Introduction
Lung cancer is a frequently diagnosed malignant cancer and the leading cause of cancer-related death in the world (1). Non-small cell lung cancer (NSCLC) accounts for 75–80% of lung cancer cases worldwide (2). Although great progress has been made in the development of various common treatments, including surgical resection, chemotherapy, radiotherapy and targeted drugs, considering the low survival rate, the improvement of the prognosis and treatment of NSCLC is still urgently needed (3). Therefore, it is crucial to further study the molecular mechanisms underlying NSCLC carcinogenesis and to identify biomarkers and targets for early detection, intervention and treatment.
Long non-coding RNA (lncRNA) is a common class of non-coding small RNA with a length >200 nucleotides (4,5). Although most of lncRNA molecules do not encode proteins or peptides, they can modulate the expression and function of a given gene epigenetically, transcriptionally and post-transcriptionally by sponging microRNA (miRNA/miR) (6–8). Moreover, lncRNA has been implicated in numerous pathological and physiological processes, such as cell proliferation, apoptosis, tissue differentiation, angiogenesis, carcinogenesis, migration, invasion, anti-tumor drug resistance and energy metabolism (9,10). Therefore, lncRNA holds potential for the development of biomarkers for the early diagnosis of NSCLC.
lncRNA forkhead box D3 antisense 1 (FOXD3-AS1) is an antisense transcript of the protein-coding gene FOXD3, which is located on chromosome 1p31.3 in humans. Upregulation of FOXD3-AS1 is associated with poor prognosis in glioma, thyroid cancer, breast cancer, hepatocellular carcinoma, colorectal cancer and malignant melanoma (11–15). For example, high levels of FOXD3-AS1 result in poor prognosis in patients with glioma, while FOXD3-AS1 silencing suppresses glioma cell proliferation, migration and invasion (11). Additionally, FOXD3-AS1 regulates thyroid cancer progression by regulating miR-296-5p/TGF-β1/Smad (Smad2/3, 4 and 7) axis (16). In hepatocellular carcinoma cells, inhibition of FOXD3-AS1 through regulation of miR-335 and rapamycin-insensitive companion of mTOR expression reduced cell proliferation, colony formation, migration and invasion. In recent studies, FOXD3-AS1 has also been confirmed to play an important role in NSCLC. For instance, FOXD3-AS1 promotes cisplatin chemoresistance of NSCLC cells by directly acting on the miR-127-3p/MDM2 axis (17). High expression levels of FOXD3-AS1 promote NSCLC progression by regulating the miR-127-3p/mediator complex subunit 28 (MED28) axis (18). Therefore, FOXD3-AS1 might be a crucial regulator involved in NSCLC tumorigenesis, although the underlying mechanisms of FOXD3-AS1 in NSCLC remain elusive. The present study demonstrated that FOXD3-AS1 expression was increased in both NSCLC tissue and cell lines. Furthermore, an axis involving NSCLC, miR-135a-5p/CDK6 was identified as essential to NSCLC progression. These findings may provide potential novel insights for therapeutics in patients with NSCLC.
Materials and methods
Cell culture and patient samples
The NSCLC tissue and adjacent non-cancerous tissue samples (distance from tumor tissues >5 cm.) were collected between May 2016 and December 2019 at the Affiliated Nanhai Hospital, Southern Medical University. A total of 30 patients with lung cancer (age range, 38–75 years; mean age, 62±1.5 years; male/female ratio, 7:8) were selected because they met the following inclusion criteria: Pathological examination confirmed primary lung cancer by biopsy samples; and no anticancer treatments were given before surgery. All participants provided written informed consent and were aware that their samples would be used for research, according to the Declaration of Helsinki. The clinicopathological characteristics of the 30 patients are presented in Table I. This study was approved by The Institutional Board of The Affiliated Nanhai Hospital, Southern Medical University (People's Hospital of Nanhai District; approval no. KY-E-2019-6-17).
 |
Table I.
Relationship between the expression of FOXD3-AS1 and the clinicopathological characteristics of patients with non-small cell lung cancer (n=15).
|
Table I.
Relationship between the expression of FOXD3-AS1 and the clinicopathological characteristics of patients with non-small cell lung cancer (n=15).
| |
FOXD3-AS1 expression |
|
| |
|
|
| Variable |
Low, n |
High, n |
P-value |
| Age, years |
|
|
|
| <60 |
7 |
5 |
0.456 |
| ≥60 |
8 |
10 |
|
| Sex |
|
|
|
| Male |
6 |
8 |
0.464 |
| Female |
9 |
7 |
|
| Tumor stage |
|
|
|
| I+II |
10 |
4 |
0.028 |
| III+IV |
5 |
11 |
|
| Differentiation |
|
|
|
| Well-differentiated |
12 |
5 |
0.010 |
| Moderate-poor |
3 |
10 |
|
| Lymph node metastasis |
|
|
|
| Negative |
11 |
5 |
0.028 |
| Positive |
4 |
10 |
|
| Smoking status |
|
|
|
| Yes |
7 |
6 |
0.713 |
| No |
8 |
9 |
|
The NSCLC cell lines (CALU3, HCT116, A549 and H1229) and the normal human bronchial epithelial cells (16HBE) were purchased from the American Type Culture Collection (ATCC). These cells were cultured in Dulbecco's modified Eagles medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) in a humidified atmosphere containing 5% CO2 at 37°C.
