Introduction
Prostate cancer (PCa) is the second most leading cause of mortality and morbidity among men worldwide, with an estimated 248,530 new cases and 34,130 deaths in 2021 (1). For the past decade, standard therapy has comprised chemotherapy, radiation, secondary hormonal and targeted therapy; although a multitherapy approach has been followed in PCa, the heterogeneity of the disease prompts poor treatment outcomes among the majority of patients (2). Large-scale genomic rearrangements and extensive copy number alterations involving multiple chromosomes within PCa cells contribute to failure or resistance to therapy among a number of patients (3). Intratumor heterogeneity renders choosing the best treatment for a specific patient a clinical challenge (4). Therefore, it is crucial to establish novel and more reliable in vitro models to understand the heterogeneity and progression of PCa, identify new therapeutic targets, and empirically test personalized treatment strategies in a pre-clinical setting.
Various in vitro and in vivo model systems have been established to understand the biological mechanisms and molecular alterations that underly PCa pathology; however, these models are relatively scarce, clinically unreliable and have limitations (5); for example, they include cell lines, such as PC-3, DU-145 and LnCAP, and other in vivo experimental models. PCa cell lines are difficult to culture due to heterogeneity (6–11). Over the past 10 years, organoids have been used following the development of an intestinal organoid culture in 2009. This biotechnological advancement was considered a breakthrough in stem cell research. This method uses endogenous stem cell niche components to create three-dimensional (3D), long term and near-physiological culture systems that mimic the geno-/phenotypic features of human disease (12–16). To date, organoids have been established from a variety of organs, including the prostate (13,15,17–29).
Karthaus et al (30) and Gao et al (31) successfully adapted this culture method to PCa and cultured normal human and murine prostate epithelial cells without genetic manipulation in an in vitro 3D system that modelled prostate glandular structure (30,31). However, this system has been optimized to use human metastatic-rather than primary-prostate specimens to generate new organoid lines that express previously identified common genetic alterations in advanced PCa, such as PTEN, TP53 and TMPRSS2 (31,32). Using this methodology, it has been shown that patient-derived PCa organoids mimic the tumor of origin both genetically and phenotypically (30,31). The advantage of this culture medium is that it allows highly efficacious and unlimited proliferation of both normal and tumor prostate cells without substantial genetic aberrations (30,32). Nonetheless, culturing prostate organoids with a high success rate remains a challenge as there is a lack of knowledge in this area.
To the best of our knowledge, there are few studies reporting successful establishment of patient-derived organoids from PCa specimens (13,33–37), cell lines (38–40) or transformed primary normal prostate cells (39,41). The reported success rate using patient-derived PCa specimens is <20% and the capacity for long-term maintenance is variable and limited (31). Also, patient-derived organoids have primarily been established from metastatic biopsies or circulating tumor cells from patients with advanced PCa and rarely from the primary tumor (31). Richards et al (34) developed and characterized PCa organoids from primary tumors highlighting the role of stromal cells in this process of organoid establishment via a co-culture model that incorporated the prostate stroma. The importance of modeling PCa from primary tissue, particularly from treatment-naïve samples, is that the latter comprises a reservoir of extensively heterogeneous and resistant clones (42,43). Subsets in primary malignancy that may be masked in secondary lesions are the seeds for tumor progression and metastasis and represent a key target to develop efficacious therapies (44–46).
The aim of the present study was to derive organoids from treatment-naïve patients with PCa undergoing radical prostatectomy and to characterize organoids at cellular and molecular levels. The study also aimed to assess response to different regimen of treatments, including hormonal-, chemo- and radiotherapy. It was hypothesized that this methodology may provide a cost- and time-effective approach to understand the mechanisms involved in PCa development and progression. In addition, the study aims included characterizing these PCa organoid models through next-generation and RNA sequencing (seq) to capture the wide spectrum of heterogeneity of PCa and compare patient-derived organoids with parental tissue to demonstrate the accuracy of this model (Fig. 1).
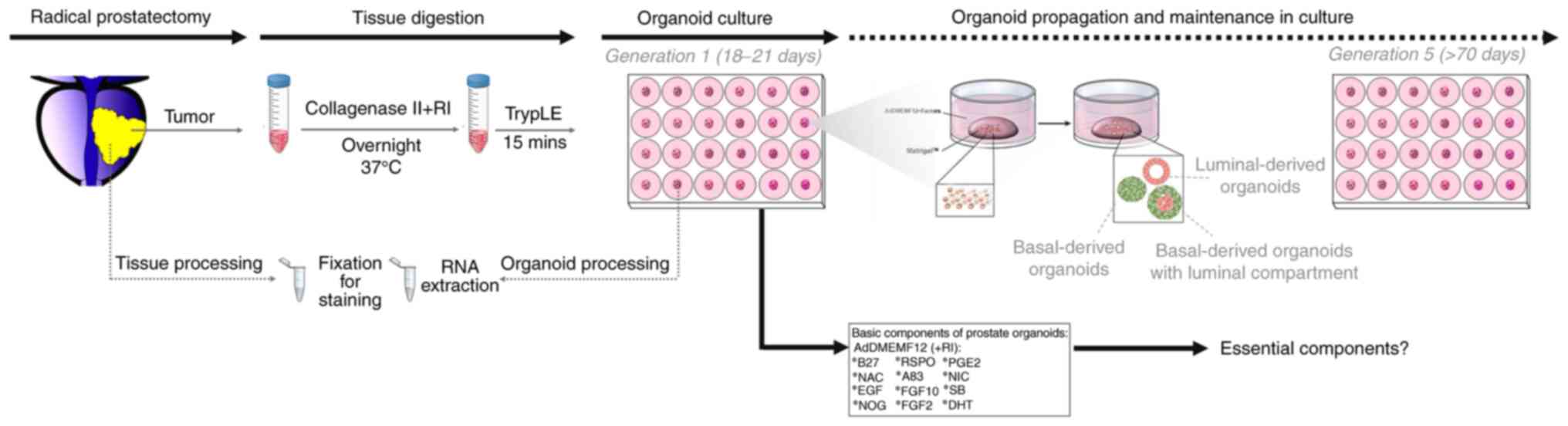 |
Figure 1.
Schematic diagram of the establishment and characterization of patient-derived prostate organoids.
|
Materials and methods
Patients
A total of 70 fresh tissue samples (two per patient) from distinct stages of human prostate adenocarcinoma were obtained from consenting treatment-naïve patients undergoing radical prostatectomy at the American University of Beirut-Medical Center (AUB-MC, Beirut, Lebanon). Primary tissue samples were collected (between July 2016 and June 2019) only if it would not compromise the sample for diagnosis or staging. A core biopsy was taken from the area most likely to be involved with cancer according to a pathologist's recommendation guided by imaging reports of the prostate. A total of 35 primary tumor and 35 paired healthy prostate specimens from patients aged 52–75 years were included in the present study. Inclusion criteria: i) Proven diagnosis of prostate cancer or undergoing a biopsy/prostate surgery for diagnostic, treatment or follow-up purposes; and ii) written, dated and signed informed consent. Exclusion criteria: i) Patients who were or had suffered from any other malignancy within the last 5 years, except completely excised basal cell carcinoma of the skin; and ii) patients with clinically significant co-morbidities that would be a contraindication to tumor biopsy.
Ethics statement
The present study was approved by the Institutional Review Board of AUB-MC prior to commencement. All protocols were performed in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving human subjects. Written informed consent was obtained from all patients and confidentiality was maintained.
Collection and dissection of PCa tissue specimens
The collection and dissection of fresh tissue specimens was performed as described previously (47). In brief, tissues were placed in human prostate growth medium (Table I) and maintained at 4°C. The time between surgical resection and tissue processing was <6 h to maximize the reliability of organoid generation. Prostate tissue samples were 3–5 mm in size.
 |
Table I.
Components and concentrations of human prostate growth medium.
|
Table I.
Components and concentrations of human prostate growth medium.
| Component |
Stock concentration |
Solvent |
Final concentration |
| B27 |
50.0X |
– |
1.00X |
| Nicotinamide |
1.0 M |
PBS |
10.00 mM |
| NAC |
500.0 mM |
PBS |
1.25 mM |
| EGF |
500.0 µg/mla |
PBS |
10.00 ng/mla |
| A83 |
5.0 mM |
DMSO |
500.00 nM |
| NOG |
100.0 µg/mla |
PBS + 0.1% BSAa |
50.00 ng/mla |
| RSPO |
500.0 µg/mla |
PBS + 0.1% BSAa |
250.00 ng/mla |
| DHT |
10.0 µM |
Ethanol |
1.00 nM |
| FGF2 |
100.0 µg/mla |
PBS + 0.1% BSAa |
6.00 ng/mla |
| FGF10 |
0.1 mg/ml |
PBS + 0.1% BSA |
10.00 ng/ml |
| PGE2 |
10.0 mM |
DMSO |
1.00 µM |
| SB |
10.0 mM |
DMSO |
10.00 µM |
| Y-27632 |
10.0 mM |
PBS + 0.1% BSA |
10.00 µM |
Prostate tumor tissue fragments were minced using sterile scalpel blades into 0.1–0.5 mm diameter pieces and washed with human prostate growth medium to remove cellular debris. The majority of minced fragments was employed for organoid culturing; remaining fragments were used for RNA extraction or transferred directly to 4% methanol-free paraformaldehyde (PFA) for histological examination.
