Introduction
Hepatocellular carcinoma (HCC) is the sixth most prevalent type of cancer and the third most common cause of cancer-related mortality (1,2). Considering that morbidity and mortality rates increase each year, HCC has become a growing public health concern worldwide (3), accounting for >80% of all liver cancer cases. Currently, early-stage HCC can be treated by surgical resection, liver transplantation and radiofrequency ablation (4). However, patients with early-stage HCC are asymptomatic and 70–80% of cases are diagnosed at an advanced stage (5). However, the effects of treatments, such as transcatheter arterial chemoembolization, radiofrequency ablation and liver transplantation are rather limited for advanced HCC. Notably, immunotherapy has become a hotspot in a number of cancers, including liver cancer. Certain immune checkpoint molecules, such as programmed cell death protein-1 (PD-1), PD-1 ligand 1 (PD-L1), cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4) are often the targets of immunotherapy (6,7). However, the majority of immune checkpoint inhibitors (ICIs) are effective in only a portion of HCC patients. For instance, nivolumab and pembrolizumab (PD-1 inhibitors) exhibit a response rate of 16–20% of in patients with advanced HCC (8,9). Therefore, the exploration of novel immunotherapy targets is of utmost urgency.
HCC is a typical inflammation-driven cancer, with almost 90% of the cases developing from chronic non-resolving inflammation, including chronic viral hepatitis, hepatic fibrosis and cirrhosis (10,11). Chronically unresolved inflammation can remodel the tumor microenvironment (TME) from immunoactivation to immunosuppression, through a variety of mechanisms that induce immune tolerance and enable tumor cells to evade immune surveillance, ultimately promoting tumor proliferation, invasion and metastasis (12). In addition, the liver is an organ with immune privilege properties (13), in which most antigens suffer immune tolerance. Approximately only 30% of early-stage HCC cases exhibit genomic evidence of immune activation (14). The dysfunction of immunity results in HCC development and progression.
The TME consists of the stromal and immune cells, which infiltrate a given tumor. The main function of these immune cells, particularly macrophages, dendritic cells (DCs) and other innate immune cells, within a TME of immune activation, would have been to identify and eliminate foreign pathogens, including tumor cells (15), whereas the immunosuppressive TME promotes tumor progression (16). Moreover, a number of studies have demonstrated that the TME can affect the therapeutic efficacy of ICIs (17,18). However, the underlying mechanisms through which HCC progresses through TME remodeling, thereby evading the innate immune surveillance and acquiring resistance to ICIs remain unclear (12,19). Therefore, further investigations into the mechanisms of immunosuppression in HCC are required, with the identification of effective approaches for immunotherapy as the main goal.
Herein, integrated bioinformatics analyses were performed, including differential gene expression, Gene Ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, gene set enrichment analyses (GSEA), survival analysis, multivariate Cox regression model analysis and Pearson's correlation analysis; the tumor immune dysfunction and exclusion (TIDE) algorithm was also used. Data were retrieved from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) database. The results of the present study indicated that collectin-12 (COLEC12), matrix metalloproteinase-12 (MMP12) and mucin-12 (MUC12) were the key innate immune-related genes (IIRGs) involved in HCC immunosuppression, possibly providing novel hypotheses for HCC immunotherapy.
Materials and methods
Sources of data
HCC mRNA expression profiles with clinical information of 374 liver hepatocellular carcinoma (LIHC) and 50 normal control samples were downloaded from TCGA database (https://portal.gdc.cancer.gov/, RNA-seq, Illumina) (20). In addition, high-sequence data of GSE17548 (21) and GSE83148 (22) (GPL570, Affymetrix Human Genome U133 Plus 2.0 Array) were retrieved from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). GSE17548 contained data for 37 liver tissues (17 HCC tissues and 20 liver cirrhosis tissues). Furthermore, six samples from healthy liver tissues in the GSE83148 dataset were selected to act as normal controls. The clinical information and sequencing data were acquired in accordance with the requirements of TCGA and the GEO databases. Thus, no approval from an ethics committee or consent procedure was needed.
Human subjects and ethical compliance
Surgical samples of HCC tissues (T1: male, 46 years old, stage II, BMI 24; T2: female, 50 years old, stage II, BMI 25; T3: male, 56 years old, stage III, BMI 23; and T4: female, 49 years old, stage III, BMI 27) and their corresponding adjacent normal liver tissues (N1, N2, N3 and N4) from 4 patients with HCC were obtained from the Biological Specimen Banks of the Second Affiliated Hospital of Nanchang University, China, following the approval of the Ethics Committee of the hospital (approval no. 20190913). Written informed consents were acquired from all study subjects, and all experiments were performed in compliance with the relevant guidelines and regulations. The surgical tissue samples were subjected to reverse transcription-quantitative PCR (RT-qPCR) and western blot analysis.
Identification of differentially expressed mRNAs (DEMs), GO annotation and KEGG pathway analyses
Firstly, all TCGA raw count data were normalized using the transcripts per million (TPM) method and transformed using the log2 approach. Subsequently, the DEMs between the HCC samples and normal liver tissues were screened using the ‘edgeR’ package downloaded from Bioconductor (23) in R (RStudio, Inc.; version 3.5.1; 64-bit; http://www.r-project.org/). Only the genes with an absolute value of log2 fold change |logFC|>2 and a corrected P-value <0.05 were defined as DEMs. A heatmap using ‘pheatmap’ R package was created, based on the expression value of some specific genes. In order to reveal the functions and signal transduction process, and analyze the biological significance of DEMs, GO annotation and KEGG pathway enrichment analyses were conducted. Biological processes (BP), cellular components (CC) and molecular functions (MF) are all included in the GO annotation. The Database for Annotation, Visualization, and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/; date accessed, November 20, 2020; species, human) (24) was used to conduct GO annotation and KEGG pathway enrichment analyses, which were visualized using the R package ‘ggplot2’ (25).
GSEA
The potential molecular mechanisms responsible for tumor immunosuppression were explored using GSEA (26), by analyzing the DEM matrix between HCC tissues and normal liver tissues. Innate immune signature gene sets were examined in Molecular Signatures Database v7.2 of GSEA (Broad Institute, Inc.), using the key words ‘innate immune’ and ‘Homo sapiens’. A nominal value of P<0.05 and false discovery rate (FDR) <0.25 were considered to indicate statistically significant differences.
Survival analysis and development of the innate immune-related risk signature
Survival analysis was conducted using the R packages ‘survival’ and ‘survminer’ based on TCGA dataset. The HCC samples were divided into two groups, according to individual gene expression as follows: High expression (TPM values greater than the median) and low expression (TPM values less than the median). Subsequently, Kaplan-Meier survival curves, in combination with a two-sided log-rank test were used to screen the survival-related IIRGs for patients with HCC with P<0.05. An innate immune-related risk signature was then developed. The individualized risk scores were calculated with coefficients and patients were divided into a high- and low-risk groups, according to the risk score's median cutoff. The risk score was established using the following formula: Risk score = (βmRNA1 × expression level of mRNA1) + (βmRNA2 × expression level of mRNA2) + (βmRNA3 × expression level of mRNA3) + … + (βmRNAn × expression level of mRNAn) (27). The Kaplan-Meier survival curve was drawn and the differences in overall survival (OS) between the high- and the low-risk group were compared. The time-dependent receiver operating characteristic (ROC) curves were drawn and the area under the curves (AUC) was calculated, in order to evaluate the predictive value of the signature for the 1-, 3- and 5-year OS, using the R package ‘survivalROC’ (28,29). Clinical features, including age, sex, body mass index (BMI), α-fetoprotein (AFP) levels, pathological stage, were collected from the TCGA database. Subsequently, univariate and multivariate Cox regression analyses were both performed, to verify whether the signature can predict HCC patient prognosis independently from these clinical features. A value of P<0.05 was considered to indicate a statistically significant difference.
Validation in GEO and the Human Protein Atlas
The mRNA expression of the immunosuppressive genes (COLEC12, MMP12 and MUC12) screened from HCC tissues was also validated using the GSE17548 and GSE83148 datasets. The protein expression levels of these genes were subsequently explored in the Human Protein Atlas (https://www.proteinatlas.org/) online database (30).
