis. GAPDH, ACTB, TBP, 18S rRNA, HPRT1 and
TUBB are commonly used as reference genes in RT-qPCR
(13,14). However, previous studies have
reported numerous putative reference genes for a wide variety of
human tissues and human cell lines under different experimental
conditions or environmental factors (15–19). For
example, mRNA levels of GAPDH in liver cancer are not always
constant, and may vary based on changes in pathology, treatment, or
environmental conditions among different tissues or cell lines
(20–25). Furthermore, liver cancer is
heterogeneous, and therefore, an accurate and precise protocol is
required for biomarker validation. When performing RT-qPCR
analysis, the selection of the internal reference gene is arguably
the most important step. To date, studies determining suitable
reference genes for gene expression analysis in serum samples from
patients with HCC have been insufficient (20,26,27).
Therefore, the aim of the present study was to identify valid
internal control genes for the normalization of RT-qPCR studies in
both human HCC tissues and blood samples.
Materials and methods
Data processing and expression
analysis for reference genes in HCC
The gene expression profiles of the GSE114564
dataset were obtained from the Gene Expression Omnibus database
(www.ncbi.nlm.nih.gov.libproxy.ajou.ac.kr/geo/);
gene expression profiles were analyzed with the GEO2R tool, using
high-throughput sequencing to investigate the expression of 14
candidate reference genes in patients with different liver disease
statuses. A heatmap of the reference genes was generated using the
heatmap visualization tool Morpheus (https://software.broadinstitute.org/morpheus/).
Suitable reference gene candidates for analyzing gene expression in
HCC were identified using the list of housekeeping genes at
genomics-online (https://www.genomics-online.com/resources/16/5049/housekeeping-genes);
the gene accession numbers were obtained through the NCBI BLAST
database (Table I). Kruskal-Wallis
(non-parametric) followed by Dunn's post hoc test was used to
determine statistical significance between non-tumor (normal,
chronic hepatitis and liver cirrhosis) and HCC groups (early and
advanced HCC). P<0.05 was considered to indicate a statistically
significant difference.
 | Table I.Known mammalian housekeeping
genes. |
Table I.
Known mammalian housekeeping
genes.
| Gene symbol | Gene | Gene accession
number |
|---|
| ACTB | Actin, beta | NM_001101.3 |
| B2M |
Beta-2-microglobulin | NM_004048.3 |
| GAPDH |
Glyceraldehyde-3-phosphate
dehydrogenase | NM_002046.6 |
| GUSB | Glucuronidase,
beta | NM_001293105.1 |
| HMBS | Hydroxymethylbilane
synthase | NM_000190.4 |
| HPRT1 | Hypoxanthine
phosphoribosyltransferase 1 | NM_000194.3 |
| PGK1 | Phosphoglycerate
kinase 1 | NM_000291.4 |
| PPIA | Peptidylprolyl
isomerase A | NM_001300981.2 |
| RPLP0 | Ribosomal protein,
large, P0 | NM_001002.3 |
| RPL13A | Ribosomal protein
L13a | NM_012423.4 |
| SDHA | Succinate
dehydrogenase complex, subunit A, flavoprotein (Fp) | NM_004168.4 |
| TBP | TATA box binding
protein | NM_003194.5 |
| TFRC | Transferrin
receptor | NM_001128148.3 |
| YWHAZ | Tyrosine
3-monooxygenase/tryptophan5-monooxygenase activation protein, zeta
polypeptide | NM_145690.3 |
The Exocarta database (http://www.exocarta.org) is a manually curated
web-based overview of exosomal proteins, RNA and lipids. Exocarta,
which is used to evaluate corresponding data, such as exosome
characterization and molecular properties, was used to identify
reference genes expressed in exosomes (28).
Samples
Sera and tissue samples were collected from the
Biobank of Ajou University Hospital, a member of the Korea Biobank
Network, between April 2015 and July 2019. Written informed consent
was obtained from all study participants. Serum samples were
collected from 20 healthy controls and 20 patients with HCC; 20
pairs of HCC tissues with 20 corresponding non-tumor tissue samples
were also obtained from patients undergoing tumor resection
surgery. These samples were immediately frozen in liquid nitrogen
until use. Healthy controls were subjects 18 years of age or older
without a history of viral hepatitis or alcoholic liver disease who
visited the Ajou University Hospital for the purpose of regular
health checkups. HCC was diagnosed based on the American
Association for the Study of Liver Diseases practice guideline
(29) or histopathologic findings.
Subjects were excluded if they exhibited any evidence of other
malignancy except HCC or viral coinfections with the human
immunodeficiency virus. The patient clinical characteristics are
presented in Table SI. All
experiments were performed according to the Declaration of Helsinki
and the study protocol was approved by the Institutional Review
Board of Ajou University Hospital, Suwon, South Korea (approval no.
AJRIB-BMR-KSP-16-365 and AJIRB-BMR-SMP-17-189).
Cell culture
To evaluate exosomes, Huh7 cells from the Korean
Cell Line Bank were cultured in Dulbecco's modified Eagle's medium
(GenDEPOT, LLC) supplemented with 10% fetal bovine serum
(Invitrogen; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (GenDEPOT, LLC). The cells were incubated
at 37°C in a humidified atmosphere containing 5%
CO2.
Separation of blood sera
Blood samples (5 ml each) were collected from 20
patients directly into serum collection tubes. The whole blood
samples were centrifuged at 1,800 × g at room temperature for 10
min, and the resultant sera were aliquoted into 1.5 ml tubes. The
samples were then centrifuged at 3,000 × g at 4°C for 15 min to
remove cell debris prior to use.
Exosome isolation
Exosomes were isolated from human serum samples
using ExoQuick (System Biosciences, LLC) according to the
manufacturer's instructions (2).
Transmission electron microscopy
(TEM)
Exosome presence and size were confirmed using TEM.
Serum exosome samples were fixed with 2% glutaraldehyde and 4%
paraformaldehyde for 2 h at room temperature, and then treated with
0.4% uranyl acetate at 4°C for 10 min. Thereafter, the exosomes
were observed using a Sigma 500 electron microscope (Zeiss GmbH),
and further examined using a NanoSight NS300 instrument (Malvern
Panalytical Ltd.) equipped with a 405-nm laser, to determine the
size and quantity of the isolated particles. A 60-sec video was
generated at a frame rate of 30 frames/s, and particle movement was
analyzed using NTA software (version 3.0, Malvern Panalytical,
Ltd.). Each sample was analyzed three times and the average number
of counts were used.
