Introduction
Digestive system tumors (DSTs), such as colorectal cancer (CRC), hepatocellular carcinoma (HCC), gastric cancer (GC) and esophageal carcinoma, are the most common tumors in the world (1). DSTs also constitute a primary proportion of cancer-related deaths worldwide (2). As estimated by GLOBOCAN 2018 (2), there were ~1,000,000 new GC cases and 783,000 associated deaths worldwide in 2018; therefore, GC ranks 3rd in terms of cancer-related deaths, followed by liver cancer, which causes ~782,000 deaths annually. The prognosis of DSTs mainly depends on the cancer stage at diagnosis; however, a number of patients are not diagnosed until the advanced stage. Currently, the available treatment options for advanced DSTs are limited, and the 5-year survival rate remains low at <20% (3,4). Tumor metastasis contributes to high mortality; however, several biomarkers associated with tumor metastasis have been identified recently (5,6). Therefore, more knowledge concerning biomarker regulation and the underlying network behavior might help provide superior treatment options for DSTs.
Circular RNAs (circ/circRNAs) are long non-coding (lnc) endogenous RNAs in the form of covalently closed loops with no 3′ polyadenylated tail or 5′ cap (7). circRNAs are reported to be sponges for microRNAs (miR/miRNAs) in the cytoplasm, and they facilitate the translation of proteins and interact with the RNA-binding proteins (RBPs) to regulate transcription (8,9). Recently, the focus of research has been on the circRNA-related molecular pathways participating in tumor metastasis and progression (5,10). circRNAs may serve as novel tumor biomarkers.
Exosomes are vesicles with a diameter of 30–150 nm. Since they are released outside the cells, they are extensively present in a variety of body fluids such as plasma/serum, urine and saliva (11). Exosomes have been found to contain proteins, DNA, miRNAs and lncRNAs (12). Typically, exosomes eliminate the redundant cellular components. circRNAs have been identified in exosomes (13). When exosomes are released, they are absorbed by distant or neighboring cells, and those containing circRNAs interfere with the processes that regulate the tumor microenvironment (TME), and may therefore, facilitate cancer occurrence, development and metastasis.
Biogenesis of exosomes
Exosomes were initially identified in the mature reticulocytes of sheep (14). Exosomes seem cup-shaped when observed using a cryo-electron and transmission electron microscope (15).
Three distinct processes are required to release exosomes (Fig. 1). Firstly, the selected cytosolic factors, such as proteins, are required to accumulate on the endosomal membrane for exosome biogenesis, to control the lipid bilayer invagination of endosomes to their luminal cavity, thus forming the intraluminal vesicle (ILV) (16). The protein sorting by ILVs is recognized as a highly regulated mechanism and it is dependent on the endosomal sorting complex required for transport (ESCRT) mechanism (17). Specifically, 4 complexes, namely ESCRT-0, -I, -II and -III, make up the ESCRT apparatus. This process is repeated to produce the ILVs that fill the multivesicular body (MVB). The MVB then fuses with the cell membrane using an adenosine triphosphate-dependent process, and exosomes are released into the extracellular space (18).
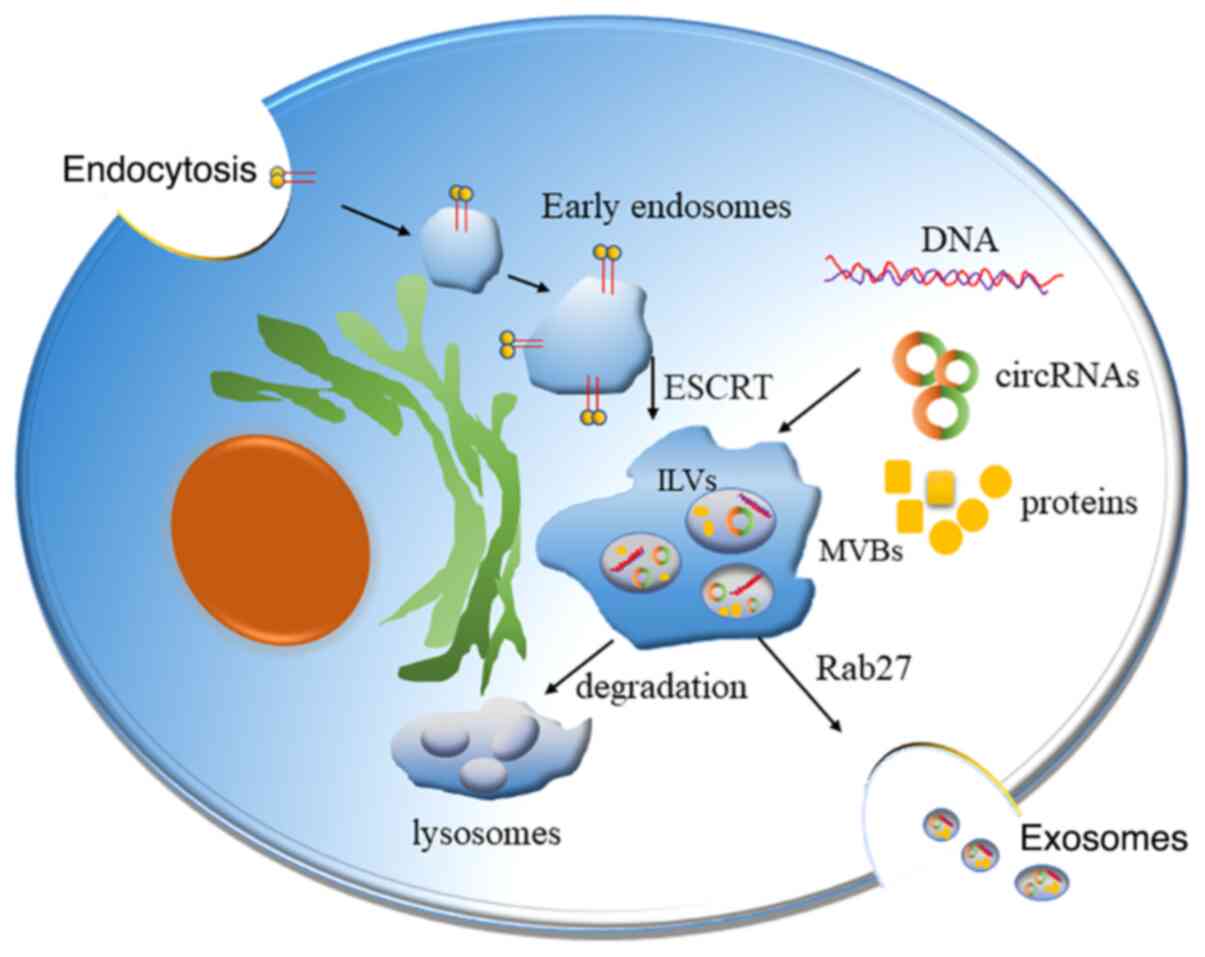 |
Figure 1.
Function and processing of exosomes. Substances are encapsulated using endocytosis forming the early endosomes, which further mature into ILV-containing MVBs. The formation of ILVs is mediated by mechanisms that depend on the ESCRT. ILVs subsequently merge with the cell membrane via an adenosine triphosphate-dependent process, thus releasing exosomes outside the cells. These exosomes travel through biological fluids. circRNA, circular RNA; ILV, intraluminal vesicle; MVB, multivesicular body; ESCRT, endosomal sorting complex required for transport.
|
A protein complex comprising nucleic acids (miRNA, mRNA and DNA), lipids, extracellular matrix (ECM) proteins, transcription factors, enzymes and receptors is present either in the interior or on the exterior of the exosomes. These components contribute to the signal transmission between the recipient and the donor cells, and affect the recipient cells (19). An analysis of the exosomal protein composition revealed that certain proteins are present in specific cell- and tissue-derived exosomes, while others are commonly seen in all the exosomes (20). Lipids are involved in the biogenesis of exosomes and also maintain homeostasis in the recipient cells. For example, lipids with a high density, such as lyosbisphosphatidic acid (LBPA), on the inner MVB membrane lead to the formation of ILV and subsequent exosomes (21). LBPA interacts with the apoptosis-linked gene-2-interacting protein X, which contributes to the inward sprouting of the MVB membrane (22).
Role of exosomes in tumor metastasis
Several studies (23,24) have verified that tumor metastasis is related to various oncogenes and their diverse oncogenic pathways (24). Yang et al (25) found that FOXP3 can act as a co-activator to facilitate the Wnt-b-catenin signaling pathway, inducing epithelial-mesenchymal transition (EMT), tumor growth and metastasis in non-small cell lung cancer. It was suggested that the cell-to-cell communication in cancer together with the adjacent stroma facilitates tumor metastasis. Therefore, the present review further elaborates on the effect of the exosomes on tumor metastasis.
Promotion of tumor migration
When tumor cells metastasize, they need to break the basement membrane to penetrate the blood vessels. Such processes and tumor growth at the primary and metastatic sites are closely related to the TME. The exchange of information between the tumor cells and TME via the tumor-derived exosomes (TDEs) plays an important role in the promotion of cancer metastasis (26,27).
Mediators of EMT
Zomer et al (28) demonstrated for the first time that exosomes derived from highly invasive tumor cells enhance the invasiveness of lowly invasive tumor cells. Tight junctions are the main components in the adhesion complex of endothelial and epithelial cells. The natural barrier limits the metastasis of tumor cells; however, this natural barrier is destroyed by TDEs, which promote penetration of blood vessels by tumor cells. It was confirmed through in vivo experiments in mice that TDEs with high miR-105 expression increase the incidence rates of brain and lung metastases, accompanied by the downregulation of zonula occludens-1 (ZO-1) in endothelial cells and an increase in vascular permeability (29).