RNA isolation and reverse transcription-quantitative (RT-qPCR)
Total RNA was isolated from tissue samples or cultured cells using RNAiso Plus reagent (Invitrogen; Thermo Fisher Scientific, Inc.). A NanoDrop® 2000 spectrophotometer (Thermo Fisher Scientific, Inc.) was used to check the concentration and purity of RNA, and the A260/A280 ratio of all samples was between 1.9–2.1. After measuring concentration, RNA was reverse transcribed into cDNA using the PrimeScript™ II 1st RT reagent kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's protocol. qPCR was performed using the SYBR Premix Ex Taq II kit (Takara Biotechnology Co., Ltd.). The reaction mixture was incubated at 95°C for 2 min. The thermocycling program consisted of holding at 94°C for 2 min, followed by 30 cycles of 30 sec at 94°C, 30 sec at 56°C, and 60 sec at 72°C. The expression levels of the small nucleolar RNA U6 and GAPDH served as an endogenous control. The relative expression levels were calculated using the 2−ΔΔCq method (19). The sequences of the primers used for FOXD3-AS1, miR-135a-5p, GAPDH and U6 were as follows: i) FOXD3-AS1 forward, 5′-TTTCCGTAGCTCCACAGCTT-3′ and reverse, 5′-CAGAGCAAAGGAGGTTCCTG-3′; ii) miR-135a-5p forward, 5′-ATACCGAAAAATAAGGATACACT-3′ and reverse 5′-GCGCGCGTAACAGTCTACAGC-3′; iii) CDK6 forward, 5′-TCTTCATTCACACCGAGTAGTGC-3′ and reverse, 5′-TGAGGTTAGAGCCATCTGGAAA-3′; GAPDH forward, 5′-TGCACCACCAACTGCTTAGC-3′ and reverse, 5′-GGCATGCACTGTGGTCATGAG-3′; and iv) U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
Cell transfection
To construct the sh-FOXD3-AS1 vector, the effective shRNA was subcloned into the lentivirus vector (pHBLV-U6-MCS-CMV-ZsGreen-PGKPuro; HanBio Biotechnology Co, Ltd.) sh-NC was used as the negative control. The virus was packaged with three-plasmid system. Briefly, the FOXD3-AS1 or sh-NC were carried by 1 ng lentiviral vector and two packaging vectors (psPAX2, 1 ng and pMD2.G, 1 ng; HanBio Biotechnology Co., Ltd.) and were transiently transfected into 293T (ATCC) using Lipofectamine 3000 (Sigma Aldrich; Merck KGaA) according to the manufacturer's instructions at 37°C for 15 min. Following transfection, the viral supernatants were collected at 48 and 72 h and filtered through a 0.45 µm filter. Subsequently, a 40 ml ultracentrifuge tube was used to centrifuge the viral supernatants (4°C, 58,700 × g, 120 min). The viral supernatants (sh-FOXD3-AS1 and sh-NC) were used to infect A549 and H1299 cells, with a MOI to infect the cells of 20–40. Subsequently, the virus was screened with puromycin and the expression level of FOXD3-AS1 was detected by RT-qPCR. miR-135a-5p inhibitor, miR-135a-5p mimics, miR negative control (NC) mimics and miR-NC-inhibitor were carried out by Shanghai GeneChem Co., Ltd. The open reading frame of CDK6 was inserted into the expression vector pcDNA3.1+ (Sigma-Aldrich; Merck KGaA) to overexpress CDK6 by Genechem Shanghai GeneChem Co., Ltd. The empty pcDNA3.1+ vector was used as the negative control for CDK6 overexpression. The NSCLC cells were transfected with the indicated constructs (20 nM) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 15 min following the manufacturer's protocol. Cells were collected at 48 h after transfection for subsequent experimentation. The sequences used were as follows: i) sh-FOXD3-AS1#1, 5′-GCUACUUGGAGUUGUUAAAUUCAAGAGACGAUGAACCUCAACAAUUUUUUUUU-3′; ii) sh-FOXD3-AS1#2, 5′-GGAGUUGUUAAACGACUUAUUCAAGAGACCUCAACAAUUUGCUGAAUUUUUUU-3′; iii) sh-NC, 5′-CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTTG-3′; iv) miR-NC-mimics, 5′-GUCGUAUCCAGUGCAGGGUCC-3′; v) miR-NC-inhibitor, 5′-CGAACGUGUCACGUTT-3′; vi) miR-135a-5p mimics, 5′-AUACCGAAAAAUAAGGAUACACU-3′; and vii) miR-135a-5p inhibitor, 5′-UGUAUCCUUAUUUUUCGGUAU-3′. Following transfection, RT-qPCR was performed to determine the transfection efficiency.
Cell Counting Kit-8 (CCK-8) assay
CCK-8 (Dojido) assays were performed to assess cell proliferation. After transfection, cells were cultured in 96-well plates and used for the assay at the 0, 24, 48 and 72-h time-points. After incubation with CCK-8 reagent for 2 h at 37°C, cell viability was evaluated according to the manufacturer's protocol.
Isolation of cell cytosolic/nuclear fractions
The Cytoplasmic Nuclear RNA Purification kit (Norgen Biotek Corp.) was used to separate the A549 and H1299 cell nuclear and cytosolic fractions according to the manufacturer's protocol. The RNA was extracted from the cytosolic as well as nuclear fractions. Subsequently, the expression level of FOXD3-AS1 in both fractions of NSCLC cell was determined using RT-qPCR.
Dual luciferase reporter assay
Dual luciferase assay was performed using a customized pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega). Briefly, wild-type (WT) containing putative binding sites or mutant (Mut) reporter containing all mutated putative binding sites of FOXD3-AS1 and CDK6 were inserted into the pmirGLO basic vector. For the luciferase reporter assay, cells with the reporter vector (Pmir-GLO-NC, Pmir-GLO-FOXD3-AS1-WT/Mut or Pmir-GLO-CDK6 WT/Mut; Promega) were co-transfected with negative control and miR-135a-5p mimics at the final concentration of 20 nM for each construct using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After transfection for 36 h, the luciferase activity was measured using the Dual-Lucy Assay Kit (Promega) according to the manufacturer's protocol. Luciferase activity was first normalized to that of Renilla luciferase from the same sample.
RNA immunoprecipitation (RIP) assay
RIP assay was utilized to explore the interaction between FOXD3-AS1 and miR-135a-5p. The EZMagna RIP kit (Merck KGaA) was used for RIP assay according to the manufacturer's protocol. A549 and H1299 cells were collected by centrifugation at 800 × g for 5 min at 4°C and lysed using complete Lysis buffer (Beyotime Institute of Biotechnology) containing RNase (Sigma-Aldrich; Merck KGaA) and protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA). Cell extracts (100 µl) were co-immunoprecipitated with protein G Sepharose beads (included in the kit) pre-coated with the conjugated Argonaute2 (Ago2) antibodies (1:150; cat. no. 2897S; Cell Signaling Technology, Inc.) or IgG (1:150; cat. no. 3900S; Cell Signaling Technology, Inc.) according to the manufacturer's protocol. Following incubation for 2 h at 4°C, the precipitated RNA was isolated from the beads and quantified by RT-qPCR.
Apoptotic assay
Annexin V-FITC apoptosis detection kit (Beyotime Institute of Biotechnology) was used to determine A549 and H1299 cell apoptosis. Annexin V-FITC and propidium iodide (PI) (Beyotime Institute of Biotechnology) were used to stain A549 and H1299 cells at room temperature in the dark for 20 min to assess apoptotic cells following the manufacturer's instructions. After staining, the percentage of apoptotic cells was quantified using flow cytometry (FACSCalibur; BD Biosciences) following the supplier's instructions. A negative control was used to confirm the correct gate. A549 and H1299 cell apoptosis was determined using FlowJo 7.6.1 software (FlowJo LLC).
Cell cycle assay
Cell cycle progression was analyzed by fixing the cells overnight in cold 70% alcohol at 4°C followed by washing and staining with PI (Beijing Solarbio Science & Technology Co., Ltd.) containing RNase A. After incubation with an RNA enzyme containing PI (40%; Sigma-Aldrich; Merck KaA) for 30 min at 4°C, the cells were analyzed within 1 h. The percentages of cells in each cell cycle phase (G0/G1, S and G2/M) were determined. The data are presented as a histogram.
Western blot analysis
Proteins were extracted using RIPA lysis buffer (Thermo Fisher Scientific, Inc.). BCA protein assay kit (Beyotime Institute of Biotechnology) was used to determine the protein concentration. Proteins (40 µg) were separated by 10% SDS-PAGE and transferred onto PVD membranes (EMD Millipore). Subsequently, the membranes were blocked in 5% non-fat milk and incubated with primary antibodies against CDK6 (1:1,000; cat. no. 3136S; Cell Signaling Technology, Inc.) and GAPDH (1:500; cat. no. ab8245; Abcam) overnight at 4°C. The following day, the membranes were blocked with appropriate HRP-conjugated secondary antibodies (1:1,000; cat. no. 7076S; Cell Signaling Technology, Inc.) at 37°C for 1 h. Lastly, the bands were visualized using a chemiluminescence detection kit (Thermo Fisher Scientific, Inc.). ImageJ 1 software (National Institutes of Health) was used for the quantification of band densities. GAPDH protein was used as the internal reference.