Western blotting
To validate the expression of exosomal protein
markers, serum exosomes and Huh7 cell lysates were lysed in RIPA
lysis buffer (Thermo Fisher Scientific, Inc.) containing the Halt
protease inhibitor cocktail (Thermo Fisher Scientific, Inc.). Total
protein concentration was quantified using the bicinchoninic acid
assay method (Thermo Fisher Scientific, Inc.); equal amounts (10
µg) of protein sample were separated with 10% gel, and then
transferred onto polyvinylidene difluoride membranes
(MilliporeSigma). The membranes were blocked with 5% non-fat milk
(in Tris-buffered saline and 0.1% Tween-20) for 1 h at room
temperature, and then incubated with the following primary
antibodies: Mouse anti-Alix (1:1,000; cat. no. sc53538; Santa Cruz
Biotechnology, Inc.), mouse anti-CD81 (1:250; cat. no. 10630D;
Invitrogen; Thermo Fisher Scientific, Inc.), rabbit anti-CD9
(1:2,000; cat. no. ab92726; Abcam) and mouse anti-BiP/GRP78
(1:1,000; cat. no. 610979; BD Biosciences). The resulting immune
complexes were then probed using secondary horseradish
peroxidase-conjugated anti-rabbit (cat. no. BR170-6515; Bio-Rad
Laboratories, Inc.) or anti-mouse (cat. no. BR170-6516; Bio-Rad
Laboratories, Inc.) antibodies. Luminescence was observed using the
ChemiDoc™ Imaging System (Bio-Rad Laboratories, Inc.).
Primer design
The NCBI BLAST database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for
primer design. All primers were designed with target amplicons
<200 bp in length. The primer sequences are listed in Table II. The specificity of these primer
sets was confirmed using melting curve analysis (Fig. S1A).
 | Table II.Details of primer sequences of
candidate reference genes. |
Table II.
Details of primer sequences of
candidate reference genes.
| Gene symbol | Gene accession
number | Forward primer
sequence | Reverse primer
sequence | Size, bp |
|---|
| ACTB | NM_001101.3 |
ACCCAGATCATGTTTGGACCT |
GAGTCCATCACGATCCAGT | 108 |
| GAPDH | NM_002046.6 |
AGTATGACAACAGCCTCAAG | TCATGAGTCCTTCCA
CGATA | 111 |
| HMBS | NM_000190.4 |
GGAGGGCAGAAGGAAGAAAACAG | CACTGTCCGTCTGTA
TGCGAG | 91 |
| PPIA | NM_001300981.2 |
GCTGTGAGGAGGTACTGCTTG |
CCTGAGAAACCAAGTCCTTAGTG | 145 |
| RPLP0 | NM_001002.3 |
TGGTCATCCAGCAGGTGTTCG A |
ACAGACACTGGCAACATTGCGG | 119 |
RNA extraction and cDNA synthesis
Total RNA from the selected tissue samples was
isolated using QIAzol reagent (Qiagen GmbH), and serum RNA was
extracted from the selected blood samples using the
TRIzol® LS reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Exosomal RNA was isolated from serum using the
SeraMir™ Exosome RNA Amplification kit (System Bioscience, LLC)
according to the manufacturer's instructions. RNA concentration was
quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher
Scientific, Inc.). Following the manufacturer's instructions, serum
RNA (500 ng) was reverse transcribed into cDNA using the
PrimeScript™ RT Master Mix (Takara Bio, Inc.), and exosomal RNA (50
ng) was reverse transcribed using the miScript II RT kit (Qiagen
GmbH).
qPCR
qPCR was performed using the amfiSure qGreen Q-PCR
Master Mix (GenDEPOT, LLC) according to the manufacturer's
instructions, on the CFX Connect Real-Time PCR Detection System
(Bio-Rad Laboratories, Inc.). Each sample was prepared in a total
volume of 10 µl, containing 4 µl diluted cDNA template, 5 µl
amfiSure qGreen Q-PCR Master Mix (GenDEPOT, LLC), and 500 nM of
each primer. The PCR conditions were as follows: 95°C for 2 min, 40
cycles of 95°C for 15 sec, 58°C or 60°C for 34 sec, and 72°C for 30
sec, followed by a dissociation stage of 95°C for 10 sec, 65°C for
5 sec, and 95°C for 5 sec. Relative gene expression levels were
calculated using the 2−ΔΔCq method (30). All PCR reactions were performed in
triplicate.
Analysis of reference gene expression
stability
The stability of candidate reference gene expression
was evaluated using the Excel-based software BestKeeper (https://www.gene-quantification.de/bestkeeper.html).
All data processing was based on crossing point (CP). The stability
rankings of the individual genes were determined according to the
lowest standard deviations.
Statistical analysis
All experiments were performed independently in
triplicate. Results are presented as the mean ± standard deviation
or standard error of the mean. Statistical differences between
groups were analyzed using paired Student's t-test for the tissue
samples or Welch's t-test for the serum and serum exosome samples.
All statistical analyses were performed using GraphPad Prism 5.0
(GraphPad Software Inc.) and P<0.05 was considered to indicate a
statistically significant difference.