EMT involves the loss of intercellular adhesion and polarity, which exacerbates the processes of invasion and metastasis of various cancer cell types, such as GC, HCC and CRC (30). By contrast, EMT facilitates tissue remodeling and is regarded as a prerequisite for tumor metastasis (31). Under the regulation of the TME, epithelial tumor cells undergo EMT, characterized by low proliferation, high invasion and metastasis. When tumor cells reach the metastatic microenvironment in distant organs, a mesenchymal-epithelial transition occurs, thus recovering their high proliferation status, which is beneficial for the growth of metastatic tumors (30). TDEs have been proposed as the conduits to initiate EMT signaling. For instance, in the bladder cancer 5637 cell line with high expression of lncRNA-urothelial carcinoma-associated 1 (UCA1), EMT was used to transport lncRNA-UCA1 to low-expression sites through exosomes to promote tumor invasion and growth (32). Moreover, in the exosomes collected from nasopharyngeal carcinoma (NPC) cells infected with Epstein-Barr virus, high levels of hypoxia-inducible factor 1 (HIF1) and latent membrane protein 1 are detectable; HIF1 interacts with the Snail pathway to upregulate Twist, which triggers EMT in NPC cells (33). Co-culturing NPC cells with exosomes isolated from these cells induced EMT in an autocrine manner, which was supported by upregulated vimentin and N-cadherin levels, and downregulated E-cadherin levels (34). miR-23a in exosomes enhance the effect of TGF-1 to promote EMT by suppressing the synthesis of E-cadherin in melanoma and lung carcinoma cells (35). Yang et al (4) reported that hepatoma-derived exosomal miR-92a-3p plays a critical role in EMT progression and the promotion of metastasis by inhibiting PTEN and activating Akt/Snail signaling.
Involvement in angiogenesis
In tumorigenesis, exosomes activate neovascularization and deliver more nutritious blood to avoid tumor necrosis. TDEs mediate the exchange of information between the tumor cells and the TME, which also plays important roles in promoting angiogenesis (36). The NPC-derived exosomes contain miR-23a, which acts on endothelial cells, regulates target gene expression of testis-specific 10 and promotes angiogenesis (37). On the other hand, TDEs containing miR-25-3p enter endothelial cells to target Kruppel-like factor (KLF)2 and KLF4, regulate the expression of vascular endothelial growth factor (VEGF) receptor 2, ZO-1 and atrein, improve vascular permeability and promote neovascularization (38). Further animal experiments confirmed that miR-25-3p in exosomes markedly increased the occurrence rates of liver and lung metastases in mice (38).
Involvement in immunosuppression
TDEs suppress both cellular and humoral immune responses to maintain tumor cell proliferation, and are linked to most immune cells. Specifically, they interfere with the functions of immune cells through a variety of pathways, including suppressing proliferation of immune cells, transmitting tolerance signals to immunocytes, suppressing natural killer (NK) cell activities, mediating the apoptosis of CD8+ T cells and interfering with the differentiation of monocytes (39). For example, exosomes derived from epithelial ovarian cancer cells inhibit the cytotoxicity of NK cells by downregulating the natural killer group 2 member D receptor on NK cells (40).
Assisting tumor cells in escaping immune cell attack
The circulating tumor cells in the blood must escape the surveillance of the immune system for survival. Platelets help in protecting the tumor cells from the immune cells; they adhere to the circulating tumor cell surface to assist it to escape the destruction by obstructing the interactions between NK cells and tumor cells (41).
In addition, platelets also enhance the aggressiveness of tumor cells. Pang et al (42) demonstrated that the thrombin-activated platelets induced tumor cells to become more aggressive, and that the platelet membrane proteins p-selectin and glycoprotein IIb/IIIa were beneficial for the tumor cells, as they helped them to exude from blood vessels. Moreover, activated platelet-derived microparticles (PMPs) also contain p-selectin and glycoprotein IIb/IIIa. Co-culturing PMPs with tumor cells increased the proliferation, invasion and metastasis abilities of tumor cells, such as prostate and gastric cancer cells (43,44). The upregulation of PMPs is related to a poor prognosis and cancer development (45).
Participation in the establishment of a pre-metastatic microenvironment
Organotropic metastasis indicates that the metastatic site is a result of the complicated interaction between tumor and stroma within the host organ, rather than being selected at random. Paget first explained this by proposing the ‘seed and soil hypothesis’ in 1889. The ‘seed and soil hypothesis’ suggested that metastases only formed when the seeds (circulating tumor cells) and the soil (host organ) were compatible (46). Based on this, the establishment of a microenvironment before metastasis is essential for tumor cells to colonize the metastatic site. For example, TDEs containing miR-122 act on the non-cancerous cells in the pre-metastatic microenvironment, downregulating the expression of alanine kinase in the non-cancerous cells, decreasing glucose uptake and utilization, and thereby elevating nutritional support for the tumor cells (47). TDEs from pancreatic ductal adenocarcinoma induce the formation of the microenvironment before liver metastasis in mice and elevate the occurrence of liver metastasis (48).
Classification and biological functions of circRNAs
circRNAs are generated through splicing events during the maturation of the corresponding precursor mRNAs subject to RNA polymerase II transcription (49). According to the parental gene components, circRNAs are classified into 3 subtypes (50): Exon-intron circRNAs (elciRNAs), circular intronic RNAs and exonic circRNAs (ecircRNAs). Among them, only ecircRNAs contain back-spliced exon sequences, which are mainly distributed in the cytoplasm and function as miRNA or RNA-binding protein sponges. Today, the term circRNA is commonly applied to describe ecircRNAs, in which downstream donor-exons splice to the upstream acceptor-exons (51). Further studies have uncovered the functions of circRNAs, and the biological functions of each circRNA subtype are suggested to be different across different types of cells (52,53).
circRNAs play a role in the processes regulating human physiology and pathology. Several biological functions of circRNAs have been demonstrated; for instance, they act as miRNA sponges and affect the downstream target genes of miRNAs, regulate parental gene transcription, modulate alternative splicing and interact with RBPs (Fig. 2). The present study attempts to provide an overview of the molecular mechanisms underlying the regulation of various biological processes by circRNAs.
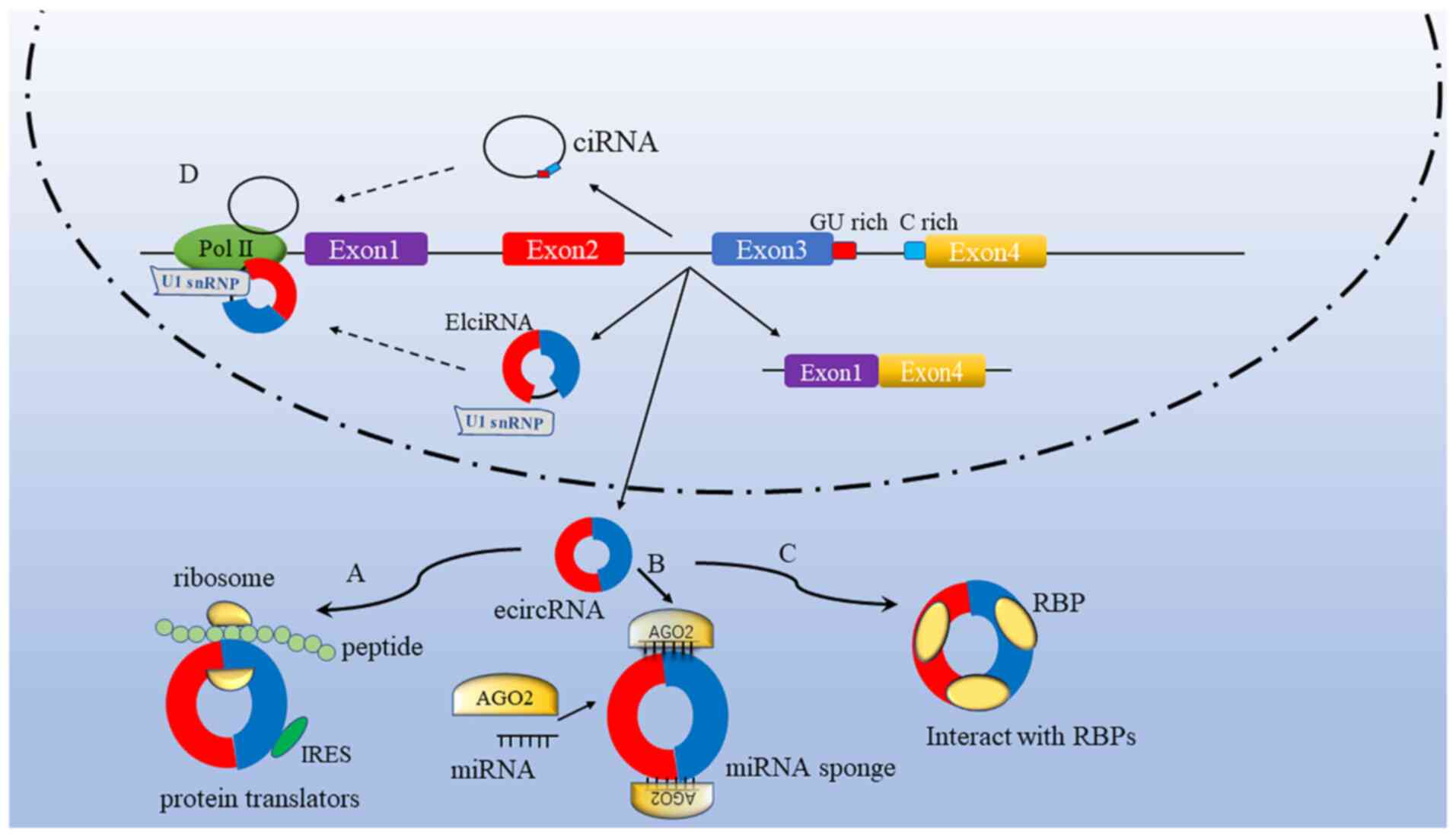 |
Figure 2.