Western blot analysis
Western blot analysis was performed as previously described (31). Total protein of HCC and corresponding adjacent normal liver tissues was extracted using RIPA lysis buffer (Beyotime Institute of Biotechnology) with 1% PMSF. Next, the protein concentration was measured by Pierce BCA Protein assay kit (Thermo Fisher Scientific, Inc.). Briefly, 40 µg protein obtained from each HCC sample and their corresponding adjacent normal specimens were resolved using 10% SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. Following membrane blocking with 5% milk powder diluted in PBS for 1 h at room temperature, antigens were detected using the following primary antibodies: Anti-COLEC12 (1:1,000; ab-DF10165; Affinity Biosciences, Ltd.), anti-MUC12 (1:500; sc-377269; Santa Cruz Biotechnology, Inc.), anti-MMP12 (1:500; ab137444; Abcam) and anti-β-actin (1:1,000; M01263-2; Boster Biological Technology) at 4°C overnight. The membrane was then incubated with horseradish peroxidase-conjugated anti-rabbit (1:3,000; AS014; ABclonal Biotech Co., Ltd.) or anti-mouse IgG (1:1,000; AS003; ABclonal Biotech Co., Ltd.) secondary antibodies for 1.5 h at 25°C. All data were detected using the ChemiDoc™ Touch Imaging System and analyzed using the Image Lab 6.1 software (Bio-Rad Laboratories, Inc.). Moreover, COLEC12, MUC12 and MMP12 gene protein levels were respectively normalized against the level of β-actin protein.
RT-qPCR
Total RNA was extracted from frozen HCC tissues and their adjacent normal liver tissues using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, 40 µl of the isolated RNA were reverse transcribed into cDNA using the EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (cat. no. AE311-02; TransGen Biotech Co., Ltd.), followed by quantitative PCR (qPCR) with the PerfectStart™ Green qPCR SuperMix kit (cat. no. AQ601-01; TransGen Biotech Co., Ltd.) in accordance with a previously described protocol (28). Relative mRNA expression was normalized to β-actin mRNA, which was used as an internal control. The primer sequences for gene amplification were as follows: COLEC12 forward, 5′-AATCCTTCGGTTACAAGCGGT-3′ and reverse, 5′-ACTGTGATTGTTAGCAAGGCAC-3′; MMP12 forward, 5′-CATGAACCGTGAGGATGTTGA-3′ and reverse, 5′-GCATGGGCTAGGATTCCACC-3′; MUC12 forward, 5′-CCTGGAAACCTTAGCACCAG-3′ and reverse, 5′-GACAGACGCATTGTTTTCCAT-3′; and β-actin forward, 5′-GCACCACACCTTCTACAATG-3′ and reverse, 5′-TGCTTGCTGATCCACATCTG-3′. The mRNA levels of COLEC12, MUC12 and MMP12 were calculated using the 2−ΔΔCq method (32).
Pearson's correlation analysis and validation of immunotherapy response
A Pearson's correlation analysis of the genes between HCC tissues and normal liver tissues on the online Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/index.html) was conducted, based on TCGA-LIHC (33). The correlation coefficient (R) was used to indicate the strength of the connection between the target gene and immune checkpoint molecules or immunosuppressive cytokines genes. Subsequently, the TIDE algorithm was used to calculate the clinical efficacy of immune checkpoint inhibitors between different groups (34). The grouping method was performed according to the individual gene expression (COLEC12, MMP12 and MUC12) in the HCC samples of TCGA database, as follows: High expression group (TPM values greater than the median) and low expression group (TPM values less than the median). The immune checkpoint blockade (ICB) response scores were calculated by the TIDE algorithm, based on gene relative expression levels. The graphics were visualized using the R packages ‘ggplot2’ and ‘ggpubr (0.4.0)’.
Statistical analyses
Statistical analyses were performed and visualized using SPSS version 22.0 (IBM Corp.), R software (RStudio, Inc.; version 3.5.1) and GraphPad Prism v7.0 (GraphPad Software, Inc.). Continuous variables are presented as the mean ± SE or median, whereas categorized variables are presented as frequency/number (n) and proportion (%). Quantitative variables were analyzed using paired t-tests or a non-parametric Wilcoxon rank-sum test/Mann-Whitney U test. Univariate and multivariate Cox regression analyses were performed to estimate the predictive power of the innate immune risk signature and clinical features. P-values of P<0.05 and FDR <0.25 were considered to indicate statistically significant differences.
Results
Identification of DEMs and functional enrichment
A flow chart illustrating how the study was conducted is presented in Fig. 1. A total of 3,676 genes were identified as DEMs with a P-value <0.05 and |logFC| ≥2.0 when the HCC tissues (n=374) were compared with the normal liver tissues (n=50). Among the 3,676 genes, 3,392 were significantly upregulated, while 284 genes were significantly downregulated in the tumor tissues (P<0.05). ‘Extracellular region’, ‘extracellular space’, ‘integral component of plasma membrane’ and ‘blood microparticle’ were the main enriched terms in the GO-CC category, according to GO analysis (Fig. 2A; P<0.05). ‘Proteolysis’, ‘immunological response’, ‘oxidation-reduction process’ and ‘mitotic nuclear division’ were the enriched terms in the GO-BP annotations (Fig. 2B; P<0.05). Moreover, DEMs in the GO-MF category were mainly enriched in ‘serine-type endopeptidase activity’, ‘antigen binding’, ‘heme binding’ and ‘iron ion binding’ (Fig. 2C; P<0.05). Conversely, KEGG pathway analysis revealed that the DEMs were enriched in some KEGG pathways, including ‘retinol metabolism’, ‘Metabolism of xenobiotics by cytochrome P45’, ‘chemical carcinogenesis’, ‘mineral absorption’, ‘steroid hormone biosynthesis’, ‘Staphylococcus aureus infection’, ‘prion diseases’, ‘PPAR signaling pathway’, ‘PI3K-Akt signaling pathway’ and ‘p53 signaling pathway’, among others (Fig. 2D; P<0.05). The aforementioned results suggested that some of these DEMs may be involved in the immune response.
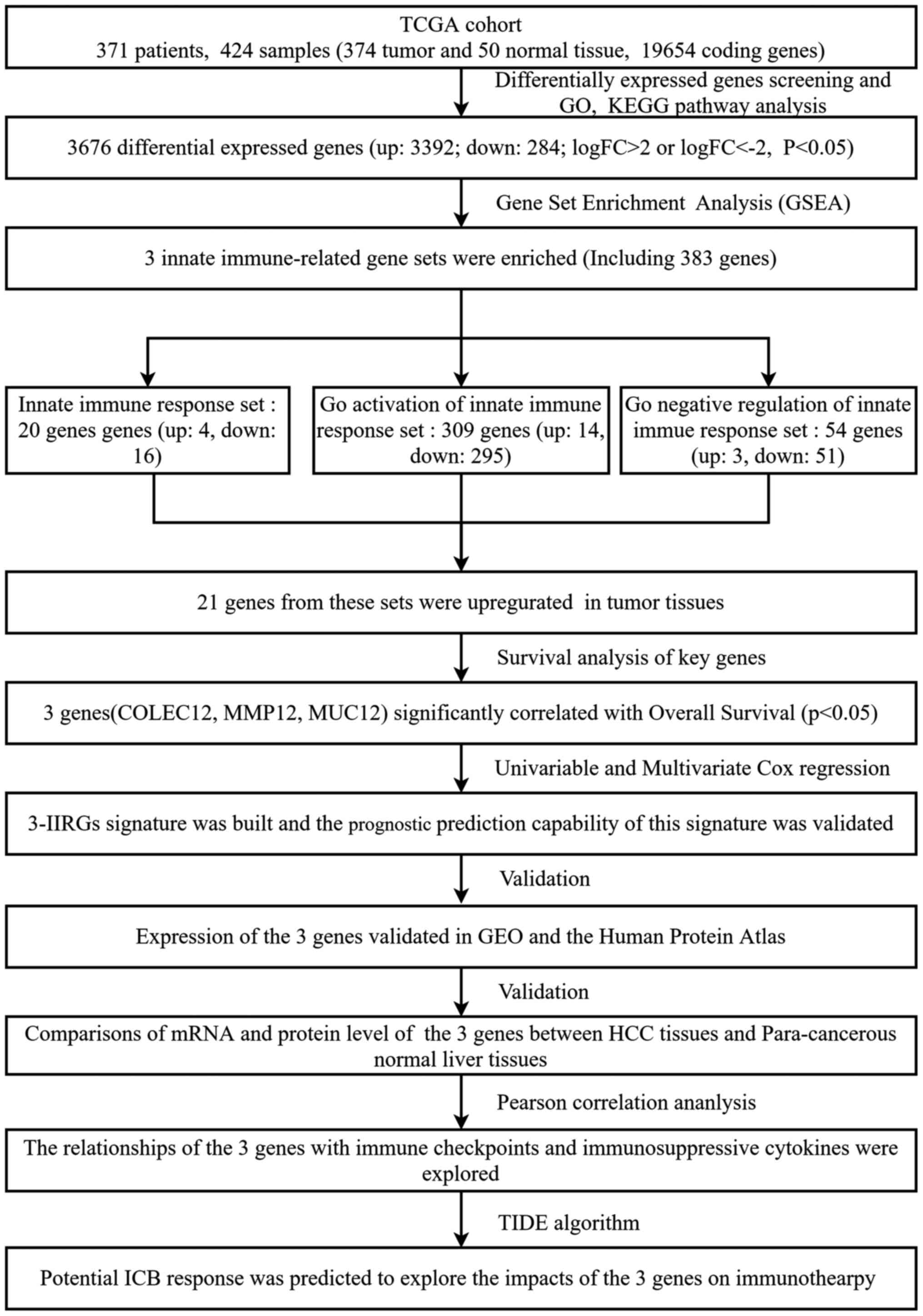 |
Figure 1.