Results
Selection of candidate reference genes
for HCC marker studies
Expression levels of the 14 selected reference
genes, analyzed using the next-generation sequencing multistage HCC
RNA seq dataset GSE114564, are represented as a heat map based on
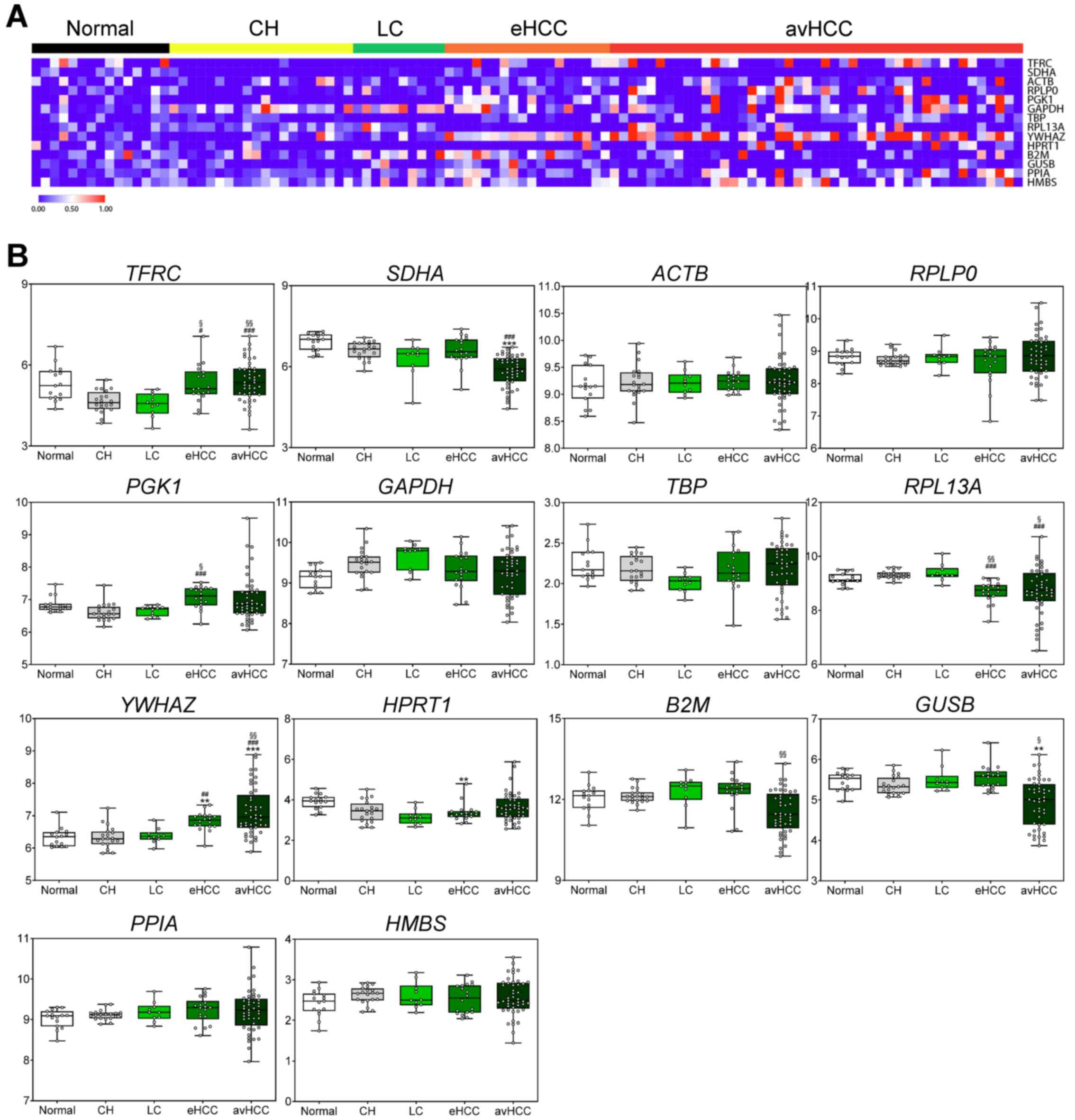
liver disease status (Fig. 1A and
Table SII). Differences in
expression levels between the control group and the HCC group were
identified in patients with different liver disease statuses. From
the 14 genes, ACTB, GAPDH, HMBS, PPIA, RPLP0 and TBP
were selected, as they did not show a statistically significant
difference between the control and HCC groups (Fig. 1B and Table SII). For the exosome samples, the
Exocarta database (http://www.exocarta.org/) was used to identify
suitable reference genes from the five selected genes. TBP,
which is not registered in the Exocarta database, was excluded from
the final selected candidates.
The ACTB gene performs key functions of the
cytoskeleton, such as cell motility and contraction (31). The GAPDH gene has
glyceraldehyde-3-phosphate dehydrogenase and nitrosylase
activities, and is involved in glycolysis and nuclear function. It
also regulates the organization and assembly of the cytoskeleton
(32,33). The HMBS gene supports the
generation of hydroxymethylbilane synthase, and is indirectly
involved in the production of heme (34). The PPIA gene catalyzes the
cis-trans isomerization of proline imidic peptide bonds in
oligopeptides, and is involved in apoptosis signaling through
NF-κΒ, AKT1 and BCL2 upregulation (35,36). The
RPLP0 gene encodes a ribosome, an organelle that catalyzes
protein synthesis, is composed of a small 40S and a large 60S
subunit, and is associated with pathologies including Chagas
disease (37). Based on these
results, the expression levels of 6 genes exhibited no statistical
significance between the control and HCC groups. Among them, 5
genes were expressed in exosomes using the Exocarta database. The
present study subsequently identified the molecular characteristics
of those 5 candidate reference genes.
Primer specificity of candidate
reference genes
Following primer design using NCBI BLAST, and
confirmation of specificity using melting curve analysis, all
primers were observed as a single peak (Fig. S1A). The most suitable annealing
temperature and mean Cq values were then selected (Fig. S1B).
RT-qPCR Cq values of candidate
reference genes
Pure exosomes were identified by isolation from
serum samples and characterization using TEM analysis (Fig. S2A). Furthermore, positive and
negative protein markers of extracellular vesicles were confirmed
through western blotting (Fig.