Multiple functions of circRNAs. (A) A subset of circRNAs are involved in protein coding. Protein coding by circRNAs is also possible with an IRES. (B) A majority of circRNAs function as miRNA sponges that interact with miRNA-Ago2 (representative protein) complexes to suppress the functions of miRNAs. (C) circRNAs bind with RBPs to form RNA-protein complexes, thus affecting their functions and translocation. (D) For transcriptional regulation, elcircRNAs and ciRNAs induce the transcription of parental genes by binding transcription complexes to the host promoters. circRNA, circular RNA; IRES, internal ribosome entry site; miRNA, microRNA; Ago2, argonaute 2; RBP, RNA-binding protein; elciRNA, exon-intron circRNA; ciRNA, circular intronic RNA; ecircRNA, exonic circRNA; snRNP, small nuclear ribonucleoprotein; Pol II, RNA polymerase II.
|
Competitive endogenous RNA and miRNA sponges
Some RNAs with corresponding miRNA response elements competitively bind to miRNA sites to affect their target genes, thereby affecting tumor behavior. This process is referred to as ‘miRNA sponging’. Additionally, circRNAs have also been shown to competitively bind to miRNA sites, and affect the expression of miRNA and downstream target genes (54,55). Furthermore, circRNAs serve as tumor suppressors or oncogenes in tumorigenesis as they act as miRNA sponges through response elements. circRNAs also affect cancer development. As early as 1993, Capel et al (56) showed through use of sequence analysis that circRNA sex-determining region Y (Sry), which was derived from the sex-determining region of the mouse Y chromosome, possessed 16 binding sites for miR-138, and it inhibited the expression of miR-138 and its downstream target genes. In 2013, Hansen et al (8) found that circRNA ciRS-7, which is also known as the circRNA sponge for miR-7, possessed >70 miR-7-specific binding sites. The miR-7 sponge suppressed miR-7 activity, thus increasing miR-7 expression and that of its downstream target genes. At present, bioinformatics analysis has helped identify tens of thousands of circRNAs capable of miRNA sponging (57,58).
Participation in protein translation
Although circRNA is defined as a non-coding RNA, some studies (59,60) report that circRNAs possess open reading frames, which are presented in the form of proteins or polypeptides. In 1995, Chen and Sarnow (61) found that circRNAs containing internal ribosome entry sites (IRESs) might potentially induce the recruitment of ribosomes together with the initiation of translation, but no protein was encoded in the IRES-lacking circRNAs.
Regulation of transcription and alternative splicing
circRNAs are abundant in the nuclei and can bind to RNA polymerase II, thus regulating the process of transcription. For example, Ci-ankrd52 is mainly located near the transcription site and is a positive regulator of the RNA polymerase II (Pol II) complex, which exerts the cis-regulation of its parent gene elciRNA in combination with U1 small nuclear ribonucleoprotein (snRNP) to form the elciRNA-U1snRNP complex using a special binding method. Specifically, this complex binds to Pol II to enhance the transcription of the parent gene through cis-regulation (62). circRNAs are stably expressed in cells, display tissue specificity and are present predominantly in the cytoplasm. Studies have shown that circRNAs also participate in alternative splicing (63). circMBL is a circRNA formed from exon 2 of the muscleblind gene that encodes the splicing factor MBL. The flanking introns of circMBL possess the conservative binding sites of muscleblind and display strong and specific MBL binding. Regulating the MBL content potently impacts the biosynthesis of circMBL, which depends upon the binding sites of MBL (64).
Interaction with RBPs
RBPs are capable of binding to circRNA, and they also participate in nearly all cellular processes, such as differentiation, apoptosis and proliferation. circRNAs act as protein sponges, which promote the regulatory effects of RBPs on protein expression. For instance, human and drosophila circMBL binds to the MBL protein at a variety of binding sites (64). Also, Forkhead box O3 (Foxo3) is a protein encoded by the FOXO3 gene, while circFoxo3 is one of the most researched circRNAs at present, and has been found to serve as an adapter to connect cyclin-dependent kinase 2 (CDK2) with p21 (65). circFoxo3 also induces p21 to release CDK2, and p21 phosphorylates cyclin E and cyclin A while promoting the cell cycle. Furthermore, circFoxo3 promotes the combination of p53 and the oncogene murine double minute 2, and accelerates p53 degradation (66).
Exosomes and circRNAs
circRNAs in exosomes are mainly 200–600 bp in length. Li et al (13) performed circRNA sequencing analyses using the MHCC-LM3 HCC cell line and exosomes secreted by these cells, and discovered 5,484 and 6,751 circRNAs in the cells and exosomes, respectively. The circRNA content in the exosomes was nearly twice as high as that in the cells. The sequencing analyses on the normal human serum exosomes identified a total of 1,215 circRNAs, indicating the abundance of circRNAs in serum exosomes. Tumor tissue also continuously releases exosomes during its growth (67). circRNAs are stable in exosomes, which are also stable at room temperature. Therefore, the detection of circRNAs in humoral exosomes is easy and facilitates the diagnosis and prognosis of patients. In addition, Li et al (13) performed circRNA sequencing analyses on patients with CRC and found that, compared with that in healthy individuals, 67 types of circRNA were missing from the exosomes of patients, and an additional 257 types of circRNA were detected. Most of the circRNAs are housekeeping genes. PCR results showed that circ-kelch domain containing 10 can be used to distinguish between tumor patients and the non-tumor population, serving as a potential tumor biomarker. Exosomes are secreted by most cells in the body and contain multiple endogenous active substances (13). Exosomes can be stably stored in peripheral body fluids, making it easy to obtain and store exosomes (68). Importantly, they reflect the physiological and pathological changes in cells or tissues from which they are derived. When taken up by the recipient cells, exosomes impact the functionality of the recipient cell in a variety of physiological ways, especially since miRNA can mediate RNA interference (69). Numerous studies (70–72) have been performed on exosomal proteins, nucleic acids and other biologically active substances, aimed at finding new potential biomarkers, but there are still few studies on the circRNAs in exosomes. Among the thousands of circRNAs discovered using sequencing, microarray analysis and other methods, only a small proportion have been validated using PCR; therefore, further research to check whether the circRNAs in plasma exosomes can be used as biomarkers is warranted.
The exosomes secreted by cells come in contact with recipient cells and fuse to release their contents into recipient cells, thereby achieving the transfer of intracellular proteins, RNA and lipids (73). In such exosome-mediated non-contact cell communication, the TDEs play a dual regulatory role in either suppressing or promoting tumors using the paracrine and endocrine pathways. Moreover, exosomes derived from non-tumor cells also participate in tumor growth (74). Typically, the differences in the functions of exosomes are ascribed to the different types of cells and their contents. Generally, exosomes derived from tumor cells promote tumors (27), in contrast to those obtained from non-tumor cells; for instance, exosomes obtained from the immune cells mainly play a role in tumor inhibition (75). In general, exosomes affect tumor initiation and progression. In recent years, it has been suggested that exosome-derived circRNAs are closely related to tumor metastasis. For example, Li et al (76) found that the expression of circ_0044516 in exosomes from patients with prostate cancer was upregulated and inhibition of expression could inhibit the metastasis of tumor cells. In the current review, studies on the novel mechanisms and functions of exosomal circRNAs for regulating the initiation of DSTs are summarized. In addition, the association between the dysregulation of exosomal circRNAs and DST metastasis is also assessed.
Role of exosomal circRNAs in tumor proliferation
Exosomal circRNAs and HCC
HCC is the seventh most frequently occurring cancer in the world and the second most common cause of cancer mortality (77); it has a high degree of malignancy and a 5-year survival rate of <10% (78). HCC is a primary tumor originating in hepatocytes and is mainly caused by chronic cirrhosis and the gradual progression of hepatitis B or C. The pathogenesis of HCC is not fully elucidated yet, but it is characterized by rapid progression, a poor prognosis, a high mortality rate and early intrahepatic metastasis (79). Therefore, early detection and early intervention are particularly important to improve the prognosis and long-term survival rate of patients with HCC.
Zhang et al (80) demonstrated that the exosomal circRNAs released by adipocytes promoted HCC growth by sponging miR-34a and activated the ubiquitin-specific protease 7 (USP7)/Cyclin A2 pathway. Also, exosomes derived from adipose tissue upregulated USP7, which decreased oncogene ubiquitination.
Exosomal circRNAs also exist that suppress HCC cell proliferation. Chen et al (81) reported that circ0051443 level was significantly lower in the plasma and tissue exosomes in patients with HCC compared with that in healthy controls. circ_0051443 was transmitted from normal cells to HCC cells via exosomes and suppressed malignant behavior by promoting cell apoptosis and arresting the cell cycle. Consequently, exosome circ_0051443 could be used as a predictor and potential therapeutic target of HCC.
Exosomal circRNAs and CRC
Studies have shown that the pathology of CRC is characterized by multiple steps and stages, from benign polyps to invasive adenocarcinoma and distant metastasis (82). Such pathological changes are caused by the inactivation or abnormal activation of the protein-encoding oncogenes and tumor suppressor genes.
circ-formin 2 (circFMN2) may serve as a new treatment target and biomarker for CRC. Li et al (83) demonstrated for the first time that exosomal circFMN2 promoted the proliferation of CRC by directly binding with miR-1182 and subsequently decreasing the inhibition of human telomerase reverse transcriptase. A dual-luciferase reporter assay was performed to further validate this result and it suggested that the knockdown of circFMN2 inhibited the growth of CRC in vivo. In addition, circFMN2 expression was detectable in the extracted serum exosomes derived from patients with CRC, and circFMN2 expression was inversely correlated with miR-1182 in serum exosomes (83).
Role of exosomal circRNAs in tumor metastasis
Exosomal circRNAs are involved in the regulation of tumor metastasis. This review section emphasizes the functions of various aberrantly expressed exosomal circRNAs during tumor metastasis (Table I).
 |
Table I.
Exosomal circRNAs involved in tumor metastasis: Signaling pathways and functions.
|
Table I.
