Introduction
Neuroblastoma (NB) is the most common extracranial malignancy in children but can also occur in the adult population, with 1 case per 10 million adults per year diagnosed in adulthood (1). NB accounts for 8–10% of all childhood tumors and ~15% of all cancer-associated mortality in children (2). Despite progress in treatment of NB in low and intermediate-risk patients, particularly in patients <1 year old, the 5-year overall survival rate for high-risk patients remains poor (3).
With the development of radiation therapy, autologous stem cell transplants and immunotherapy, among other treatment modalities, the 5-year survival rate of high-risk patients with NB is now approaching 50% (4); however, treatments frequently fail due to recurrence and development of chemoresistance. Hence, there is an urgent need for novel targeted therapies to improve the outcomes of such patients.
TP53 is a well-known tumor suppressor that serves as ‘the guardian of the genome’. TP53 is activated in response to DNA damage and other cellular stresses, subsequently inducing cell cycle arrest, apoptosis and senescence (5–7). Additionally, it is a key regulator of numerous signaling pathways, including the regulation of insulin-like growth factor I-AKT-mTOR signal transduction pathway (8) and the regulation of exosome secretion (9). Inactivation of TP53 is a common event in tumorigenesis. However, only ~50% of tumors harbor TP53 mutations and the remaining 50% tumor cells with wild-type (wt) TP53 are suggested to harbor mutations that lie either up- or downstream of p53 and may also lead to inactivation of the p53 pathway, which suggests that alternative mechanisms are used by tumor cells to impair TP53 function (10,11).
Murine double minute2 homolog (MDM2), a well-known negative regulator of TP53, is amplified in several types of human malignancy, including NB (12–17). As an E3 ubiquitin ligase, MDM2 exerts its inhibitory effect by directly binding to the p53 transactivation domain (TAD), shuttling p53 to the cytoplasm and mediating its poly-ubiquitination and degradation (18), thus impeding the activity of TP53. De novo somatic mutations of TP53 or MDM2 are rarely detected in NB (19), occurring in <2% of primary cases of NB and 14% of relapsed cases (18,20). Thus, the intact MDM2/p53 axis provides an opportunity for targeted therapy in NB treatment.
Numerous studies have shown that targeting the MDM2/p53 negative feedback loop using small molecule inhibitors allows for restoration of the tumor suppressive function of p53 (21–25). At present, several MDM2 antagonists have been designed to target the MDM2/p53 interaction and to restore the tumor suppressive function of p53, such as SAR405838 (MI-773), RITA (26), MI-219 (21), Nutlin-3 (27), MI-63 (12), RG7388 (28), ALRN-6924 (29) and APG-115 (30); some of these have shown promising antitumor efficacy in vitro. For example, MI-63 inhibits the proliferation of cancer cell lines with wt p53 expression by binding to MDM2 and decreasing ubiquitin degradation of p53 (31–33). MI-219 and Nutlin-3 dose-dependently induce p53 accumulation and upregulation of p53-target gene products (MDM2, p21 and Bcl2 binding component 3) in wt p53 NB cell lines, leading to apoptosis of tumor cells (16). Based on promising antitumor effects via modulation of the MDM2/p53 axis, novel MDM2 antagonists with high specificity and efficacy should be further investigated.
MI-773 is a highly potent and selective inhibitor that binds to MDM2 (23,24). The antitumor effects of MI-773 in wt p53 NB have been reported in previous studies (24,34). Meanwhile, its ability to augment the cytotoxic effects of doxorubicin highlight its potential as an effective adjuvant to current chemotherapeutic regimens for NB treatment (34). In our previous study, the antitumor effects of MI-773 were evaluated using high-throughput screening of small molecule inhibitors in NB cells (35). In the present study, the molecular mechanisms modulated by MI-773 in different NB cell lines were further evaluated by in vitro functional experiments and RNA microarray assay, which may provide evidence that MI-773 could serve as a novel strategy for NB treatment.
Materials and methods
Cell culture
The NB cell lines with wt p53 (IMR-32 and SK-N-SH) were cultured in MEM (Thermo Fisher Scientific, Inc.) supplemented with 15% heat-inactivated FBS (Biological Industries; Sartorius AG), while NGP, LAN-1 and SH-SY5Y were cultured in DMEM/High glucose with 10% FBS (Biological Industries), 1% penicillin-streptomycin (MilliporeSigma) and 0.1% Ciprofloxacin (Heowns Biochem Technologies LLC) in a humidified incubator with 5% CO2 at 37°C and routinely assessed for mycoplasma contamination. The p53 mutant NB cell line (KELLY) was cultured in RPMI medium (Biological Industries; Sartorius AG) supplemented with 10% heat-inactivated FBS and 1% penicillin-streptomycin. All cell lines were purchased from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences within the past 5 years and verified by short tandem repeat analysis between 2018 and 2019.
Cell proliferation and viability assays
A total of 2×104 NB cells (IMR-32, SH-SY5Y, SK-N-SH, LAN-1 and NGP) were seeded into 96-well plates and allowed to adhere overnight at 37°C. Subsequently, cells were treated with different concentrations (0.05–20.00 µM) MI-773 (Selleck Chemicals) for 24 h at 37°C. The control group was treated with an equivalent volume of DMSO [final DMSO volume, <0.1% of the cell culture fluid (v/v) and toxicity <0.1%]. Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.) was used to assess cell viability and the absorbance at 450 nm after 2-h incubation at 37°C was measured using a scanning multi-well spectrophotometer (Bio-Rad Model 550; Bio-Rad Laboratories, Inc.). The half-maximal inhibitory concentration (IC50) values and relative survival rates of NB cells treated with MI-773 were calculated using GraphPad Prism 8.4.3 (GraphPad Software, Inc.). Treatment with each drug concentration was replicated three times and the background reading of the medium from each well was subtracted to standardize the results.
Colony formation assay
A total of 2×103 NB cells/well (SH-SY5Y, SK-N-SH and KELLY) was seeded into a 6-well plate and cultured at 37°C for 24 h. Treated with serial concentrations (0 µM, the same volume of DMSO, 0.5, 1, 5 or 10 µM) of MI-773, NB cells were incubated at 37°C for 2 weeks. Subsequently, the adherent cells in the 6-well plate were fixed with 100% methanol for 15 min and stained with 1X Giemsa staining solution (Beijing Solarbio Science & Technology Co., Ltd.; 1 ml 10X Giemsa stock solution diluted in 9 ml 0.01 M PBS at PH 6.8) at room temperature for 1 h. The plates were scanned and the number of colonies (diameter ≥1 mm) was counted manually.
Cell cycle analysis
Cell cycle analysis by propidium iodide (PI) staining was performed as previously described (24). NB cells (IMR-32 and SH-SY5Y) were washed with cold PBS and then fixed with 70% ethanol at 4°C overnight. The following day, fixed cells were permeabilized with 0.5% Triton X-100 and stained with PI (1.5 µmol/l; cat. no. P4170; Sigma-Aldrich; Merck KGaA) and 25 µg/ml RNase A at 37°C in the dark for 1 h. Cell cycle distribution was assessed using a Beckman Gallios™ Flow Cytometer (Immunotech; Beckman Coulter, Inc.). The proportion of cells in different phases of the cell cycle was analyzed using MultiCycle AV DNA analysis software (version 328; Verity Software House, Inc.).
Cell apoptosis analysis
Cell apoptosis analysis was performed as previously described (35). Briefly, ~1×105 NB cells/well (IMR-32 and SH-SY5Y) were treated with MI-773 at the predetermined concentrations of 0.0, 0.5, 1.0, 5.0 and 10.0 µM in a 6-well plate at 37°C. Following 48-h incubation, cells were collected and washed with cold PBS. Apoptosis analysis was performed using a FITC Annexin V apoptosis detection kit (cat. no. 556420; BD Biosciences, Inc.). Cell apoptosis was analyzed using a flow cytometry as aforementioned.