S2B). Next, RT-qPCR analysis was used to evaluate the
expression levels of the selected genes in the control and HCC
groups. All samples were analyzed in triplicate, and Welch's t-test
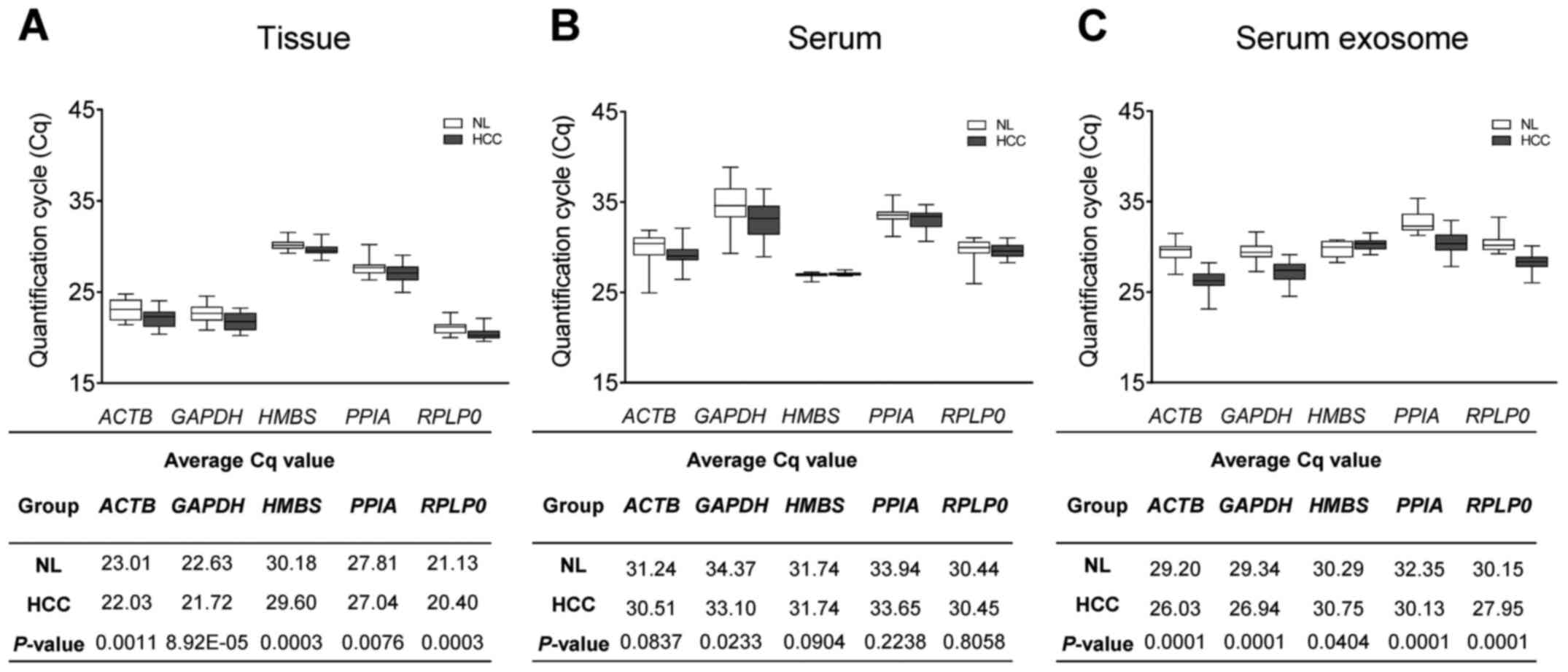
was performed with the average Cq values for each group. First, Cq
values of the five selected reference genes were calculated in 20
healthy and 20 HCC tissues. The expression levels of PPIA
(P=0.0076) showed the lowest significant difference between the
control and HCC tissue groups, and the expression levels of
ACTB (P=0.0011), GAPDH (P=8.92E-05), HMBS
(P=0.0003), and RPLP0 (P=0.0003) indicated a more
significant difference. (Fig. 2A).
Next, Cq values of the selected reference genes in serum and serum
exosome samples were estimated. Unlike the tissue samples, the
expression levels of ACTB (P=0.0837), HMBS
(P=0.0904), PPIA (P=0.2238) and RPLP0 (P=0.8058)
showed no significant difference. However, similar to the tissue
the samples, GAPDH (P=0.0233) indicated a significant
difference in expression level between the control and HCC groups
(Fig. 2B). Finally, the expression
levels of the five reference genes were confirmed in exosomal RNA
isolated from patient serum. Of these five genes, HMBS
(P=0.0404) exhibited the least significantly different expression
between the control and HCC serum exosome groups; however, the
expression of ACTB (P=0.0001), GAPDH (P=0.0001),
PPIA (P=0.0001) and RPLP0 (P=0.0001) indicated a
substantially significant difference (Fig. 2C). Therefore, among the five
reference genes identified, HMBS exhibited the least
significant difference in expression between the control and HCC
groups for blood samples (both serum and serum exosome).
Identification of the most suitable
reference genes in HCC studies
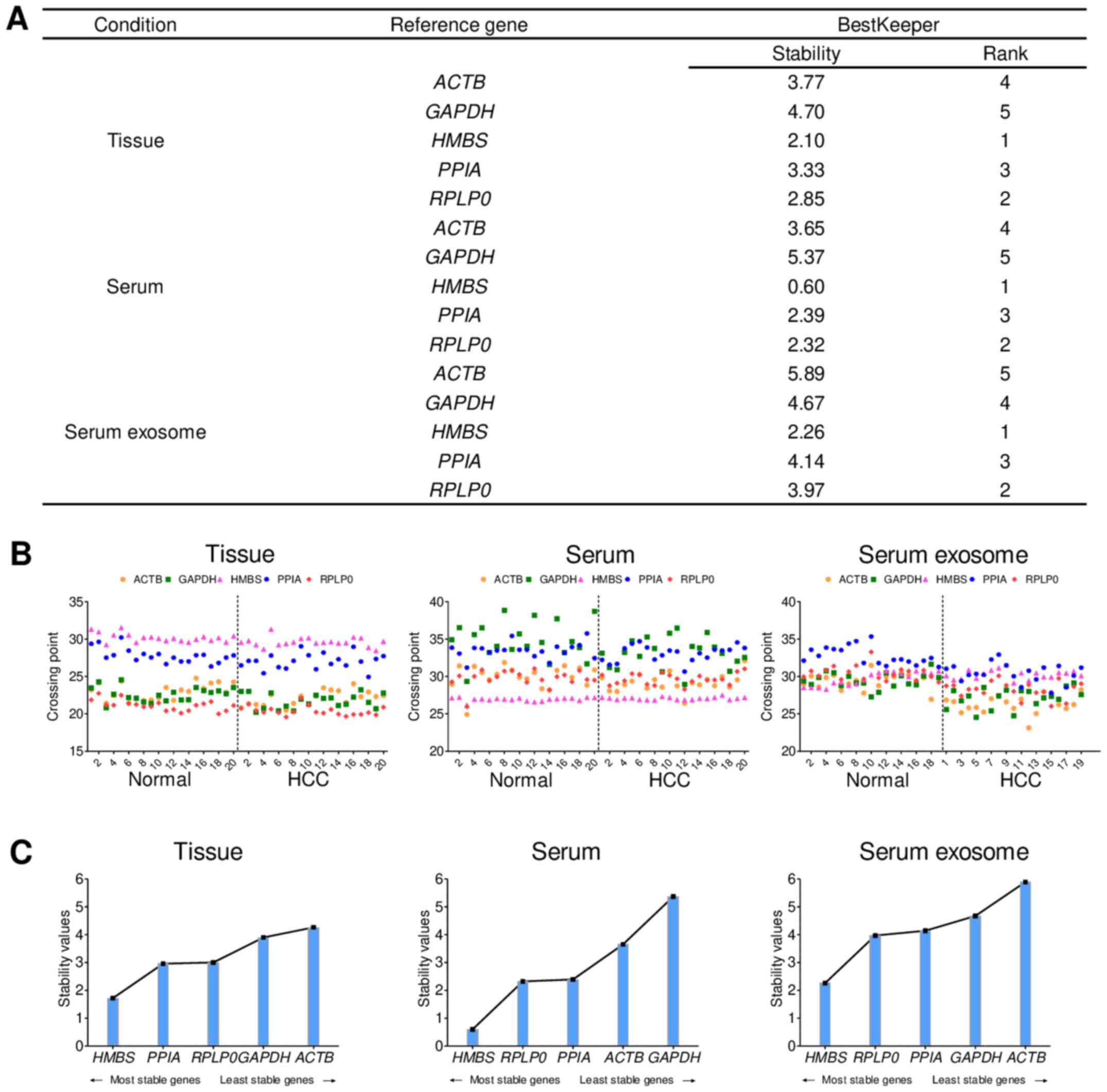
BestKeeper analyses of the tissue, blood and serum
samples were performed to investigate the stability of the five
reference genes. Descriptive statistics of the derived CPs were
calculated for each reference gene. CPs are direct results obtained
from the threshold line crosses fluorescence plots for each of the
samples. All CP data for all groups were compared throughout the
study (38). Stability rankings for
each sample were evaluated according to the coefficient of variance
values of the BestKeeper analyses. As such, the most stable
reference gene was identified to be HMBS. GAPDH,
which is a commonly used reference gene, was found to be the least
stable (Fig. 3A). In all 40 tissues
and blood samples, HMBS had the most consistent CP values
among five reference genes (Fig.
3B). The stability values obtained from the BestKeeper analyses
are represented in Fig. 3C. Also,
when performing NormFinder analysis (another tool for calculation
of stability), HMBS exhibited the highest stability in
tissue samples among the five candidate reference genes (data not
shown). In conclusion, HMBS was selected as the most stable
reference gene in tissue, serum and serum exosomes based on
Bestkeeper, a software that identifies the suitable reference gene
(39). Additionally, NormFinder
analysis revealed that HMBS is the most stable reference
gene for tissue samples.
In the present study, we found that HMBS is the most
suitable reference gene for blood and tissue samples in HCC. This
study will be helpful for future studies by finding suitable