In vivo experiment
A total of 12 male BALB/c nude mice aged 6–8 weeks (20-25 g) were obtained from Shanghai Animal Laboratory Center. The mice were raised under a 12L/12D photoperiod at 18–22°C and 50–60% humidity. Drinking water was continuously provided and food was supplemented 3 times a week. The behavior and food intake of the mice were monitored every day to maintain their health. After transfection, 5×106 A549 cells were injected into the BALB/c nude mice subcutaneously. Tumor size was monitored using a caliper and tumor volumes were calculated every week for 28 days after injection. The mice were separately sacrificed at 7, 14, 21 and 28 days. In strict accordance with the principles of animal welfare, anesthesia is used to relieve pain during euthanasia. The mice were euthanized using sodium pentobarbital injected intraperitoneally at a dose of 150 mg/kg, and death was confirmed by the cessation of the heartbeat. All animal protocols were approved by The Animal Ethics Committee of Affiliated Nanhai Hospital, Southern Medical University (People's Hospital of Nanhai District; approval no. KY-E-2019-7-05).
Bioinformatics analysis
The Cancer Genome Atlas (TCGA, http://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) and Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) were used to explore the clinical characteristics of FOXD3-AS1 in patients with NSCLC. The LncBASE-DIANA database (http://bigd.big.ac.cn/databasecommons/database/id/336) was used to predict FOXD3-AS1 binding sites to miR-135a-5p. The Starbase miRNA target prediction website (https://www.lncrnablog.com/tag/starbase-v2-0/) was mined to explore the potential miR-135a-5p binding sites to CDK6.
Statistical analysis
The results are presented as the mean ± SEM. The data were statistical analyzed using GraphPad Prism 7.0 software (GraphPad Software, Inc.). All statistical analysis was conducted via SPSS 13.0 (SPSS Statistics, Inc.). All the experiments were performed three times. The data were normalized to the mean value of the control group and are presented as fold change, unless otherwise indicated. The differences between two groups were analyzed using unpaired Student's t-test. The differences between multiple groups were compared using one-way ANOVA with Bonferroni correction. The χ2 test was used to analyze the differences in the clinicopathological characteristics of the patients with NSCLC. P<0.05 was considered to indicate a statistically significant difference.
Results
FOXD3-AS1 is highly expressed in NSCLC tissues and cell lines
To investigate the clinical significance of FOXD3-AS1 in NSCLC tissue samples, TCGA data were analyzed, which demonstrated that FOXD3-AS1 expression was significantly upregulated in lung squamous cell carcinoma and lung adenocarcinoma samples relative to normal samples (Fig. 1A). Moreover, FOXD3-AS1 expression levels in 30 pairs of human NSCLC tissues and matched adjacent normal tissues were determined using RT-qPCR. FOXD3-AS1 expression was significantly upregulated in NSCLC tissue compared with normal adjacent tissue (Fig. 1B). In addition, the expression levels of FOXD3-AS1 were also determined in four NSCLC cell lines (CALU3, HCT116, A549 and H1229) and normal human bronchial epithelial cells (16HBE). Echoing the expression pattern found in NSCLC tissue, FOXD3-AS1 expression was significantly increased in NSCLC cells compared with 16HBE cell (Fig. 1C).
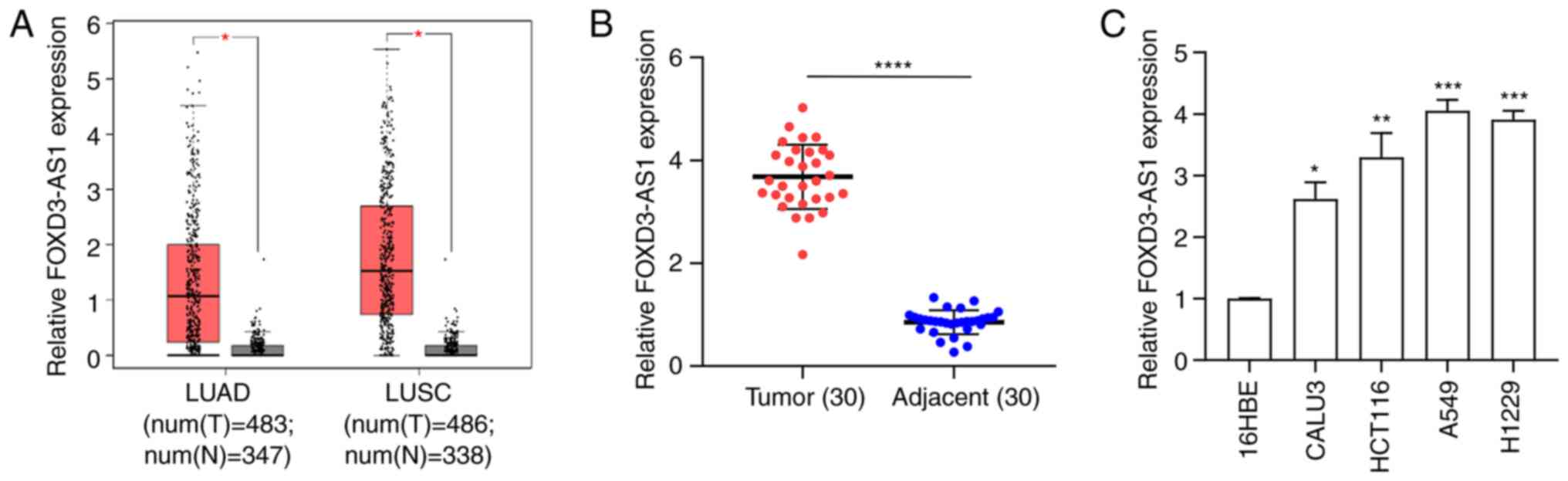 |
Figure 1.
FOXD3-AS1 is highly expressed in NSCLC tissue samples and cell lines. (A) FOXD3-AS1 was overexpressed in LUAD and LUSC samples from The Cancer Genome Atlas. (B) Relative FOXD3-AS1 expression in 30 pairs of NSCLC and matched adjacent normal tissue samples. (C) Relative expression level of FOXD3-AS1 in four NSCLC cell lines (CALU3, HCT116, A549 and H1229) and normal human bronchial epithelial cells (16HBE). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. NSCLC, non-small cell lung cancer; FOXD3-AS1, forkhead box D3 antisense 1; LUAD, lung adenocarcinoma; LUSC, lung squamous carcinoma; num(T), number of tumor samples; num(N), number of normal samples.
|
The 30 patients with NSCLC were then divided equally into high- and low-expression groups using the median FOXD3-AS1 expression value as the cut-off. FOXD3-AS1 levels were positively associated with NSCLC tumor differentiation, lymph node metastasis and TNM stage (Table I; P<0.05). Collectively, these data suggested that FOXD3-AS1 expression was significantly increased, both in NSCLC tissue and cell lines.
FOXD3-AS1 promotes proliferation and inhibits apoptosis of NSCLC cells in vitro and tumor growth in vivo
In order to clarify the function of FOXD3-AS1 in NSCLC, FOXD3-AS1 expression was silenced in the A549 and H1229 cell lines using a shRNA specifically targeting FOXD3-AS1. FOXD3-AS1 expression levels significantly decreased in the cells transfected with sh-FOXD3-AS1#1 and #2 compared with sh-NC (Fig. 2A). Additionally, CCK-8 and colony formation assays indicated that FOXD3-AS1 enhanced the proliferation of both A549 and H1229 cells (Fig. 2B and C). Furthermore, the effect of FOXD3-AS1 on the apoptosis in A549 and H1229 cells was also examined by flow cytometry. The results suggested that cell apoptosis was significantly increased following transfection with sh-FOXD3-AS1 compared with sh-NC (Fig. 2D). In order to evaluate the effect of FOXD3-AS1 on tumor growth in vivo, a subcutaneous tumor xenograft model was established. The weight of the tumors was smaller in the sh-FOXD3-AS1 group compared with the sh-NC group (Fig. 2E). In the tumor tissue, the expression levels of FOXD3-AS1 significantly decreased in the sh-FOXD3-AS1 group in comparison with the sh-NC group (Fig. 2F). Taken together, the results indicated that FOXD3-AS1 increased NSCLC cell proliferation whilst inhibiting apoptosis.