Western blotting
Western blotting was performed as described previously (35). Cells (IMR-32 and SH-SY5Y) were lysed using RIPA lysis buffer (Beyotime Institute of Biotechnology) containing protease and phosphatase inhibitors (Roche Diagnostics) and ultrasonication on ice for 10 min. The supernatant protein collected by centrifugation at 14,000 × g for 15 min at 4°C was quantified using a BCA assay kit (Thermo Fisher Scientific, Inc.). A total of 25–50 µg denatured protein was resolved using 4–20% gradient SDS-PAGE (Genscript) and then transferred onto a PVDF membrane (MilliporeSigma). Membranes were blocked in 5% skimmed milk in 0.1% TBS-Tween-20 (TBST) for 1 h at room temperature, then incubated with primary antibodies at 4°C overnight. Primary antibodies were as follows: Cleaved caspase-3 (cat. no. 9661S), poly(ADP-ribose) polymerase 1 (PARP; cat. no. 9542S), p53 (cat. no. 2524), CDKN1A (p21; cat. no. 2947), MDM2 (all 1:1,000; cat. no. 86934; all Cell Signaling Technology, Inc.), GAPDH (cat. no. AP0063; 1:5,000; Bioworld Technology Inc.) and insulinoma-associated 1 (INSM1; cat. no. ab170876; 1:1,000; Abcam). The following day, membranes were washed with TBST three times, then incubated with goat anti-rabbit (cat. no. 111-035-003) or anti-mouse IgG (H+L; cat. no. 115035-003) HRP-conjugated secondary antibodies (both 1:3,000; both Jackson ImmunoResearch Laboratories, Inc.) at room temperature for 1 h. Finally, the protein bands were visualized with an ECL ultra-sensitive luminescent fluid (Thermo Fisher Scientific, Inc.) using LAS 4010 (Cytiva) imaging system and ImageQuant TL 8.1 software (Cytiva).
RNA microarray and data analysis
Arraystar Human LncRNA Microarray version 4.0 (Arraystar, Inc.) allows global profiling of human long non-coding (lnc)RNAs and protein-coding transcripts and is capable of detecting ~40,173 lncRNAs and 20,730 coding transcripts. RNA isolation, purification, microarray hybridization, array scan and normalization of gene expression were performed by Kangchen BioTech Co., Ltd. The Arraystar Human LncRNA 8×60 k version 3.01 microarray was used to identify mRNA and lncRNA expression profiles in 5 µm MI-773-treated at 37°C for 48 h SH-SY5Y cells compared with an untreated control group. Microarray analysis and original data have been submitted to the Gene Expression Omnibus (GEO) database (accession no. GSE174450). The Bioconductor Limma package 3.46 (34) was used to identify differentially expressed genes. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (35) and gene set enrichment analysis (GSEA) were performed using the clusterProfiler package 3.18.1 (36) in R 4.0.5.
Reverse transcription-quantitative PCR (RT-qPCR)
RT-qPCR was performed as described previously (37). Total RNA from SH-SY5Y was isolated using a RNeasy Mini kit (cat. no. 74104; Qiagen GmbH). A total of 2 µg total RNA was reversed transcribed into cDNA with 200 U M-MLV reverse transcriptase (Promega Corporation), 20 U RNase inhibitor (Thermo Fisher Scientific, Inc.) and 500 ng random primers (Promega Corporation), which were mixed into a total volume of 25 µl and underwent RT on an ABI PCR platform (Applied Biosystems; Thermo Fisher Scientific, Inc.). The reaction conditions were as follows: Initial denaturation at 70°C for 5 min, denaturation for 60 min at 37°C and annealing/extension for 10 min at 85°C. LightCycler® 480 SYBR Green I MasterMix (cat. no. 04707516001; Roche Diagnostics GmbH) was used for qPCR analysis on a LightCycler 480 Real-Time system (Roche Diagnostics GmbH). The qPCR amplification was performed in a LightCycler® 480 (Roche) using universal thermal cycling parameters (initial denaturation at 95°C for 10 min and 45 cycles of 15 sec at 95°C, 15 sec at 60°C, 60 sec at 72°C, melting curve for 10 sec at 95°C, 60 sec at 65°C and 30 min at 40°C). Gene expression quantification was determined using the 2−∆∆Cq method (38), using GAPDH as the housekeeping gene. The sequences of the primers are listed in Table SI.
Preparation of short hairpin (sh)RNA INSM1
The shRNA sequence targeting INSM1 in the PLKO.1 lentiviral vector was designed and constructed by IGE Biotechnology, Ltd. The inserted INSM1 shRNA sequence and locus were confirmed by sequencing (Table SII). For lentivirus preparation, envelop plasmid and packaging plasmid was purchased from Addgene, Inc. (pMD2.G, cat. no. 12259; psPAX2, cat. no. 12260). Lentiviral packaging was performed as previously described (39). 293FT cells were allowed to grow to ~70% confluence before transfection. The packaging plasmid and expression plasmid were mixed at the ratio of 1:1. Then, 15 µl of 1 mg/ml PEI (Polysciences) was mixed with the plasmid and incubated at room temperature for 20 min. Subsequently, the transfection complex was added dropwise to the cell culture dish and incubated at 37°C. The medium containing the transfection complex was replaced with fresh culture medium 6 h post-transfection. Lentivirus were harvested 48 h after changing the medium and filtered with 0.45 µm, syringe filter (Millipore). NB cells were transfected with prepared lentivirus at the multiplicity of infection of 10. Stably transfected NB cells were established by selection with 10 µg/ml puromycin (Invitrogen; Thermo Fisher Scientific, Inc.) for one week. The lentivirus operation was performed in a biological safety cabinet, and all operations complied with the operating safety standards for use of lentiviruses.
Public database
The expression level of INSM1 mRNA in different types of tumor cell lines was obtained from the Cancer Cell Line Encyclopedia (http://www.broadinstitute.org/ccle). Kaplan Meier analysis of the prognostic relevance of MDM2 and INSM1 expression in NB patients was generated from R2: Genomics Analysis and Visualization Platform (r2platform.com), and P-values were calculated using the log rank test. Scan or Median cut-off mode provided on the R2 platform was used to stratify the NB patients into high- and low-expression groups.
Statistical analysis
Statistical analysis was performed using GrapdPad Prism 8.4.3 (GraphPad Software, Inc.). Data are presented as the mean ± SD. All experiments were performed independently at least three times. Differences between multiple groups were compared using one-way ANOVA. Fisher's Least Significant Difference post hoc test was used following ANOVA to determine the significance for pairwise comparison. All tests were two-tailed and P<0.05 was considered to indicate a statistically significant difference.
Results
MI-773 exhibits potent anticancer effects on NB cells
In our previous study, the anticancer effect of 33 inhibitors (Table SIII) in eight NB cell lines was screened by high throughput screening (35). These inhibitors were developed against multiple targets, including histone deacetylase 1, polo-like kinase 1 (PLK1), CDK4/6 and enhancer of zeste 2 polycomb repressive complex 2 subunit (Fig. 1A). Amongst these, inhibitors targeting PLK1 and p21 (RAC1) activated kinase 4 showed potent favorable anticancer effects on NB cells as reported in our previous study (35,40). In the present study, MI-773, a novel inhibitor of MDM2, was assessed in more detail as seven of the NB cell lines were sensitive to this agent.
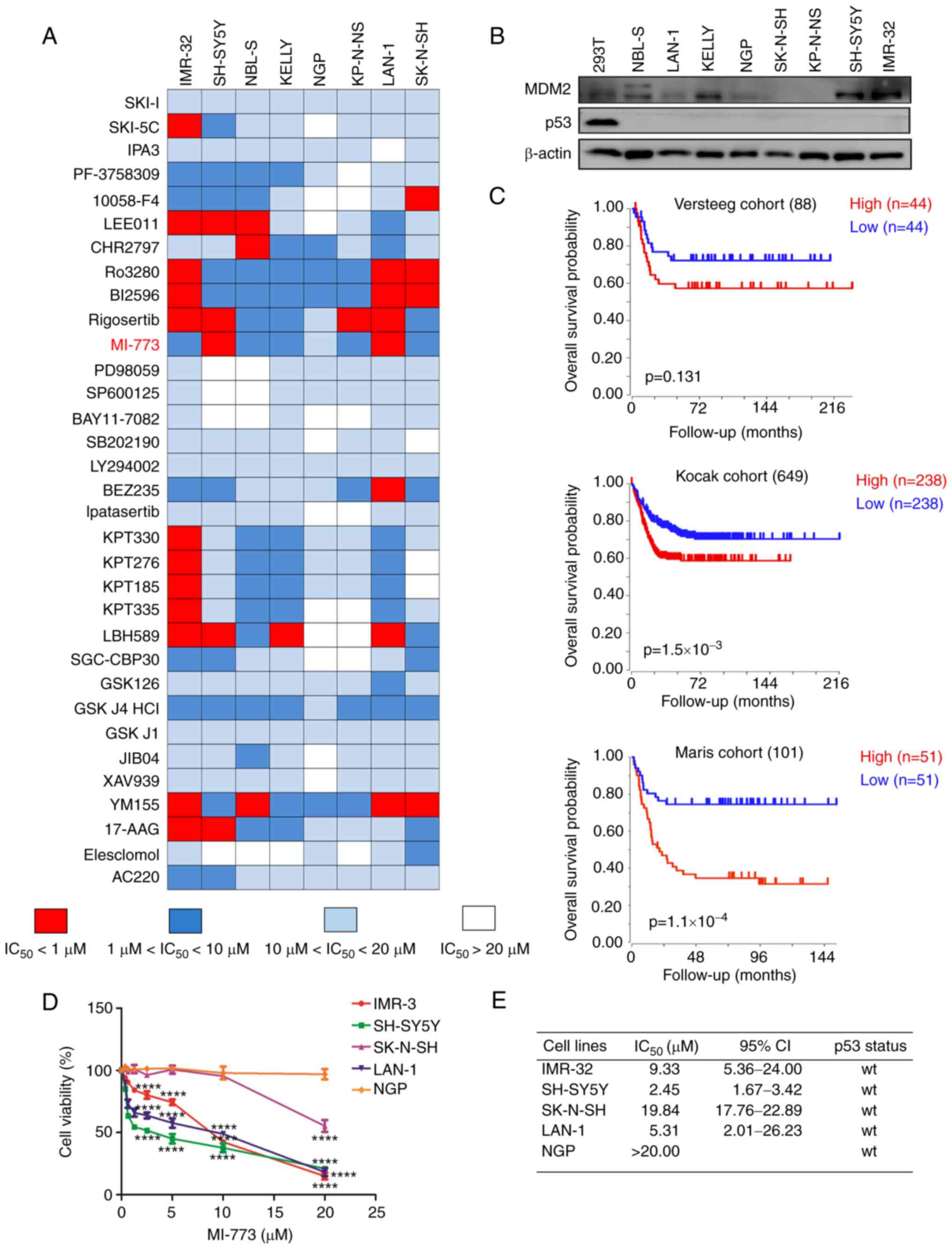 |
Figure 1.