Processing of PCa tissue specimens and establishment of patient-derived PCa organoids
Processing of the PCa tissue specimens was performed as described by Drost et al (13). Minced fragments for organoid culturing were digested in 2 ml 5 mg/ml collagenase type II (Gibco; Thermo Fisher Scientific, Inc.; cat. no. #17101-015) in Advanced DMEM-F12 (adDMEM/F12; Gibco; Thermo Fisher Scientific, Inc.; cat. no. #12634-010) with rho-associated coiled-coil containing protein kinase (ROCK) inhibitor (Y-27632; Santa Cruz Biotechnology, Inc.; cat. no. #sc-281642A) overnight at 37°C in a humidified incubator containing 5% CO2. The next day, the pellet was digested using TrypLE™ (Thermo Fisher Scientific, Inc.; cat. no. #12605-010), resuspended in adDMEM/F12 and passed through a 40-µm cell strainer to remove tissue debris and obtain single-cell suspensions. A specific cell count (~5,000 cells) was frozen in fetal bovine serum (FBS; Sigma-Aldrich; Merck KGaA; cat. no. #F9665) + 10% dimethyl sulfoxide (DMSO; Scharlab, SL; cat. no. #SU01571000) as passage zero cells to serve as a stock of patient-derived cells for later use. All cells were maintained as a stock in liquid nitrogen. The required cell count (20,000 cells) was resuspended in Matrigel (Corning Life Sciences; cat. no. #354230) at a ratio of 20,000 cells/40 µl 90% Matrigel. The derived cell count was estimated using trypan blue and cells were resuspended in growth factor-reduced Matrigel in a master mix at a ratio of 20,000 cells/40 µl 90% Matrigel. From this master mix, droplets were plated in the center of a 24-well culture plate at one droplet (40 µl) per well. The plate was then placed upside down in a 37°C incubator for 30 min to allow the Matrigel to solidify. Pre-warmed (37°C) human prostate or modified growth medium plus ROCK inhibitor (Y-27632; 10 µM) was added into each well.
Propagation of patient-derived PCa organoids
Harvesting and propagation of PCa organoids at each generation (G) was performed after 18–22 days or more depending on the sample. In brief, propagation of organoids was performed by adding ice-cold adDMEM/F12 without factors, collecting the organoids with a pipette tip, dissociating organoids enzymatically using TrypLE, splitting cells at a ratio of 1:2 or 1:3, resuspending cells in 90% Matrigel and seeding as aforementioned.
PCa organoid proliferation and growth analysis
To measure the growth of PCa organoid over time, organoids were visualized and counted manually under a Zeiss AG Axiovert inverted light microscope, as previously described (48). The size and shape of organoids were assessed using Carl Zeiss AG ZEN 2013 image software. A total of ≥50 organoids/condition/experiment were analyzed to calculate mean diameter. The organoid formation count (OFC; number of organoids per 20,000 cells seeded) was calculated by counting the number of organoids formed, starting with the same number of input cells in all conditions (20,000 cells) and in every generation (G; from G1 until >G7).
Human prostate growth medium components
The human prostate growth medium included adDMEM/F12 containing 1% Penicillin/Streptomycin (v/v; Biowest; cat. no. #L0022-100), 0.2% Gentamicin/Amphotericin B (v/v; Thermo Fisher Scientific, Inc.; cat. no. #R01510), 0.2% plasmocin prophylactic (v/v; InvivoGen; cat. no. #ant-mpp), 10 mM HEPES (cat. no. #15630-056) and 2 mM GlutaMAX (cat. no. #35050-061; both Gibco; Thermo Fisher Scientific, Inc.). Components specified in Table I were added fresh on a weekly basis. For the first week after plating, ROCK inhibitor (Y-27632) was added fresh to the culture medium on the same days that medium was changed (every 2–3 days).
Immunohistochemistry (IHC)
To embed organoids, droplets of Matrigel containing organoids were fixed in pre-warmed (to 37°C) 4% PFA (v/v) for 30 min to keep the Matrigel intact, then the droplet of Matrigel was collected carefully using a spatula. Histogel (Thermo Fisher Scientific, Inc.; cat. no. #HG-4000-012) was liquefied at 60°C and used to surround the droplet of Matrigel, then allowed to solidify on ice for 10 min before transferring into a tissue cassette to be incubated in 10% formalin (v/v) overnight at 37°C. Finally, organoids and corresponding tissue specimens were processed for paraffin embedding, sliced into 4-µm sections using a cryostat (Leica Biosystems; cat. no. #CM1850) and melted onto charged slides [Star Frost Microscope Slides Adhesive with frosted end (76×26 mm); Servoprax GmbH; cat. no. #B4 0401].
Paraffin embedding, microtome sectioning and standard hematoxylin and eosin (H&E) staining were all performed by the Histology Laboratory at the Diana Tamari Sabbagh building; all steps were performed at room temperature. For IHC staining, organoid and tissue sections were first outlined with a hydrophobic pen (Vector Laboratories, Inc; Maravai Life Sciences), washed with tris-buffered saline (TBS) containing 0.1% Tween-20 (Sigma-Aldrich; Merck KGaA; cat. no. #P1379; v/v; TBS-T), and deparaffinized. Antigen retrieval was performed in a citrate buffer (for 30 min in steamer at 95–98°C). Then, the Novolink™ polymer detection system (Leica Biosystems; cat. no. #RE7150-K) was employed per the manufacturer's instructions; sections were incubated overnight at 4°C with primary antibodies diluted in TBS-T with 3% bovine serum albumin (BSA; v/v; Amresco, LLC; cat. no. #0332-100G), 2% normal goat serum (NGS; v/v; Thermo Fisher Scientific, Inc.; cat. no. #16210064) and 0.1% Triton X-100 (v/v; Sigma-Aldrich; Merck KGaA; cat. no. #T9284). The sections were incubated at 37°C with Post Primary and Novolink Polymer for 30 min each, before developing the peroxidase activity using ImmPACT DAB peroxidase substrate (Vector Laboratories, Inc.; cat. no. #SK-4105) for 1–10 min. Finally, sections were counterstained with hematoxylin, dehydrated and mounted using Permount medium (Invitrogen; Thermo Fisher Scientific, Inc.; cat. no. #P36934). Images were acquired using an Olympus CX41 light microscope and processed using Carl Zeiss AG ZEN 2013 image software.
Immunofluorescence (IF) and morphological analysis of organoids
Indirect IF analysis was used to characterize prostate epithelial lineage and stem cell markers expressed by organoids. IF analysis was performed on both paraffin-embedded organoids and organoids in suspension to preserve the 3D architecture. To perform IF in suspension, organoids were collected when they reached the appropriate size and confluency for passaging (18–22 days after plating). Matrigel was dissolved using ice-cold medium as aforementioned. The pellet was fixed in 4% PFA for 30 min at room temperature. For paraffin-embedded organoids, section thickness was 4–5 µm. Antigen retrieval was performed in citrate buffer (for 30 min in steamer at 95–98°C). Thereafter, the slides were left at room temperature for 30 min to cool down, and then rinsed twice in distilled water. Then, both paraffin-embedded organoids and organoids in suspension were permeabilized with 0.5% Triton X-100 (v/v) for 30 min at room temperature, followed by incubation with blocking buffer [0.10 BSA, 0.20 Triton X-100, 0.05 Tween-20 and 10.00% NGS (all v/v) in PBS] for 1 h at room temperature. Organoids were then incubated overnight with primary antibodies at 4°C. Following gentle washing with PBS containing 0.1% Tween-20 (v/v), organoids were incubated with corresponding secondary antibodies for 1 h at room temperature, the pellet was then washed gently and mounted with anti-fade reagent Fluoro-gel II with DAPI (Electron Microscopy Sciences; cat. no. #17985-51). Indirect IF microscopic analysis was performed using Carl Zeiss AG Axio Observer. Z1 and LSM710 laser scanning confocal microscopes using 40X oil objective. Images were acquired and analyzed using Carl Zeiss AG ZEN 2013 image software.
Antibodies
Antibodies used in IF staining were as follows: Mouse monoclonal anti-cytokeratin (CK)8 (1:200; cat. no. #MMS-162P; Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-CK5 (1:200; cat. no. #PRB-160P; BioLegend, Inc.), anti-CK14 (1:200; cat. no. sc-17104; Santa Cruz Biotechnology, Inc.), anti-vimentin (VIM; 1:50; cat. no. #sc-5565; Santa Cruz Biotechnology, Inc.), mouse monoclonal anti-CD44 (1:50; cat. no. sc-7297; Santa Cruz Biotechnology, Inc.), rat monoclonal anti-CD49f (1:50; cat. no. 555734; BD Pharmingen; BD Biosciences), Alexa 568-conjugated goat anti-mouse IgG (cat. no. #A-11004) and anti-rabbit IgG (both Thermo Fisher Scientific, Inc.; cat. no. #A-11011), Alexa 488-conjugated goat anti-rabbit (cat. no. A-11034), and anti-mouse IgG (cat. no. A-11001; both Thermo Fisher Scientific, Inc.). All secondary Alexa Fluor antibodies were used at 1:200. Fluorogel II with DAPI (Electron Microscopy Sciences) was used for mounting. Primary antibodies used in IHC staining included rabbit monoclonal anti-androgen receptor (AR; 1:50; cat. no. ab52615; Abcam), anti-prostate-specific antigen (PSA; 1:200; Abcam; cat. no. ab224799), mouse monoclonal anti-p63 (dilution 1:50; Leica Biosystems-Bond™; cat. no. P63-L-CE), rabbit polyclonal anti c-MYC (dilution 1:50; cat. no. #9402; Cell Signaling Technology) and rabbit monoclonal anti-ETS transcription factor ERG (dilution 1:50; cat. no. ab92513; Abcam;).