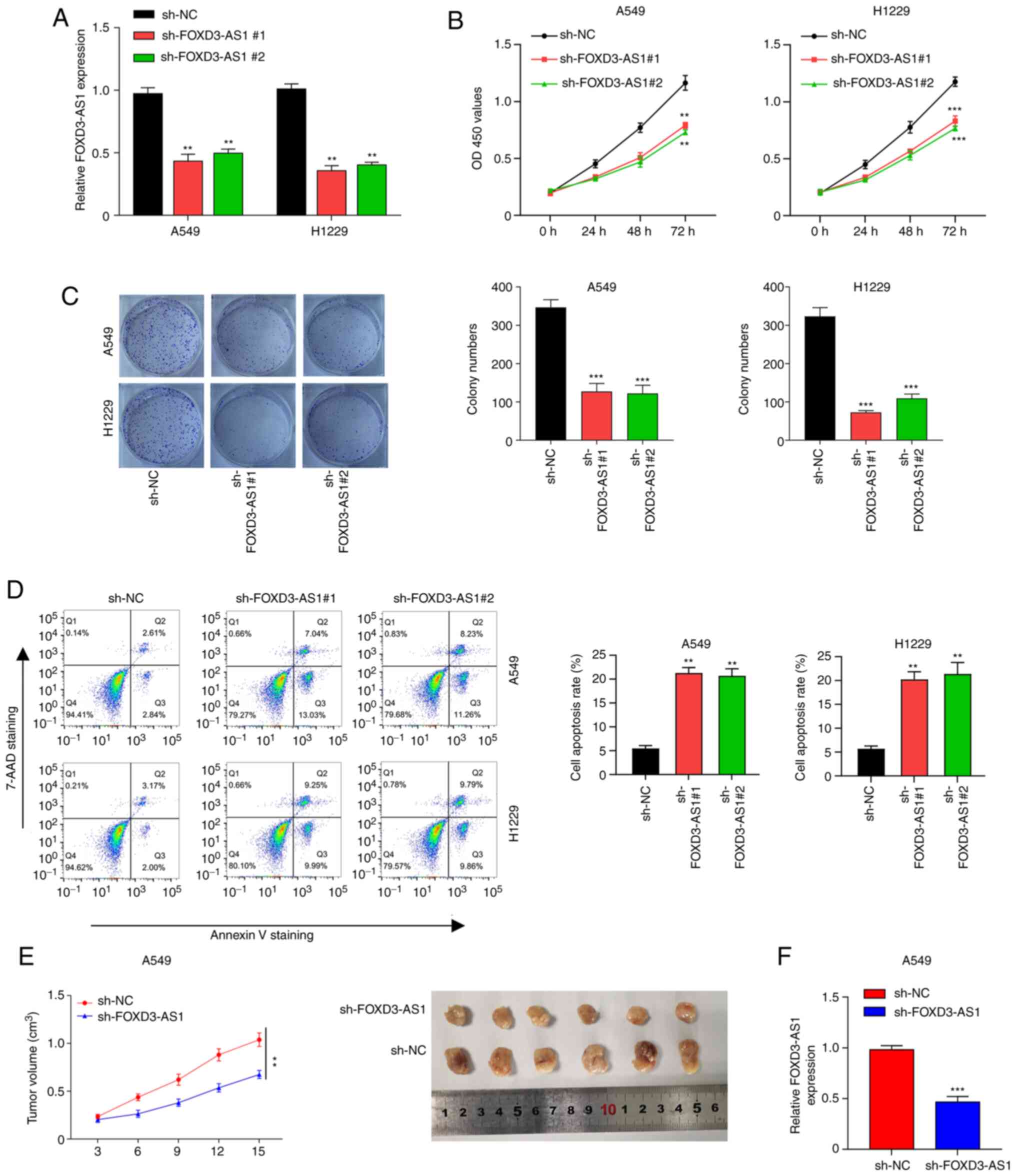 |
Figure 2.
FOXD3-AS1 promotes proliferation and inhibits apoptosis of non-small cell lung cancer cells in vitro and tumor growth in vivo. (A) Knockdown efficiency of FOXD3-AS1 was examined in A549 and H1229 cells. (B) Effects of FOXD3-AS1 on A549 and H1229 cell proliferation. (C) Effects of FOXD3-AS1 on the viability of A549 and H1229 cells. (D) Flow cytometric analysis of the effects of FOXD3-AS1 knockdown on the apoptosis of A549 and H1229 cells. (E) Tumor weight in nude mice injected subcutaneously with A549 cells transfected with sh-NC and sh-FOXD3-AS1#1. (F) Relative expression levels of FOXD3-AS1 in tumor tissue from the mice. **P<0.01, ***P<0.001. FOXD3-AS1, forkhead box D3 antisense 1; sh, short hairpin RNA; NC, negative control; OD, optical density; 7-AAD, 7-aminoactinomycin D.
|
FOXD3-AS1 promotes CDK6 expression by sponging miR-135a-5p
Subsequently, it was further demonstrated that FOXD3-AS1 was mainly localized in the cytoplasm (Fig. 3A). Since miRNA is a well-documented downstream target of lncRNA (20), an online database lncBASE-DIANA to predict miRNA candidates with putative binding sites for FOXD3-AS1 (Fig. 3B). This database suggested that miR-135a-5p may be sponged by FOXD3-AS1. Before performing validation experiments, we first ensured that miR-135a-5p mimics was successfully transfected in cells. RT-qPCR was used to detect the expression of miR-135a-5p in A549 and H1229 cells transfected with miR-135a-5p mimics (Fig. S1A). The results indicated that in cells transfected with miR-135a-5p mimics, the levels of miR-135a-5p were significantly increased compared with miR-NC-mimics, indicating a successful transfection. A luciferase reporter assay was then performed to confirm whether FOXD3-AS1 could bind to miR-135a-5p. The results demonstrated that the relative luciferase activity was significantly decreased in 293T cells which miR-135a-5p co-transfected with FOXD3-AS1-WT, whereas the luciferase activity of cells transfected with FOXD3-AS1-MUT showed no obvious difference (Fig. 3B). To further confirm that FOXD3-AS1 could sponge miR-135a-5p, the enrichment of FOXD3-AS1 was tested in A549 and H1229 using RIP and RT-qPCR experiments. The results revealed a higher expression of FOXD3-AS1 in the Ago2 group than that in the IgG group (Fig. 3C). Furthermore, RT-qPCR demonstrated that miR-135a-5p was upregulated following transfection with sh-FOXD3-AS1 compared with sh-NC (Fig. 3D). These results suggested that FOXD3-AS1 could directly bind to miR-125a-5p and regulate its expression.