Flow chart illustrating the protocols used in the present study. TCGA, The Cancer Genome Atlas; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; HCC, hepatocellular carcinoma; COLEC12, collectin-12 gene; MMP12, matrix metalloproteinase-12 gene; MUC12, mucin-12 gene; GEO, Gene Expression Omnibus; TIDE, tumor immune dysfunction and exclusion; ICB, immune checkpoint blockade.
|
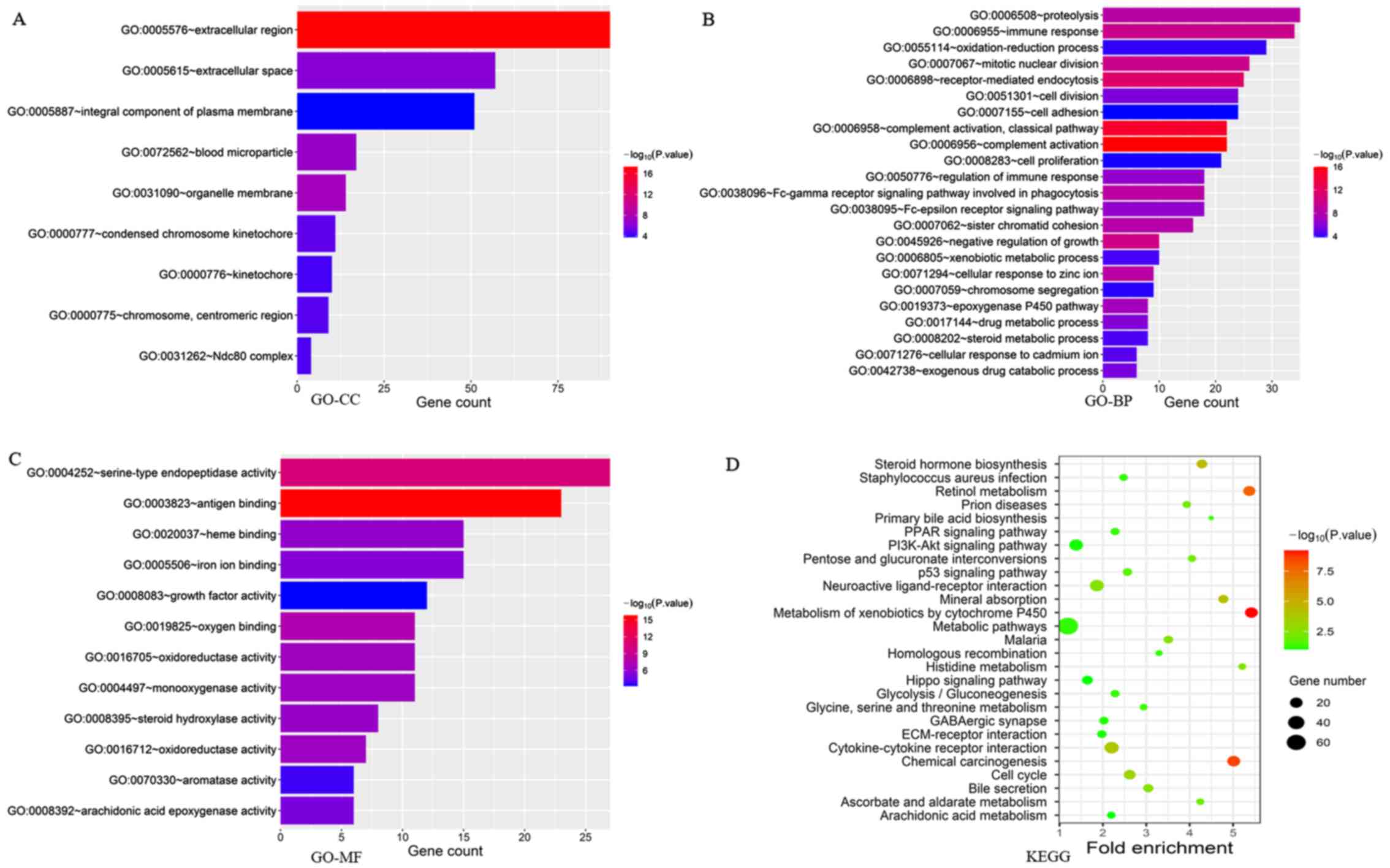 |
Figure 2.
Main GO terms associated with cellular components. (A) GO cellular components; (B) GO biological processes; (C) GO molecular functions; and (D) KEGG pathway analysis of DEMs between HCC (n=374) and normal liver tissues (n=50) in TCGA-LIHC dataset. GO, Gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; HCC, hepatocellular carcinoma; DEMs, differentially expressed mRNAs; TCGA, The Cancer Genome Atlas; LIHC, liver hepatocellular carcinoma.
|
Screening three genes based on survival analysis and development of the innate immune-related gene risk signature
DEMs were analyzed using GSEA to compare the HCC tissues to normal liver tissues at innate immune signature gene sets, in order to elucidate the underlying molecular mechanisms of immunosuppression. The results of the present study indicated that three innate immune signature gene sets, including the innate immune response gene set [enrichment score (ES), −0.48; normalized enrichment score (NES), −1.52; nominal P-value, P=0.037; FDR=0.134], GO activation of innate immune response gene set (ES, −0.28; NES, −1.27; nominal P-value, P=0.039; FDR=0.232) and the GO negative regulation of innate immune response gene set (ES, −0.33; NES, −1.17; nominal P-value, P=0.043; FDR=0.227) were enriched in the normal liver tissue group (Fig. 3A). However, no set was enriched in the HCC tissue group. Notably, there were 383 IIRGs in the three sets. Among these, 362 IIRGs were highly expressed in the normal liver tissues, while 21 IIRGs [BPI fold containing family B, member 1 (BPIFB1), Trem-like transcript 4 (TREML4), apolipoprotein A-IV (APOA4), S100 calcium-binding protein A1 (S100A1), hypothetical protein LOC55765 gene (INAVA), proteasome subunit α8 (PSMA8), regenerating islet-derived protein 3γ (REG3G), deleted in malignant brain tumors 1 (DMBT1), pyrin domain-containing protein 1 gene (PYDC1), MMP12, MUC15, proton-coupled folate transporter gene (SLC46A2), MUC13, MUC5B, Serpin B4 gene (SERPINB4), DC-STAMP Domain Containing 1 (DCST1), p21-activated kinase 2 (PAK3), COLEC12, MUC12, mucin-like protein 1 (MUCL1) and MUC5AC] were highly expressed in the HCC tissues. It was hypothesized that the proteins encoded by the 21 IIRGs can probably favor suppression of the innate immune response. The heatmap depicted the expression levels of these 21 IIRGs between the normal and HCC tissues in TCGA dataset (Fig. 3B).