Exosomal circRNAs involved in tumor metastasis: Signaling pathways and functions.
| First author, year |
Exosomal circRNA |
Cancer type |
Regulation |
Signaling pathway |
Function/clinical association |
(Refs.) |
| Wang et al, |
circPTGR1 |
HCC |
Upregulated |
circPTGR1/ |
Increases invasive and |
(86) |
| 2019 |
|
|
|
miR-449a/ |
migratory capacities in |
|
| |
|
|
|
MET |
poorly metastatic and |
|
| |
|
|
|
|
non-metastatic cells |
|
| Liu et al, |
circMMP2 |
HCC |
Upregulated |
circMMP2/ |
Promotes tumor metastasis. |
(87) |
| 2020 |
|
|
|
miR-136-5p |
Associated with low overall |
|
| |
|
|
|
|
survival of patients with |
|
| |
|
|
|
|
HCC |
|
| Huang et al, |
circRNA _100338 |
HCC |
Upregulated |
Not |
The metastatic ability of |
(88) |
| 2020 |
|
|
|
mentioned |
HCC cells can be enhanced |
|
| |
|
|
|
|
by influencing the |
|
| |
|
|
|
|
proangiogenic activity |
|
| |
|
|
|
|
by regulating angiogenesis |
|
| Li et al, |
circZNF652 |
HCC |
Upregulated |
circZNF652/ |
Contributes to HCC cell |
(89) |
| 2020 |
|
|
|
miR-29a-3p/ |
proliferation, migration, |
|
| |
|
|
|
GUCD1 |
invasion and glycolysis |
|
| Li et al, |
circPDE8A |
Pancreatic |
Upregulated |
circPDE8A/ |
Plays a vital role during |
(92) |
| 2018 |
|
cancer |
|
miR-338/ |
cancer metastasis. Serves as |
|
| |
|
|
|
MACC1/ |
an efficient biomarker for |
|
| |
|
|
|
MET/ERK |
predicting the prognosis and |
|
| |
|
|
|
or AKT |
diagnosis of pancreatic |
|
| |
|
|
|
|
cancer |
|
| Li et al, |
circIRAS |
Pancreatic |
Downregulated |
circIRAS/ |
Decreases ZO-1 and |
(93) |
| 2018 |
|
cancer |
|
miR-122/ZO-1/ |
miR-122 expression, |
|
| |
|
|
|
RhoA/F-actin |
increases and the |
|
| |
|
|
|
|
permeability of the |
|
| |
|
|
|
|
endothelial monolayer, |
|
| |
|
|
|
|
enhances tumor metastasis |
|
| |
|
|
|
|
and invasion |
|
| Wang et al, |
circ_ 0000284 |
Cholangio |
Upregulated |
circ_0000284/ |
Upregulates the circ-0000284 |
(100) |
| 2019 |
|
carcinoma |
|
miR-637/ |
level and stimulates cell |
|
| |
|
|
|
LY6E |
proliferation, invasion and |
|
| |
|
|
|
|
migration |
|
| Zhang et al, |
circNRIP |
GC |
Upregulated |
circNRIP1/ |
Sponges miRNA and |
(102) |
| 2019 |
|
|
|
miR-149-5p/ |
promotes the proliferation, |
|
| |
|
|
|
AKT1/mTOR |
migration and invasion of |
|
| |
|
|
|
|
GC cells |
|
| Xie et al, |
circSHKBP1 |
GC |
Upregulated |
circSHKBP1/ |
Suppresses HSP90 |
(103) |
| 2020 |
|
|
|
miR-582-3p/ |
degradation and promotes |
|
| |
|
|
|
HUR/VEGF |
cell proliferation, migration, |
|
| |
|
|
|
|
invasion and angiogenesis |
|
| Lu et al, |
circRanGAP1 |
GC |
Upregulated |
circRanGAP1/ |
Enhances GC cell |
(104) |
| 2020 |
|
|
|
miR-877-3p/ |
migration and invasion. |
|
| |
|
|
|
VEGFA |
Suppresses tumor growth |
|
| |
|
|
|
|
and metastasis |
|
| Zhong et al, |
circ_ 0032821 |
GC |
Upregulated |
circ_0032821/ |
Boosts the proliferation, |
(107) |
| 2021 |
|
|
|
miR-515-5p/ |
migration, and invasion |
|
| |
|
|
|
SOX9 |
|
|
| Yao et al, |
circPVT1 |
GC |
Upregulated |
circPVT1/ |
Modulates autophagy, |
(108) |
| 2021 |
|
|
|
miR-30a-5p/ |
invasion, and apoptosis |
|
| |
|
|
|
YAP1 |
|
|
| Liu et al, |
circ_0026611 |
ESCC |
Upregulated |
Not |
Is significantly upregulated |
(109) |
| 2021 |
|
|
|
mentioned |
in ESCC with lymph node |
|
| |
|
|
|
|
metastasis and is a |
|
| |
|
|
|
|
predictor of ESCC |
|
| |
|
|
|
|
prognosis |
|
| Zang, |
circ_0000337 |
ESCC |
Upregulated |
circ_0000337/ |
Promotes DDP resistance |
(110) |
| 2021 |
|
|
|
miR-377-3p/ |
and metastasis in |
|
| |
|
|
|
JAK2 |
DDP-sensitive esophageal cancer cells |
|
| Zhao et al, |
circABCC1 |
CRC |
Upregulated |
circABCC1/ |
Mediates cell stemness and |
(112) |
| 2020 |
|
|
|
β-catenin/ |
metastasis |
|
| |
|
|
|
Wnt pathway |
|
|
| Yang et al, |
circ_133 |
CRC |
Upregulated |
circ_133/ |
Transports to normoxic |
(113) |
| 2020 |
|
|
|
miR-133a/ |
cancer cells and promotes |
|
| |
|
|
|
GEF-H1/ |
cell migration |
|
| |
|
|
|
RhoA |
|
|
| Shang et al, |
circPACRGL |
CRC |
Upregulated |
circPACRGL/ |
Promotes cell proliferation, |
(114) |
| 2020 |
|
|
|
miR-142-3p/ |
migration and invasion. |
|
| |
|
|
|
miR-506-3p- |
Plays an oncogenic role in |
|
| |
|
|
|
TGF-β1 |
CRC proliferation and |
|
| |
|
|
|
|
metastasis |
|
Exosomal circRNAs and HCC
Several studies have reported the effect of circRNAs on HCC (84,85), and the presence of circRNAs in exosomes has also been verified. However, the effects of exosomal circRNAs on highly metastatic HCC cells are rarely investigated. Wang et al (86) found that exosomal circ-prostaglandin reductase 1 (circPTGR1) enhanced the metastasis of HCC through the miR-449a-MET signaling pathway. In the study, exosomal circRNAs obtained from highly metastatic (LM3), poorly metastatic (97L) and non-metastatic (HepG2) cells were sequenced. The highly metastatic cells transferred their contents to the poorly metastatic and non-metastatic cells via exosomes, leading to increased invasive and migratory capacities in the recipient cells. During this process, circPTGR1 affected the poorly metastatic recipient cells by decreasing the interaction between miR-449a and MET within, destroying TME homeostasis and promoting HCC metastasis.
Liu et al (87) speculated that exosomal circMMP2 may be a novel biomarker for HCC treatment. The study found that circMMP2 was delivered by 97H- or LM3-secreted exosomes to L02 and HepG2 cells, and promoted metastasis in HCC by sponging miR-136-5p to enhance MMP2 expression. A high level of circMMP2 and a low level of miR-136-5p was associated with low overall survival rates in patients with HCC.
Huang et al (88) showed for the first time that circ_100338 was highly expressed in both metastatic HCC cells and their secreted exosomes. The study reported that the overexpression or knockdown of exosomal circ_100338 significantly enhanced or decreased the invasive abilities of the HCC cells, respectively. The results suggested that the metastatic ability of HCC cells could be enhanced by transferring exosomal circRNA-100338 to receptor human umbilical venous endothelial cells (HUVECs) and influencing their proangiogenic activity by regulating angiogenesis.
In addition to the aforementioned studies, Li et al (89) explored the function of exosomal circ-zinc finger protein 652 (circZNF652) in HCC. The study found that exosomal circZNF652 was upregulated in the serum and HCC cells of patients with HCC. Exosomal circZNF652 contributed to HCC cell proliferation, migration, invasion and glycolysis via the miR-29a-3p/guanylyl cyclase domain containing 1 axis.
Exosomal circRNAs and pancreatic cancer
Pancreatic cancer has the worst prognosis among the gastrointestinal system cancers, and it ranks fourth in the world in terms of number of cancer-related deaths (90). Metastasis is a primary cause of death among patients with pancreatic cancer (91). Pancreatic cancer occurrence, development and metastasis involve intricate biological processes comprising several signaling pathways and target genes. Therefore, it is important to identify and verify the specific gene and its molecular mechanism that shows a causal relationship with pancreatic cancer metastasis in the diagnosis and treatment of pancreatic cancer.
circ-phosphodiesterase 8A (circPDE8A), which was discovered using microarray analyses of pancreatic ductal adenocarcinoma (PDAC) cells in liver metastasis, plays an important role in tumor invasion. Li et al (92) discovered that exosomal circPDE8A enhanced metastasis through the miR-338/MET transcriptional regulator MACC1/MET signaling pathway in pancreatic cancer (Fig. 3). In addition, circPDE8A was identified to be a miR-338 sponge and regulated MET (one of the tyrosine kinase receptors). miR-338 is a critical oncogene in the epithelial tumor subset. Li et al (92) discovered an association between circPDE8A upregulation and lymphatic metastasis, poor prognosis and high TNM stage in patients suffering from pancreatic cancer. Furthermore, circPDE8A secreted by the tumor was introduced to the circulation via exosomes, and the exosomal circPDE8A in the plasma showed an association with metastasis and a poor prognosis in PDAC cases. Therefore, the aforementioned results suggested that exosomal circRNA potentially serves as an efficient and suitable biomarker for PDAC.