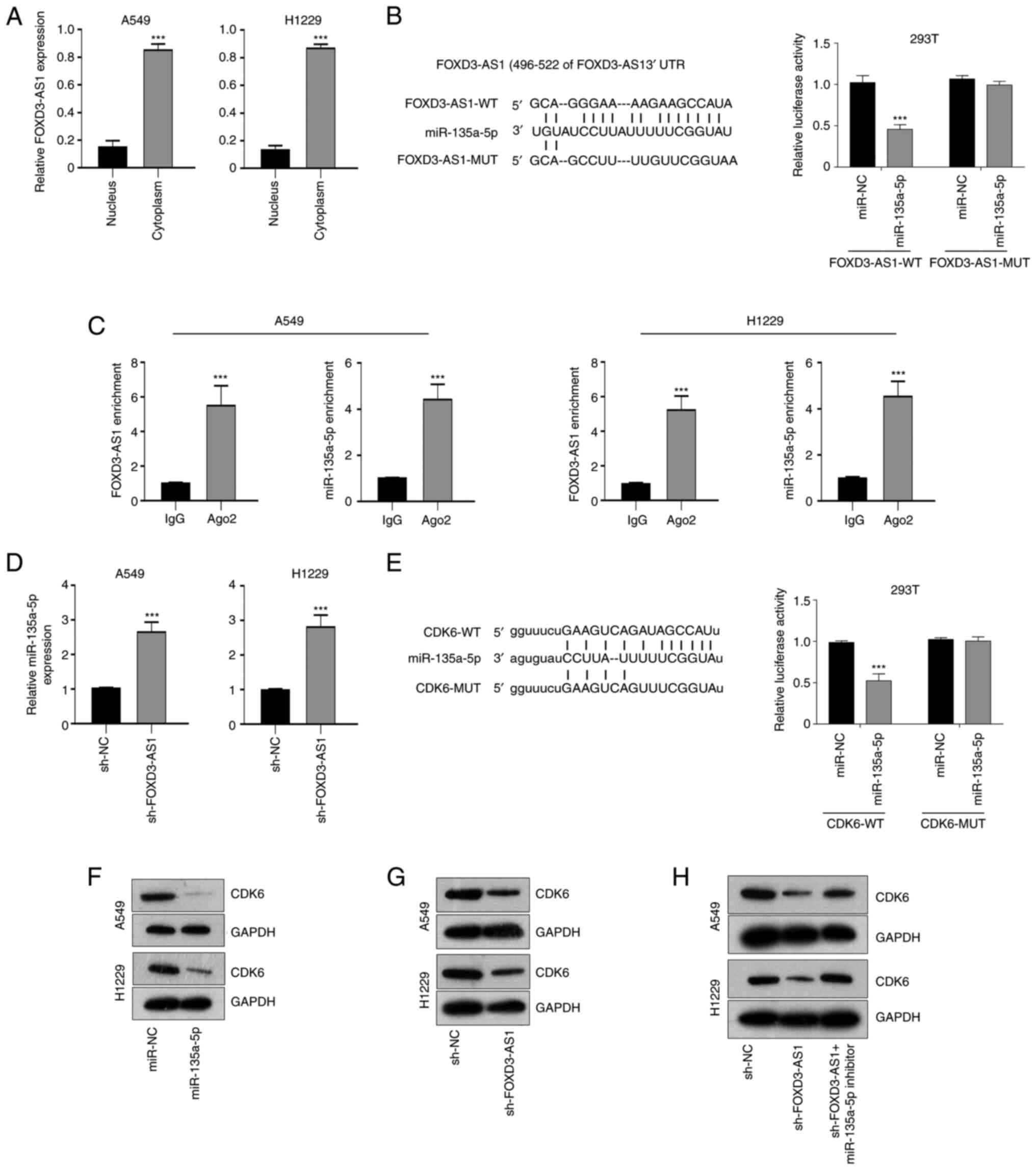 |
Figure 3.
FOXD3-AS1 facilitates CDK6 expression by sponging miR-135a-5p. (A) Localization of FOXD3-AS1 was determined in A549 and H1229 cells using subcellular localization experiment. (B) Potential binding sites of FOXD3-AS1 in miR-135a-5p and relative luciferase activity following co-transfection with miR-135a-5p mimics and FOXD3-AS1-WT or MUT. (C) Enrichment of FOXD3-AS1 following RNA immunoprecipitation. (D) Relative miR-135a-5p expression levels in A549 and H1229 cells transfected with sh-NC or sh-FOXD3-AS1#1. (E) Potential binding sites of CDK6 in miR-135a-5p. Luciferase reporter assay was used to analyzed the relative luciferase activity in cells co-transfected with luciferase vectors containing WT or MUT CDK6 and miR-135a-5p. (F) Protein expression levels of CDK6 were determined in A549 and H1229 cells transfected miR-135a-5p mimics or miR-NC. (G) Protein levels of CDK6 were determined in A549 and H1229 cells transfected with sh-FOXD3-AS1#1 or sh-NC. (H) Protein levels of CDK6 were determined in A549 and H1229 cells transfected with sh-FOXD3-AS1#1 alone or with miR-135a-5p inhibitor. ***P<0.001. FOXD3-AS1, forkhead box D3 antisense 1; CDK6, cyclin-dependent kinase 6; miR, microRNA; WT, wild-type; MUT, mutant; UTR, untranslated region; NC, negative control; Ig, immunoglobulin; Ago2, argonaute 2; sh, short hairpin RNA.
|
Subsequently, in order to further characterize the axis through which FOXD3-AS1 promotes the cancer progression, the downstream target of miR-125a-5p was examined. Using the Starbase software, it was identified that CDK6 contained putative binding sites for miR-135a-5p. A luciferase reporter assay indicated that the miR-135a-5p mimics reduced the relative luciferase activity in cells co-transfected with CDK6-WT, while no significant change was observed in cells transfected with CDK6-MUT (Fig. 3E). In A549 and H1229 cells, transfection with miR-135a-5p mimics downregulated the expression of CDK6 compared with miR-NC (Fig. 3F). Moreover, in sh-FOXD3-AS1-transfected cells, the expression of CDK6 was downregulated compared with sh-NC (Fig. 3G). However, whether miR-135a-5p inhibitor could influence CDK6 expression should be confirmed, and we also ensured the successful transfection. RT-qPCR was used to detect the expression of miR-135a-5p in A549 and H1229 cells transfected with miR-135a-5p inhibitor (Fig. S1B). The results indicated that the expression level of miR-135a-5p was significantly decreased in cells transfected with miR-135a-5p inhibitor compared with miR-NC-inhibitor. As shown in Fig. 3H, in sh-FOXD3-AS1-transfected cells, the expression of CDK6 was downregulated compared with sh-NC; however, in A549 and H1229 cells co-transfected with sh-FOXD3-AS1 and miR-135a-5p inhibitor, the CDK6 expression levels did not change compared with sh-NC (Fig. 3H). These results suggested that FOXD3-AS1 could promote CDK6 expression by sponging miR-135a-5p in NSCLC cells.