MI-773 shows potent anticancer effect in NB cells. (A) High-throughput drug screening of 33 small molecule inhibitors was performed on NB cells. Data are presented as the mean ± standard deviation of three independent experiments (n=3-6/group). (B) Western blot analysis of MDM2 and p53 expression in NB cell lines. (C) Overall survival curve using public cohorts from R2 Genomics Analysis and Visualization Platform (r2.amc.nl). NB patients were categorized into high or low MDM2 expression groups using median value of MDM2 expression as a cutoff point. Kaplan-Meier survival analysis revealed shorter survival time in the patients with high MDM2 expression levels in tumors. (D) Viability of NB cells treated with serial concentrations of MI-773 for 72 h. The cell viability rate was calculated as a percentage of DMSO-treated control. ****P<0.0001. (E) IC50 values of MI-773 in each cell line were calculated based on cell viability assay. TP53 status in NB cell lines was also shown. NB, neuroblastoma; MDM2, murine double minute 2 homolog; IC50, half-maximal inhibitory concentration; wt, wild-type.
|
The expression levels of p53 and MDM2 in NB cells were assessed. Western blotting showed that TP53 was expressed at low levels in NB cell lines (Fig. 1B). MDM2 was highly expressed in IMR-32, SH-SY5Y, but was low in LAN-1, NGP, SK-N-SH and KP-N-NS.
The prognostic significance of MDM2 in patients with NB was investigated using R2: Genomics Analysis and Visualization Platform (r2platform.com). Kaplan-Meier curves were generated from three public NB expression datasets derived from GEO (accession nos. GSE16476, GSE45547 and GSE49710). The median value of MDM2 expression was used as a cutoff value to stratify the data into high- and low-expression groups. High MDM2 expression was associated with unfavorable outcomes in patients with NB (Fig. 1C), indicating that MDM2 may serve as a promising therapeutic target for NB. Thus, the findings provided strong support for the use of MDM2 inhibitor MI-773 as a potential drug candidate for the treatment of NB.
CCK-8 assay was used to identify the short-term cytotoxicity of MI-773 on NB cells. Following treatment with increasing concentrations of MI-773 treated for 72 h, a notable dose-dependent inhibitory effect of MI-773 on the viability of NB cells was observed (Fig. 1D). The IC50 of MI-773 was analyzed in five NB cell lines (IMR-32 IC50, 9.33 µM; SH-SY5Y IC50, 2.45 µM; SK-N-SH IC50, 19.84 µM; LAN-1 IC50, 5.31 µM and NGP IC50, >20.00 µM). Of these, three wt p53 NB cell lines (IMR-32, SH-SY5Y and LAN-1) that exhibited high expression of MDM2 were sensitive to MI-773 and the cell viability decreased in a dose- and time-dependent manner.
Inhibitory effect of MI-773 on proliferation of NB cells
After treating cells with increasing concentrations of MI-773 for 72 h, cells exhibited abnormal morphological features, with cell clustering, weakened adhesion and an increase in the number of floating cells (Fig. 2A). This effect was attenuated in SK-N-SH and NGP cells, which expressed low levels of MDM2. Colony formation assay was used to evaluate the long-term effect of MI-773 on proliferation in wt p53 NB cells. The results showed a significant decrease in colony formation ability in wt p53 cell lines (SK-N-SH and SH-SY5Y) following MI-773 treatment (Fig. 2B). Colony numbers were counted in each group (Fig. 2C and D), showing that MI-773 attenuated the proliferation of the wt p53 NB cells in a time- and dose-dependent manner. By contrast, this was not observed in the p53 mutant KELLY cell line. Collectively, these data showed that MI-773 exhibited a potent antiproliferative effect on wt p53 NB cell lines.
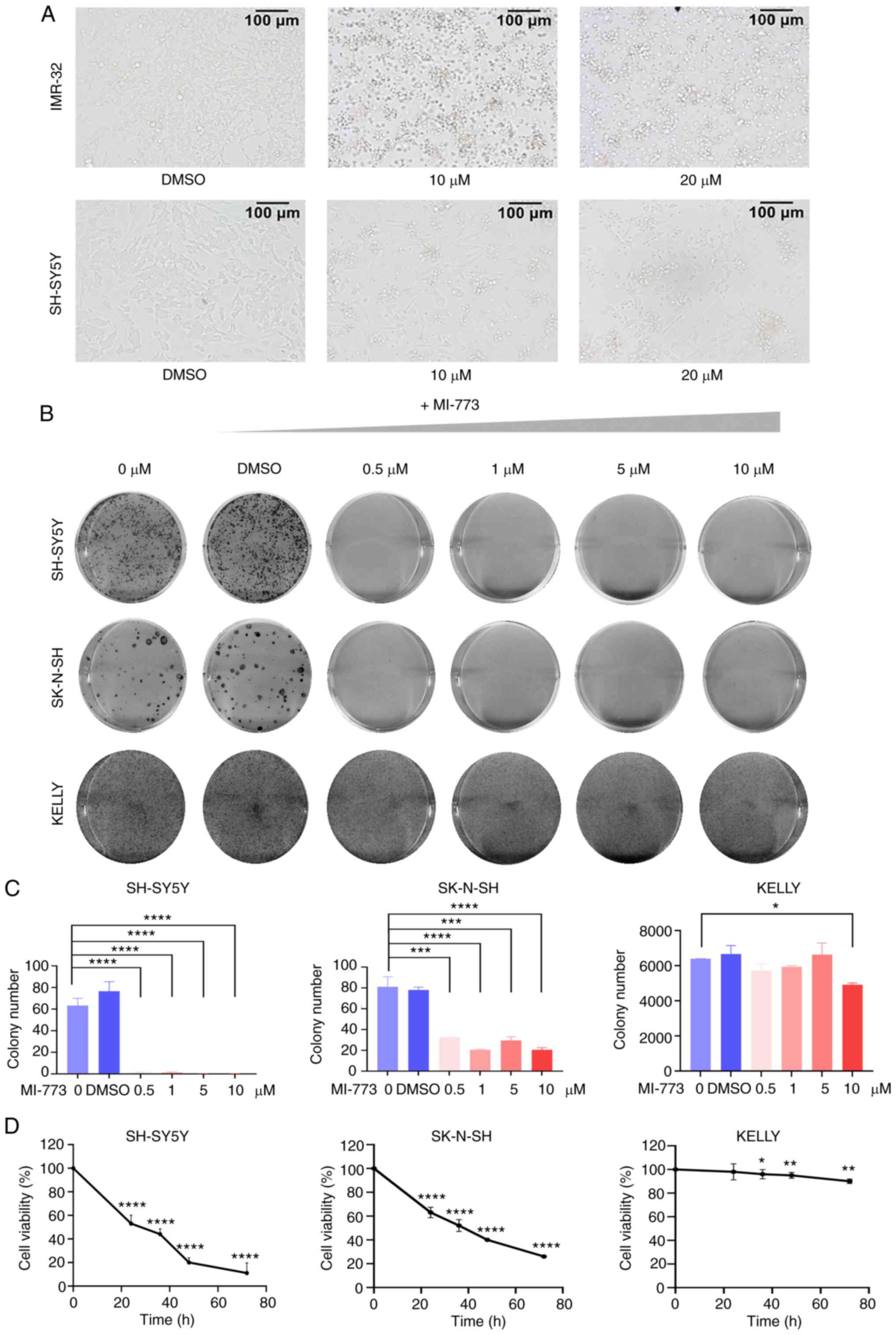 |
Figure 2.