RNA isolation and purification
Tissues and organoids were washed once with 1 ml PBS then total RNA extraction was performed using TRIzol® reagent (Thermo Fisher Scientific, Inc.) and RNAeasy Mini kit (Qiagen GmbH) as per the manufacturer's instructions. Then, 0.2 ml chloroform was added and centrifugation at 8,000 × g was performed for 15 min at 4°C to isolate total RNA. The upper aqueous phase containing isolated RNA was mixed with equal volume of 70% ethanol, then RNA purification was performed using RNAeasy Mini spin column (Qiagen GmbH) according to the manufacturer's protocol. NanoDrop 2000™ (Thermo Fisher Scientific, Inc.) and BioAnalyzer 2100™ (Agilent Technologies, Inc.) were used to determine the concentration and RNA integrity number (RIN) of isolated RNA, respectively.
RNA-seq library preparation and sequencing of patient-derived PCa organoids and corresponding parental tissue
Total concentrations of >0.5 ng/µl RNA samples from two patients (Table SI) were determined using NanoDrop™ 2000/2000c spectrophotometer (Thermo Scientific™), whereas RNA integrity number (RIN) of >8 were determined with the 2100 Bioanalyzer system (Agilent) using Bioanalyzer RNA 6000 Nano assay chips (Agilent). An amount of 100 ng/µl of total RNA with RIN >8 was used for RNA-seq library preparation using Illumina TruSeq Stranded mRNA prep kit (Illumina, Inc.; cat. no. RS-122-2101). RNA-Seq libraries preparations, multiplex and sequencing were all performed with 100 bp pair-end after barcoding using Illumina Hi-Seq 2500 sequencing platform system at NYU Abu Dhabi (NYUAD) Genomic Core Facility (Abu Dhabi, UAE).
Transcriptome data computational analysis
Raw count reads aligned to the reference genome (49) were estimated via DESeq2 computational pipeline [(Bioconductor version 3.13; http://bioconductor.org/packages/release/bioc/html/DESeq2.html) and R project-based package software (R version 4.1.1; http://www.r-project.org/)]. NYUAD High Performance Computing server platform Dalma was used to run the computing methods on a Linux-based command system. Principal component analysis (PCA) was performed using RNA-Seq Shiny Transcriptome Analysis Resource Tool application via the NYUAD Center of Genomic and Systems Biology Bioinformatics Online Analysis and Visualization Portal (https://tsar.abudhabi.nyu.edu/) (50). Data counts were deposited at National Center for Biotechnology Information Gene Expression Omnibus (51) (accession no. GSE148937; ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE148937).
Gene array data analysis
Differentially expressed gene (DEGs) were subjected to Gene Ontology (GO) term and gene set enrichment analysis (GSEA) using Cytoscape (version 3.8.0 by The Cytoscape Consortium) and EnrichmentMap (Cytoscape plugin version 3.3) bioinformatics tools (52). The database of pathway gene sets used for pathway enrichment analysis was downloaded from baderlab.org/GeneSets and included eight data sources: MSigDB (C2 collection) (53), National Cancer Institute (54), Institute of Bioinformatics, NetPath (55), HumanCyc (56), Reactome (57), GO (58), MSigDB (C3 collection; Specialty enrichment map gene set microRNAs, transcription factors) (53) and Panther (59).
Treatments of patient-derived PCa organoids
Cell suspensions derived from fresh digested tissue were plated in Matrigel™ as aforementioned. Then, human prostate growth medium with classical components (Table I) was added to each well in the presence or absence of different drugs (two replicates each). The organoid culture was maintained at 37°C in a humidified incubator (5% CO2). The culture medium was changed every 2–3 days and organoids were counted after 18–22 days as aforementioned. All drugs were dissolved in DMSO to a final concentration of 0.1% (v/v). The control in all experiments was human prostate growth medium with classical components + DMSO. Drugs tested were as follows: Bicalutamide (Sigma-Aldrich; Merck KGaA; cat. no. #B9061-50MG), AR signaling pathway, 1st generation antiandrogen, 1 and 10 µM within the range of IC50 values; concentrations were used as previously described (60,61); Enzalutamide (Selleck Chemicals; cat. no. #MDV3100), AR signaling pathway, 2nd generation antiandrogen, 1 and 10 µM within the range of IC50 values, concentrations were used as previously described (62,63); and Docetaxel (Sigma-Aldrich; Merck KGaA; cat. no. #01885-25MG-F), common chemotherapeutic agent used in the treatment of PCa; 1 and 50 nM within the range of IC50 values, concentrations were used as previously described (64).
In addition, combinatorial therapy of Enzalutamide (1 µM) + Docetaxel (1 nM) was evaluated. The drugs were added at the same time under the same conditions and for the same duration as aformentioned before.
Organoids were subjected to radiation therapy. The irradiator was a 225 kV Precision X-Ray (Pxi) X-RAD 225. Irradiation was performed at a rate of 3 Gy/min and a 1.5 mm aluminum filter was used. To assess the effect of irradiation on organoids, irradiation was performed at 2 Gy (a clinically achievable dose) on day 1 of plating cell suspensions in Matrigel. Corresponding controls were sham irradiated, where cultured cells were subjected to the same irradiation protocol without turning on the irradiator. After 18–22 days, the effect of drugs or irradiation on cell proliferation and survival was analyzed. Diameter and number of organoids were assessed. A total of ≥50 organoids was analyzed to calculate the mean diameter using Carl Zeiss AG ZEN 2013 image software. Total number of organoids was counted manually under bright-field light microscopy (20X objective). OFC was estimated by plating 20,000 cells/well in duplicate and counting the total number of organoids formed per well after 18–22 days.
Optimization of PCa organoid culture conditions
Cell suspensions derived from fresh digested tissue or fresh-frozen P0 cells were plated in Matrigel, as aforementioned. To assess the importance of each of 12 components (Table I) in the human prostate growth medium, different factors were removed to generate media as follows: Control (CTRL), all 12 components; -N-acetylcysteine (NAC), all except NAC; -noggin (NOG), all except NOG; -R-spondin (RSPO), all except RSPO; -A83-01 (A83), all except A83; -prostaglandin E2 (PGE2), all except PGE2; -dihydrotestosterone (DHT), all except DHT; -fibroblast growth factor (FGF)10, all except FGF10; -FGF2, all except FGF2; -EGF, all except EGF; and -SB202190 (SB), all except SB. Experiments were performed on tumor samples derived from 3 random PCa patients with similar clinical manifestations [grade group 2; Gleason score, 7 (3+4); 2 replicates/condition/patient].
For follow-up experiments, essential components, including NOG, NAC, A83, B27 and nicotinamide were designated ‘5F’. Human prostate growth medium was modified as follows: CTRL, all 12 components; 5F, essential components only; 5F + RSPO; 5F + SB; 5F + PGE2; 5F + FGF2; 5F + FGF10 and 5F + EGF. Experiments were performed using cells freshly isolated from tumor tissue of one random patient [patient 29; Gleason Score 7 (3+4)] and were frozen as P0. Cells were then thawed and organoids were plated (two replicates/condition).
In both sets of experiments, at the end of G1 and before propagation to G2 (after 18–22 days), size and number of organoids were assessed to determine the effect of different media on organoid proliferation and survival. To calculate the mean diameter, ≥50 organoids were analyzed using Carl Zeiss AG ZEN 2013 image software. Total number of PCa organoids was counted manually and OFC was calculated.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 7 software (GraphPad Software, Inc.). One-way ANOVA followed by post hoc Bonferroni's multiple comparisons was used to analyze treatment response to chemotherapy, androgen-deprivation therapy and irradiation in vitro, single factor effects and the effect of withdrawal of individual factors from human prostrate growth medium on organoid growth and maintenance (propagation). P<0.05 was considered to indicate a statistically significant difference.
Results
Establishment and characterization of patient-derived PCa organoids and corresponding tissue
Fresh radical prostatectomy tumor specimens examined by expert pathologists were obtained from treatment-naïve patients and organoids were successfully established (Fig. 2A). For certain patients, organoids were not successfully established. Out of 70 specimens, >90% were successfully established as organoids (Fig. S1). For most unestablished samples, primary viable cells (P0) were not successfully derived following initial tissue digestion, indicating that the quality of tissue may be a factor that affects organoid derivation. The OFC ranged from 142 (0.71%) to 607 (3.03%; Fig. 2B) and the diameter of PCa organoids ranged from 61.8 to 106.0 µm (Fig. 2C).