reference genes for RT-qPCR, used to detect gene expression widely.
In this respect, we suggest that the expression stability of
reference genes should be validated to obtain accurate and reliable
results.
Discussion
Various studies have suggested biomarkers for liver
cancer, and the effort to identify additional markers is ongoing
(40). Numerous methods, including
immunohistochemistry, ELISA and western blotting, have previously
been used in such studies (41,42).
However, these methods are relatively time-consuming and expensive.
Similarly, droplet digital PCR technology (which can be used for
extremely low target quantitation) and microarrays (that can
measure the absolute expression of genes in cells or tissues so
that can perform precise analyses) are widely-used newer
technologies, but the cost of the associated instruments and
reagents is higher (43,44). In this respect, RT-qPCR has been the
most cost-effective and widely-used technique for biomarker
analysis studies, and can be used for analyzing both tissue and
blood samples (45). However,
despite the uncertainty surrounding gene normalization, RT-qPCR is
still used as one of the most accurate methods for transcript
quantification, and since liver cancer is heterogenous, a suitable
reference gene is required for this method (25,46).
Therefore, an accurate protocol for the validation of biomarker
studies needs to be developed.
The selection of an internal control gene for
normalizing target gene expression is an important consideration
for RT-qPCR. In particular, since exosomes are presently and
commonly used to identify biomarkers in cancer, the identification
of a suitable reference gene for exosome detection is also required
(47). Gorji-Bahri et al
(48) validated reference genes in
pooled cancer exosomes, and Dai et al (49) revealed that GAPDH, YWHAZ and
UBC were the most stably expressed reference genes in
exosomal RNA isolated from liver and breast cancer cell lines.
However, reference genes in HCC tissues and blood were not
evaluated by in vitro experiments and another software
analysis to determine stable housekeeping genes. The aim of the
present study was to identify the most reliable reference genes in
HCC tissue and blood samples using RT-qPCR. Therefore, 14
candidate, commonly used reference genes, were selected through a
systematic literature search.
Previous studies have reported that ACTB is
upregulated in liver cancer tissues and is therefore unsuitable for
the normalization of RT-qPCR (50).
Furthermore B2M was expressed at different levels depending on
hepatitis infection status (25).
Barber et al (51) indicated
that normalization is unstable for a single gene, as the
between-tissue variation for GAPDH can be substantial
(51). GUSB was not suitable
as a reference gene in RT-qPCR study for lung squamous-cell
carcinoma (52). Furthermore,
HMBS has been verified as suitable for the normalization of
gene expression data among tumor tissues in HCC (23). In addition, and as reported by Ceelen
et al (53), gene expression
stability level was analyzed in the human HepaRG cell line using
three algorithms (geNorm, BestKeeper, NormFinder). The results
revealed that TBP and HMBS exhibited the highest
stability (53). Also, in tumor
tissues from male HCC patients with hepatitis B infection and
cirrhosis, CTBP1 was the most stable reference gene, and
HMBS ranked third (24).