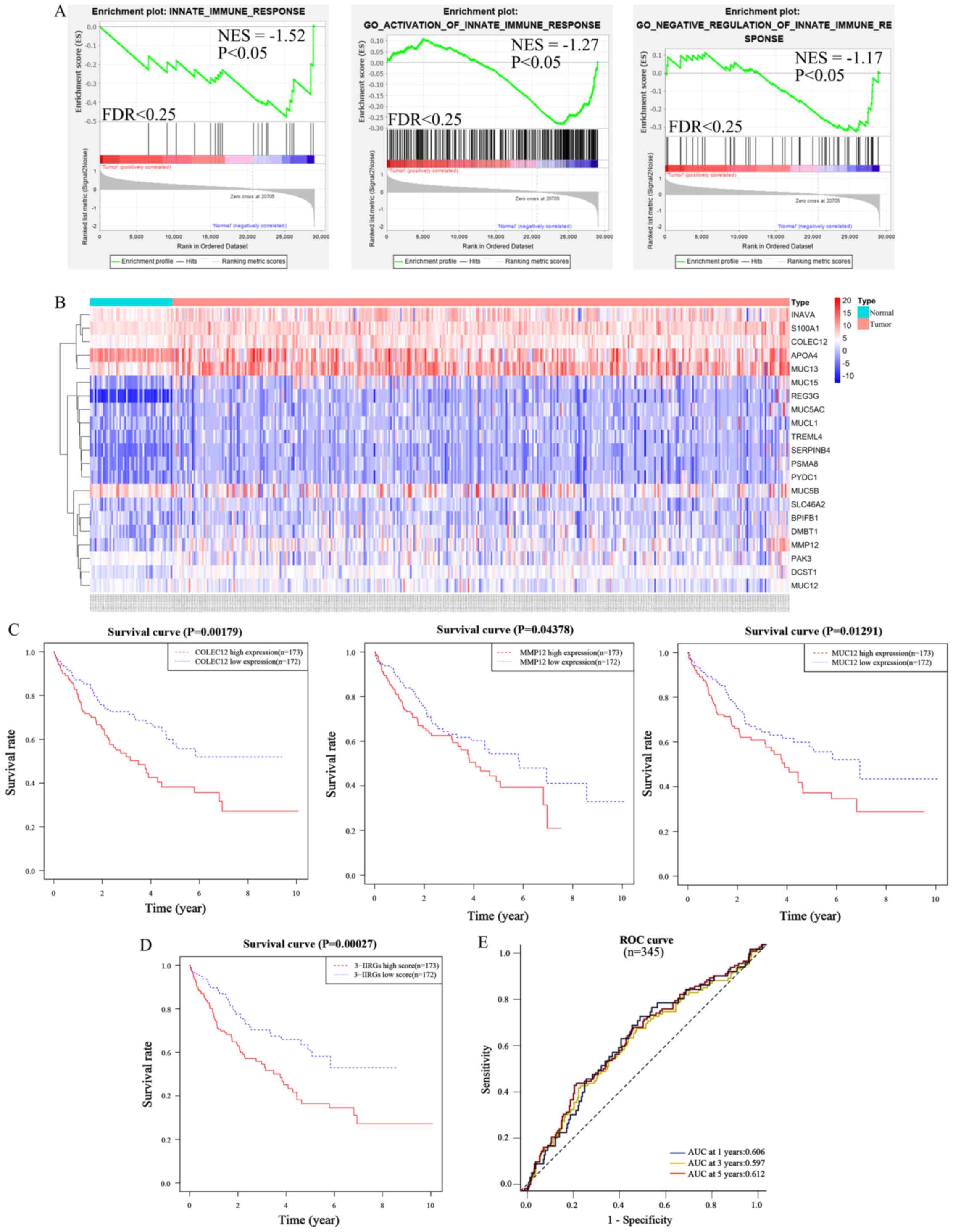 |
Figure 3.
COLEC12, MMP12 and MUC12 were screened from innate immune gene sets based on survival analysis and an innate immune risk signature was developed based on the three IIRGs. (A) Gene set enrichment plots of DEMs between HCC (n=374) and normal liver tissues (n=50) in TCGA-LIHC dataset: Innate immune response, GO activation of innate immune response and GO negative regulation innate immune response sets were enriched in normal liver tissues. (B) Heatmap of the 21 genes expression in HCC tissues (pink plot, n=374) and normal liver tissues (azure plot, n=50) from TCGA-LIHC dataset. (C) Kaplan-Meier OS curves according to the expression levels of COLEC12, MMP12, and MUC12 in HCC patients (n=345) from TCGA-LIHC dataset. (D) Kaplan-Meier OS curves according to the risk scores of the three IIRGs. (E) Time-ROC curve of the three-IIRG risk signature. COLEC12, collectin-12 gene, MMP12, matrix metalloproteinase-12 gene; MUC12, mucin-12 gene; GO, Gene Ontology; HCC, hepatocellular carcinoma; DEMs, differentially expressed mRNAs; TCGA, The Cancer Genome Atlas; LIHC, liver hepatocellular carcinoma; OS, overall survival; IIRGs, innate immune-related genes; ROC, receiver operating characteristic; NES, normalized enrichment score; FDR, false discovery rate.
|
A survival analysis on each of the aforementioned 21 IIRGs was performed, in order to identify the prognostic immune-related genes among them. A total of 345 patients with a follow-up period >1 month were included for these survival analyses. Kaplan-Meier survival curves were drawn between the high expression group (n=173) and low expression group (n=172) of individual genes among the 21 IIRGs, based on TCGA-HCC dataset samples. The results demonstrated that COLEC12 (cut-off value, 376.022), MMP12 (cut-off value, 8.507) and MUC12 (cut-off value, 1.515) significantly affected the OS (P<0.05; Fig. 3C). Patients expressing higher levels of COLEC12, MUC12, or MMP12 had a significantly shorter OS rate than those with a lower expression.
To establish an innate immune-related risk signature based on the expression of the three IIRGs and their corresponding coefficient obtained from multivariate Cox regression (Table I). The formula included a risk score of three IIRGs = (0.406 × MMP12) + (0.521 × MUC12) + (0.484 × COLEC12). Subsequently, the risk score was computed for each patient, based on TCGA-HCC samples (n=345) and the median of the risk score was defined to classify patients into a high-risk group (n=173) or a low-risk group (n=172). Patients in the low-risk group presented a significantly improved OS in comparison with those in the high-risk group (Fig. 3D; P<0.01 log-rank test). ROC curve analysis based on the three-IIRG risk signature demonstrated an acceptable discrimination with an AUC of 0.606, 0.597 and 0.612 in predicting the 1-, 3-, 5-year OS, respectively (Fig. 3E). This finding indicated that the three-IIRG risk signature resulted in a robust and efficient prediction of the prognosis of patients with HCC.
 |
Table I.
Coefficients and multivariable Cox model results of 3 innate immune-related signature genes.
|
Table I.
Coefficients and multivariable Cox model results of 3 innate immune-related signature genes.
| Gene |
LogFC |
Coefficient |
HR (95% CI) |
P-value |
| COLEC12 |
2.454 |
0.484 |
1.622 (1.117–2.355) |
0.011 |
| MMP12 |
4.220 |
0.406 |
1.500 (1.028–2.190) |
0.036 |
| MUC12 |
2.527 |
0.521 |
1.683 (1.163–2.436) |
0.006 |
Moreover, a univariate Cox analysis was performed in order to analyze the association between the three-IIRG risk signature, clinical variables [including age, sex, body mass index (BMI), pathological stage and AFP levels] and the OS of patients in TCGA-HCC database. The high-risk group (high vs. low) presented a strong association with a worse OS in the database and the observed hazard ratio (HR) was 1.579 [95% confidence interval (CI), 1.028–2.426; P<0.05; Table II]. In the multivariate Cox regression model, following the multivariable adjustment by clinical features, the risk scores could independently predict OS in the set (HR, 1.701; 95% CI, 1.006–2.875; P<0.05; Table II). These results suggested that the three-IIRG signature may be a robust and independent risk factor for HCC.
 |
Table II.
Univariate and multivariate analysis of clinical features and risk score with Cox proportional hazard model.
|
Table II.