FOXD3-AS1 promotes the cell cycle progression and apoptosis of NSCLC cells by regulating miR-135a-5p/CDK6 axis
To further confirm that FOXD3-AS1 promoted NSCLC progression by regulating the miR-135a-5p/CDK6 axis, western blotting was used to quantify the protein levels of CDK6 following FOXD3-AS1 knockdown. First of all, successful transfection with OE-CDK6 was verified, CDK6 protein expression was detected by western blotting in A549 and H1229 cells transfected with OE-CDK6 (Fig. S1C). The results demonstrated that, following OE-CDK6 transfection, CDK6 expression was increased significantly compared with empty vector. Compared with sh-NC, CKD6 protein expression levels were reduced in cells transfected with sh-FOXD3-AS1, but not in cells co-transfected with sh-FOXD3-AS1 and miR-135a-5p inhibitor. Similar results were also observed in A549 and H1229 cells co-transfected with sh-FOXD3-AS1 and CDK6 (Fig. 4A). Furthermore, in cells transfected with sh-FOXD3-AS1, cell proliferation was significantly inhibited compared with the sh-NC group. However, proliferation was significantly increased in either the sh-FOXD3-AS1+ miR-135a-5p inhibitor group or the sh-FOXD3-AS1 + CDK6 group compared with sh-FOXD3-AS1 alone (Fig. 4B). Since CDK6 is an important cell cycle regulator, it was hypothesized that cell cycle progression in A549 and H1229 cells transfected with sh-FOXD3-AS1 could be affected. Indeed, following FOXD3-AS1 knockdown >60% of the cells were in the G0/G1 and 20% in S phases, indicating that cell cycle progression was inhibited. A possible explanation for this observation is that FOXD3-AS1 knockdown resulted in the downregulation of CDK6, thereby inhibiting cell cycle progression (Fig. S2). In addition, flow cytometry assays suggested that cell apoptosis significantly increased following transfection with sh-FOXD3-AS1 compared with sh-NC. However, this effect was reversed following miR-135a-5p inhibitor transfection or CDK6 overexpression (Fig. 4C). These results indicated that FOXD3-AS1 promoted the cell cycle progression and apoptosis of NSCLC cells by regulating the miR-135a-5p/CDK6 axis.
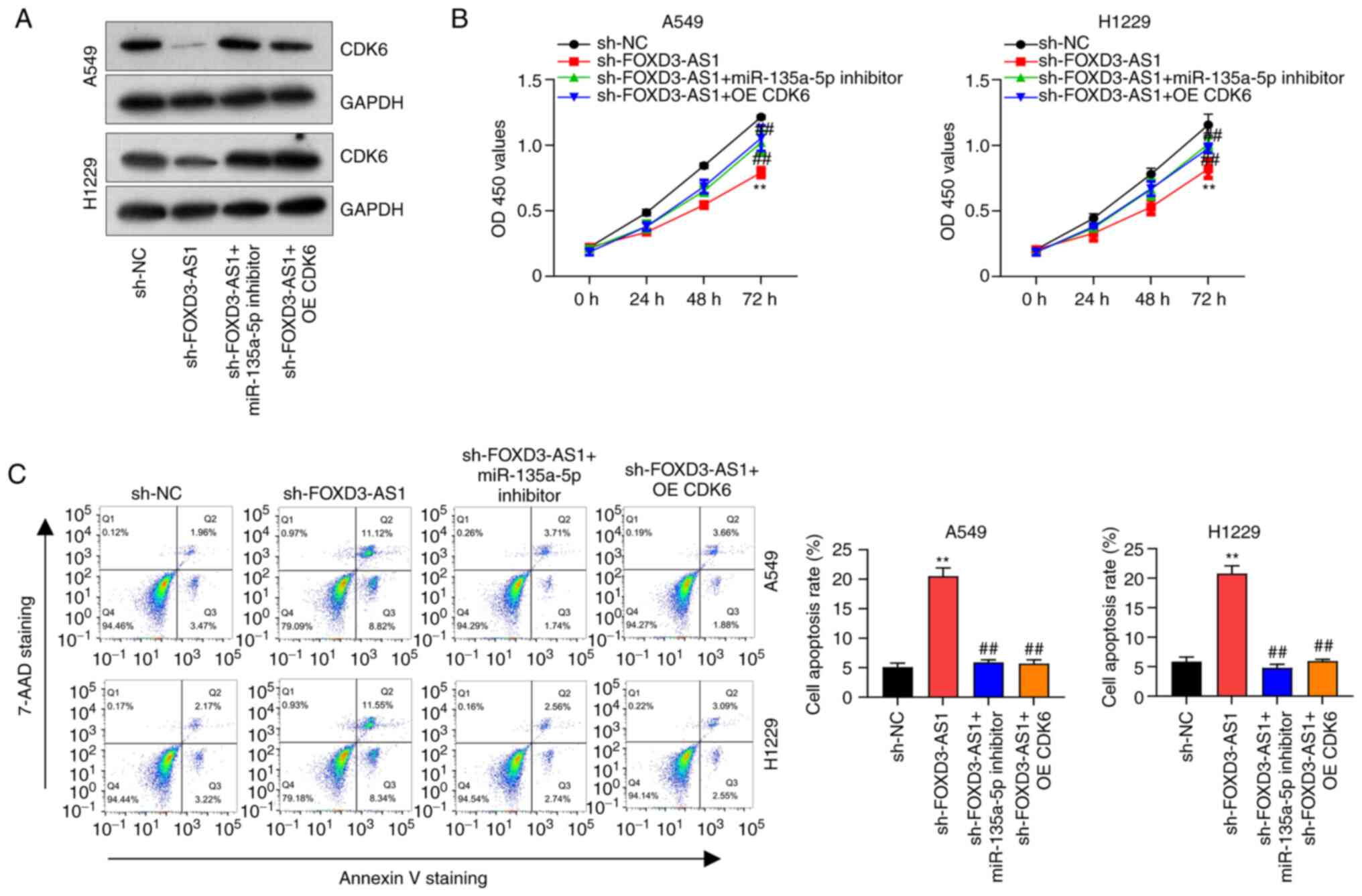 |
Figure 4.
FOXD3-AS1 promotes non-small cell lung cancer progression by regulating the miR-135a-5p/CDK6 axis. (A) Relative protein expression levels of CDK6 in A549 and H1229 cells transfected with sh-FOXD3-AS1, sh-FOXD3-AS1 + miR-135a-5p inhibitor or sh-FOXD3-AS1 + CDK6. (B) Proliferation in A549 and H1229 cells transfected with sh-FOXD3-AS1, sh-FOXD3-AS1 + miR-135a-5p inhibitor and sh-FOXD3-AS1 + CDK6. (C) Flow cytometric analysis of cell apoptosis in A549 and H1229 cells after transfection with sh-FOXD3-AS1, sh-FOXD3-AS1 + miR-135a-5p inhibitor and sh-FOXD3-AS1 + CDK6. **P<0.01 vs. sh-NC group; ##P<0.01 vs. sh-FOXD3-AS1 group. FOXD3-AS1, forkhead box D3 antisense 1; CDK6, cyclin-dependent kinase; sh, short hairpin RNA; NC, negative control; miR, microRNA; OD, optical density; 7-AAD, 7-aminoactinomycin D.
|
Discussion
Growing evidence suggests that lncRNA acts as a sponge for miRNA in NSCLC progression. For example, the lncRNA muscleblind like splicing regulator 1-AS1 suppresses NSCLC progression by inhibiting cell proliferation, cell cycle progression and metastasis, as well as increasing cell apoptosis by sponging miR-135a-5p (21). In contrast, lncRNA AWPPH and small nucleolar RNA host gene 4 were found to accelerate cell proliferation, migration, invasion and epithelial-to-mesenchymal transition by sponging miR-204 and miR-98-5p respectively in NSCLC (22,23). Recent evidence also indicates that FOXD3-AS1 is closely associated with poor prognosis in patients with NSCLC and plays an oncogenic role. Indeed, FOXD3-AS1 is involved in the pathological cell activities and chemo-resistance of NSCLC by sponging miR-127-3p to modulate MED28 or MDM2 expression, respectively (17,18). However, the role of FOXD3-AS1 in NSCLC remains poorly understood. In the present study, FOXD3-AS1 silencing reduced cell proliferation while inducing apoptosis, suggesting that FOXD3-AS1 might be a new potential biomarker for NSCLC.