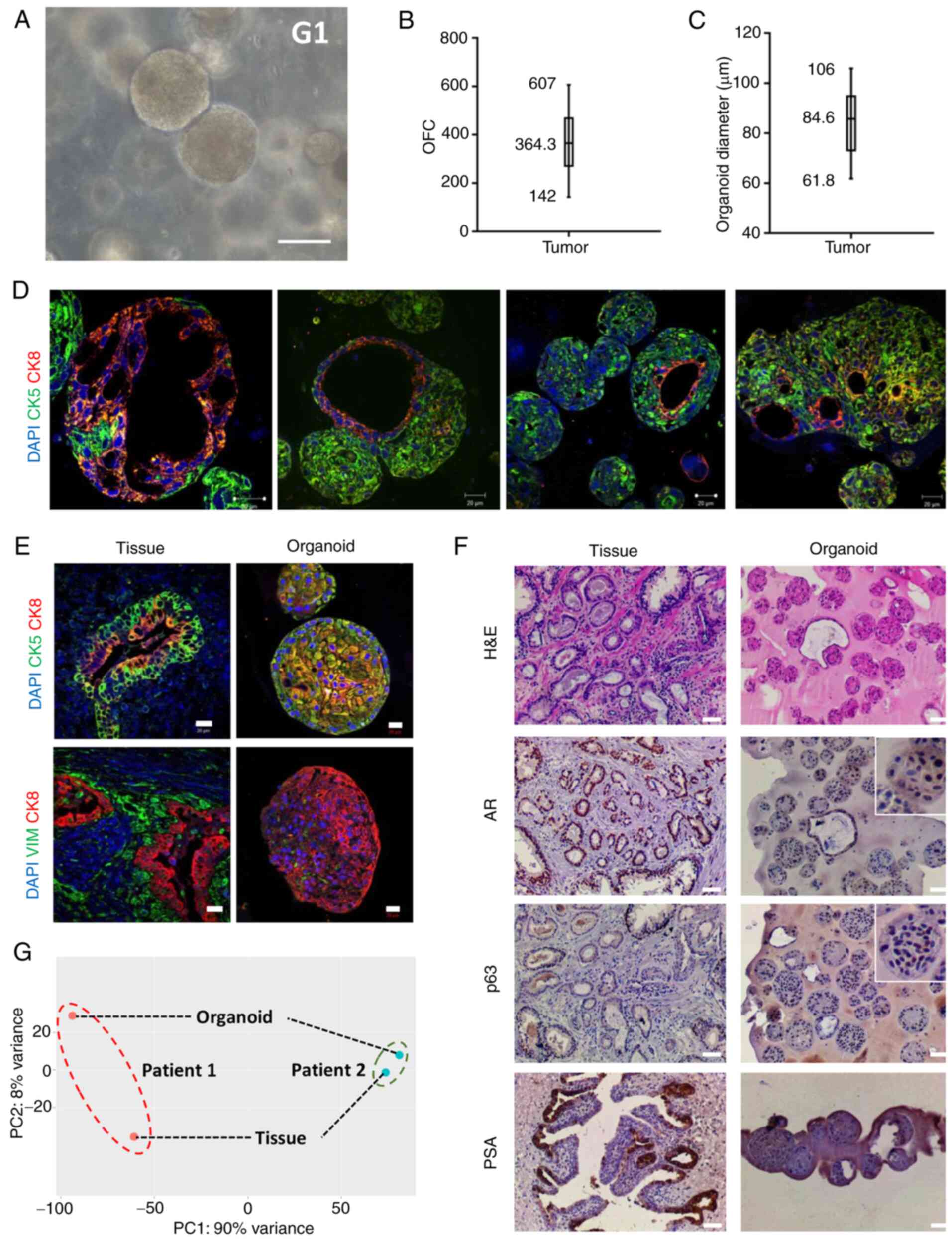 |
Figure 2.
Establishment and characterization of patient-derived PCa organoids and corresponding tissue. (A) Representative bright-field image showing established PCa organoids (G1) grown in culture. Scale bar, 100 µm. Mean, minimum and maximum (B) OFC and (C) diameter of patient-derived PCa organoids (n=10). (D) Immunofluorescent images of organoids [patient 22; grade group 5; Gleason score, 9 (5+4)] stained with prostate lineage epithelial markers CK8 and CK5 revealing the presence of both prostate epithelial lineages in the established organoid cultures, with organoids expressing luminal- or basal-only or luminal and basal double-positive cells. (E) Immunofluorescent images of organoids and corresponding tissue stained with prostate lineage epithelial markers CK8 and CK5 [patient 8; grade group 1; Gleason score, 6 (3+3)] and mesenchymal marker VIM [patient 25; grade group 3; Gleason score 7 (4+3)]. The nuclei were stained with anti-fade Fluorogel II with DAPI. Representative confocal microscopy images were acquired using a Zeiss LSM 710 laser scanning confocal microscope and processed using Carl Zeiss ZEN 2013 image software. (F) Immunohistochemistry images of organoids and corresponding tissue [patient 7; grade group 2; Gleason score 7 (3+4)] stained with H&E and prostate lineage epithelial markers p63, AR and PSA. Scale bar, 50 µm. Representative microscopy images were acquired using an Olympus CX41 light microscope (×10 magnification). Inset magnification, ×2.5. (G) RNA-seq gene expression principal component analysis plot of PCa organoids and corresponding tissue for two patients [patient 1, grade group 1/Gleason score 6 (3+3); patient 2, grade group 2/Gleason score 7 (3+4)]. PCa, prostate cancer; G, generation; OFC, organoid formation count; CK, cytokeratin; VIM, vimentin; H&E, hematoxylin and eosin; AR, androgen receptor; PSA, prostate-specific antigen.
|
Patient-derived PCa organoids and corresponding tissue were characterized by assessing the expression of prostate epithelial lineage markers using IHC and IF staining. The sections were probed for different markers, including luminal (CK8), basal (p63 and CK5) and mesenchymal markers (VIM), as well as AR and PSA. The expression of luminal CK8 and basal CK5 markers confirmed the presence of both prostate epithelial lineages in the established organoid cultures (Fig. 2D and E). Moreover, luminal CK8-positive cells were detected around a lumen-like gland and surrounded by basal CK5-positive cells in parental tissue and organoids (Fig. 2E). In addition, an intermediate cell population, referred to as intermediate or transit amplifying cells, co-expresses luminal and basal markers (65,66). This population was also detected in organoids, including cells that co-expressed luminal CK8 and basal CK5 (Fig. 2E). The results revealed strong p63 and nuclear expression of AR in PCa organoids and corresponding tumor tissue and high levels of PSA in the original tissue (Fig. 2F). PSA was detected in a limited number of PCa organoids (Fig. 2F). Strong expression of c-MYC and ERG was noted in PCa organoids as well as corresponding tumor tissue (Fig. S2).
Whole-transcriptome seq analysis of PCa organoids vs. corresponding tissue
To understand the variance between patients and established PCa organoids and tissue counterparts at the genome wide level, RNA was isolated. RNA-seq libraries were prepared and the association between samples was assessed via PCA. Comparison of tissue samples and established organoids revealed high variance primarily at the cellular structural level at PC1=90% and PC2=8%, indicating variability between patients (Fig. 2G).
To assess the transcriptomic features that signify PCa organoids vs. corresponding tissue, paired-end (100 bp) RNA-seq was performed using the Hi-Seq 2500 Illumina platform to identify DEGs between patient-derived PCa organoids and corresponding tissue. Using P-adj<0.05 cut-off, 3,134 transcripts were significantly differentially expressed between PCa organoids and corresponding tissue (792 up- and 2,342 downregulated; Table SII). The DEG expression volcano plot is presented in Fig. S3A. Heatmap and hierarchical cluster analysis demonstrated that the identified DEGs were able to effectively distinguish PCa organoids and corresponding tissue (Fig. S3B).
GSEA identifies enrichment of cell cycle and epithelial lineage-associated signalling pathways in PCa organoids relative to tissue
Enrichment maps were used to evaluate DEGs and associated pathways (Fig. S4) using Cytoscape 3.7.2 software (EnrichmentMap). GSEA identified significantly altered pathways in PCa organoids relative to corresponding tissue (Table SIII). Results indicated significant differences (FDR<0.01, NOM P-value <0.05) in enrichment of gene sets. The 20 most significantly enriched signalling pathways were selected based on normalized enrichment score (Fig. S5). Results indicated the PCa organoids were enriched for: i) Cell cycle pathways such as ‘cell_cycle_reactome’, ‘cell_cycle_checkpoints_reactome’, and ‘m_phase_reactome’ among others; ii) e2f signaling such as ‘hallmark_e2f_targets’; iii) mitosis reactions, such as ‘mitotic_cell_cycle’, ‘mitotic_anaphase_reactome’, and ‘sister_chromatid_segregation’, among others; and iv) epithelial differentiation pathways, such as ‘epidermis development’, ‘keratinization’, ‘keratinocyte differentiation’, and ‘skin development’ among others.
Differential treatment response on established organoid growth
Drug response was assessed in vitro. The therapeutic response was evaluated by quantifying the total number (OFC) and average size of organoids. The culture medium (with or without drugs) was changed every 2–3 days and organoid growth was assessed after 18–22 days.
G1 PCa organoids from 3 random patients with different clinical gradings were maintained under different treatment regimens. Patients 21 and 32 both exhibited International Society of Urological Pathology grade group 3 [Gleason score, 7 (4+3)]; patient 22 exhibited grade group 5 [Gleason score, 9 (5+4)]. The effect on the count (OFC) and size of cultured organoids was observed following drug treatment (Fig. 3A). While both Enzalutamide (10 µM) in patient 22 and Bicalutamide (10 µM) in patients 21 and 22, significantly decreased the size and the count of organoids, no effect was observed on OFC of patient 32 for both drugs (Fig. 3B-G). The chemotherapeutic drug Docetaxel exhibited an inhibitory effect on organoid formation in patients 21 and 22 at a concentration of 1 µM (though less prominent than the effect of androgen deprivation drugs in both patients), but no effect was observed in patient 32. Combinatorial treatment of Enzalutamide (1 µM) + Docetaxel (1 µM) elicited a statistically significant inhibition on both OFC and size of organoids in all three patients.