MI-773 suppresses proliferation in NB cells. (A) Morphology of IMR-32 and SH-SY5Y cells incubated with MI-773 at indicated concentrations for 72 h. (B) Clone formation assay showed increasing doses of MI-773 inhibited clonal formation ability in SH-SY5Y, SK-N-SH and KELLY cells. (C) Colony formation analysis of three NB cell lines treated with increasing concentrations of MI-773. ****P<0.0001; *P<0.05. (D) Cell viability analysis of SH-SY5Y, SK-N-SH and KELLY cells treated with 10 µm MI-773 at various times. *P<0.05, **P<0.01 and ****P<0.0001. One-way ANOVA was performed. Data are shown as the mean ± SD and of three independent experiments. NB, neuroblastoma.
|
MI-773 induces apoptosis and cell cycle arrest in NB cells
The results of flow cytometry analysis showed that NB cells treated with different concentrations of MI-773 for 72 h exhibited increased apoptosis compared with the control. The proportion of apoptotic cells increased ~3-fold in the MI-773 treated group compared with the control, suggesting that MI-773 treatment significantly induced apoptosis in the cell lines (Fig. 3A).
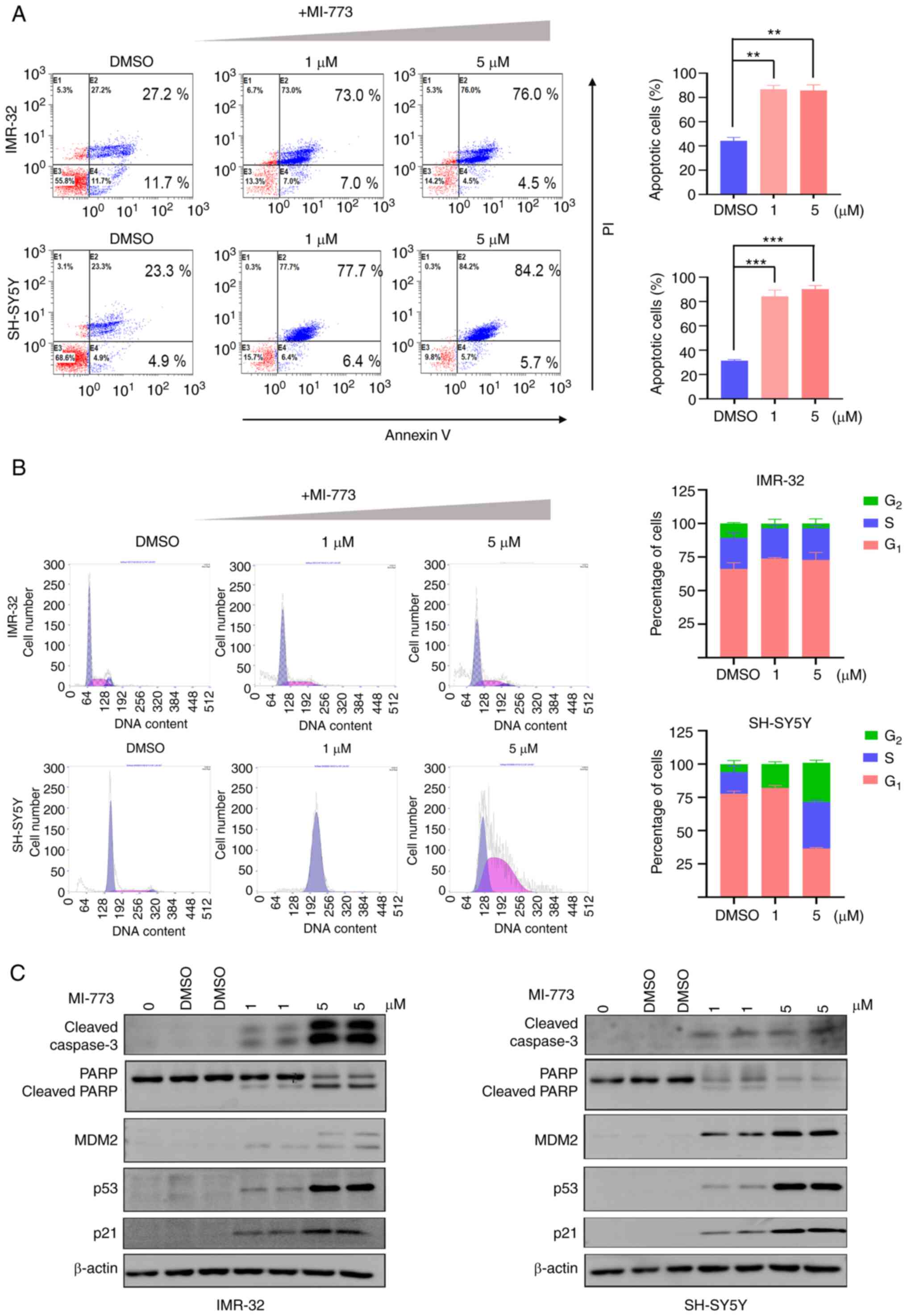 |
Figure 3.
MI-773 inhibits NB cells via induction of apoptosis and cell cycle arrest. (A) Annexin V/PI staining analysis showed increased proportion of apoptotic cells in NB cells treated with MI-773. **P<0.01, ***P<0.001. (B) Cell cycle analysis showed MI-773 altered G1 and G2/M phase ratio in NB cells. (C) Western blot analysis showed that MI-773 increased levels of p53, Caspase-3, MDM2 and p21 and decreased PARP expression in NB cells. One-way ANOVA was performed. Data are shown as the mean ± SD and of three independent experiments. NB, neuroblastoma; MDM2, murine double minute 2 homolog; PARP, poly(ADP-ribose) polymerase 1.
|
The effect of MI-773 on cell cycle progression was analyzed in two NB cell lines (SH-SY5Y and IMR-32) treated with increasing concentrations of MI-773 for 72 h (Fig. 3B). The results showed that the population of G1 phase cells increased in both NB cell lines treated with 1 µM MI-773, while a higher dose of MI-773 (5 µM) induced a notable increase in the proportion of cells in the G1 phase. These findings suggested that MI-773 induced G2/M phase cell cycle arrest.
Next, the levels of p53 downstream effectors CDKN1A (p21), MDM2 and the apoptosis-associated proteins PARP and caspase-3 were assessed using western blotting (Fig. 3C). The results showed that expression of p53 increased when NB cells were treated with MI-773, suggesting a p53 stabilization resulting from inhibition of MDM2-p53 interaction by MI-773. By contrast with the control group (0 µM), a notable increase in the cleavage of PARP and caspase-3 were observed in NB cell lines in a dose-dependent manner, confirming the pro-apoptosis effect of MI-773. Increased expression of MDM2 and CDKN1A (p21) protein, which were p53 targets, was shown in cells treated with MI-773. These findings indicated that MI-773 effectively induced apoptosis in NB cells and p53 stabilization.
Transcriptome analysis of MI-773 in NB cells
To determine the molecular pathways regulated by MI-773 and downstream target genes, RNA microarray analysis of MI-773-treated NB cells and the corresponding control was performed. Following data analysis, mRNA expression was clustered and plotted (Fig. 4A).