Univariate and multivariate analysis of clinical features and risk score with Cox proportional hazard model.
| Covariates |
HR (95% CI) |
P-value |
| Univariate analysis |
| Age (≥65/<65 years) |
1.58 (1.047–2.398) |
0.029 |
| Sex (male/female) |
0.732 (0.481–1.113) |
0.145 |
| BMI (≥25/<25) |
1.117 (0.713–1.751) |
0.629 |
| Pathological stage (III+IV/I+II) |
2.009 (1.315–3.069) |
0.001 |
| AFP (≥400/400) |
0.860 (0.490–1.511) |
0.600 |
| Three-IIRG score (high/low) |
1.579 (1.028–2.426) |
0.037 |
| Multivariate analysis |
| Age (≥65/<65 years) |
1.462 (0.888–2.408) |
0.136 |
| Sex (male/female) |
0.814 (0.483–1.374) |
0.442 |
| BMI (≥25/<25) |
1.016 (0.612–1.688) |
0.950 |
| Pathological stage (III+IV/I+II) |
1.900 (1.107–3.262) |
0.020 |
| AFP (≥400/400) |
0.721 (0.392–1.326) |
0.293 |
| Three-IIRG score (high/low) |
1.701 (1.006–2.875) |
0.048 |
Validation in the GEO database and the Human Protein Atlas
The GEO database indicated that COLEC12, MUC12 and MMP12 mRNA expression was significantly upregulated in HCC tissues, in comparison with liver cirrhosis or normal liver tissues (P<0.01; Fig. 4A-C), in consistency with TCGA cohort results. The protein expression of the three-IIRG signature was detected using immunohistochemical stains retrieved from the Human Protein Atlas Profiles, revealing that the COLEC12 protein level was considerably higher in the HCC tissues (tumor) than in normal liver tissues (normal) (Fig. 4A, the right two immunohistochemical pictures). However, no significant differences were observed in the MUC12 expression level between the HCC tissues (tumor) and normal liver tissues (normal) (Fig. 4C, the right two immunohistochemical pictures). Notably, the immunohistochemical stain of MMP12 was not found in the database.
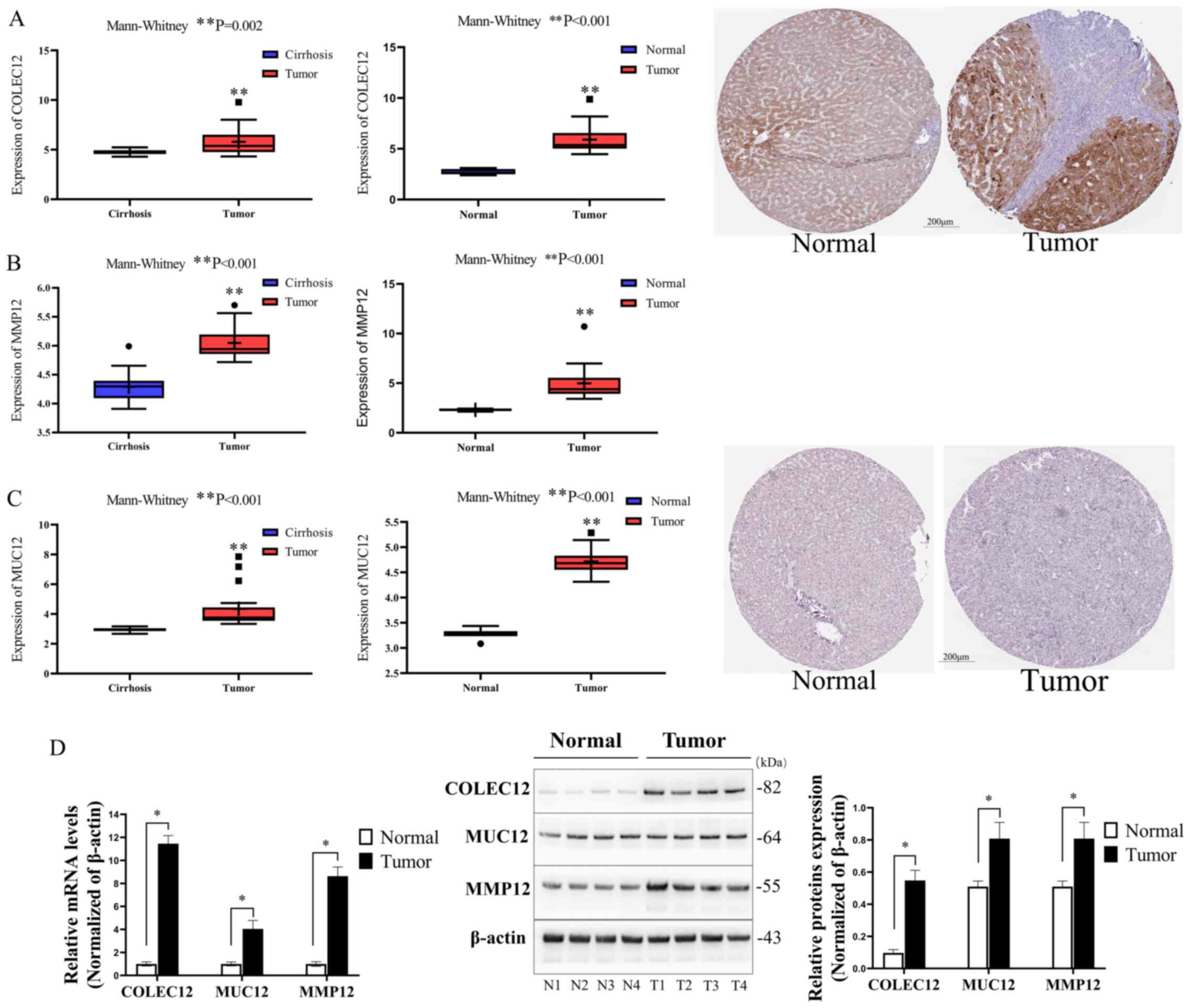 |
Figure 4.
Validation of COLEC12, MMP12 and MUC12 mRNA and protein levels. (A) Comparison of COLEC12 mRNA levels between HCC tissues (n=17) and cirrhosis (n=20) or normal liver tissues (n=6) in the GEO datasets (GSE17548 and GSE83148), and immunohistochemical staining of COLEC12 in HCC tissues and normal liver tissues downloaded from the Human Protein Atlas. (B) The comparisons of the MMP12 mRNA level between HCC tissues and cirrhosis or normal liver tissues in the GEO datasets (GSE17548 and GSE83148). (C) The comparisons of the MUC12 mRNA level between HCC tissues and cirrhosis or normal liver tissues in the GEO datasets (GSE17548 and GSE83148), and immunohistochemical staining of MUC12 in HCC tissues and normal liver tissues downloaded from the Human Protein Atlas. (D) The comparisons of the mRNA and protein expression levels of COLEC12, MUC12 and MMP12 between tumor samples (HCC tissues, n=4) and normal (corresponding adjacent normal liver tissues, n=4) samples of HCC patients from the Second Affiliated Hospital of Nanchang University. *P<0.05 and **P<0.01 vs. normal or cirrhosis, respectively. COLEC12, collectin-12 gene, MMP12, matrix metalloproteinase-12 gene; MUC12, mucin-12 gene; HCC, hepatocellular carcinoma; GEO, Gene Expression Omnibus.
|
Validation using human HCC and normal liver tissues
Protein and mRNA expression of the HCC patient tissue surgical samples (T1, T2, T3 and T4) and their corresponding adjacent normal liver tissues (N1, N2, N3 and N4) were examined using RT-qPCR and western blot analysis. Following three independent experiments, the results of RT-qPCR and western blot analysis demonstrated that both the mRNA and protein expression levels of COLEC12, MUC12 and MMP12 were significantly increased in the human HCC tissues as compared with the normal liver tissues (P<0.05; Fig. 4D).