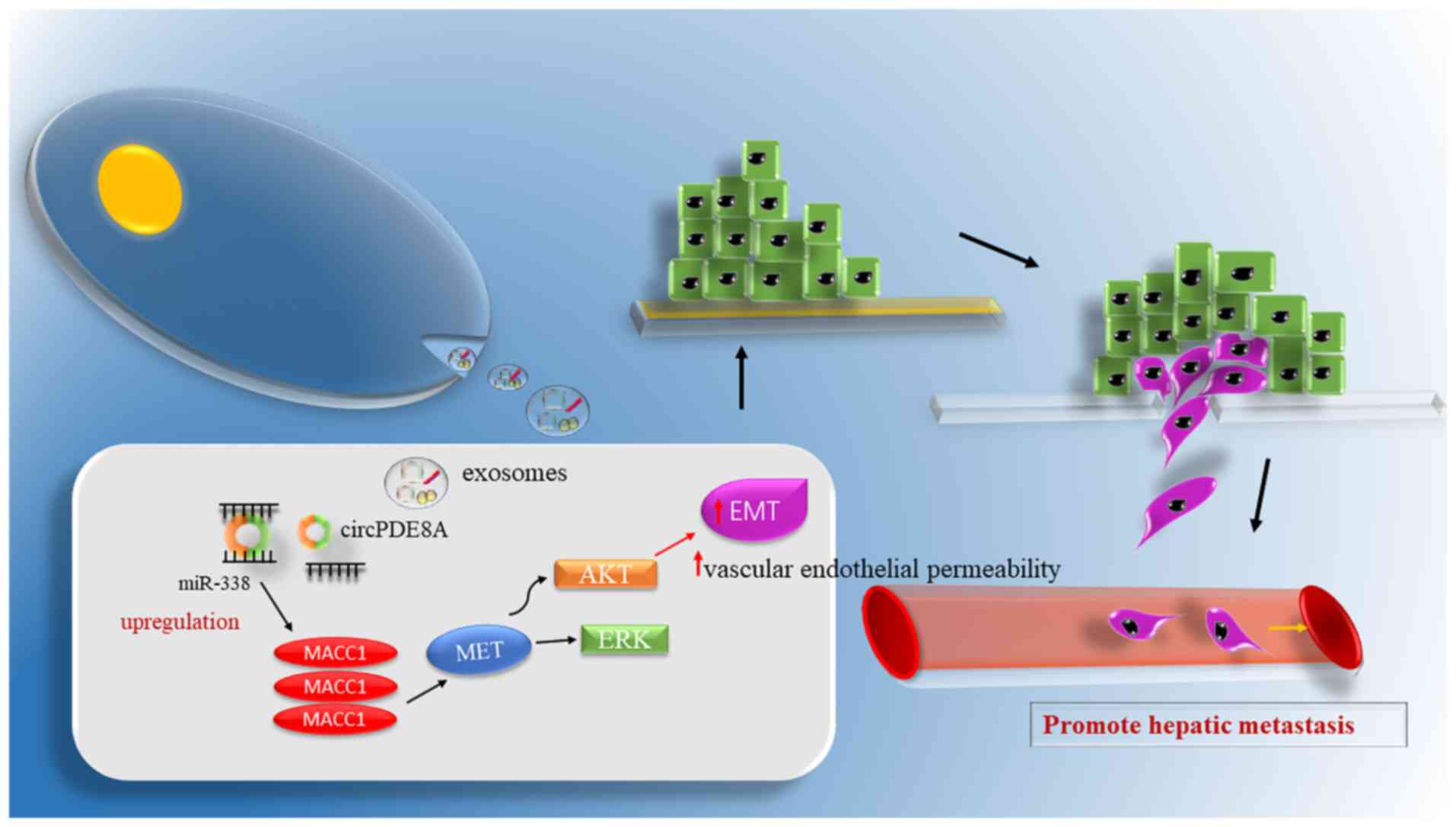 |
Figure 3.
Exosomal circRNAs affect tumor metastasis. Exosomal circPDE8A plays the role of a competing endogenous RNA, sponging miR-338 and inducing the invasive growth of tumors via the AKT, ERK, MET and MACC1 pathways. Finally, such processes result in EMT, increase the permeability of vascular endothelial cells and promote liver metastasis of pancreatic cancer. circ/circRNA, circular RNA; miR, microRNA; EMT, epithelial-mesenchymal transition; MACC1, MET transcriptional regulator MACC1; PDE8A, phosphodiesterase 8A.
|
As shown in previous studies, exosomes are rich in circRNAs and take part in intercellular communication. However, it remains unclear whether the exosomal circRNA-associated mechanisms contribute to cancer metastasis and invasion. As discovered by Li et al (93), circIARS accessed HUVECs through pancreatic cancer cell-derived exosomes, which elevated the permeability of the endothelial monolayer, thereby promoting tumor metastasis and invasion. Specifically, circIARS sponged miR-122, which elevated the activity of the downstream target gene Ras homolog family member A, upregulated F-actin levels, downregulated ZO-1 levels and increased the permeability of the endothelial monolayer. Therefore, these processes boost the metastasis and invasion of the tumor. Exosomal circRNAs serve as vital biomarkers to diagnose and predict tumor prognosis early.
Several studies have shown that exosomes derived from bone marrow mesenchymal stem cells (BM-MSCs) are crucial regulators of the progression of various tumors, such as colorectal cancer and multiple myeloma (94,95). Yao et al (96) reported for the first time that circ_0030167 derived from BM-MSCs regulated miR-338-5p, enhanced Wnt inhibitory factor 1 expression and inhibited the Wnt8/β-catenin pathway, ultimately inhibiting the invasion, migration, proliferation and stemness of pancreatic cancer cells.
Exosomal circRNAs and cholangiocarcinoma
Cholangiocar-cinoma is a cancer originating from the epithelial cells of the bile duct that has a high malignancy grade (97). The morbidity and mortality rates of cholangiocarcinoma have increased over time (98). Currently, radical surgical resection is the only curative method that achieves the long-term survival of patients with cholangiocarcinoma (99). However, as cholangiocarcinoma is not associated with chronic liver disease, the early symptoms are not obvious in patients, and diagnostic markers with high specificity are lacking. Therefore, a number of patients are affected by intrahepatic or extrahepatic lymph node metastasis (LNM), while only 30% are eligible for radical surgery, yielding a low surgical resection rate. The postoperative recurrence rate is up to 50%, even for patients who have the chance of surgery, and the 5-year survival rate is 20–45%. Therefore, it is of great clinical significance to investigate the molecular mechanism underlying cholangiocarcinoma invasion and metastasis, and to identify novel therapeutic targets. Wang et al (100) found that the upregulation of circ_0000284 promoted cholangiocarcinoma cell proliferation, invasion and migration both in vitro and in vivo. Additionally, circ_0000284 was upregulated within the cholangiocarcinoma cell-derived exosomes. Even more unexpected was that the cholangiocarcinoma cell-derived exosomes upregulated the circ-0000284 level, and stimulated the proliferation and migration of adjacent normal cells.
Exosomal circRNAs and GC
GC has been identified as the second leading cause of cancer-related deaths in the world, and is associated with a poor prognosis, a high relapse rate, a decreased cure rate and a decreased survival rate (101). GC treatment is complicated due to heterogeneity in the tumor tissue resulting from both epigenetic and genetic alterations. Gastric cancer easily develops metastasis due to changes in the TME. Therefore, it is important to understand the mechanism of genetic links in GC metastasis. The functions and mechanisms of exosomal circRNA in GC are not known.
Zhang et al (102) reported for the first time that exosomal communication across GC cells was able to transmit circ-nuclear receptor interacting protein 1 (circNRIP1). Specifically, circNRIP1 was significantly upregulated within human GC tissues, where it promoted GC cell invasion, proliferation and migration by acting as a miR-149-5p sponge. The knockdown of exosome-derived circNRIP1 blocked GC cell invasion, migration and proliferation, and downregulated AKT serine/threonine kinase 1. In addition, by injecting a luciferase label into circNRIP1-containing exosomes in nude mice through the tail vein, Zhang et al (102) also proved that exosomal circNRIP1 enhanced tumor metastasis. circNRIP1 expression was higher in GC clinical samples compared with that in adjacent normal tissues. The study verified that circNRIP1 transmitted by exosomes upregulated EMT markers to promote GC metastasis via EMT.
Xie et al (103) demonstrated that exosomal circ-SH3KBP1 binding protein 1 (circSHKBP1) regulated the miR-582-3p/HUR/VEGF pathway, suppressed heat shock protein 90 degradation, and promoted GC cell proliferation, migration, invasion and angiogenesis in vitro and in vivo, while the suppression of circSHKBP1 had the opposite effect. Liquid biopsy of serum exosomes targeting circSHKBP1 helped diagnose and predict the prognosis of GC. Lu et al (104) identified that circ-Ran GTPase activating protein 1 (circRanGAP1) was markedly upregulated in exosomes from the plasma of patients with GC. Plasma exosomes in these patients enhanced GC cell migration and invasion. The study found that silencing of circRanGAP1 markedly suppressed tumor growth and metastasis of GC in vivo. In terms of mechanism, circRanGAP1 sponged miR-877-3p to upregulate VEGFA expression and promote GC progression.
Several studies have reported that exosome-mediated circRNAs are associated with tumor drug resistance (105,106). Zhong et al (107) reported that circ_0032821 contributed to oxaliplatin (OXA) resistance in gastric cancer cells. circ_0032821 was highly expressed in exosomes secreted by OXA-resistant GC cells. circ_0032821-containing exosomes secreted by OXA-resistant GC cells boosted the proliferation, migration and invasion of OXA-sensitive GC cells by acting as an miR-515-5p sponge to regulate SOX9 expression. Yao et al (108) highlighted a novel mechanism for the development of cisplatin (DDP) resistance in GC cells. It was discovered that exosomal circPVT1 accelerated DDP resistance by modulating autophagy, invasion and apoptosis via the miR-30a-5p/yes-associated protein 1 axis in GC cells. It was hypothesized that these exosomal circRNAs are promising diagnostic biomarkers for GC treatment.