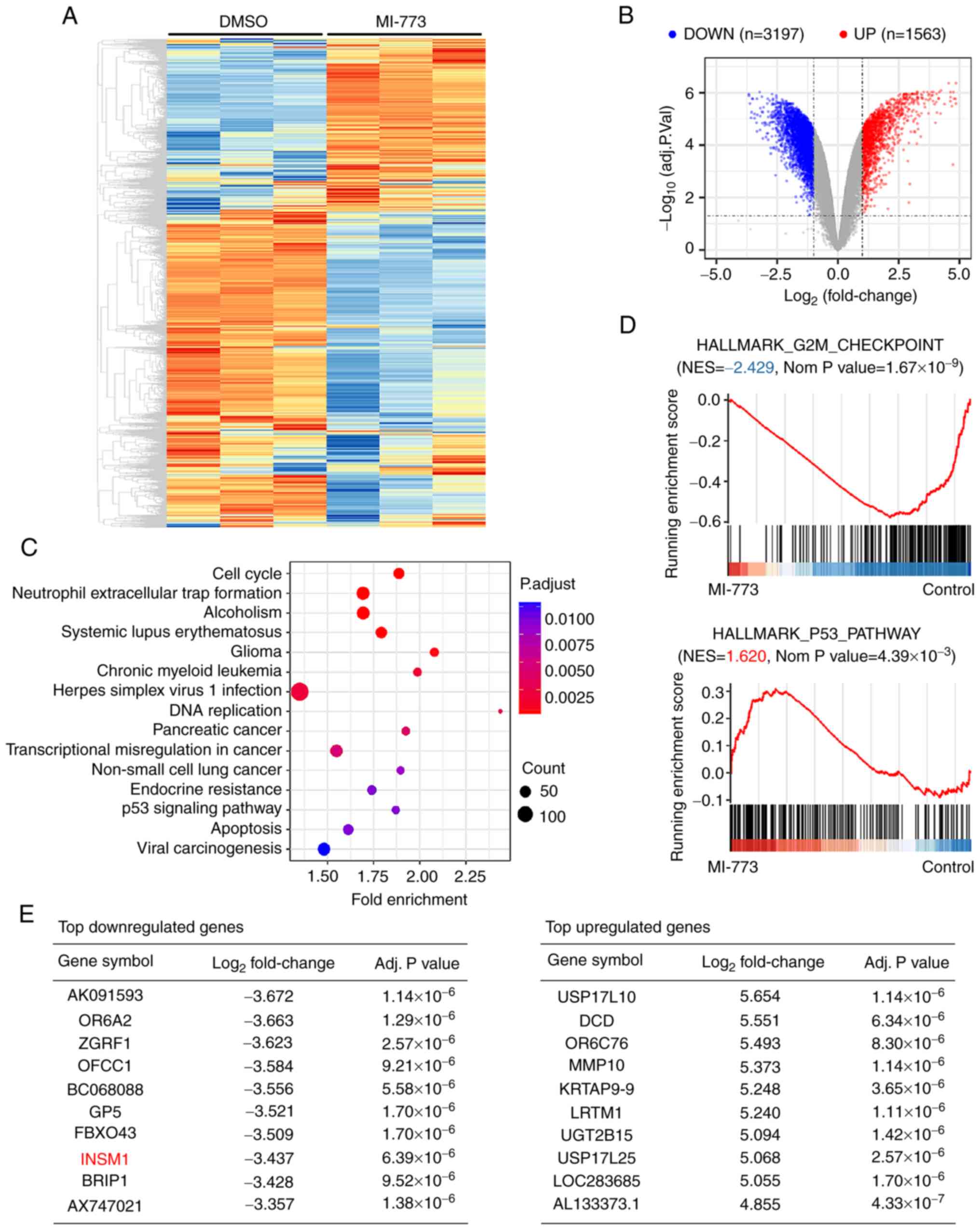 |
Figure 4.
Molecular mechanism analysis of MI-773 in NB cells. (A) SH-SY5Y cells were treated with MI-773 for 48 h. A total of three independent samples were used in the microarray analysis. mRNA expression data were clustered and plotted using the heatmap package in R. Red, upregulation; blue, downregulation. (B) Volcano plot to identify the differentially expressed genes with absolute (log2FoldChange)>1.0 and adjusted P-value <0.05. (C) Kyoto Encyclopedia of Genes and Genomes and (D) gene set enrichment analysis of differentially expressed genes was performed to investigate the function of MI-773 in gene regulation. (E) Top differentially expressed genes in SH-SY5Y following MI-773 treatment.
|
To identify the differentially expressed genes following MI-773 treatment, a volcano plot of all expressed genes was plotted with absolute (log2FoldChange)>1.0 and adjusted P-value <0.05 (Fig. 4B). A total of 1,563 up- and 3,197 downregulated genes were identified between the MI-773-treated and control group.
To investigate the role of MI-773 in gene regulation, the function of differentially expressed genes was annotated and enriched using KEGG pathway enrichment analysis (Fig. 4C). The results indicated that MI-773 was associated with regulation of ‘cell cycle’, ‘DNA replication’, ‘p53 signaling pathway’, ‘apoptosis’ and several other tumor-associated processes and activities. Moreover, GSEA revealed significant downregulation of genes belonging to the G2/M checkpoint pathway and upregulation of hallmark genes associated with the p53 pathway (Fig. 4D).
The top differentially expressed genes are displayed in Fig. 4E. One candidate gene was INSM1. The INSM1 gene was significantly downregulated by 3-fold following MI-773 treatment in SH-SY5Y cells based on data from RNA microarray analysis. Although the function of INSM1 in NB cells remains unclear, the role of INSM1 in NB cells was further investigated based on its reported function in neuronal differentiation (41).
INSM1 is a novel target of MI-773 in NB cells
First, INSM1 mRNA expression levels in different types of tumor were analyzed using the Cancer Cell Line Encyclopedia (Fig. 5A). The results showed that INSM1 had notable cancer type specificity and was highly expressed in NB and small cell lung cancer, both of which are hypothesized to originate from sympathetic nerve cells (39). The prognostic association of INSM1 in NB was analyzed using 476 samples in the GSE45547 and 102 samples in the GSE3446 dataset from GEO (42) and 249 samples from the TARGET website (Fig. 5B). The results showed that patients with NB with higher INSM1 expression were associated with a shorter overall survival time.
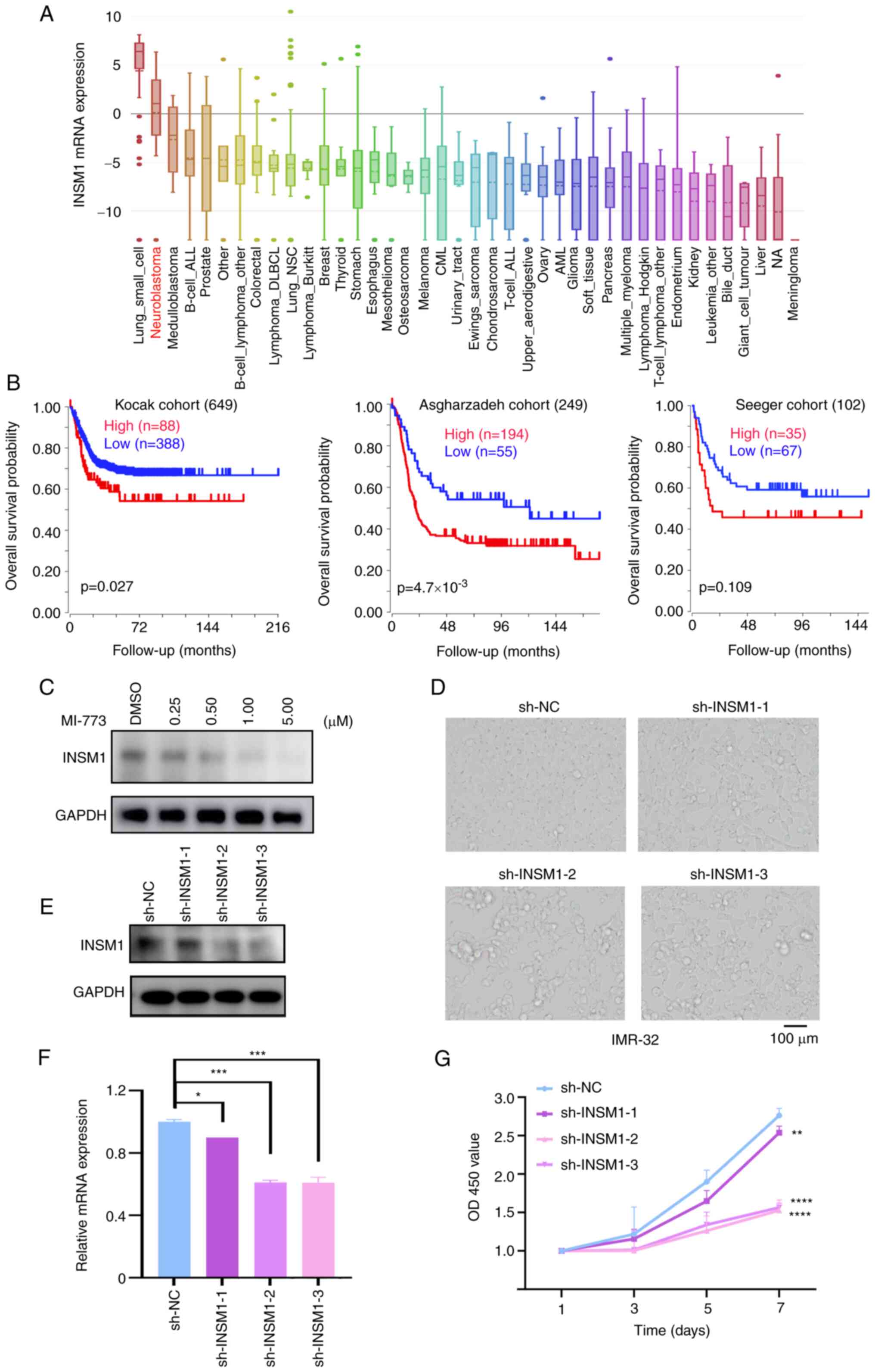 |
Figure 5.