Pearson's correlation analyses of the three genes with immunosuppressive cytokines and immune checkpoints, and ICB response prediction
Pearson's correlation analyses were performed to further confirm whether the expression of the three IIRGs correlated with that of immunosuppressive cytokines and immune checkpoints (n=374). The present results revealed that: i) The correlations of COLEC12 with PDL1 (R=0.64, P<0.01) and with IL-10 (R=0.78, P<0.01) were strong; ii) the correlation of MMP12 with CTLA-4 (R=0.39, P<0.01) was moderate; iii) the correlations of COLEC12 with IL23-p19 (R=0.21, P<0.05), and MMP12 with PDL1 (R=0.26, P<0.05), IL10 (R=0.12, P<0.05) and IL23-p19 (R=0.21, P<0.05) were weak. However, the correlations of COLEC12 with CTLA-4, and MUC12 with CTLA-4, PDL1, IL10 and IL23-p19 were not statistically significant (P>0.05) (Fig. 5A-C).
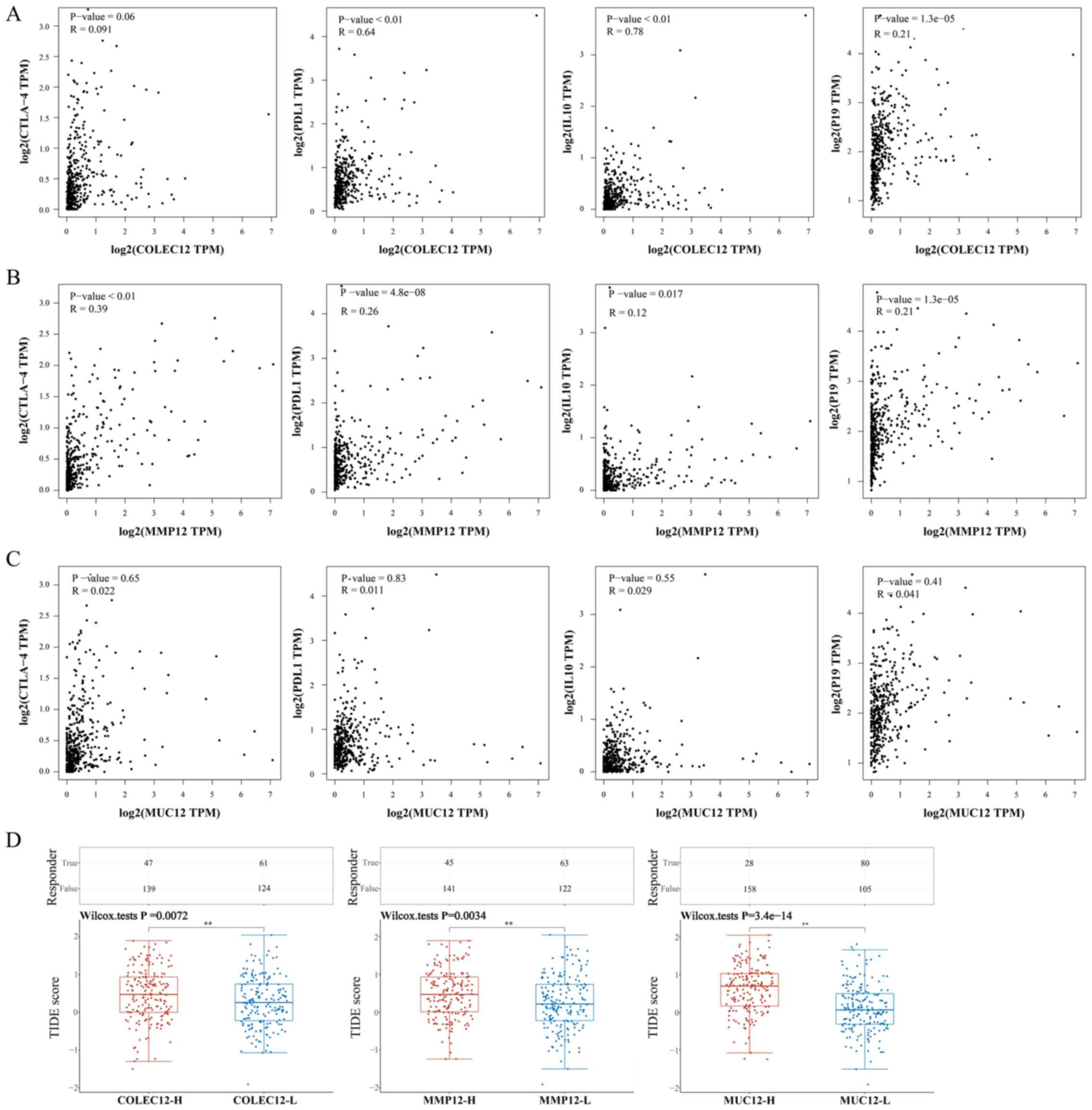 |
Figure 5.
Pearson's correlation analysis of COLEC12, MMP12 and MUC12 with immune checkpoints and immunosuppressive cytokines (n=374), and ICB response prediction using the TIDE algorithm (TCGA-LIHC dataset, n=371). (A) Correlations of COLEC12 with CTLA-4, PDL1, IL10 and IL23-P19 (P<0.05). (B) Correlations of MMP12 with CTLA-4, PDL1, IL10, and IL23-P19 (P<0.05). (C) Correlations of MUC12 with CTLA-4, PDL1, IL10, or IL23-P19 (P>0.05). (D) Statistical table of immune response of samples in different groups in the prediction results and the corresponding distribution of immune response scores in the different groups in the prediction results (COLEC12-H, high level of COLEC12 group, n=186; COLEC12-L, low level of COLEC12 group, n=185; MMP12-H, high level of MMP12 group, n=186; MMP12-L, low level of MMP12 group, n=185; MUC12-H, high level of MUC12 group, n=186; MUC12-L, low level of MUC12 group, n=185). **P<0.01. COLEC12, collectin-12 gene, MMP12, matrix metalloproteinase-12 gene; MUC12, mucin-12 gene; ICB, immune checkpoint blockade; TCGA, The Cancer Genome Atlas; LIHC, liver hepatocellular carcinoma; PD-L1, PD-1 ligand 1; CTLA-4, cytotoxic T lymphocyte-associated antigen 4.
|
Furthermore, in order to validate the effect of the three IIRGs on immunotherapy, the TIDE algorithm was applied. Raw counts of HCC patient mRNA and corresponding clinical information obtained from TCGA dataset were used (n=371). The ICB response scores were calculated using the TIDE algorithm. As the results demonstrated, the high expression levels in the COLEC12 group (COLEC12-H, n=186) had a higher TIDE score than the low expression levels in the COLEC12 group (COLEC12-L, n=185) (P<0.01; Fig. 5D). The same results were found when comparing the high expression level in the MMP12 group (MMP12-H, n=186) with the low expression level in the MMP12 group (MMP12-L, n=185), and the high expression level in the MUC12 group (MUC12-H, n=186) with the low expression level in the MUC12 group (MUC12-L, n=185) (Fig. 5D). These findings indicated that the high expression levels of the three IIRGs were associated with a poor ICB response of patients with HCC, suggesting that the three genes probably affect the ICB of patients with HCC. Therefore, considering the aforementioned results, the mechanisms of the three IIRGs inducing tumor immune evasion do not only rely on immune checkpoint factor expression.
Discussion
Recently, several immunotherapies have been increasingly used in patients with HCC; however, their overall efficacy remains unsatisfactory. For example, PD-1/PD-L1 inhibitor, a well-known anticancer drug, has been shown to present only a 20% objective response rate in patients with HCC in spite of showing promising results in patients with lung cancer and other cancers (35). This may be attributed to specificity of the liver, creating an immunotolerant tumor microenvironment due to its broad exposure to gut-derived antigens (36). Although the immune system can monitor and eliminate cancer cells, HCC develops and progresses via pathological mechanisms, including immunotolerance and immunosuppression, ultimately evading immune response. The innate immune system, as the first line of defense, has two effects. Firstly, the innate immune system can identify and phagocytize antigens, triggering acquired immunity and ultimately clearing the pathogen. Furthermore, the consistent non-resolving inflammation induces immune fatigue or immunosuppression when the immune response is not strong enough to clear pathogens (37). In the present study, integrated bioinformatics analysis was performed, in order to identify IIRGs that are involved in the immunosuppressive mechanism, with the overarching goal of providing critical evidence for the exploration of novel putative targets for HCC treatment.