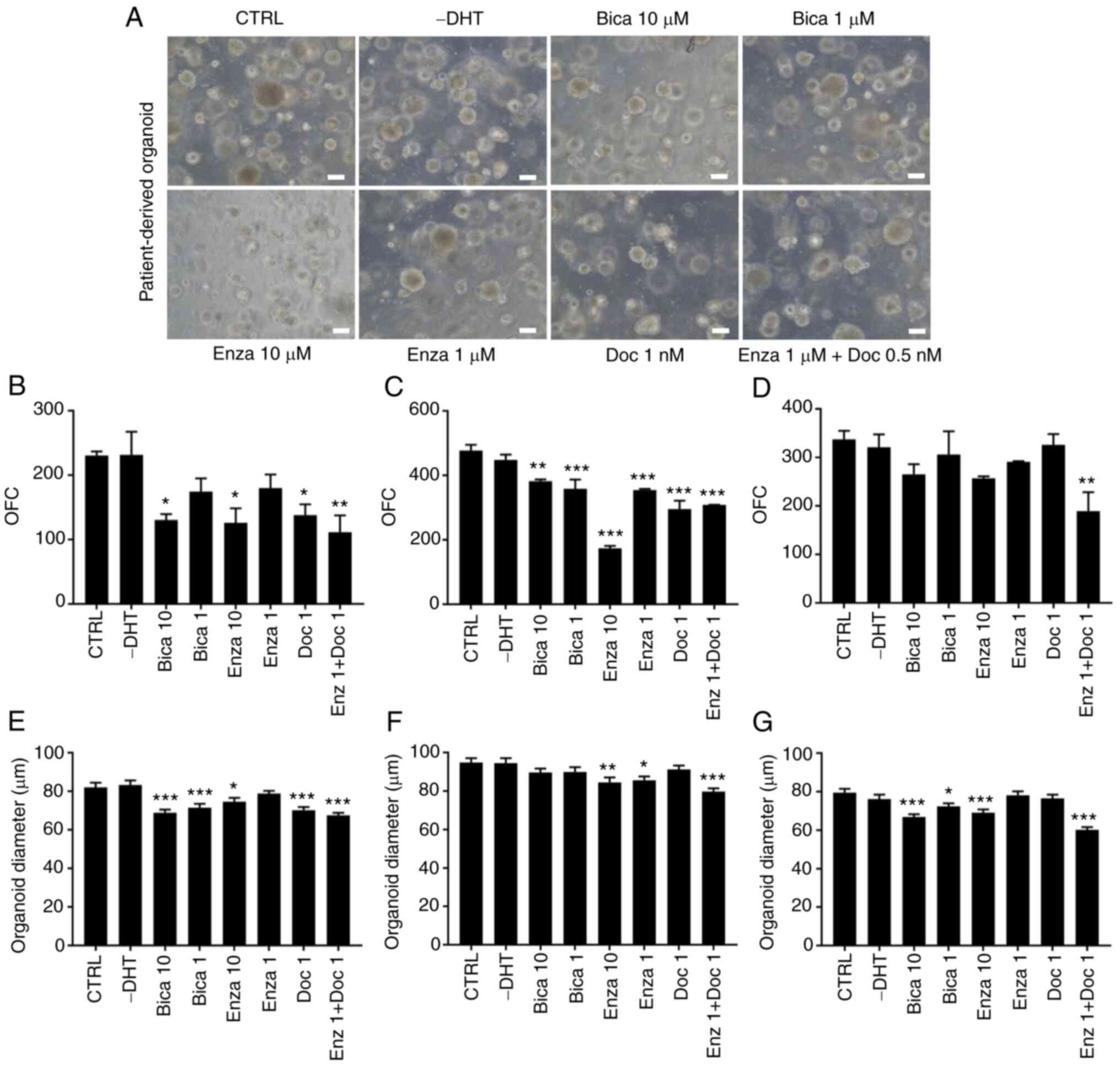 |
Figure 3.
Differential treatment response of chemotherapy and androgen-deprivation therapy on G1 patient-derived PCa organoid growth. (A) Representative brightfield images of G1 PCa organoids [patient 22; grade group 5; Gleason score, 9 (5+4)] grown in the presence or absence of different drugs. Scale bar, 100 µm. (B) OFC for patients 21 [grade group 3; Gleason score, 7 (4+3); treatment F sampling distribution (8,9)=10.1, P=0.0011], (C) 22 [grade group 5; Gleason score, 9 (5+4); treatment F(8,9)=129.7, P<0.0001] and (D) 32 [grade group 3; Gleason score, 7 (4+3); treatment F(7,8)=6.454, P=0.0087]. Data are presented as the mean ± SD of duplicate experiments. Average diameter of ≥100 organoids from patients (E) 21 [treatment F(9,950)=175.8, P<0.0001], (F) 22 [treatment F(7,792)=5.515, P<0.0001] and (G) 32 [treatment F(7,792)=13.2, P<0.0001]. Data are presented as the mean ± SEM of duplicate experiments. One-way ANOVA followed by Bonferroni multiple comparisons. *P<0.05; **P<0.01; ***P<0.001 vs. CTRL. G, generation; PCa, prostate cancer; OFC, organoid formation count; CTRL, control; DHT, dihydrotestosterone; Bica, Bicalutamide; Enza, Enzalutamide; Doc, Docetaxel.
|
After propagating organoids from G1 to G2, dissociated PCa organoids from two patients were maintained under different irradiation regimens (1, 2 and 4 Gy; Fig. 4A). Patients 6 and 13 both exhibited grade group 2 [Gleason score, 7 (3+4)]. As with chemotherapeutic treatment, irradiation showed differential effects between patients. For patient 6, increasing irradiation dose led to a significant decrease in organoid size while no effect on OFC was observed (Fig. 4B and C). For patient 13, however, both PCa organoid size and OFC decreased significantly with increasing doses of irradiation from 1 to 4 Gy (Fig. 4D and E). Although both patients exhibited similar tumor grade [Gleason score. 7 (3+4)], there was a notable difference in response to irradiation. This variability in treatment effect may be due to underlying genetic and epigenetic alterations in each patient.
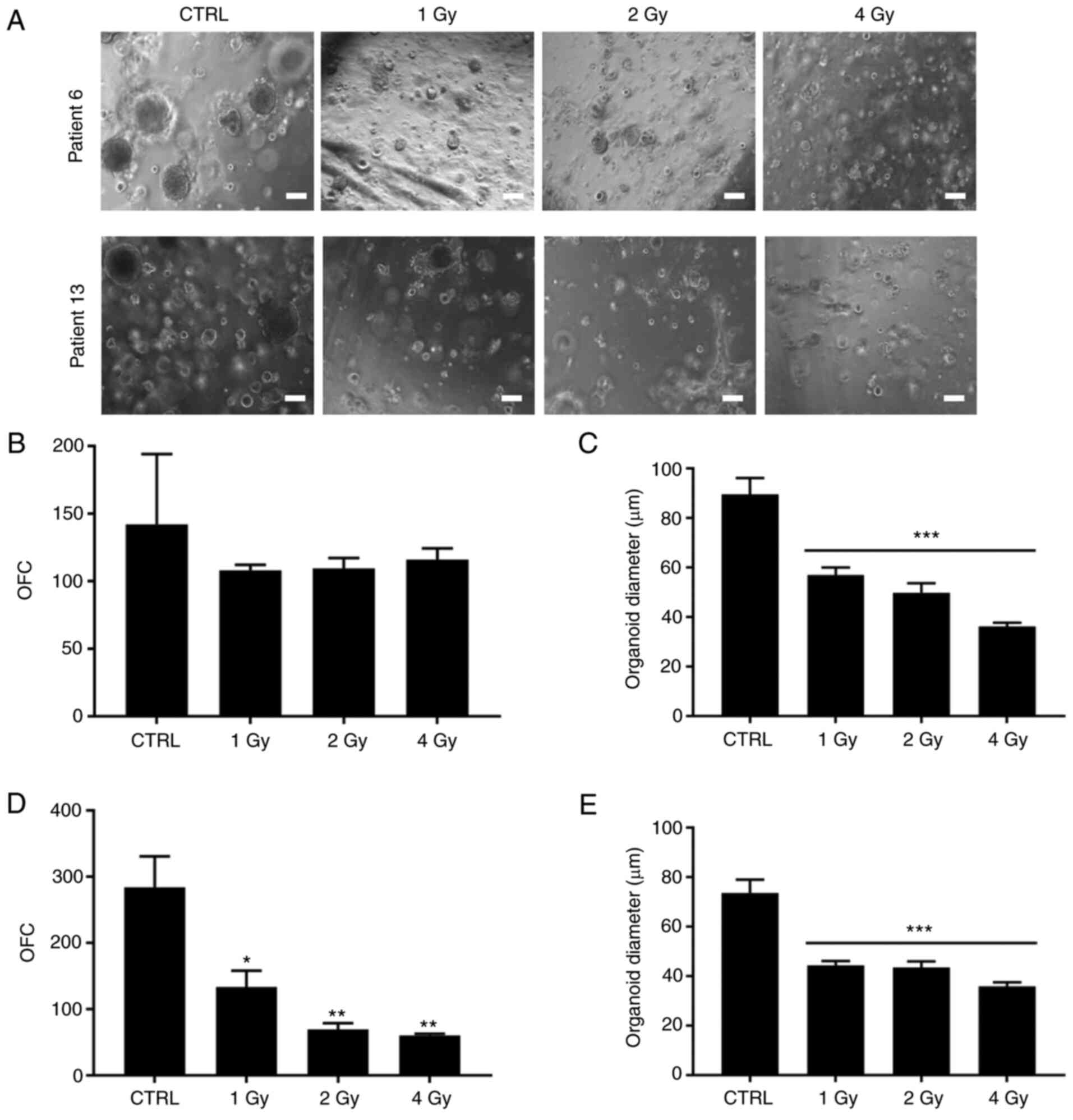 |
Figure 4.