CDK6 plays a key role in cell cycle progression in solid tumors such as colorectal cancer and NSCLC (24,25). For example, CDK6 overexpression rescues cell cycle arrest at the G1 phase following lncRNA NR2F2-AS1 silencing in colorectal cancer (24). A growing number of studies have revealed that miRNA plays crucial roles in promoting cancer tumorigenesis and progression via direct interaction with the 3′-UTRs of target genes (26–28). For example, miR-218 directly targets the CDK6 mRNA 3′-UTR and inhibits NSCLC cell proliferation by negatively regulating CDK6 expression (25). In NSCLC, miR-149-5p mimics co-transfected with CDK6 3′-UTR WT markedly inhibits the luciferase activities of H1563 and SK-MES-1 cells, suggesting that CDK6 is the direct target of miR-149-5p (29). In the present study, CDK6 could bind to miR-135a-5p. Additionally, CDK6 expression was negatively associated with miR-135a-5p expression, and restoring CDK6 expression completely rescued the inhibitory effect of FOXD3-AS1 knockdown in A549 and H1299 cells. These results demonstrated that CDK6 was directly regulated by miR-135a-5p.
miR-135a-5p has been implicated in lung cancer, bladder cancer, prostate cancer, glioma, colorectal cancer, gastric cancer and ovarian cancer (30–36). Previously, Zhang et al suggested that miR-135a-5p played an oncogenic role in lung adenocarcinoma, promoting cell proliferation, migration, invasion and inhibiting apoptosis by targeting lysyl oxidase-like 4 (37). By contrast, miR-135a-5p could act as a tumor suppressor in colorectal cancer, NSCLC, ovarian cancer, as well as other cancer types (38–41). For example, miR-135a-5p reduces glioblastoma malignancy by targeted regulation of syndecan binding-protein (42). In addition, miR-135a-5p overexpression suppresses tumor cell progression by regulating Kruppel-like factor 8 expression in glioma cell lines (43). The varied expression and function of target genes may result in different functions of miR-135a-5p in human tumors. Nevertheless, the present study demonstrated that miR-135a-5p inhibits cell proliferation while inducing apoptosis in NSCLC cells by targeting CDK6 in NSCLC.
Although FOXD3-AS1 has been studied in NSCLC as an oncogene in the present study, the regulatory mechanism of FOXD3-AS1 in NSCLC has also been reported by Zeng et al (17). This previous study reported that FOXD3-AS1 could promote chemo-resistance of NSCLC cells via directly acting on miR-127-3p/MDM2 axis. However, the volume of subcutaneous tumors is slightly larger than the tumor volume in our study. Moreover, Zeng et al also suggested that FOXD3-AS1 promoted NSCLC through the miR-127-3p/MED28 axis, with slightly larger tumor volume than those reported in our study (18). Ji et al (44) also reported that FOXD3-AS1 was significantly downregulated in NSCLC tissue and suppressed the progression of NSCLC by regulating the miR-150/SRC kinase signaling inhibitor 1 axis. The results from the above studies were different from those from the present study, suggesting that this study may have a theoretical basis and may be innovative.
In conclusion, the present findings demonstrated that FOXD3-AS1 played an oncogenic role in NSCLC, promoting cell proliferation and inhibiting apoptosis by regulating the expression of CDK6 via sponging miR-135a-5p. This study identifies a new cancer cell regulatory pathway involving FOXD3-AS1, miR-135a-5p and CDK6, providing novel insights for therapeutics in NSCLC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HG and MH conceived and designed the study. SL and ZG performed the experiments. HG wrote the paper. MH reviewed and edited the manuscript. JX and JZ performed the literature review. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved. HG and MH confirm the authenticity of all the raw data.
Ethics approval and consent to participate
The present study was approved by The Institutional Board of Affiliated Nanhai Hospital, Southern Medical University (People's Hospital of Nanhai District) (approval no. KY-E-2019-6-17). Written informed consent was obtained from all the participants prior to the start of the study. All animal protocols were approved by The Animal Ethics Committee of Affiliated Nanhai Hospital, Southern Medical University (People's Hospital of Nanhai District) (approval no. KY-E-2019-7-05).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Mao Y, Yang D, He J and Krasna MJ: Epidemiology of lung cancer. Surg Oncol Clin N Am. 25:439–445. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fujimoto J and Wistuba II: Current concepts on the molecular pathology of non-small cell lung carcinoma. Semin Diagn Pathol. 31:306–313. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL and Paz-Ares L: Lung cancer: Current therapies and new targeted treatments. Lancet. 389:299–311. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chi Y, Wang D, Wang J, Yu W and Yang J: Long non-coding RNA in the pathogenesis of cancers. Cells. 8:10152019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ponting CP, Oliver PL and Reik W: Evolution and functions of long noncoding RNAs. Cell. 136:629–641. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xiong W, Qin J, Cai X, Xiong W, Liu Q, Li C, Ju Y, Wang Q, Li Y and Yang Y: Overexpression LINC01082 suppresses the proliferation, migration and invasion of colon cancer. Mol Cell Biochem. 462:33–40. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Si Y, Yang Z, Ge Q, Yu L, Yao M, Sun X, Ren Z and Ding C: Long non-coding RNA Malat1 activated autophagy, hence promoting cell proliferation and inhibiting apoptosis by sponging miR-101 in colorectal cancer. Cell Mol Biol Lett. 24:502019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rinn JL and Chang HY: Genome regulation by long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kong S, Tao M, Shen X and Ju S: Translatable circRNAs and lncRNAs: Driving mechanisms and functions of their translation products. Cancer Lett. 483:59–65. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Akhade VS, Pal D and Kanduri C: Long noncoding RNA: Genome organization and mechanism of action. Adv Exp Med Biol. 1008:47–74. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen ZH, Hu HK, Zhang CR, Lu CY, Bao Y, Cai Z, Zou YX, Hu GH and Jiang L: Down-regulation of long non-coding RNA FOXD3 antisense RNA 1 (FOXD3-AS1) inhibits cell proliferation, migration, and invasion in malignant glioma cells. Am J Transl Res. 8:4106–4119. 2016.PubMed/NCBI
|
|
12
|