The GO-CC results of the present study demonstrated that DEMs were mainly focused on extracellular matrix and cell membranes, while GO-BP results indicated that DEMs were mainly focused on immune response. Moreover, GO annotation under MF was mostly enriched in genes associated with antigen binding. Among the results of KEGG pathway analysis for DEMs, the pathway ‘Staphylococcus aureus infection’ was enriched. Oncogenesis is well known for triggering the recruitment of innate immune cells such as granulocytes, monocytes, macrophages and natural killer cells, which mainly accumulate in the extracellular matrix and stroma (38). Therefore, the results of the present study strongly suggested that the differences between HCC tissues and the normal liver tissues in terms of immune response were significant. The present study focused on DEMs involved in the innate immune response, since the innate immune response is the first line of defense in humans. Two sets of C5 (GO activation of innate immune response set and GO negative regulation of innate immune response set) and 1 set of C7 (innate immune response set) were enriched in the normal liver tissues, as opposed to HCC tissues. This indicated that innate immunity was attenuated in tumors, which is consistent with the results of a previous study on HCC and liver cancer (39). Notably, both the GO activation of the innate immune response set and GO-negative regulation of the innate immune response set were enriched in normal liver tissues. This finding confirmed that the proper immune response rapidly reverted to its original inactive state after effectively eradicating pathogens. Although the majority of the genes in the three sets were highly expressed in normal liver tissues, 21 IIRGs were highly expressed in HCC tissues. It was thus hypothesized that the activation of these 21 IIRGs is insufficient for generating an effective antitumor immune response. Alternatively, they cause immunosuppression, allowing the tumor to evade immune surveillance and prompting tumor proliferation and metastasis. Furthermore, three prognostic IIRGs were screened (COLEC12, MUC12 and MMP12) among the 21 IIRGs. It is worth noting that all the three IIRGs were linked to a short survival time. An innate immune-related risk signature was developed based on the expression of the three IIRGs. The use of 3-IIRGs risk signature resulted in a robust and efficient prediction of HCC patient prognosis. The results of the univariate and multivariate Cox analysis further confirmed that the risk group (high levels of COLEC12, MUC12 and MMP12 relative to low levels of the three genes) based on the three-IIRG risk signature was a robust and independent prognostic factor. The expression of the three IIRGs was also confirmed through external validation. Only COLEC12 and MMP12 were correlated with immunosuppressive cytokines and immune checkpoints. However, the scores of TIDE algorithm predicted that each of the three IIRGs was associated with resistance to ICIs. These results further indicated that the mechanisms through which the three IIRGs induce tumor immunosuppression do not only depend on affecting the expression of immune checkpoints.
COLEC12 is a member of the C-type Lectin receptor family, which can influence the adaptive immune response through the carbohydrate-recognition domain through the recognition of the complex glycan structures of pathogens (35,40). Li et al (41) reported a higher COLEC12 expression in osteosarcoma tissues in comparison with normal tissues, and that a higher expression of COLEC12 was associated with a worse outcome in patients with osteosarcoma. Chang et al (42) demonstrated that the expression of COLEC12 in gastric stromal cells mediated immunosuppression and promoted the development of gastric cancer. MUC12 is a member of mucins (MUCs), which are high molecular-weight glycoproteins that constitute the first line of innate immunity (43). MUCs can be divided into two subtypes: Secretory and membrane-bound. MUC12 is a membrane-bound MUC, whose function is epithelial cell protection (44). MMP12 is a member of MMPs, a family of zinc- and calcium-dependent endopeptidases that can degrade almost all extracellular matrix components. This behavior is consistent with the role of MMPs in enhancing tumorigenesis, tumor angiogenesis and tumor invasiveness (45). Conversely, MMP-mediated proteolysis can also inhibit tumor growth (46). These contradictory findings have caused the role of MMPs in cancer to remain controversial.
Notably, COLEC12 is a bacterial lipopolysaccharide (LPS) binding receptor, suggesting the possibility for bacterial infection in HCC tissue. Previous studies have reported that the products of intestinal flora metabolism are transported to the liver via the hepatic portal vein (47,48). The close association between the gut and liver has been referred to as the ‘gut-liver axis’ (49). The changes in the types and quantities of intestinal flora may result in increased intestinal permeability and absorption of bacterial endotoxin (LPS). Kupffer cells and hepatic stellate cells are then activated, releasing a series of inflammatory factors ultimately causing hepatitis. When hepatitis persists, the liver develops fibrosis and cirrhosis due to exhaustion of the immune system. Eventually, HCC may occur due to the unsolved inflammation and attenuated immune system (50). In addition to HCC, bacterial infection also exists in other tumors. Nejman et al (51) analyzed the tumor microbiome in 1,526 tumors and their adjacent normal tissues from seven cancer types including breast, lung, ovary, pancreas, melanoma, bone and brain tumors. In that study, it was reported that each tumor type had a distinct microbiome composition, suggesting that changing the tumor microbiome can affect tumor immunity and immunotherapy response. However, the role of intra-tumor bacteria in tumor development has not yet been fully elucidated. Thus, it is possible that COLEC12 is one of the key factors that induce immunosuppression following bacterial infection, which could explain the results of the present study.
In spite of the integral bioinformatics analysis conducted, the present study has several limitations. Firstly, the sample size used to verify the mRNA and protein expression level of key genes was limited, due to time and geographical constraints. Secondly, there was no follow-up in the present study. Finally, the exact mechanisms through which the three IIRG genes affect immunity were not elucidated. Therefore, further studies are required to explore the underlying mechanisms of the genes involved in immunosuppression, in order to provide a new direction for HCC treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HH, LG and YH collected the data, including performing the experiments. HH analyzed the data and drafted the manuscript. HH and ZW designed the study. ZW critically revised the manuscript. HH and YH confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All research protocols in the present study were approved by the Ethics Committee of the Second Affiliated Hospital of Nanchang University (approval no. 20190913). Written informed consent was acquired from all study subjects, and all experiments were performed in compliance with the relevant guidelines and regulations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
IIRGs
|
innate immune-related genes
|
|
OS
|
overall survival
|
|
PD-1
|
programmed cell death protein-1
|
|
PD-L1
|
PD-1 ligand 1
|
|
CTLA-4
|
cytotoxic T-lymphocyte-associated antigen 4
|
|
COLEC12
|
collectin-12 gene
|
|
MMP12
|
matrix metalloproteinase-12 gene
|
|
MUC12
|
mucin-12 gene
|
|
ICIs
|
immune checkpoint inhibitors
|
|
TME
|
tumor microenvironment
|
|
TIDE
|
tumor immune dysfunction and exclusion
|
|
RT-qPCR
|
reverse transcription-quantitative PCR
|
|
TCGA
|
The Cancer Genome Atlas
|
|
GEO
|
Gene Expression Omnibus
|
|
DEMs
|
identification of differentially expressed mRNAs
|
|
GO
|
Gene Ontology
|
|
BP
|
biological processes
|
|
CC
|
cellular components
|
|
MF
|
molecular functions
|
|
GSEA
|
gene set enrichment analyses
|
|
KEGG
|
Kyoto Encyclopedia of Genes and Genomes
|
|
TPM
|
transcripts per million
|
|
DAVID
|
Database for Annotation, Visualization and Integrated Discovery
|
|
ROC
|
receiver operating characteristic
|
|
AUC
|
the area under the curves
|
|
GEPIA
|
Gene Expression Profiling Interactive Analysis
|
|
ICB
|
immune checkpoint blockade
|
|
BMI
|
body mass index
|
References
|
1
|