Treatment response to different doses of irradiation of G2 patient-derived PCa organoid growth. (A) Representative brightfield images of G2 PCa organoids [patients 6 and 13; both grade group 2; both Gleason score, 7 (3+4)]. Scale bar, 100 µm. (B) OFC was calculated in duplicate wells per treatment [patient 6; treatment F(3,4)=0.6917, P=0.6032]. Bonferroni post hoc analysis was performed to determine simple factor effects. (C) Quantification of the average diameter was calculated using ≥30 organoids from duplicate wells/treatment [patient 6; treatment F(3,136)=22.11, P<0.0001]. (D) OFC was calculated in duplicate wells/treatment [patient 13; treatment F(3,4)=29.7, P=0.0034]. (E) Quantification of the average diameter of ≥30 organoids from duplicate wells/treatment (patient 13). One-way ANOVA followed by Bonferroni multiple comparisons [treatment F(3,136)=18.89, P<0.0001]. The data are reported as mean ± SD of duplicate experiments. *P<0.05; **P<0.01, ***P<0.001 vs. CTRL. G, generation; PCa, prostate cancer; CTRL, control.
|
Withdrawal of NAC, NOG and A83 from human prostate growth medium significantly decreases organoid growth
To assess the components of human prostate growth medium that are are essential to culture PCa organoids, the effect of withdrawal of each component from the culture medium on the formation of PCa organoids was assessed (Fig. 5). Organoid growth was detected and evaluated by quantifying the number of PCa organoids formed (OFC), calculating the average diameter and assessing maintenance (propagation of organoids) in culture (days). Preliminary data (not shown) indicated that B27 and nicotinamide were essential and thus were not excluded from medium. Each of the 10 components (NAC, EGF, NOG, RSPO, A83, FGF10, FGF2, PGE2, SB and DHT) were removed from the culture medium one at a time and the effect on organoid formation efficiency was assessed. Of these components, three (NAC, NOG and A83) induced a robust inhibitory effect on organoid growth following removal from the human prostate growth medium (Fig. 5). This effect was notable in all six samples from three patients (Fig. S6). For patient 1, withdrawal of the three aforementioned components significantly decreased OFC from 405.00±14.14 organoids in the CTRL to 227.00±19.10 (−NAC), 259.00±10.61 (−NOG), and 328.00±23.34 (−A83; Fig. 5). A similar effect was observed on the size of cultured organoids (Fig. 5) following withdrawal of NAC, NOG and A83. Patients 2 and 3 demonstrated comparable effects, although results were not consistent for all factors (Fig. S6). For example, withdrawal of FGF2 affected the count and size of patient 2 organoids, but not those of patient 3 (Fig. S6). Despite inter-patient variability, removal of PGE2 resulted in an increase in both diameter and organoid count for all three patient samples (Fig. S6) but did not enhance maintenance beyond G3 (Fig. 5D). In addition, SB removal increased the diameter of organoids derived from 2 out of 3 patient samples (Fig. S6). Consequently, it was hypothesized that these components were undesirable factors, and that their effect was further assessed in combination with B27 and nicotinamide. It was also hypothesized that these factors (NAC, NOG, A83, B27 and nicotinamide) would be sufficient to establish patient-derived prostate organoids. In our previous study on the role of EGF in the maintenance of prostate epithelial cells isolated from patient-derived organoids, results demonstrated a significant decrease in cell viability and proliferation when EGF was removed from the medium of 2D cultured cells (47). Here, there was enhanced growth and long-term maintenance of PCa organoids in culture reaching G7 following withdrawal of EGF. It was hypothesized that this effect was due to survival mechanisms and long-term stem cell maintenance of luminal prostate epithelial cells within organoids, similar to what has been previously found in mammary organoids where removal of EGF results in an increase in the relative proportion of mature luminal cells with a concomitant decrease in basal cells (67).
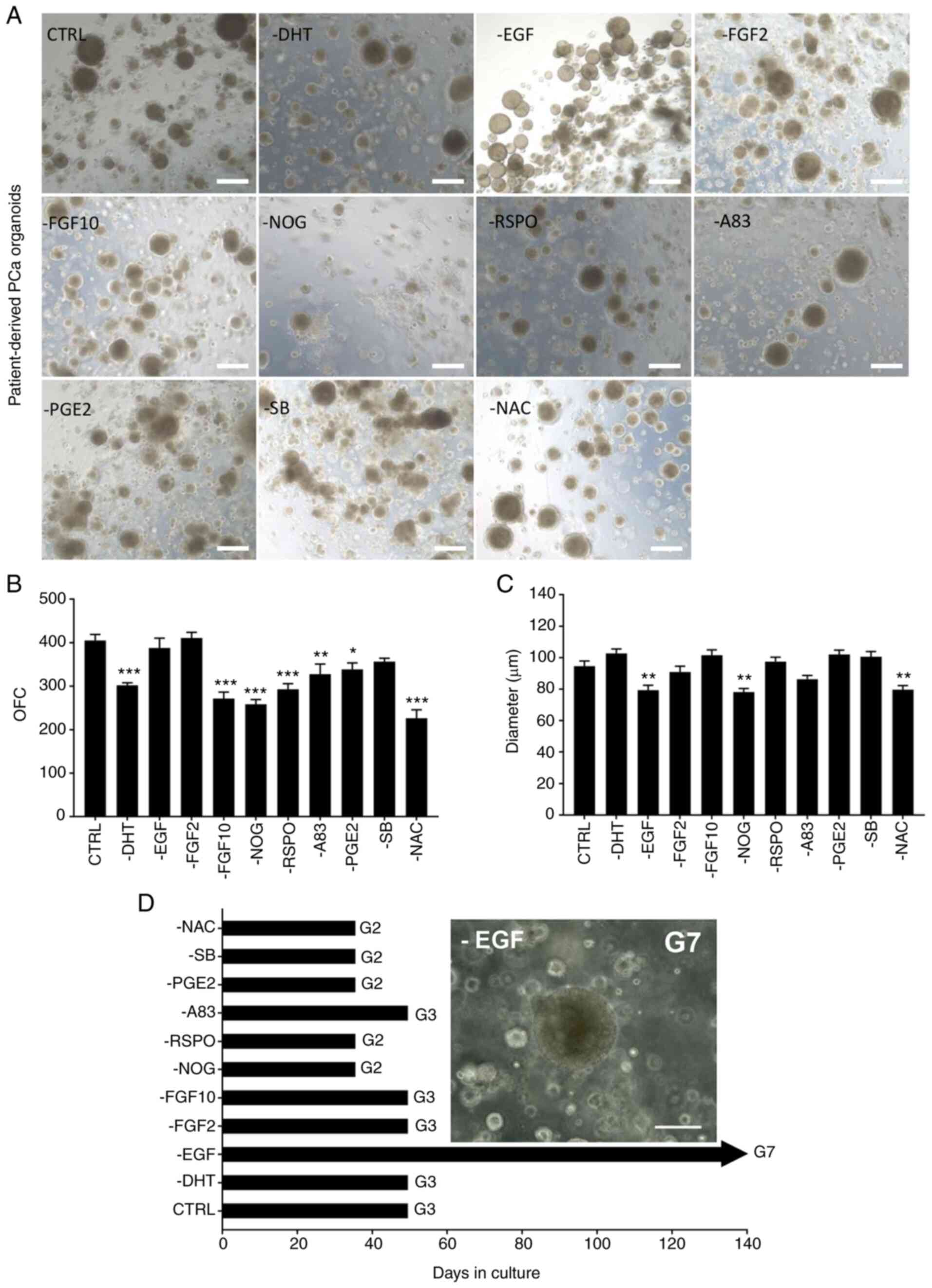 |
Figure 5.
Withdrawal of individual factors from human prostate growth medium boosts organoid growth and maintenance. (A) Representative bright-field images of G2 PCa organoids [patient 29; grade group 2; Gleason score, 7 (3+4)]. Organoids were grown in adDMEM/F12 including all 12 components (CTRL) or in the absence of one component at a time. Scale bar, 200 µm. (B) OFC [condition F(10,11)=31.83, P<0.0001]. Data are presented as the mean ± SD of duplicate experiments. *P<0.05; **P<0.01; ***P<0.001 vs. CTRL. (C) Quantification of the average diameter of ≥50 G1 PCa organoids from duplicate wells/condition using Carl Zeiss ZEN 2013 image software. One-way ANOVA followed by Bonferroni multiple comparisons: Condition F(10,539)=10.71, P<0.0001. Data are presented as the mean ± SEM. **P<0.01 vs. CTRL. (D) Withdrawal of EGF from media results in maintenance of PCa organoids beyond G3 and up to G7 (equivalent to 5 months). Scale bar, 100 µm. G, generation; PCa, prostate cancer; ad, advanced; CTRL, control; OFC, organoid formation count; EGF, epidermal growth factor; DHT, dihydrotestosterone; FGF, fibroblast growth factor; NOG, noggin; RSPO, R-spondin; PGE2, prostaglandin E2; SB, SB202190; A83, A83-01; NAC, N-acetylcysteine.
|
Culturing in 5F medium increases efficiency of organoid propagation
To confirm the hypothesis whether patient-derived organoids can be cultured and maintained using only 5F (NAC, NOG, A83, B27 and nicotinamide), frozen P0 patient 1 cells were thawed and cultured using this minimal medium. The present study validated the role of each of the remaining factors (EGF, RSPO, FGF2, FGF10, PGE2 and SB) in combination with 5F (Fig. 6). Under 5F, organoid diameter and count increased significantly compared with the CTRL containing all 12 components (Fig. 6B and C). Addition of RSPO, FGF10 and FGF2 each boosted organoid growth. The presence of 5F alone also enhanced the establishment of prostate organoids: OFC increased from 207.0±17.6 organoids in the CTRL to 394.0±19.8 in the 5F group (Fig. 6B; P<0.0001). A similar effect was observed on the size of cultured organoids, as the diameter significantly increased in the 5F group compared with the CTRL (Fig. 6C). Organoids at G5 in the presence of FGF10 reached 350 µm while organoids growing under 5F only reached a maximum size of 150 µm (Fig. 6D). As FGF10 is an essential growth factor for prostate development (68), it was hypothesized that FGF10 was a key component for long-term maintenance beyond G5.