Exosomal circRNAs and esophageal carcinoma
Esophageal carcinoma is an aggressive disease, with 5-year survival rates ranging from 15–25%. Due to this aggressiveness, 30% of esophageal tumors have already invaded adjacent tumors at the time of diagnosis (90). Therefore, early detection and diagnosis are important.
Recently, Liu et al (109) elaborated on the diagnostic and prognostic value of serum exosome circ_0026611 in esophageal squamous cell carcinoma (ESCC). The study found that the expression of circ_0026611 in serum exosomes from cases of ESCC with LNM was higher than that for ESCC without LNM. This suggested that circ_0026611 is a predictor of ESCC prognosis.
Exosome-mediated transfer of circRNAs is reported to be related to drug resistance in esophageal cancer. Zang et al (110) found that circ_0000337-containing exosomes secreted by DDP-resistant esophageal cancer cells promoted DDP resistance and metastasis in DDP-sensitive esophageal cancer cells in vitro through the miR-377-3p/Janus kinase 2 axis.
Exosomal circRNAs and CRC
CRC is one of the most prevalent types of cancer globally, causing multiple cancer-related deaths (111). A growing body of evidence suggests that circRNAs play a regulatory role in the initiation and development of cancer.
Zhao et al (112) attempted to explore the regulatory role of circ-ATP binding cassette subfamily C member 1 (circABCC1) in CRC for the first time. Exosomes from CD133+ cells are known to regulate tumor progression in cancer types such as breast cancer and glioblastoma. A study found that exosomes from CD133+ cells carrying circABCC1 mediated cell stemness and metastasis in CRC. circABCC1 was shown to bind with β-catenin in the cell nucleus, which activated the Wnt pathway.
Hypoxia is one of the important characteristics of solid tumors. The metastatic potential of hypoxic tumor cells is different from that of normoxic tumor cells. Yang et al (113) observed that hypoxia-derived exosomes transported circ-133 to normoxic cancer cells and promoted cell migration via the miR-133a/GEF-H1/RhoA axis in CRC.
Shang et al (114) found that circ-Parkin coregulated like (circPACRGL) was significantly upregulated in CRC cells after treatment with TDEs. Moreover, circPACRGL promoted CRC cell proliferation, migration and invasion, and the differentiation of N1 to N2 neutrophils via the miR-142-3p/miR-506-3p-TGF-β1 axis. The study suggested that cancer-derived exosomal circPACRGL plays an oncogenic role in CRC proliferation and metastasis.
Conclusions and future perspectives
Tumor metastasis is a crucial factor when accounting for the high mortality rate in cancer patients. The main purpose of therapy for cancer patients is to block tumor metastasis and enhance the survival rate. Continuous efforts have been made to explore the mechanism of tumor metastasis. It is now known that, before the metastatic cells reach the metastatic site, the environment at the metastatic site, including the cell state, blood supply and ECM, needs to be altered to host and help proliferation of the metastatic cells (115). Similarly, the instability of the tumor cell genome is recognized as the driving force for tumor metastasis; although this is still a hypothesis, it is supported by most researchers in the field.
With rapid developments in biotechnology, particularly in RNA sequencing technology, circRNAs have now been confirmed to act as sponges of miRNAs. Researchers can control circRNAs to regulate miRNAs, and thereby further modulate the occurrence and development of gastrointestinal tumors. circRNAs are stable in serum exosomes, and consequently, circRNA sampling has the advantages of reduced trauma for patients, compared with surgery to remove tissues, and ease of access. Also, circRNAs are highly stable due to their special loop structures, and are widely distributed in various tissues and organs. Tissue expression is temporally and spatially different, which regulates tumor proliferation and metastasis. Thus, circRNAs are expected to serve as biomarkers for the early diagnosis of DSTs. Currently, clinical diagnosis of DSTs is mainly performed using imaging and biopsy. Further studies are required to demonstrate whether DSTs can be diagnosed by detecting the levels of circRNAs.
At present, an increasing number of studies are being performed to elucidate the role of circRNAs as tumor biomarkers, providing a clearer picture of their role in physiology. However, research on circRNAs in exosomes and their role as tumor biomarkers is lacking. Exosomes carry a large number of circRNAs, and they can transfer information from one cell to another. Exosomes also promote tumor metastasis by improving the invasion capacity of tumor cells, establishing the TME before metastasis and improving the tendency of organs to guide tumor metastasis. Serum circRNA levels are different in patients at various stages of cancer compared with in healthy patients, which increases the number of biomarkers available for diagnosis, treatment and evaluation of prognosis. The exploration of the mechanism of tumor occurrence and development is a long-term project, and exosomes represent a key method for the transmission of information for tumor cells; therefore, greater efforts should be made to further explore the limited information available, which will help to provide strong guidance for tumor diagnosis and treatment strategies in the next few years.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
BS searched the literature and wrote the manuscript. KS searched the literature and revised the manuscript. All authors have read and approved the final manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Nagtegaal ID, Odze RD, Klimstra D, Paradis V, Rugge M, Schirmacher P, Washington KM, Carneiro F and Cree IA; WHO Classification of Tumours Editorial Board, : The 2019 WHO classification of tumours of the digestive system. Histopathology. 76:182–188. 2020. View Article : Google Scholar
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar
|
|
3
|
Hu JX, Zhao CF, Chen WB, Liu QC, Li QW, Lin YY and Gao F: Pancreatic cancer: A review of epidemiology, trend, and risk factors. World J Gastroenterol. 27:4298–4321. 2021. View Article : Google Scholar
|
|
4
|
Yang L, Ying X, Liu S, Lyu G, Xu Z, Zhang X, Li H, Li Q, Wang N and Ji J: Gastric cancer: Epidemiology, risk factors and prevention strategies. Chin J Cancer Res. 32:695–704. 2020. View Article : Google Scholar
|
|
5
|
Jiang K, Dong C, Yin Z, Li R, Mao J, Wang C, Zhang J, Gao Z, Liang R, Wang Q and Wang L: Exosome-derived ENO1 regulates integrin α6β4 expression and promotes hepatocellular carcinoma growth and metastasis. Cell Death Dis. 11:9722020. View Article : Google Scholar
|
|
6
|
Sexton RE, Al Hallak MN, Diab M and Azmi AS: Gastric cancer: A comprehensive review of current and future treatment strategies. Cancer Metastasis Rev. 39:1179–1203. 2020. View Article : Google Scholar
|
|
7
|
Salzman J, Gawad C, Wang PL, Lacayo N and Brown PO: Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 7:e307332012. View Article : Google Scholar
|
|
8
|
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function as efficient microRNA sponges. Nature. 495:384–388. 2013. View Article : Google Scholar
|
|
9
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar
|
|
10
|
Zhu Z, Huang J, Li X, Xing J, Chen Q, Liu R, Hua F, Qiu Z, Song Y, Bai C, et al: Gut microbiota regulate tumor metastasis via circRNA/miRNA networks. Gut Microbes. 12:17888912020. View Article : Google Scholar
|
|
11
|
Boriachek K, Islam MN, Möller A, Salomon C, Nguyen NT, Hossain MSA, Yamauchi Y and Shiddiky MJA: Biological functions and current advances in isolation and detection strategies for exosome nanovesicles. Small. 14:17021532018. View Article : Google Scholar
|
|
12
|
Lakshmi S, Hughes TA and Priya S: Exosomes and exosomal RNAs in breast cancer: A status update. Eur J Cancer. 144:252–268. 2021. View Article : Google Scholar
|
|
13
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 25:981–984. 2015. View Article : Google Scholar
|
|
14
|
Johnstone RM, Adam M, Hammond JR, Orr L and Turbide C: Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 262:9412–9420. 1987. View Article : Google Scholar
|
|
15
|
Braicu C, Tomuleasa C, Monroig P, Cucuianu A, Berindan-Neagoe I and Calin GA: Exosomes as divine messengers: Are they the hermes of modern molecular oncology? Cell Death Differ. 22:34–45. 2015. View Article : Google Scholar
|
|
16
|
Larios J, Mercier V, Roux A and Gruenberg J: ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J Cell Biol. 219:e2019041132020. View Article : Google Scholar
|
|
17
|
Elia N, Sougrat R, Spurlin TA, Hurley JH and Lippincott-Schwartz J: Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci USA. 108:4846–4851. 2011. View Article : Google Scholar
|
|
18
|
Wei D, Zhan W, Gao Y, Huang L, Gong R, Wang W, Zhang R, Wu Y, Gao S and Kang T: RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 31:157–177. 2021. View Article : Google Scholar
|
|
19
|
Chen WX, Liu XM, Lv MM, Chen L, Zhao JH, Zhong SL, Ji MH, Hu Q, Luo Z, Wu JZ and Tang JH: Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS One. 9:e952402014. View Article : Google Scholar
|
|
20
|
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, Wang G, Wu P, Wang H, Jiang L, et al: Exosomal circRNAs: Biogenesis, effect and application in human diseases. Mol Cancer. 18:1162019. View Article : Google Scholar
|
|
21
|
Gruenberg J: Life in the lumen: The multivesicular endosome. Traffic. 21:76–93. 2020. View Article : Google Scholar