Forner A, Reig M and Bruix J: Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Villanueva A: Hepatocellular carcinoma. N Engl J Med. 380:1450–1462. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lechel A and Gougelet A: Early HCC treatment: A future strategy against interferon/miR-484 axis to revert precancerous lesions? Gut. 65:1073–1074. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kudo M: Early detection and curative treatment of early-stage hepatocellular carcinoma. Clin Gastroenterol Hepatol. 3 (Suppl 2):S144–S148. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thomas MB, Jaffe D, Choti MM, Belghiti J, Curley S, Fong Y, Gores G, Kerlan R, Merle P, O'Neil B, et al: Hepatocellular carcinoma: Consensus recommendations of the National Cancer Institute Clinical Trials Planning Meeting. J Clin Oncol. 28:3994–4005. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Philips GK and Atkins M: Therapeutic uses of anti-PD-1 and anti-PD-L1 antibodies. Int Immunol. 27:39–46. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rowshanravan B, Halliday N and Sansom DM: CTLA-4: a moving target in immunotherapy. Blood. 131:58–67. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu AX, Finn RS, Edeline J, Cattan S, Ogasawara S, Palmer D, Verslype C, Zagonel V, Fartoux L, Vogel A, et al KEYNOTE-224 investigators, : Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): A non-randomised, open-label phase 2 trial. Lancet Oncol. 19:940–952. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kambhampati S, Bauer KE, Bracci PM, Keenan BP, Behr SC, Gordan JD and Kelley RK: Nivolumab in patients with advanced hepatocellular carcinoma and Child-Pugh class B cirrhosis: Safety and clinical outcomes in a retrospective case series. Cancer. 125:3234–3241. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu LX, Ling Y and Wang HY: Role of nonresolving inflammation in hepatocellular carcinoma development and progression. NPJ Precis Oncol. 2:62018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ringelhan M, Pfister D, O'Connor T, Pikarsky E and Heikenwalder M: The immunology of hepatocellular carcinoma. Nat Immunol. 19:222–232. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han Q, Zhao H, Jiang Y, Yin C and Zhang J: HCC-derived exosomes: Critical player and target for cancer immune escape. Cells. 8:5582019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tanimine N, Ohira M, Tahara H, Ide K, Tanaka Y, Onoe T and Ohdan H: Strategies for deliberate induction of immune tolerance in liver transplantation: From preclinical models to clinical application. Front Immunol. 11:16152020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sia D, Jiao Y, Martinez-Quetglas I, Kuchuk O, Villacorta-Martin C, Castro de Moura M, Putra J, Camprecios G, Bassaganyas L, Akers N, et al: Identification of an Immune-specific Class of Hepatocellular Carcinoma, Based on Molecular Features. Gastroenterology. 153:812–826. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xiao Y and Yu D: Tumor microenvironment as a therapeutic target in cancer. Pharmacol Ther. 221:1077532021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ngambenjawong C, Gustafson HH and Pun SH: Progress in tumor-associated macrophage (TAM)-targeted therapeutics. Adv Drug Deliv Rev. 114:206–221. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Barry KC, Hsu J, Broz ML, Cueto FJ, Binnewies M, Combes AJ, Nelson AE, Loo K, Kumar R, Rosenblum MD, et al: A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat Med. 24:1178–1191. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, Bassez A, Decaluwé H, Pircher A, Van den Eynde K, et al: Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 24:1277–1289. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu XN, Cui DN, Li YF, Liu YH, Liu G and Liu L: Multiple ‘Omics’ data-based biomarker screening for hepatocellular carcinoma diagnosis. World J Gastroenterol. 25:4199–4212. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al: The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401–404. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yildiz G, Arslan-Ergul A, Bagislar S, Konu O, Yuzugullu H, Gursoy-Yuzugullu O, Ozturk N, Ozen C, Ozdag H, Erdal E, et al: Genome-wide transcriptional reorganization associated with senescence-to-immortality switch during human hepatocellular carcinogenesis. PLoS One. 8:e640162013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou W, Ma Y, Zhang J, Hu J, Zhang M, Wang Y, Li Y, Wu L, Pan Y, Zhang Y, et al: Predictive model for inflammation grades of chronic hepatitis B: Large-scale analysis of clinical parameters and gene expressions. Liver Int. 37:1632–1641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Robinson MD, McCarthy DJ and Smyth GK: edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 4:32003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ito K and Murphy D: Application of ggplot2 to pharmacometric graphics. CPT Pharmacometrics Syst Pharmacol. 2:e792013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al: Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, Yuan A, Cheng CL, Wang CH, Terng HJ, Kao SF, et al: A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 356:11–20. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lorent M, Giral M and Foucher Y: Net time-dependent ROC curves: A solution for evaluating the accuracy of a marker to predict disease-related mortality. Stat Med. 33:2379–2389. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Blanche P, Dartigues JF and Jacqmin-Gadda H: Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med. 32:5381–5397. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pontén F, Jirström K and Uhlen M: The Human Protein Atlas--a tool for pathology. J Pathol. 216:387–393. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang B, Su X, Xie Z, Ding H, Wang T, Xie RY and Wen Z: Inositol-requiring kinase 1 regulates apoptosis via inducing endoplasmic reticulum stress in colitis epithelial cells. Dig Dis Sci. 66:3015–3025. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tang Z, Li C, Kang B, Gao G, Li C and Zhang Z: GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, Li Z, Traugh N, Bu X, Li B, et al: Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 24:1550–1558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Iñarrairaegui M, Melero I and Sangro B: Immunotherapy of hepatocellular carcinoma: Facts and hopes. Clin Cancer Res. 24:1518–1524. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rizvi S, Wang J and El-Khoueiry AB: Liver cancer immunity. Hepatology. 73 (Suppl 1):86–103. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Finn OJ: Cancer immunology. N Engl J Med. 358:2704–2715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jeong JY, Kim TB, Kim J, Choi HW, Kim EJ, Yoo HJ, Lee S, Jun HR, Yoo W, Kim S, et al: Diversity in the extracellular vesicle-derived microbiome of tissues according to tumor progression in pancreatic cancer. Cancers (Basel). 12:23462020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rohr-Udilova N, Klinglmüller F, Schulte-Hermann R, Stift J, Herac M, Salzmann M, Finotello F, Timelthaler G, Oberhuber G, Pinter M, et al: Deviations of the immune cell landscape between healthy liver and hepatocellular carcinoma. Sci Rep. 8:62202018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
McGreal EP, Miller JL and Gordon S: Ligand recognition by antigen-presenting cell C-type lectin receptors. Curr Opin Immunol. 17:18–24. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li GZ, Deng JF, Qi YZ, Liu R and Liu ZX: COLEC12 regulates apoptosis of osteosarcoma through Toll-like receptor 4-activated inflammation. J Clin Lab Anal. 34:e234692020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chang LL, Hsu WH, Kao MC, Chou CC, Lin CC, Liu CJ, Weng BC, Kuo FC, Kuo CH, Lin MH, et al: Stromal C-type lectin receptor COLEC12 integrates H. pylori, PGE2-EP2/4 axis and innate immunity in gastric diseases. Sci Rep. 8:38212018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Moehle C, Ackermann N, Langmann T, Aslanidis C, Kel A, Kel-Margoulis O, Schmitz-Madry A, Zahn A, Stremmel W and Schmitz G: Aberrant intestinal expression and allelic variants of mucin genes associated with inflammatory bowel disease. J Mol Med (Berl). 84:1055–1066. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Andrianifahanana M, Moniaux N and Batra SK: Regulation of mucin expression: Mechanistic aspects and implications for cancer and inflammatory diseases. Biochim Biophys Acta. 1765:189–222. 2006.PubMed/NCBI
|
|
45
|
Kataoka H, Uchino H, Iwamura T, Seiki M, Nabeshima K and Koono M: Enhanced tumor growth and invasiveness in vivo by a carboxyl-terminal fragment of alpha1-proteinase inhibitor generated by matrix metalloproteinases: A possible modulatory role in natural killer cytotoxicity. Am J Pathol. 154:457–468. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shapiro SD and Senior RM: Matrix metalloproteinases. Matrix degradation and more. Am J Respir Cell Mol Biol. 20:1100–1102. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jirillo E, Caccavo D, Magrone T, Piccigallo E, Amati L, Lembo A, Kalis C and Gumenscheimer M: The role of the liver in the response to LPS: Experimental and clinical findings. J Endotoxin Res. 8:319–327. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Haque TR and Barritt AS IV: Intestinal microbiota in liver disease. Best Pract Res Clin Gastroenterol. 30:133–142. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wiest R, Albillos A, Trauner M, Bajaj JS and Jalan R: Targeting the gut-liver axis in liver disease. J Hepatol. 67:1084–1103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jiang JW, Chen XH, Ren Z and Zheng SS: Gut microbial dysbiosis associates hepatocellular carcinoma via the gut-liver axis. Hepatobiliary Pancreat Dis Int. 18:19–27. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E, et al: The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 368:973–980. 2020. View Article : Google Scholar : PubMed/NCBI
|



