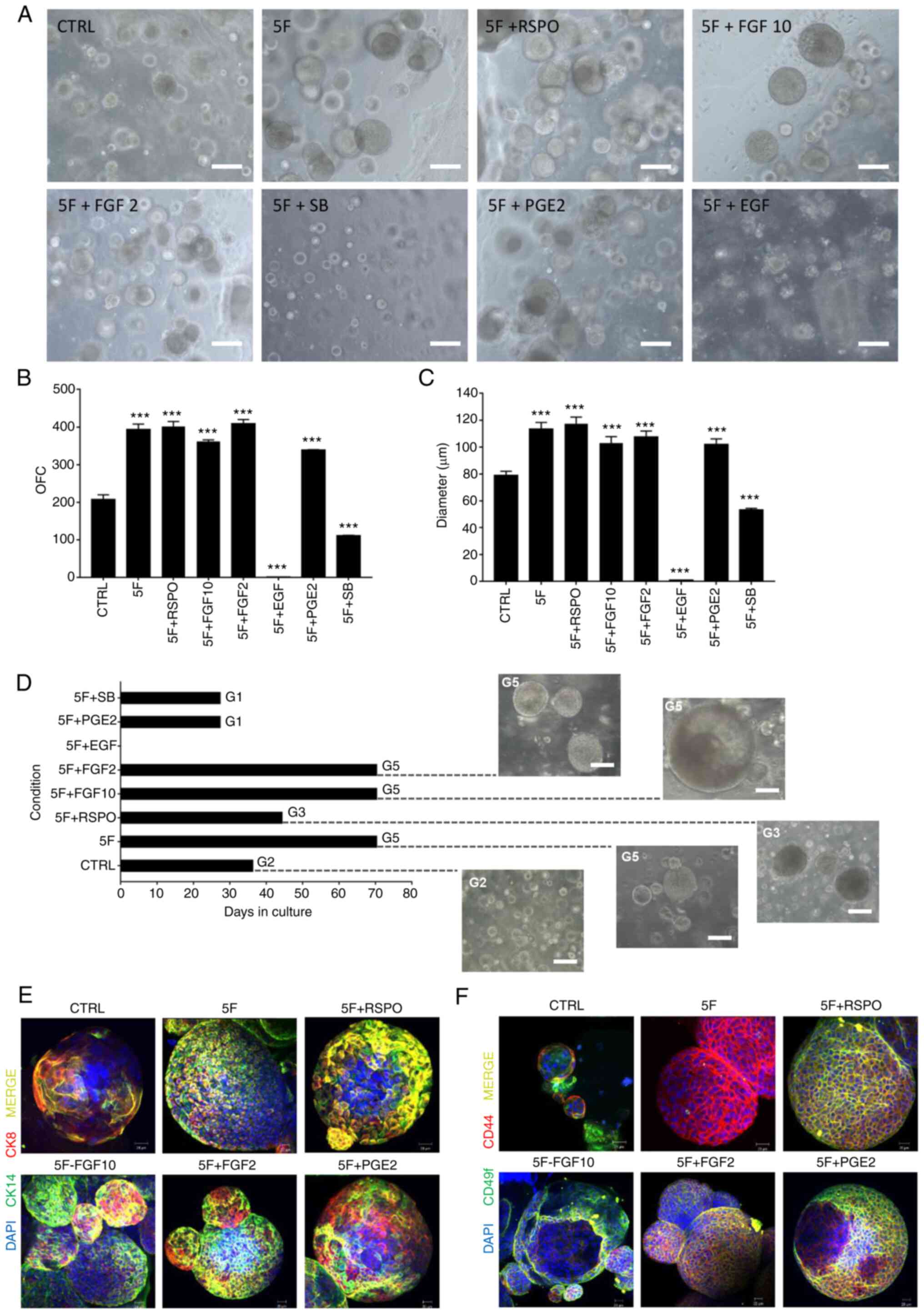 |
Figure 6.
Addition of individual components (EGF, RSPO, FGF2, FGF10, PGE2 and SB) to minimal essential 5F components (NAC, NOG, A83, B27 and nicotinamide) effect on organoid growth. (A) Representative bright-field images of PCa organoids [patient 29; grade group 2; Gleason score, 7 (3+4)] grown in adDMEM/F12 including 5F vs. CTRL (all 12 components) and individual components. Scale bar, 100 µm. (B) OFC [treatment F(8,9)=271.6, P<0.0001]. The data are reported as mean ± SD of duplicate experiments. (C) Quantification of the average diameter of P0 G1 PCa organoids. One-way ANOVA followed by Bonferroni multiple comparisons: Treatment F(8,441)=98.57, P<0.0001. Data are presented as the mean ± SEM of triplicate experiments. ***P<0.001 vs. CTRL. (D) Maintenance of PCa organoids (patient 29) in the presence of 5F vs. CTRL and individual components. Representative bright-field images of the PCa organoids at G2, G3 and G5 were visualized by Axiovert inverted microscope (20× magnification). Scale bar, 100 µm. Immunofluorescent images of P0 G1 PCa organoids (patient 29) stained with (E) prostate lineage epithelial markers CK8 and CK14 and (F) stem cell markers CD44 and CD49f. The nuclei were stained with anti-fade Fluorogel II with DAPI. Scale bar, 40 µm. Representative confocal microscopy images were acquired using a Zeiss LSM 710 laser scanning confocal microscope at 40× oil objective and processed using Carl Zeiss ZEN 2013 image software. G, generation; PCa, prostate cancer; ad, advanced; CTRL, control; OFC, organoid formation count; EGF, epidermal growth factor; DHT, dihydrotestosterone; FGF, fibroblast growth factor; NOG, noggin; RSPO, R-spondin; PGE2, prostaglandin E2; SB, SB202190; A83, A83-01; NAC, N-acetylcysteine; CK, cytokeratin.
|
Organoids were derived from frozen cells of patient 1. Efficiency of organoid formation decreased significantly under CTRL; OFC decreased from >300 organoids with a fresh sample to <150 with frozen cells derived from the same patient. Culturing with 5F medium salvaged organoid growth from a frozen sample and the OFC was restored to >300 organoids (data not shown).
The addition of EGF, PGE2, and SB, previously predicted as undesirable components, 5F medium decreased OFC and diameter significantly compared with CTRL (P<0.0001) (data not shown). These results further confirmed the hypothesis about their undesirable effect on the culture medium.
To characterize the prostate epithelial lineages in the established organoid cultures, organoids were stained for luminal-(CK8) and basal-specific markers (CK14). The expression of luminal CK8 and basal CK14 markers confirmed the presence of both prostate epithelial lineages in the established organoid cultures, with organoids expressing luminal- or basal-only or luminal and basal double-positive cells (Fig. 6E). Co-expression of both stem cell markers, CD44 and CD49f, has been shown to identify putative prostate stem-like cells (69,70). Positive staining of CD44 and CD49f stem cell markers further demonstrated the existence of stem-like cells within the bulk of patient-derived organoids (Fig. 6F).
Withdrawal of EGF from human prostate growth medium enhances organoid growth and maintenance
Following withdrawal of EGF, OFC of G1 PCa organoids increased (Figs. 5 and S6). To validate this, organoids of patient 29 were cultured in the absence of single components and compared with the CTRL (full components). Only organoids grown in the absence of EGF survived beyond G3, reaching G7 (equivalent to 5 months; Fig. 5D). Hence, EGF may be a key negative regulatory factor for long-term growth of organoids.
Discussion
Despite published protocols (13,27,30,31,33,34,38) describing the establishment of PCa organoids, the successful culture and maintenance of those organoids in culture remains a challenge. The present study obtained treatment-naïve patient-derived PCa organoids from fresh radical prostatectomy specimens. These PCa organoids mimicked the heterogeneity of corresponding parental tumor tissue; histopathological analysis demonstrated similar tissue architecture and cellular morphology, as well as consistent IHC marker expression. The majority of PCa organoids consisted of solid 3D structures, with a minority having luminal-like hollow structures as detected by bright-field microscopy and H&E staining of sectioned organoids. Gao et al (2014) (31) demonstrated that human organoid cultures mostly comprise solid basal cell-derived organoids. The existence of prostate luminal and basal lineages was confirmed in the established cultures by positive expression of luminal- (CK8, AR and PSA) and basal-specific markers (CK5, CK14 and p63).