INSM1 is a novel target of MI-773 in neuroblastoma cells. (A) INSM1 mRNA expression level in a range of tumors (generated from Broad Cancer Cell Line Encyclopedia: portals.broadinstitute.org/ccle). (B) Overall survival curve using public cohorts generated from R2 Genomics Analysis and Visualization Platform (r2.amc.nl). NB patients were categorized into high- or low-expression by using the scan cut-off mode. (C) Western blot analysis showed that the expression of INSM1 in IMR-32 cells treated with different concentrations of MI-773. (D) Cell morphology of IMR-32 cells following transfection with sh-INSM1 knockdown plasmids for 8 days. (E) Western blotting showed the effect knockdown of INSM1 expression by sh-INSM1 lentivirus in IMR-32 cells. (F) Reverse transcription-quantitative PCR analysis showed that transfection with INSM1 knockdown plasmids suppressed INSM1 mRNA expression in IMR-32 cells. *P<0.05 and ***P<0.001. (G) Proliferation rate of IMR-32 cells transfected with sh-INSM1 or sh-NC. **P<0.01; ****P<0.0001. One-way ANOVA was performed. Data are shown as the mean ± SD and of three independent experiments. INSM1, insulinoma-associated 1; sh, short hairpin; NC, negative control; OD, optical density.
|
Western blotting was performed on IMR-32 cells treated with increasing concentrations of MI-773; the results showed that the protein expression levels of INSM1 decreased in a dose-dependent manner, which was consistent with the results of RNA microarray (Fig. 5C). To investigate the role of INSM1 in NB cells, an interfering vector plasmid containing shRNA targeting the INSM1 gene was produced and transfected into IMR-32 cells. Knockdown of INSM1 was detected using western blotting (Fig. 5E) and RT-qPCR (Fig. 5F). Compared with the sh-negative control group, expression levels of INSM1 (sh-INSM1-2 and sh-INSM1-3) were significantly decreased by ~40% at the mRNA and protein level. Cell proliferation rate was significantly decreased following INSM1 knockdown (Fig. 5F). The cell viability rate on day 3 and 7 of the sh-INSM1-2 group decreased to 18.0 and 44.6%, respectively, compared with the sh-NC group (Fig. 5G). These results suggested that INSM1 served a role in regulating proliferation and may be a novel target of MI-773 in NB cells.
Discussion
The p53 pathway is key in maintaining genomic integrity in the face of cellular stress signals. It is hypothesized that nearly all cancers have a compromised p53 pathway (43). Tumor cells inactivate the tumor suppressive function of TP53 via different mechanisms. The most common mechanism is the acquired somatic TP53 mutation, which occurs in ~50% of tumor cells (44). The remaining 50% tumor cells with wt TP53 are suggested to harbor mutations that lie either up- or downstream of p53 and may also lead to the inactivation of the p53 pathway (43). p53 pathway regulators include MDM2 and 4 (10,11), p21 (45), ADP ribosylation factor (46) and tripartite motif-containing (47). A common result of in non-mutation-dependent inactivation of p53 is the upregulation of MDM2. MDM2 functions as a E3 ubiquitin ligase of p53, promoting degradation of p53 and blocking the TAD and tumor suppressive function of p53. Mutations in p53 rarely occur in NB, with <2% of cases at diagnosis and ~15% of cases at relapse possessing p53 mutations. However, amplification of the MDM2 oncogene is prevalent in NB; interfering with the interaction of MDM2 and p53 and restoring the tumor suppressive function of p53 is a potential therapeutic strategy for NB treatment (48–51).
A number of MDM2 antagonists have been developed and their antitumor efficacy has been investigated in a range of tumors. MI-219, belonging to the spiro-oxindole family, which has been shown to be well tolerated in extensive toxicity evaluations in animals, can disrupt the MDM2-p53 interaction (52). Nutlin-3 is a cis-imidazoline analogue that binds MDM2 at the p53 binding site (27,32,33). MI-63, a novel, non-peptide small-molecule inhibitor, is more effective and highly selective in blocking p53/MDM2 interactions compared with the traditional MDM2 inhibitors (12,22). However, although these inhibitors have been effective in in vitro experiments in early studies, the majority of do not exhibit favorable effects in in vivo studies or cannot be used in clinical applications due to high cytotoxicity, poor absorption and/or acquired chemoresistance (22,24,36,37,40,49,53).
MI-773, an optimized and potent MDM2 antagonist, has high selectivity and affinity in binding to MDM2 and shows tumor suppressive activity in different types of tumor, including adenoid cystic carcinomas (54) and mucoepidermoid carcinoma (55). The clinical safety of MI-773 was demonstrated in patients with solid tumors in a Phase I trial (trial no. NCT01636479) (56). MI-773 has also been demonstrated to augment the cytotoxic effects of doxorubicin in NB cell lines and may serve as an effective adjuvant (34).
In the present study, CCK-8 and clonogenic assay demonstrated the impact of MI-773 on short- and long-term cell viability, respectively. The results showed potent antiproliferative effects of MI-773 on NB cells with wt p53, with IC50 values in the micromolar concentration range (IMR-32 IC50, 9.33 µM; SH-SY5Y IC50, 2.45 µM; SK-N-SH IC50, 19.84 µM; LAN-1 IC50, 5.31 µM and NGP IC50, >20.00 µM). However, these effects were not observed in the KELLY cells, which possess mutant p53.
MDM2 antagonists have been reported to exert their antitumor effects by inducing cell cycle arrest, apoptosis, senescence or differentiation of NB cells (12,57). Mechanistically, MDM2 antagonists block the p53 binding site of MDM2 and prevent its interaction with p53, allowing the reactivation of p53 function in cells with intact p53 (58). The present study showed that MI-773 treatment induced apoptosis and cell cycle arrest in a dose-dependent manner. Additionally, the results confirmed that MI-773 blocked ubiquitination-mediated degradation of p53 as p53 protein accumulated following MI-773 treatment. The upregulation of downstream effector genes MDM2 and p21 in the wt p53 NB cells suggests the reactivation of p53. The discrepancy of basal MDM2 protein expression levels in Figs. 1B and 3C is likely due to the differences in auto-exposure time from each blot, although the loaded protein amount was the same (20 µg/lane). Since the expression of MDM2 protein significantly increased in MI-773-treated IMR-32 and SH-SY5Y cells, the imaging system automatically adjusts exposure time to avoid overexposure based on the high expression level in the experimental group, which makes the basal MDM2 expression appear undetectable. Therefore, the band only reflects the relative expression, not the quantitative level, of protein of interest.
The underlying molecular mechanisms and pathways involved in MI-773-induced proliferation arrest in wt p53 NB cells were investigated. The significantly enriched pathways were involved in ‘cell cycle regulation’, ‘neutrophil extracellular trap formation’, ‘glioma’ and ‘chromic myeloid leukemia’. Differentially expressed genes were clustered in the G2/M phase checkpoint and p53 pathway genes, which was consistent with the antitumor effects of MI-773 in NB.
In particular, the INSM1 gene was identified, shown to be significantly downregulated following MI-773 treatment and served a role in NB cell proliferation. The INSM1 gene, an established cytoplasmic marker of neuroendocrine tumors (57), also serves as a transcription factor via an autoregulatory mechanism (59). In NB, INSM1 forms a positive-feedback loop with N-Myc and stimulates NB cell proliferation, which highlights INSM1 signaling as a potential target for future drug development (43,60). The present study demonstrated that knockdown of the INSM1 gene exhibited an inhibitory effect on proliferation and induced apoptosis in NB cells. The available data above suggest that INSM1 is a downstream effector of the MDM2/p53 pathway in NB cells and may serve as a novel therapeutic target for the treatment of NB.