|
|
22
|
Pattanakitsakul SN, Poungsawai J, Kanlaya R, Sinchaikul S, Chen ST and Thongboonkerd V: Association of Alix with late endosomal lysobisphosphatidic acid is important for dengue virus infection in human endothelial cells. J Proteome Res. 9:4640–4648. 2010. View Article : Google Scholar
|
|
23
|
Huang D, Sun W, Zhou Y, Li P, Chen F, Chen H, Xia D, Xu E, Lai M, Wu Y and Zhang H: Mutations of key driver genes in colorectal cancer progression and metastasis. Cancer Metastasis Rev. 37:173–187. 2018. View Article : Google Scholar
|
|
24
|
Asem M, Young AM, Oyama C, Claure De La Zerda A, Liu Y, Yang J, Hilliard TS, Johnson J, Harper EI, Guldner I, et al: Host Wnt5a potentiates microenvironmental regulation of ovarian cancer metastasis. Cancer Res. 80:1156–1170. 2020. View Article : Google Scholar
|
|
25
|
Yang S, Liu Y, Li MY, Ng CSH, Yang SL, Wang S, Zou C, Dong Y, Du J, Long X, et al: FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer. 16:1242017. View Article : Google Scholar
|
|
26
|
Fang T, Lv H, Lv G, Li T, Wang C, Han Q, Yu L, Su B, Guo L, Huang S, et al: Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 9:1912018. View Article : Google Scholar
|
|
27
|
Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu Y, Zhang Z, Cai S, Xu Y, Li X, et al: Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol. 13:1562020. View Article : Google Scholar
|
|
28
|
Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, Beerling E, Schiffelers RM, de Wit E, Berenguer J, Ellenbroek SIJ, et al: In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 161:1046–1057. 2015. View Article : Google Scholar
|
|
29
|
Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O'Connor ST, Chin AR, et al: Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 25:501–515. 2014. View Article : Google Scholar
|
|
30
|
Pastushenko I and Blanpain C: EMT transition states during tumor progression and metastasis. Trends Cell Biol. 29:212–226. 2019. View Article : Google Scholar
|
|
31
|
Shu DY, Butcher E and Saint-Geniez M: EMT and EndMT: Emerging roles in age-related macular degeneration. Int J Mol Sci. 21:42712020. View Article : Google Scholar
|
|
32
|
Xue M, Chen W, Xiang A, Wang R, Chen H, Pan J, Pang H, An H, Wang X, Hou H and Li X: Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol Cancer. 16:1432017. View Article : Google Scholar
|
|
33
|
Aga M, Bentz GL, Raffa S, Torrisi MR, Kondo S, Wakisaka N, Yoshizaki T, Pagano JS and Shackelford J: Exosomal HIF1α supports invasive potential of nasopharyngeal carcinoma-associated LMP1-positive exosomes. Oncogene. 33:4613–4622. 2014. View Article : Google Scholar
|
|
34
|
Zhou Y, Xia L, Lin J, Wang H, Oyang L, Tan S, Tian Y, Su M, Wang H, Cao D and Liao Q: Exosomes in nasopharyngeal carcinoma. J Cancer. 9:767–777. 2018. View Article : Google Scholar
|
|
35
|
Kim J, Kim TY, Lee MS, Mun JY, Ihm C and Kim SA: Exosome cargo reflects TGF-β1-mediated epithelial-to-mesenchymal transition (EMT) status in A549 human lung adenocarcinoma cells. Biochem Biophys Res Commun. 478:643–648. 2016. View Article : Google Scholar
|
|
36
|
Olejarz W, Kubiak-Tomaszewska G, Chrzanowska A and Lorenc T: Exosomes in angiogenesis and anti-angiogenic therapy in cancers. Int J Mol Sci. 21:58402020. View Article : Google Scholar
|
|
37
|
Bao L, You B, Shi S, Shan Y, Zhang Q, Yue H, Zhang J, Zhang W, Shi Y, Liu Y, et al: Metastasis-associated miR-23a from nasopharyngeal carcinoma-derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene. 37:2873–2889. 2018. View Article : Google Scholar
|
|
38
|
Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, Zhou K, Liu X, Ren X, Wang F, et al: Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 9:53952018. View Article : Google Scholar
|
|
39
|
Kugeratski FG and Kalluri R: Exosomes as mediators of immune regulation and immunotherapy in cancer. FEBS J. 288:10–35. 2021. View Article : Google Scholar
|
|
40
|
Labani-Motlagh A, Israelsson P, Ottander U, Lundin E, Nagaev I, Nagaeva O, Dehlin E, Baranov V and Mincheva-Nilsson L: Differential expression of ligands for NKG2D and DNAM-1 receptors by epithelial ovarian cancer-derived exosomes and its influence on NK cell cytotoxicity. Tumour Biol. 37:5455–5466. 2016. View Article : Google Scholar
|
|
41
|
Maurer S and Ferrari de Andrade L: NK cell interaction with platelets and myeloid cells in the tumor milieu. Front Immunol. 11:6088492020. View Article : Google Scholar
|
|
42
|
Pang JH, Coupland LA, Freeman C, Chong BH and Parish CR: Activation of tumour cell ECM degradation by thrombin-activated platelet membranes: Potentially a P-selectin and GPIIb/IIIa-dependent process. Clin Exp Metastasis. 32:495–505. 2015. View Article : Google Scholar
|
|
43
|
Gay LJ and Felding-Habermann B: Contribution of platelets to tumour metastasis. Nat Rev Cancer. 11:123–134. 2011. View Article : Google Scholar
|
|
44
|
Haemmerle M, Stone RL, Menter DG, Afshar-Kharghan V and Sood AK: The platelet lifeline to cancer: Challenges and opportunities. Cancer Cell. 33:965–983. 2018. View Article : Google Scholar
|
|
45
|
Helley D, Banu E, Bouziane A, Banu A, Scotte F, Fischer AM and Oudard S: Platelet microparticles: A potential predictive factor of survival in hormone-refractory prostate cancer patients treated with docetaxel-based chemotherapy. Eur Urol. 56:479–484. 2009. View Article : Google Scholar
|
|
46
|
Paget S: The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 8:98–101. 1989.
|
|
47
|
Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J, Chow A, O'Connor ST, Li S, Chin AR, et al: Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 17:183–194. 2015. View Article : Google Scholar
|
|
48
|
Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, et al: Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 17:816–826. 2015. View Article : Google Scholar
|
|
49
|
Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ and Xu RH: Circular RNA: Metabolism, functions and interactions with proteins. Mol Cancer. 19:1722020. View Article : Google Scholar
|
|
50
|
Patop IL, Wüst S and Kadener S: Past, present, and future of circRNAs. EMBO J. 38:e1008362019. View Article : Google Scholar
|
|
51
|
Ragan C, Goodall GJ, Shirokikh NE and Preiss T: Insights into the biogenesis and potential functions of exonic circular RNA. Sci Rep. 9:20482019. View Article : Google Scholar
|
|
52
|
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019. View Article : Google Scholar
|
|
53
|
Chen LL: The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 21:475–490. 2020. View Article : Google Scholar
|
|
54
|
Zhang Y, Liu Q and Liao Q: CircHIPK3: A promising cancer-related circular RNA. Am J Transl Res. 12:6694–6704. 2020.
|
|
55
|
Zeng Z, Xia L, Fan S, Zheng J, Qin J, Fan X, Liu Y, Tao J, Liu Y, Li K, et al: Circular RNA CircMAP3K5 Acts as a MicroRNA-22-3p sponge to promote resolution of intimal hyperplasia via TET2-mediated smooth muscle cell differentiation. Circulation. 143:354–371. 2021. View Article : Google Scholar
|
|
56
|
Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P and Lovell-Badge R: Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 73:1019–1030. 1993. View Article : Google Scholar
|
|
57
|
Panda AC: Circular RNAs Act as miRNA sponges. Adv Exp Med Biol. 1087:67–79. 2018. View Article : Google Scholar
|
|
58
|
Liu Z, Yu Y, Huang Z, Kong Y, Hu X, Xiao W, Quan J and Fan X: CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 10:9002019. View Article : Google Scholar
|
|
59
|
Zang J, Lu D and Xu A: The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J Neurosci Res. 98:87–97. 2020. View Article : Google Scholar
|
|
60
|
Huang A, Zheng H, Wu Z, Chen M and Huang Y: Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics. 10:3503–3517. 2020. View Article : Google Scholar
|
|
61
|
Chen CY and Sarnow P: Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 268:415–417. 1995. View Article : Google Scholar
|
|
62
|
Okholm TLH, Sathe S, Park SS, Kamstrup AB, Rasmussen AM, Shankar A, Chua ZM, Fristrup N, Nielsen MM, Vang S, et al: Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Med. 12:1122020. View Article : Google Scholar
|
|
63
|
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL and Yang L: Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 26:1277–1287. 2016. View Article : Google Scholar
|
|
64
|
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 56:55–66. 2014. View Article : Google Scholar
|
|
65
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P and Yang BB: Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44:2846–2858. 2016. View Article : Google Scholar
|
|
66
|
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z and Yang BB: Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 24:357–370. 2017. View Article : Google Scholar
|
|
67
|
Whiteside TL: Tumor-derived exosomes and their role in cancer progression. Adv Clin Chem. 74:103–141. 2016. View Article : Google Scholar
|
|
68
|
Wu X, Zheng T and Zhang B: Exosomes in Parkinson's disease. Neurosci Bull. 33:331–338. 2017. View Article : Google Scholar
|
|
69
|
Srinivasan S, Vannberg FO and Dixon JB: Lymphatic transport of exosomes as a rapid route of information dissemination to the lymph node. Sci Rep. 6:244362016. View Article : Google Scholar
|
|
70
|
Lin Z, Gu Y, Zhou R, Wang M, Guo Y, Chen Y, Ma J, Xiao F, Wang X and Tian X: Serum exosomal proteins F9 and TSP-1 as potential diagnostic biomarkers for newly diagnosed epilepsy. Front Neurosci. 14:7372020. View Article : Google Scholar
|
|
71
|