Guan Y, Bhandari A, Xia E, Yang F, Xiang J and Wang O: lncRNA FOXD3-AS1 is associated with clinical progression and regulates cell migration and invasion in breast cancer. Cell Biochem Funct. 37:239–244. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu C, Zhang M, Zhao J, Zhu X, Zhu L, Yan M, Zhang X and Zhang R: LncRNA FOXD3-AS1 Mediates AKT pathway to promote growth and invasion in hepatocellular carcinoma through regulating RICTOR. Cancer Biother Radiopharm. 35:292–300. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen X, Gao J, Yu Y, Zhao Z and Pan Y: LncRNA FOXD3-AS1 promotes proliferation, invasion and migration of cutaneous malignant melanoma via regulating miR-325/MAP3K2. Biomed Pharmacother. 120:1094382019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu Q, Shi M, Meng W, Wang Y, Hui P and Ma J: Long noncoding RNA FOXD3-AS1 promotes colon adenocarcinoma progression and functions as a competing endogenous RNA to regulate SIRT1 by sponging miR-135a-5p. J Cell Physiol. 234:21889–21902. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Y, Gao H and Li Y: Inhibition of LncRNA FOXD3-AS1 suppresses the aggressive biological behaviors of thyroid cancer via elevating miR-296-5p and inactivating TGF-β1/Smads signaling pathway. Mol Cell Endocrinol. 500:1106342020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zeng Z, Zhao G, Zhu H, Nie L, He L, Liu J, Li R, Xiao S and Hua G: LncRNA FOXD3-AS1 promoted chemo-resistance of NSCLC cells via directly acting on miR-127-3p/MDM2 axis. Cancer Cell Int. 20:3502020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zeng ZL, Zhu HK, He LF, Xu X, Xie A, Zheng EK, Ni JJ, Liu JT and Zhao GF: Highly expressed lncRNA FOXD3-AS1 promotes non-small cell lung cancer progression via regulating miR-127-3p/mediator complex subunit 28 axis. Eur Rev Med Pharmacol Sci. 24:2525–2538. 2020.PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang W, Lou W, Ding B, Yang B, Lu H, Kong Q and Fan W: A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer. Aging (Albany NY). 11:2610–2627. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cao G, Tan B, Wei S, Shen W, Wang X, Chu Y, Rong T and Gao C: Down-regulation of MBNL1-AS1 contributes to tumorigenesis of NSCLC via sponging miR-135a-5p. Biomed Pharmacother. 125:1098562020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu D, Qin BY, Qi XG, Hong LL, Zhong HB and Huang JY: LncRNA AWPPH accelerates the progression of non-small cell lung cancer by sponging miRNA-204 to upregulate CDK6. Eur Rev Med Pharmacol Sci. 24:4281–4287. 2020.PubMed/NCBI
|
|
23
|
Tang Y, Wu L, Zhao M, Zhao G, Mao S, Wang L, Liu S and Wang X: LncRNA SNHG4 promotes the proliferation, migration, invasiveness, and epithelial-mesenchymal transition of lung cancer cells by regulating miR-98-5p. Biochem Cell Biol. 97:767–776. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li F, Jiang Z, Shao X and Zou N: Downregulation of lncRNA NR2F2 Antisense RNA 1 Induces G1 arrest of colorectal cancer cells by downregulating cyclin-dependent kinase 6. Dig Dis Sci. 65:464–469. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Z, Lu C, Zhao G, Han X, Dong K, Wang C, Guan JZ and Wang Z: Downregulation of miR-218 by nicotine promotes cell proliferation through targeting CDK6 in non-small cell lung cancer. J Cell Biochem. 120:18370–18377. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sokol E, Kedzierska H, Czubaty A, Rybicka B, Rodzik K, Tanski Z, Boguslawska J and Piekielko-Witkowska A: MicroRNA-mediated regulation of splicing factors SRSF1, SRSF2 and hnRNP A1 in context of their alternatively spliced 3′UTRs. Exp Cell Res. 363:208–217. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao X, Sun Z, Li H, Jiang F, Zhou J and Zhang L: MiR-135a-5p modulates biological functions of thyroid carcinoma cells via targeting VCAN 3′-UTR. Cancer Biomark. 20:207–216. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wijayakumara DD, Mackenzie PI, McKinnon RA, Hu DG and Meech R: Regulation of UDP-Glucuronosyltransferases UGT2B4 and UGT2B7 by MicroRNAs in liver cancer cells. J Pharmacol Exp Ther. 361:386–397. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu L, Chen Y, Li Q and Duan P: lncRNA HNF1A-AS1 modulates non-small cell lung cancer progression by targeting miR-149-5p/Cdk6. J Cell Biochem. 120:18736–18750. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He Q, Fang Y, Lu F, Pan J, Wang L, Gong W, Fei F, Cui J, Zhong J, Hu R, et al: Analysis of differential expression profile of miRNA in peripheral blood of patients with lung cancer. J Clin Lab Anal. 33:e230032019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Luo W, Sun C, Zhou J, Wang Q, Yu L, Bian XW, Zhou X, Hua D, Wang R, Rao C, et al: MiR-135a-5p functions as a glioma proliferation suppressor by targeting tumor necrosis factor receptor-associated factor 5 and predicts patients' prognosis. Am J Pathol. 189:162–176. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Q, Zhang H, Shen X and Ju S: Serum microRNA-135a-5p as an auxiliary diagnostic biomarker for colorectal cancer. Ann Clin Biochem. 54:76–85. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Deng Z, Li X, Wang H, Geng Y, Cai Y, Tang Y, Wang Y, Yu X, Li L and Li R: Dysregulation of CircRNA_0001946 contributes to the proliferation and metastasis of colorectal cancer cells by targeting MicroRNA-135a-5p. Front Genet. 11:3572020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pan J, Xu X and Wang G: lncRNA ZFAS1 is involved in the proliferation, invasion and metastasis of prostate cancer cells through competitively binding to miR-135a-5p. Cancer Manag Res. 12:1135–1149. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zheng Y, Zheng B, Meng X, Yan Y, He J and Liu Y: LncRNA DANCR promotes the proliferation, migration, and invasion of tongue squamous cell carcinoma cells through miR-135a-5p/KLF8 axis. Cancer Cell Int. 19:3022019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang Z, Ren L, Zhao Q, Lu G, Ren M, Lu X, Yin Y, He S and Zhu C: TRPC1 exacerbate metastasis in gastric cancer via ciRS-7/miR-135a-5p/TRPC1 axis. Biochem Biophys Res Commun. 529:85–90. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang Y, Jiang WL, Yang JY, Huang J, Kang G, Hu HB and Xie S: Downregulation of lysyl oxidase-like 4 LOXL4 by miR-135a-5p promotes lung cancer progression in vitro and in vivo. J Cell Physiol. 234:18679–18687. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang J, Yang J, Zhang H, Liao Y, Xu D and Ma S: Effects of miR-135a-5p and miR-141 on proliferation, invasion and apoptosis of colorectal cancer SW620 cells. Oncol Lett. 20:914–920. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zheng C, Li X, Ren Y, Yin Z and Zhou B: Long noncoding RNA RAET1K Enhances CCNE1 expression and cell cycle arrest of lung adenocarcinoma cell by sponging miRNA-135a-5p. Front Genet. 10:13482020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Duan S, Dong X, Hai J, Jiang J, Wang W, Yang J, Zhang W and Chen C: MicroRNA-135a-3p is downregulated and serves as a tumour suppressor in ovarian cancer by targeting CCR2. Biomed Pharmacother. 107:712–720. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Guo LM, Ding GF, Xu W, Ge H, Jiang Y, Chen XJ and Lu Y: MiR-135a-5p represses proliferation of HNSCC by targeting HOXA10. Cancer Biol Ther. 19:973–983. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lin J, Wen X, Zhang X, Sun X, Yunzhi L, Peng R, Zhu M, Wang M, Zhang Y, Luo W, et al: MiR-135a-5p and miR-124-3p inhibit malignancy of glioblastoma by downregulation of syndecan binding protein. J Biomed Nanotechnol. 14:1317–1329. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Feng L, Lin T, Che H and Wang X: Long noncoding RNA DANCR knockdown inhibits proliferation, migration and invasion of glioma by regulating miR-135a-5p/BMI1. Cancer Cell Int. 20:532020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ji T, Zhang Y, Wang Z, Hou Z, Gao X and Zhang X: FOXD3-AS1 suppresses the progression of non-small cell lung cancer by regulating miR-150/SRCIN1 axis. Cancer Biomark. 29:417–427. 2020. View Article : Google Scholar : PubMed/NCBI
|


