In conclusion, the antitumor effect of the MDM2 inhibitor MI-773 was assessed, including the molecular mechanisms underlying NB cell regulation. The results showed that MI-773 potently inhibited proliferation and induced apoptosis and cell cycle arrest in NB cells. Mechanistically, MI-773 effectively rescued TP53 expression and regulated p53 pathways. Additionally, the INSM1 gene was identified as a downstream member of the MDM2/p53 pathway and was suggested to be involved in NB cell proliferation.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from Natural Science Fund for Colleges and Universities of Jiangsu Province (grant no. 18KJB320022); the National Natural Science Foundation (grant nos. 82072767, 81770145, 81702339, 81701596, 817072737, 81802499, 81872845, 81902534, 81971867 and 81902534); Natural Science Foundation of Jiangsu Province (grant nos. BK20180207, BK20180206, SBK2019021442, BK20191175 and BK20190185); Jiangsu Provincial Medical Youth Talent (grant nos. QNRC2016762, QNRC2016756, QNRC2016758 and QNRC2016768); Gusu Health Talents Program of Soochow City (grant no. 2020-104); Department of Pediatrics Clinical Center of Suzhou (grant no. Szzx201504); Applied Foundational Research of Medical and Health Care of Suzhou City (grant nos. SYS2019080, SYS2019082, SYS2018075, SYS2018074, SYS2019077 and SS201809); Accented Term of Jiangsu Provincial Health Commission (grant no. K2019005); Suzhou Special Foundation of Clinical Key Diseases Diagnosis and Therapy (grant no. LCZX201907).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. Microarray analysis and original data have been submitted to the GEO database (accession no. GSE174450).
Authors' contributions
JP and ZHL designed and directed the study. ZMZ, HRW and XLL performed statistical analysis. YLC performed the experiments, analyzed the data and wrote the paper. JJP and JJY performed experiments. HBC and RZ performed lentivirus preparation and transfection. DW, XML and GL performed western blotting, PCR and in vitro experiments. YFT, JL and FF collected clinical data. YX and SYW designed primers for PCR. CXF, XRC and YPZ performed apoptosis and cell cycle analysis. JWW and ZZ performed plasmid construction. SYH designed the study and revised the manuscript. ZZ and ZHL confirmed the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Glossary
Abbreviations
Abbreviations:
|
TAD
|
transactivation domain
|
|
PI
|
propidium iodide
|
|
CCK-8
|
Cell Counting Kit- 8
|
|
IC50
|
half-maximal inhibitory concentration
|
|
shRNA
|
short hairpin RNA
|
|
GSEA
|
gene set enrichment analysis
|
|
KEGG
|
Kyoto Encyclopedia of Genes and Genomes
|
|
GEO
|
Gene Expression Omnibus
|
References
|
1
|
Conter HJ, Gopalakrishnan V, Ravi V, Ater JL, Patel S and Araujo DM: Adult versus pediatric neuroblastoma: The M.D. Anderson Cancer Center experience. Sarcoma. 2014:3751512014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Barbieri E, De Preter K, Capasso M, Johansson P, Man TK, Chen Z, Stowers P, Tonini GP, Speleman F and Shohet JM: A p53 drug response signature identifies prognostic genes in high-risk neuroblastoma. PLoS One. 8:e798432013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lerone M, Ognibene M, Pezzolo A, Martucciello G, Zara F, Morini M and Mazzocco K: Molecular genetics in neuroblastoma prognosis. Children (Basel). 8:4562021.PubMed/NCBI
|
|
4
|
Smith V and Foster J: High-risk neuroblastoma treatment review. Children (Basel). 5:1142018.PubMed/NCBI
|
|
5
|
Donghi R, Longoni A, Pilotti S, Michieli P, Porta GD and Pierotti MA: Gene p53 mutations are restricted to poorly differentiated and undifferentiated carcinomas of the thyroid gland. J Clin Invest. 91:1753–1760. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ago K, Shibutani M, Saegusa Y, Shima T, Taniai E and Mitsumori K: A case report of a cerebellar neuroblastoma in a p53 null mutation mouse. J Vet Med Sci. 71:345–347. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bu X, Shen Z and Jin B: Expression of p53 in nephroblastoma and neuroblastoma. Zhonghua Wai Ke Za Zhi. 35:220–222. 1997.(In Chinese). PubMed/NCBI
|
|
8
|
Feng Z, Zhang H, Levine AJ and Jin S: The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 102:8204–8209. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu X, Harris SL and Levine AJ: The regulation of exosome secretion: A novel function of the p53 protein. Cancer Res. 66:4795–4801. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, de Graaf P, Francoz S, Gasparini P, Gobbi A, et al: Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 24:5835–5843. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Oliner JD, Saiki AY and Caenepeel S: The role of MDM2 amplification and overexpression in tumorigenesis. Cold Spring Harb Perspect Med. 6:a0263362016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gamble LD, Kees UR, Tweddle DA and Lunec J: MYCN sensitizes neuroblastoma to the MDM2-p53 antagonists Nutlin-3 and MI-63. Oncogene. 31:752–763. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nicolai S, Pieraccioli M, Peschiaroli A, Melino G and Raschella G: Neuroblastoma: Oncogenic mechanisms and therapeutic exploitation of necroptosis. Cell Death Dis. 6:e20102015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Isaacs JS, Saito S and Neckers LM: Requirement for HDM2 activity in the rapid degradation of p53 in neuroblastoma. J Biol Chem. 276:18497–18506. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Inomistova MV, Svergun NM, Khranovska NM, Skachkova OV, Gorbach OI and Klymnyuk GI: Prognostic significance of MDM2 gene expression in childhood neuroblastoma. Exp Oncol. 37:111–115. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fan YH, Cheng J, Vasudevan SA, Dou J, Zhang H, Patel RH, Ma IT, Rojas Y, Zhao Y, Yu Y, et al: USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis. 4:e8672013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng J, Fan YH, Xu X, Zhang H, Dou J, Tang Y, Zhong X, Rojas Y, Yu Y, Zhao Y, et al: A small-molecule inhibitor of UBE2N induces neuroblastoma cell death via activation of p53 and JNK pathways. Cell Death Dis. 5:e10792014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vogan K, Bernstein M, Leclerc JM, Brisson L, Brossard J, Brodeur GM, Pelletier J and Gros P: Absence of p53 gene mutations in primary neuroblastomas. Cancer Res. 53:5269–5273. 1993.PubMed/NCBI
|
|
19
|
Diskin SJ, Capasso M, Diamond M, Oldridge DA, Conkrite K, Bosse KR, Russell MR, Iolascon A, Hakonarson H, Devoto M and Maris JM: Rare variants in TP53 and susceptibility to neuroblastoma. J Natl Cancer Inst. 106:dju0472014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Imamura J, Bartram CR, Berthold F, Harms D, Nakamura H and Koeffler HP: Mutation of the p53 gene in neuroblastoma and its relationship with N-myc amplification. Cancer Res. 53:4053–4058. 1993.PubMed/NCBI
|
|
21
|
Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, Ding K, Wang G, Chen J, et al: Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci USA. 105:3933–3938. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Canner JA, Sobo M, Ball S, Hutzen B, DeAngelis S, Willis W, Studebaker AW, Ding K, Wang S, Yang D and Lin J: MI-63: A novel small-molecule inhibitor targets MDM2 and induces apoptosis in embryonal and alveolar rhabdomyosarcoma cells with wild-type p53. Br J Cancer. 101:774–781. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang S, Sun W, Zhao Y, McEachern D, Meaux I, Barrière C, Stuckey JA, Meagher JL, Bai L, Liu L, et al: SAR405838: An optimized inhibitor of MDM2-p53 interaction that induces complete and durable tumor regression. Cancer Res. 74:5855–5865. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bill KL, Garnett J, Meaux I, Ma X, Creighton CJ, Bolshakov S, Barriere C, Debussche L, Lazar AJ, Prudner BC, et al: SAR405838: A novel and potent inhibitor of the MDM2:p53 axis for the treatment of dedifferentiated liposarcoma. Clin Cancer Res. 22:1150–1160. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chessari G, Hardcastle IR, Ahn JS, Anil B, Anscombe E, Bawn RH, Bevan LD, Blackburn TJ, Buck I, Cano C, et al: Structure-based design of potent and orally active isoindolinone inhibitors of MDM2-p53 protein-protein interaction. J Med Chem. 64:4071–4088. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Burmakin M, Shi Y, Hedström E, Kogner P and Selivanova G: Dual targeting of wild-type and mutant p53 by small molecule RITA results in the inhibition of N-Myc and key survival oncogenes and kills neuroblastoma cells in vivo and in vitro. Clin Cancer Res. 19:5092–5103. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tesson M, Vasan R, Hock A, Nixon C, Rae C, Gaze M and Mairs R: An evaluation in vitro of the efficacy of nutlin-3 and topotecan in combination with 177Lu-DOTATATE for the treatment of neuroblastoma. Oncotarget. 9:29082–29096. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen L, Rousseau RF, Middleton SA, Nichols GL, Newell DR, Lunec J and Tweddle DA: Pre-clinical evaluation of the MDM2-p53 antagonist RG7388 alone and in combination with chemotherapy in neuroblastoma. Oncotarget. 6:10207–10221. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carvajal LA, Neriah DB, Senecal A, Benard L, Thiruthuvanathan V, Yatsenko T, Narayanagari SR, Wheat JC, Todorova TI, Mitchell K, et al: Dual inhibition of MDMX and MDM2 as a therapeutic strategy in leukemia. Sci Transl Med. 10:eaao30032018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fang DD, Tang Q, Kong Y, Rong T, Wang Q, Li N, Fang X, Gu J, Xiong D, Yin Y, et al: MDM2 inhibitor APG-115 exerts potent antitumor activity and synergizes with standard-of-care agents in preclinical acute myeloid leukemia models. Cell Death Discov. 7:902021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ding K, Lu Y, Nikolovska-Coleska Z, Wang G, Qiu S, Shangary S, Gao W, Qin D, Stuckey J, Krajewski K, et al: Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2-p53 interaction. J Med Chem. 49:3432–3435. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Aziz MH, Shen H and Maki CG: Acquisition of p53 mutations in response to the non-genotoxic p53 activator Nutlin-3. Oncogene. 30:4678–4686. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Michaelis M, Rothweiler F, Barth S, Cinatl J, van Rikxoort M, Löschmann N, Voges Y, Breitling R, von Deimling A, Rödel F, et al: Adaptation of cancer cells from different entities to the MDM2 inhibitor nutlin-3 results in the emergence of p53-mutated multi-drug-resistant cancer cells. Cell Death Dis. 2:e2432011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lu J, Guan S, Zhao Y, Yu Y, Wang Y, Shi Y, Mao X, Yang KL, Sun W, Xu X, et al: Novel MDM2 inhibitor SAR405838 (MI-773) induces p53-mediated apoptosis in neuroblastoma. Oncotarget. 7:82757–82769. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li Z, Li X, Xu L, Tao Y, Yang C, Chen X, Fang F, Wu Y, Ding X, Zhao H, et al: Inhibition of neuroblastoma proliferation by PF-3758309, a small-molecule inhibitor that targets p21-activated kinase 4. Oncol Rep. 38:2705–2716. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu G, Wang LG, Han Y and He QY: ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li ZH, Du WW, Xu LX, Zhao H, Li XL, Fang F, Wu Y, Lu J, Li YH, Du WW, et al: A novel sphingosine kinase 1 inhibitor (SKI-5C) induces cell death of Wilms' tumor cells in vitro and in vivo. Am J Transl Res. 8:4548–4563. 2016.PubMed/NCBI
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Morales La Madrid A, Campbell N, Smith S, Cohn SL and Salgia R: Targeting ALK: A promising strategy for the treatment of non-small cell lung cancer, non-Hodgkin's lymphoma, and neuroblastoma. Target Oncol. 7:199–210. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lee H, Ren J, Pesavento RP, Ojeda I, Rice AJ, Lv H, Kwon Y and Johnson ME: Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via high-throughput screening and molecular modeling. Bioorg Med Chem. 27:1981–1989. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen C and Lan MS: A promoter-driven assay for INSM1-associated signaling pathway in neuroblastoma. Cell Signal. 76:1097852020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pieters R, Schrappe M, De Lorenzo P, Hann I, Rossi GD, Felice M, Hovi L, LeBlanc T, Szczepanski T, Ferster A, et al: A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): An observational study and a multicentre randomised trial. Lancet. 370:240–250. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Timmerman DM, Remmers TL, Hillenius S and Looijenga LHJ: Mechanisms of TP53 pathway inactivation in embryonic and somatic cells-relevance for understanding (Germ Cell) tumorigenesis. Int J Mol Sci. 22:53772021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rivlin N, Brosh R, Oren M and Rotter V: Mutations in the p53 tumor suppressor gene: Important milestones at the various steps of tumorigenesis. Genes Cancer. 2:466–474. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Martin-Caballero J, Flores JM, Garcia-Palencia P and Serrano M: Tumor susceptibility of p21(Waf1/Cip1)-deficient mice. Cancer Res. 61:6234–6238. 2001.PubMed/NCBI
|
|
46
|
Zhang Y, Xiong Y and Yarbrough GW: ARF Promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 92:725–734. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu J, Zhang C, Wang XL, Ly P, Belyi V, Xu-Monette ZY, Young KH, Hu W and Feng Z: E3 ubiquitin ligase TRIM32 negatively regulates tumor suppressor p53 to promote tumorigenesis. Cell Death Differ. 21:1792–1804. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Moll UM and Petrenko O: The MDM2-p53 interaction. Mol Cancer Res. 1:1001–1008. 2003.PubMed/NCBI
|
|
49
|
Tweddle DA, Pearson ADJ, Haber M, Norris MD, Xue C, Flemming C and Lunec J: The p53 pathway and its inactivation in neuroblastoma. Cancer Lett. 197:93–98. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Goldschneider D, Horvilleur E, Plassa LF, Guillaud-Bataille M, Million K, Wittmer-Dupret E, Danglot G, de Thé H, Bénard J, May E and Douc-Rasy S: Expression of C-terminal deleted p53 isoforms in neuroblastoma. Nucleic Acids Res. 34:5603–5612. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wolter J, Angelini P and Irwin M: p53 family: Therapeutic targets in neuroblastoma. Future Oncol. 6:429–444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhao Y, Liu L, Sun W, Lu J, McEachern D, Li X, Yu S, Bernard D, Ochsenbein P, Ferey V, et al: Diastereomeric spirooxindoles as highly potent and efficacious MDM2 inhibitors. J Am Chem Soc. 135:7223–7234. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Van Maerken T, Vandesompele J, Rihani A, De Paepe A and Speleman F: Escape from p53-mediated tumor surveillance in neuroblastoma: Switching off the p14(ARF)-MDM2-p53 axis. Cell Death Differ. 16:1563–1572. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Nor F, Warner KA, Zhang Z, Acasigua GA, Pearson AT, Kerk SA, Helman JI, Filho MS, Wang S and Nör JE: Therapeutic inhibition of the MDM2-p53 interaction prevents recurrence of adenoid cystic carcinomas. Clin Cancer Res. 23:1036–1048. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Andrews A, Warner K, Rodriguez-Ramirez C, Pearson AT, Nör F, Zhang Z, Kerk S, Kulkarni A, Helman JI, Brenner JC, et al: Ablation of cancer stem cells by therapeutic inhibition of the MDM2-p53 interaction in mucoepidermoid carcinoma. Clin Cancer Res. 25:1588–1600. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
de Jonge M, de Weger VA, Dickson MA, Langenberg M, Cesne AL, Wagner AJ, Hsu K, Zheng W, Macé S, Tuffal G, et al: A phase I study of SAR405838, a novel human double minute 2 (HDM2) antagonist, in patients with solid tumours. Eur J Cancer. 76:144–151. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mahalakshmi B, Baskaran R, Shanmugavadivu M, Nguyen NT and Velmurugan BK: Insulinoma-associated protein 1 (INSM1): A potential biomarker and therapeutic target for neuroendocrine tumors. Cell Oncol (Dordr). 43:367–376. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sanz G, Singh M, Peuget S and Selivanova G: Inhibition of p53 inhibitors: Progress, challenges and perspectives. J Mol Cell Biol. 11:586–599. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Breslin MB, Zhu M, Notkins AL and Lan MS: Neuroendocrine differentiation factor, IA-1, is a transcriptional repressor and contains a specific DNA-binding domain: Identification of consensus IA-1 binding sequence. Nucleic Acids Res. 30:1038–1045. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chen C, Breslin MB and Lan MS: INSM1 increases N-myc stability and oncogenesis via a positive-feedback loop in neuroblastoma. Oncotarget. 6:36700–36712. 2015. View Article : Google Scholar : PubMed/NCBI
|



