HPRT1 has been validated as the most suitable reference gene
for heart, liver and thymus samples (54), and PGK1 is known to be
suitable in small bowel studies, while PPIA is more optimal
in large bowel studies (55).
RPLP0 expression in breast, normal and adjacent tissues was
examined using geNorm and NormFinder software, and RPLP0 was
consequently found to be the least stable gene (56). Through geNorm and BestKeeper
analyses, RPL13A was selected as the most stable gene in the
granulosa cells of healthy women, as well as those of patients with
polycystic ovarian syndrome (57),
and was suitable for both healthy breast and breast tumor tissues
(58). In addition, Ohl et al
(59) identified SDHA and
TBP as reference genes for relative gene quantification in
bladder cancer, and TFRC was reported to be one of the
optimal set of reference genes for RT-qPCR analysis in HUVECs under
oxidative stress (60). Finally,
Bruce et al (61) found that
YWHAZ was stably expressed as a reference gene in studies of
non-alcoholic fatty liver disease.
Next, the expression of the 14 reference genes was
confirmed using human multistage HCC transcriptome data. Among
them, five candidate reference genes that did not show any
statistically significant difference between the control and HCC
groups, regardless of liver disease status, were selected. Primers
were designed for the five candidate reference genes using the NCBI
BLAST database, and primer efficiency was evaluated using RT-qPCR
analysis. The five reference genes were then evaluated in tissue,
serum and serum exosome samples; the characteristics of serum
exosomes were observed using TEM, and exosome markers were
confirmed using western blotting. RT-qPCR analysis was used to
measure the Cq values of the five candidate reference genes in 40
tissue samples (20 paired healthy tissues and 20 tissues from
patients with HCC) and 40 blood samples (20 healthy controls and 20
patients with HCC). HMBS showed the least significant
difference in Cq value in each group. Moreover, BestKeeper analysis
was used to evaluate the stability of the reference genes by
calculating the standard deviation of the Cq values. The results
indicated that HMBS was the most stable reference gene in
both tissue and blood samples. Thus, an in vitro study using
RT-qPCR confirmed that HMBS maintained a constant expression
level among the five candidate reference genes in HCC blood
samples. Furthermore, for the serum exosome group, BestKeeper
analysis revealed HMBS to be the most suitable reference
gene. Based on these results, HMBS is suggested as a
suitable normalization gene for RT-qPCR in HCC studies. However,
further validation via other techniques (i.e. droplet digital PCR
or NanoString) may be required in the future, although experiments
in the present study were repeated in the same sample and validated
within a constant range. Also, the current study was limited by the
small number of samples, thus in future studies, it will be
necessary to reduce error by increasing the sample population
size.
Supplementary Material
Supporting Data
Acknowledgements
The biospecimens and data used for the present study
were provided by the Biobank of Ajou University Hospital, a member
of the Korea Biobank Network.
Funding
The present study was supported by grants from the
Korea Health Technology R&D Project through the Korea Health
Industry Development Institute, funded by the Ministry of Health
and Welfare, Republic of Korea (grant no. HR21C1003), as well as
the Bio and Medical Technology Development Program of the National
Research Foundation (grant nos. NRF-2017M3A9B6061509 and
NRF-2019R1C1C1007366), funded by the Korean government (MSIT).
Availability of data and materials
The datasets analyzed during the current study are
available in the National Center for Biotechnology Information
(NCBI) Gene Expression Omnibus (GEO) database, accession no.
GSE114564.
Authors' contributions
JWE, JYC and HRA made substantial contributions to
the conception and design of the present study. HRA and HJC
performed the in vitro experiments. JAS, MGY and GOB
acquired and analyzed the data. SSK, DY and JHY interpreted all the
datasets in the present study. HJC and SSK drafted the initial
manuscript and critically revised it for important intellectual
content. JWE and JYC confirmed the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of Ajou University Hospital, Suwon, South Korea
(approval nos. AJRIB-BMR-KSP-16-365 and AJIRB-BMR-SMP-17-189).
Written informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AFP
|
α-fetoprotein
|
|
CP
|
crossing point
|
|
Cq
|
quantification cycle
|
|
EV
|
extracellular vesicle
|
|
HCC
|
hepatocellular carcinoma
|
|
LC
|
liver cirrhosis
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
TEM
|
transmission electron microscopy
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim SS, Baek GO, Ahn HR, Sung S, Seo CW,
Cho HJ, Nam SW, Cheong JY and Eun JW: Serum small extracellular
vesicle-derived LINC00853 as a novel diagnostic marker for early
hepatocellular carcinoma. Mol Oncol. 14:2646–2659. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Song P, Feng X, Zhang K, Song T, Ma K,
Kokudo N, Dong J and Tang W: Perspectives on using
des-γ-carboxyprothrombin (DCP) as a serum biomarker: Facilitating
early detection of hepatocellular carcinoma in China. Hepatobiliary
Surg Nutr. 2:227–231. 2013.PubMed/NCBI
|
|
4
|
Schulze K, Imbeaud S, Letouzé E,
Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C,
Shinde J, Soysouvanh F, et al: Exome sequencing of hepatocellular
carcinomas identifies new mutational signatures and potential
therapeutic targets. Nat Genet. 47:505–511. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang C, Peng L, Zhang Y, Liu Z, Li W,
Chen S and Li G: The identification of key genes and pathways in
hepatocellular carcinoma by bioinformatics analysis of
high-throughput data. Med Oncol. 34:1012017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao X, Wang X and Zhang S: Bioinformatics
identification of crucial genes and pathways associated with
hepatocellular carcinoma. Biosci Rep. 38:382018. View Article : Google Scholar
|
|
7
|
Xu L, Wu LF and Deng FY: Exosome: An
Emerging Source of Biomarkers for Human Diseases. Curr Mol Med.
19:387–394. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li W, Li C, Zhou T, Liu X, Liu X, Li X and
Chen D: Role of exosomal proteins in cancer diagnosis. Mol Cancer.