Wang J, Ni J, Beretov J, Thompson J, Graham P and Li Y: Exosomal microRNAs as liquid biopsy biomarkers in prostate cancer. Crit Rev Oncol Hematol. 145:1028602020. View Article : Google Scholar
|
|
72
|
Cheng J, Meng J, Zhu L and Peng Y: Exosomal noncoding RNAs in Glioma: Biological functions and potential clinical applications. Mol Cancer. 19:662020. View Article : Google Scholar
|
|
73
|
Masaoutis C, Mihailidou C, Tsourouflis G and Theocharis S: Exosomes in lung cancer diagnosis and treatment. From the translating research into future clinical practice. Biochimie. 151:27–36. 2018. View Article : Google Scholar
|
|
74
|
Whiteside TL: Exosome and mesenchymal stem cell cross-talk in the tumor microenvironment. Semin Immunol. 35:69–79. 2018. View Article : Google Scholar
|
|
75
|
Greening DW, Gopal SK, Xu R, Simpson RJ and Chen W: Exosomes and their roles in immune regulation and cancer. Semin Cell Dev Biol. 40:72–81. 2015. View Article : Google Scholar
|
|
76
|
Li T, Sun X and Chen L: Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J Cell Biochem. 121:2118–2126. 2020. View Article : Google Scholar
|
|
77
|
McGlynn KA, Petrick JL and El-Serag HB: Epidemiology of hepatocellular carcinoma. Hepatol. 73 (Suppl 1):S4–S13. 2021. View Article : Google Scholar
|
|
78
|
Wang H, Lu Z and Zhao X: Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J Hematol Oncol. 12:1332019. View Article : Google Scholar
|
|
79
|
Villanueva A: Hepatocellular carcinoma. N Engl J Med. 380:1450–1462. 2019. View Article : Google Scholar
|
|
80
|
Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, Fan Q, Li J, Ning T, Tian F, et al: Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 38:2844–2859. 2019. View Article : Google Scholar
|
|
81
|
Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, Chen L, Liu Q, He P and Ye Y: Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 475:119–128. 2020. View Article : Google Scholar
|
|
82
|
Sepulveda AR, Hamilton SR, Allegra CJ, Grody W, Cushman-Vokoun AM, Funkhouser WK, Kopetz SE, Lieu C, Lindor NM, Minsky BD, et al: Molecular biomarkers for the evaluation of colorectal cancer: Guideline from the American society for clinical pathology, college of American pathologists, association for molecular pathology, and the American society of clinical oncology. J Clin Oncol. 35:1453–1486. 2017. View Article : Google Scholar
|
|
83
|
Li Y, Li C, Xu R, Wang Y, Li D and Zhang B: A novel circFMN2 promotes tumor proliferation in CRC by regulating the miR-1182/hTERT signaling pathways. Clin Sci (Lond). 133:2463–2479. 2019. View Article : Google Scholar
|
|
84
|
Huang G, Liang M, Liu H, Huang J, Li P, Wang C, Zhang Y, Lin Y and Jiang X: CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell Death Dis. 11:10652020. View Article : Google Scholar
|
|
85
|
Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song X, Gorshkov K, Sun Q, Lin H, Zheng X, et al: CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct Target Ther. 5:2982020. View Article : Google Scholar
|
|
86
|
Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, Zhang Y, Li H, Zhang Q, Yang Y and Chen G: Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine. 40:432–445. 2019. View Article : Google Scholar
|
|
87
|
Liu D, Kang H, Gao M, Jin L, Zhang F, Chen D, Li M and Xiao L: Exosome-transmitted circ_MMP2 promotes hepatocellular carcinoma metastasis by upregulating MMP2. Mol Oncol. 14:1365–1380. 2020. View Article : Google Scholar
|
|
88
|
Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, Zhou J and Tang ZY: Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. 39:202020. View Article : Google Scholar
|
|
89
|
Li Y, Zang H, Zhang X and Huang G: Exosomal Circ-ZNF652 promotes cell proliferation, migration, invasion and glycolysis in hepatocellular carcinoma via miR-29a-3p/GUCD1 axis. Cancer Manag Res. 12:7739–7751. 2020. View Article : Google Scholar
|
|
90
|
Siegel RL, Miller KD and Jemal A: Cancer statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar
|
|
91
|
Xu JS, Liao KL, Wang X, He J and Wang XZ: Combining bioinformatics techniques to explore the molecular mechanisms involved in pancreatic cancer metastasis and prognosis. J Cell Mol Med. 24:14128–14138. 2020. View Article : Google Scholar
|
|
92
|
Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K, Zhao X, Zhang Y, Zhen P, Zhu J and Li X: Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 432:237–250. 2018. View Article : Google Scholar
|
|
93
|
Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, Liu H, Bi H, Liu X and Li X: Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 37:1772018. View Article : Google Scholar
|
|
94
|
Li H and Li F: Exosomes from BM-MSCs increase the population of CSCs via transfer of miR-142-3p. Br J Cancer. 119:744–755. 2018. View Article : Google Scholar
|
|
95
|
Roccaro AM, Sacco A, Maiso P, Azab AK, Tai YT, Reagan M, Azab F, Flores LM, Campigotto F, Weller E, et al: BM mesenchymal stromal cell-derived exosomes facilitate multiple myeloma progression. J Clin Invest. 123:1542–1555. 2013. View Article : Google Scholar
|
|
96
|
Yao X, Mao Y, Wu D, Zhu Y, Lu J, Huang Y, Guo Y, Wang Z, Zhu S, Li X and Lu Y: Exosomal circ_0030167 derived from BM-MSCs inhibits the invasion, migration, proliferation and stemness of pancreatic cancer cells by sponging miR-338-5p and targeting the Wif1/Wnt8/β-catenin axis. Cancer Lett. 512:38–50. 2021. View Article : Google Scholar
|
|
97
|
Vaquero J, Guedj N, Clapéron A, Nguyen Ho-Bouldoires TH, Paradis V and Fouassier L: Epithelial-mesenchymal transition in cholangiocarcinoma: From clinical evidence to regulatory networks. J Hepatol. 66:424–441. 2017. View Article : Google Scholar
|
|
98
|
Bertuccio P, Malvezzi M, Carioli G, Hashim D, Boffetta P, El-Serag HB, La Vecchia C and Negri E: Global trends in mortality from intrahepatic and extrahepatic cholangiocarcinoma. J Hepatol. 71:104–114. 2019. View Article : Google Scholar
|
|
99
|
DeOliveira ML: Liver transplantation for cholangiocarcinoma: Current best practice. Curr Opin Organ Transplant. 19:245–252. 2014. View Article : Google Scholar
|
|
100
|
Wang S, Hu Y, Lv X, Li B, Gu D, Li Y, Sun Y and Su Y: Circ-0000284 arouses malignant phenotype of cholangiocarcinoma cells and regulates the biological functions of peripheral cells through cellular communication. Clin Sci Lond Engl 1979. 133:1935–1953. 2019.
|
|
101
|
Tan Z: Recent advances in the surgical treatment of advanced gastric cancer: A review. Med Sci Monit. 25:3537–3541. 2019. View Article : Google Scholar
|
|
102
|
Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019. View Article : Google Scholar
|
|
103
|
Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, Ma P, Jiang H, Wu X, Shu Y and Xu T: Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer. 19:1122020. View Article : Google Scholar
|
|
104
|
Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW, Wang JB, Lin JX, Chen QY, Cao LL, et al: Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 471:38–48. 2020. View Article : Google Scholar
|
|
105
|
Ding C, Yi X, Wu X, Bu X, Wang D, Wu Z, Zhang G, Gu J and Kang D: Exosome-mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett. 479:1–12. 2020. View Article : Google Scholar
|
|
106
|
Hon KW, Ab-Mutalib NS, Abdullah NMA, Jamal R and Abu N: Extracellular vesicle-derived circular RNAs confers chemoresistance in colorectal cancer. Sci Rep. 9:164972019. View Article : Google Scholar
|
|
107
|
Zhong Y, Wang D, Ding Y, Tian G and Jiang B: Circular RNA circ_0032821 contributes to oxaliplatin (OXA) resistance of gastric cancer cells by regulating SOX9 via miR-515-5p. Biotechnol Lett. 43:339–351. 2021. View Article : Google Scholar
|
|
108
|
Yao W, Guo P, Mu Q and Wang Y: Exosome-derived Circ-PVT1 contributes to cisplatin resistance by regulating autophagy, invasion, and apoptosis via miR-30a-5p/YAP1 axis in gastric cancer cells. Cancer Biother Radiopharm. 36:347–359. 2021. View Article : Google Scholar
|
|
109
|
Liu S, Lin Z, Rao W, Zheng J, Xie Q, Lin Y, Lin X, Chen H, Chen Y and Hu Z: Upregulated expression of serum exosomal hsa_circ_0026611 is associated with lymph node metastasis and poor prognosis of esophageal squamous cell carcinoma. J Cancer. 12:918–926. 2021. View Article : Google Scholar
|
|
110
|
Zang R, Qiu X, Song Y and Wang Y: Exosomes mediated transfer of Circ_0000337 contributes to cisplatin (CDDP) resistance of esophageal cancer by regulating JAK2 via miR-377-3p. Front Cell Dev Biol. 9:6732372021. View Article : Google Scholar
|
|
111
|
Wan ML, Wang Y, Zeng Z, Deng B, Zhu BS, Cao T, Li YK, Xiao J, Han Q and Wu Q: Colorectal cancer (CRC) as a multifactorial disease and its causal correlations with multiple signaling pathways. Biosci Rep. 40:BSR202002652020. View Article : Google Scholar
|
|
112
|
Zhao H, Chen S and Fu Q: Exosomes from CD133+ cells carrying circ-ABCC1 mediate cell stemness and metastasis in colorectal cancer. J Cell Biochem. 121:3286–3297. 2020. View Article : Google Scholar
|
|
113
|
Yang H, Zhang H, Yang Y, Wang X, Deng T, Liu R, Ning T, Bai M, Li H, Zhu K, et al: Hypoxia induced exosomal circRNA promotes metastasis of colorectal cancer via targeting GEF-H1/RhoA axis. Theranostics. 10:8211–8226. 2020. View Article : Google Scholar
|
|
114
|
Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, Chen C, Chang W, Ping Y, Ji P, et al: Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p-TGF-β1 axis. Mol Cancer. 19:1172020. View Article : Google Scholar
|
|
115
|
Bergers G and Fendt SM: The metabolism of cancer cells during metastasis. Nat Rev Cancer. 21:162–180. 2021. View Article : Google Scholar
|

