16:1452017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Corrado C, Raimondo S, Chiesi A, Ciccia F,
De Leo G and Alessandro R: Exosomes as intercellular signaling
organelles involved in health and disease: Basic science and
clinical applications. Int J Mol Sci. 14:5338–5366. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wong CH and Chen YC: Clinical significance
of exosomes as potential biomarkers in cancer. World J Clin Cases.
7:171–190. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Regev-Rudzki N, Wilson DW, Carvalho TG,
Sisquella X, Coleman BM, Rug M, Bursac D, Angrisano F, Gee M, Hill
AF, et al: Cell-cell communication between malaria-infected red
blood cells via exosome-like vesicles. Cell. 153:1120–1133. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gachon C, Mingam A and Charrier B:
Real-time PCR: What relevance to plant studies? J Exp Bot.
55:1445–1454. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wong ML and Medrano JF: Real-time PCR for
mRNA quantitation. Biotechniques. 39:75–85. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huggett J, Dheda K, Bustin S and Zumla A:
Real-time RT-PCR normalisation; strategies and considerations.
Genes Immun. 6:279–284. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kozera B and Rapacz M: Reference genes in
real-time PCR. J Appl Genet. 54:391–406. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang X, Ding L and Sandford AJ: Selection
of reference genes for gene expression studies in human neutrophils
by real-time PCR. BMC Mol Biol. 6:42005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Razavi SA, Afsharpad M, Modarressi MH,
Zarkesh M, Yaghmaei P, Nasiri S, Tavangar SM, Gholami H,
Daneshafrooz A and Hedayati M: Validation of reference genes for
normalization of relative qRT-PCR studies in papillary thyroid
carcinoma. Sci Rep. 9:152412019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zampieri M, Ciccarone F, Guastafierro T,
Bacalini MG, Calabrese R, Moreno-Villanueva M, Reale A, Chevanne M,
Bürkle A and Caiafa P: Validation of suitable internal control
genes for expression studies in aging. Mech Ageing Dev. 131:89–95.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dheda K, Huggett JF, Bustin SA, Johnson
MA, Rook G and Zumla A: Validation of housekeeping genes for
normalizing RNA expression in real-time PCR. BioTechniques.
37:112–119. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Qin Z, Cai L, Zou L, Zhao J and
Zhong F: Selection of internal references for qRT-PCR assays of
human hepatocellular carcinoma cell lines. Biosci Rep. 37:372017.
View Article : Google Scholar
|
|
21
|
Fu LY, Jia HL, Dong QZ, Wu JC, Zhao Y,
Zhou HJ, Ren N, Ye QH and Qin LX: Suitable reference genes for
real-time PCR in human HBV-related hepatocellular carcinoma with
different clinical prognoses. BMC Cancer. 9:492009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Waxman S and Wurmbach E: De-regulation of
common housekeeping genes in hepatocellular carcinoma. BMC
Genomics. 8:2432007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cicinnati VR, Shen Q, Sotiropoulos GC,
Radtke A, Gerken G and Beckebaum S: Validation of putative
reference genes for gene expression studies in human hepatocellular
carcinoma using real-time quantitative RT-PCR. BMC Cancer.
8:3502008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu S, Zhu P, Zhang L, Ding S, Zheng S,
Wang Y and Lu F: Selection of reference genes for RT-qPCR analysis
in tumor tissues from male hepatocellular carcinoma patients with
hepatitis B infection and cirrhosis. Cancer Biomark. 13:345–349.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao Q, Wang XY, Fan J, Qiu SJ, Zhou J, Shi
YH, Xiao YS, Xu Y, Huang XW and Sun J: Selection of reference genes
for real-time PCR in human hepatocellular carcinoma tissues. J
Cancer Res Clin Oncol. 134:979–986. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Benz F, Roderburg C, Vargas Cardenas D,
Vucur M, Gautheron J, Koch A, Zimmermann H, Janssen J,
Nieuwenhuijsen L, Luedde M, et al: U6 is unsuitable for
normalization of serum miRNA levels in patients with sepsis or
liver fibrosis. Exp Mol Med. 45:e422013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Li T, Qiu Y, Zhang T, Guo P, Ma
X, Wei Q and Han L: Serum microRNA panel for early diagnosis of the
onset of hepatocellular carcinoma. Medicine (Baltimore).
96:e56422017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Keerthikumar S, Chisanga D, Ariyaratne D,
Al Saffar H, Anand S, Zhao K, Samuel M, Pathan M, Jois M,
Chilamkurti N, et al: ExoCarta: A web-based compendium of exosomal
cargo. J Mol Biol. 428:688–692. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marrero JA, Kulik LM, Sirlin CB, Zhu AX,
Finn RS, Abecassis MM, Roberts LR and Heimbach JK: Diagnosis,
staging, and management of hepatocellular carcinoma: 2018 practice
guidance by the American Association for the Study of Liver
Diseases. Hepatology. 68:723–750. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Drazic A, Aksnes H, Marie M, Boczkowska M,
Varland S, Timmerman E, Foyn H, Glomnes N, Rebowski G, Impens F, et
al: NAA80 is actin's N-terminal acetyltransferase and regulates
cytoskeleton assembly and cell motility. Proc Natl Acad Sci USA.
115:4399–4404. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ercolani L, Florence B, Denaro M and
Alexander M: Isolation and complete sequence of a functional human
glyceraldehyde-3-phosphate dehydrogenase gene. J Biol Chem.
263:15335–15341. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tisdale EJ: Glyceraldehyde-3-phosphate
dehydrogenase is phosphorylated by protein kinase Ciota/lambda and
plays a role in microtubule dynamics in the early secretory
pathway. J Biol Chem. 277:3334–3341. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bung N, Roy A, Chen B, Das D, Pradhan M,
Yasuda M, New MI, Desnick RJ and Bulusu G: Human
hydroxymethylbilane synthase: Molecular dynamics of the pyrrole
chain elongation identifies step-specific residues that cause AIP.
Proc Natl Acad Sci USA. 115:E4071–E4080. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wei Y, Jinchuan Y, Yi L, Jun W, Zhongqun W
and Cuiping W: Antiapoptotic and proapoptotic signaling of
cyclophilin A in endothelial cells. Inflammation. 36:567–572. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Davis TL, Walker JR, Campagna-Slater V,
Finerty PJ, Paramanathan R, Bernstein G, MacKenzie F, Tempel W,
Ouyang H, Lee WH, et al: Structural and biochemical
characterization of the human cyclophilin family of peptidyl-prolyl
isomerases. PLoS Biol. 8:e10004392010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Remacha M, Jimenez-Diaz A, Santos C,
Briones E, Zambrano R, Rodriguez Gabriel MA, Guarinos E and
Ballesta JP: Proteins P1, P2, and P0, components of the eukaryotic
ribosome stalk. New structural and functional aspects. Biochem Cell
Biol. 73:959–968. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pfaffl MW, Tichopad A, Prgomet C and
Neuvians TP: Determination of stable housekeeping genes,
differentially regulated target genes and sample integrity:
BestKeeper - Excel-based tool using pair-wise correlations.
Biotechnol Lett. 26:509–515. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sen MK, Hamouzová K, Košnarová P, Roy A
and Soukup J: Identification of the most suitable reference gene
for gene expression studies with development and abiotic stress
response in Bromus sterilis. Sci Rep. 11:133932021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chia TS, Wong KF and Luk JM: Molecular
diagnosis of hepatocellular carcinoma: trends in biomarkers
combination to enhance early cancer detection. Hepatoma Res.
5:92019.
|
|
41
|
Matos LL, Trufelli DC, de Matos MG and da
Silva Pinhal MA: Immunohistochemistry as an important tool in
biomarkers detection and clinical practice. Biomark Insights.
5:9–20. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Manole E, Bastian AE, Popescu D,
Constantin C, Mihai S, Gaina G, Codrici E and Neagu M: Immunoassay
techniques highlighting biomarkers in immunogenetic diseases.
Immunogenetics. Nov 5–2018.(Epub ahead of print).
|
|
43
|
Li H, Bai R, Zhao Z, Tao L, Ma M, Ji Z,
Jian M, Ding Z, Dai X, Bao F, et al: Application of droplet digital
PCR to detect the pathogens of infectious diseases. Biosci Rep.
38:382018. View Article : Google Scholar
|
|
44
|
Macgregor PF and Squire JA: Application of
microarrays to the analysis of gene expression in cancer. Clin
Chem. 48:1170–1177. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sanders R, Mason DJ, Foy CA and Huggett
JF: Considerations for accurate gene expression measurement by
reverse transcription quantitative PCR when analysing clinical
samples. Anal Bioanal Chem. 406:6471–6483. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Schulze K, Nault JC and Villanueva A:
Genetic profiling of hepatocellular carcinoma using next-generation
sequencing. J Hepatol. 65:1031–1042. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Soung YH, Ford S, Zhang V and Chung J:
Exosomes in cancer diagnostics. Cancers (Basel). 9:92017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gorji-Bahri G, Moradtabrizi N, Vakhshiteh
F and Hashemi A: Validation of common reference genes stability in
exosomal mRNA-isolated from liver and breast cancer cell lines.
Cell Biol Int. 45:1098–1110. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dai Y, Cao Y, Köhler J, Lu A, Xu S and
Wang H: Unbiased RNA-Seq-driven identification and validation of
reference genes for quantitative RT-PCR analyses of pooled cancer
exosomes. BMC Genomics. 22:272021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Guo C, Liu S, Wang J, Sun MZ and Greenaway
FT: ACTB in cancer. Clin Chim Acta. 417:39–44. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Barber RD, Harmer DW, Coleman RA and Clark
BJ: GAPDH as a housekeeping gene: Analysis of GAPDH mRNA expression
in a panel of 72 human tissues. Physiol Genomics. 21:389–395. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhan C, Zhang Y, Ma J, Wang L, Jiang W,
Shi Y and Wang Q: Identification of reference genes for qRT-PCR in
human lung squamous-cell carcinoma by RNA-Seq. Acta Biochim Biophys
Sin (Shanghai). 46:330–337. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ceelen L, De Spiegelaere W, David M, De
Craene J, Vinken M, Vanhaecke T and Rogiers V: Critical selection
of reliable reference genes for gene expression study in the HepaRG
cell line. Biochem Pharmacol. 81:1255–1261. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Medrano G, Guan P, Barlow-Anacker AJ and
Gosain A: Comprehensive selection of reference genes for
quantitative RT-PCR analysis of murine extramedullary hematopoiesis
during development. PLoS One. 12:e01818812017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Krzystek-Korpacka M, Diakowska D, Bania J
and Gamian A: Expression stability of common housekeeping genes is
differently affected by bowel inflammation and cancer: Implications
for finding suitable normalizers for inflammatory bowel disease
studies. Inflamm Bowel Dis. 20:1147–1156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Majidzadeh-A K, Esmaeili R and Abdoli N:
TFRC and ACTB as the best reference genes to quantify Urokinase
Plasminogen Activator in breast cancer. BMC Res Notes. 4:2152011.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lv Y, Zhao SG, Lu G, Leung CK, Xiong ZQ,
Su XW, Ma JL, Chan WY and Liu HB: Identification of reference genes
for qRT-PCR in granulosa cells of healthy women and polycystic
ovarian syndrome patients. Sci Rep. 7:69612017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Shah KN and Faridi JS: Estrogen,
tamoxifen, and Akt modulate expression of putative housekeeping
genes in breast cancer cells. J Steroid Biochem Mol Biol.
125:219–225. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ohl F, Jung M, Radonić A, Sachs M, Loening
SA and Jung K: Identification and validation of suitable endogenous
reference genes for gene expression studies of human bladder
cancer. J Urol. 175:1915–1920. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li T, Diao H, Zhao L, Xing Y, Zhang J, Liu
N, Yan Y, Tian X, Sun W and Liu B: Identification of suitable
reference genes for real-time quantitative PCR analysis of hydrogen
peroxide-treated human umbilical vein endothelial cells. BMC Mol
Biol. 18:102017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bruce KD, Sihota KK, Byrne CD and
Cagampang FR: The housekeeping gene YWHAZ remains stable in a model
of developmentally primed non-alcoholic fatty liver disease. Liver
Int. 32:1315–1321. 2012. View Article : Google Scholar : PubMed/NCBI
|