Since patients respond differently to PCa treatment, the present study used patient-derived PCa organoids to assess their potential use in predicting patient drug response and testing personalized treatment strategies. There was a substantial variability in organoid formation efficiency between patient samples, reflected by the large deviation in both OFC and organoid size. A previous study observed that samples derived from advanced PCa exhibit inconsistent growth rate (13). While Bicalutamide and Enzalutamide induced a robust inhibitory effect on growth of PCa organoids from certain patients, a more prominent significant inhibitory effect was observed with chemotherapeutic agent Docetaxel in others. Irradiation was not equally effective in impeding PCa organoid growth in all patient samples, which is an expected effect from a clinical perspective (28). The ability to grow PCa organoids from patients enables clinical screening of drug combinations that selectively target diseased tissue and may identify more effective therapies with minimal side effects (28,36,37,71). In a previous study, Puca et al (33) used PCa organoids from metastatic lesions with a neuroendocrine phenotype for therapeutic assessment. The results showed concordance between drug response in vitro and patient response in the clinic, which corresponded to the molecular background of the tumor (33). Similarly, Beshiri et al (38) used organoids from 20 models of LuCaP metastatic castration-resistant PCa patient-derived xenograft cohort to demonstrate conserved genomic heterogeneity between PDXs and organoids. Further validation is essential, including advanced genetic ananlysis to compare in vitro and clinical drug response and their association with genetic mutation profiles (72).
To minimize the costly requirements of previously established organoid culture systems, the present study developed an assay system to assess the effect of withdrawal of individual components on formation efficiency. The present study reported the establishment of a cost-effective protocol by identifying the key components needed to grow prostate organoids with a high success rate and long-term maintenance in culture. The present results demonstrated the ability to culture 3D patient-derived PCa organoids using 5 components, instead of 12 included in the initial protocol. The decreased cost and increased efficiency widen the potential use of patient samples in understanding disease mechanisms and personalized medicine.
In 2014, Gao et al (31) reported the successful generation of fully-characterized organoid lines from six metastatic tissue biopsies and one circulating tumor cell specimen (31). The method adopted by Gao et al provided a success of 20% and maintenance in culture for 1–2 months. Here, removing EGF from culture medium enhanced survival of organoids for >G7 (equivalent to 6 months). Similarly, minimal 5F medium allowed maintenance of organoids for ≥G5 FGF10 further enhanced growth and maintenance of organoids in culture. In addition, the efficiency was doubled for PCa organoids in 5F medium compared with the previously published protocol, in which the reported formation efficiency was ~1% (30).
FGF signaling serves a key role in maintaining the stemness of prostate cells (73) and is important for enhanced maintenance of PCa organoids in culture. FGF10 is a highly expressed growth factor in the mesenchyme of the developing prostate gland, while its deletion has been shown to impede branching morphogenesis (68). The present study demonstrated the importance of FGF10 in maintenance of organoids in culture. Both luminal and basal markers were observed >G6 using a minimal and less expensive medium. In line with previously published data (71,74–77), NOG and A83-01 were key to sustaining PCa organoid cultures. Bone morphogenetic protein (BMP) antagonist NOG is essential for stem cell expansion and serves a key role during prostate development by counteracting BMP-4 inhibitory effect on cell proliferation (71,74). TGF-β signaling blocks proliferation of prostate cells and serves as a defense mechanism to suppress tumorigenesis in various types of cancer, such as pancreatic, colorectal, breast and head and neck cancers (75–77). Therefore, the Alk4/5/7 inhibitor A83-01 serves a key role for enhancing organoid growth by obstructing the TGF-β signaling pathway.
Although EGF is considered to be an essential component for establishing and maintaining prostate organoids in culture (30), the present study demonstrated successful derivation and sustained PCa organoid growth in the absence of EGF. The removal of EGF enhanced organoid plating efficiency and maintenance by preventing the dissociation of organoids and migration of epithelial cells from 3D Matrigel to 2D compartment (47). The derivation and maintenance of primary 2D cells derived from 3D organoid culture were further enhanced in the presence of EGF (47). The present results demonstrated a significant decrease in viability and proliferation of 2D cells when EGF was removed from the medium, while adding EGF supports the growth of 2D cells (47). These data are consistent with the role of EGF in stimulating cell motility and migration of epithelial cells from various types of tumor, including PCa (78,79).
The present study has several limitations. The interpatient variability hinders the ability to obtain consistent results between patient samples. The conclusion that 5F key factors are necessary to grow and maintain prostate organoids in culture was based on a holistic interpretation of OFC and organoid diameter results. In addition, immunophenotypical characterization of organoids via staining with prostate lineage epithelial markers CK8 and CK14, AR and p63 and stem cell markers CD44 and CD49f relied on subject interpretation. Advanced genetic analysis is needed to elucidate PCa organoid niche requirements and their association with genetic mutation profiles. Further genetic studies, such as copy number variations and targeted sequencing, are needed in the future to elucidate the genetic signature of PCa organoids. A prostate gland that harbors a tumor cannot be normal in any way; this was supported by assessment of phosphorylated-AKT expression-a reporter for PTEN aberrations-in G1 tumor organoids via immunofluorescent staining that demonstrated high expression (data not shown). The present study did not sort cells into luminal and basal prostate epithelial cells due to the small sample size and number of cells. The results revealed both basal and luminal CK staining, indicating a mix of basal, luminal and double-positive organoids. Cell surface marker CD49f was abundant in basal cells, which are present at the periphery of organoids as well as glands (27), hence the strong stain. Certain samples exhibited luminal organoids and cells that stained positive for AR and CK8.
It remains essential to compare patient clinical response to in vitro drug response; therefore, patients are being followed up to collect clinical data regarding disease progression and drug response in case they receive drug treatment following radical prostatectomy. Further in-depth studies are warranted to assess expression levels of different genes and proteins implicated in PCa pathogenesis and progression (including AR and transmembrane serine protease 2) before and after treatment in both patient samples and corresponding organoids. Moreover, the present method used factors such as Matrigel as ECM substitute and various growth factors and molecule inhibitors were included in the culture medium. These factors may interfere with drug response. Organoid culture systems do not mimic the in vivo microenvironment since they do not include stromal, immune or endothelial cells (80). As a result, substantial efforts are dedicated to developing co-culture systems. Although it is hypothesized that organoids can be cultured indefinitely without genetic manipulation (30), organoids could not be maintained in culture beyond G3-5.
The results of the present study demonstrated the successful establishment of 3D organoids from fresh specimens of treatment-naïve patients with PCa. The presence of prostate luminal and basal epithelial lineages was confirmed in the derived models. Notably, principal component analysis plots for clustering transcriptomic gene expression of the different organoids, and their parental tissue, revealed distinct clusters, denoting a variation between the specimens. There was a differential drug response between different patient samples. Moreover, the present study established a robust method to generate prostate organoids in a reduced 5-factor culture medium directly from fresh primary tumor specimens. The present study identified EGF as a key negative regulatory factor for the long-term growth of organoids. Establishing treatment-naïve patient-derived PCa organoid cultures from radical prostatectomy tissue specimens (from a Middle Eastern cohort) which resemble parental tissue provides a unique avenue for various future applications. In addition to their use in understanding the etiology of disease, organoids can be used for drug testing and screening of traditional and novel PCa therapeutic strategies to identify personalized treatment response, as well as for broad applications in basic and translational research. By modifying a previously established PCa organoid culture system, the present study provided an optimized cost-effective culturing platform with fewer components (5, instead of 12) that increased organoid formation efficiency and long-term maintenance in culture.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by funding from the Medical Practice Plan at the American University of Beirut Faculty of Medicine (grant no. MPP‑WAK‑18) and New York University Abu Dhabi Biology Division (grant no. AD231)
Availability of data and materials
All data generated or analyzed during this study are included in this published article. Data sets have been deposited in National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE148937; ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE148937).
Authors' contributions
KC, HFB, OH, HM, AM, FB, CD, TT, NS, AT, AEH and DM performed the experiments. KC, HFB, OH, HM, AM, FB, CD, NS, AT, MES and MAS designed the experiments. KC, HFB, MAS, AT and WAK wrote the manuscript. KC, HFB, OH, HM, AM, FB, CD, TT, NS, AT, MAS, MES, AEH, DM and WAK revised and edited the manuscript. HFB and MAS collected and analyzed data. AEH, DM and WAK supervised the study. WAK conceptualized the study and obtained funding. KC and HFB confirm the authenticity of all the raw data. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was performed according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of the American University of Beirut Medical Center (approval no. IM-DM-02). Informed consent was obtained from all subjects involved in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shoag J and Barbieri CE: Clinical variability and molecular heterogeneity in prostate cancer. Asian J Androl. 18:543–548. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Williams JL, Greer PA and Squire JA: Recurrent copy number alterations in prostate cancer: An in silico meta-analysis of publicly available genomic data. Cancer Genet. 207:474–488. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Karantanos T, Corn PG and Thompson TC: Prostate cancer progression after androgen deprivation therapy: Mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 32:5501–5511. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gao D and Chen Y: Organoid development in cancer genome discovery. Curr Opin Genet Dev. 30:42–48. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Horoszewicz JS, Leong SS, Kawinski E, Karr JP, Rosenthal H, Chu TM, Mirand EA and Murphy GP: LNCaP model of human prostatic carcinoma. Cancer Res. 43:1809–1818. 1983.PubMed/NCBI
|
|
7
|
Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF and Jones LW: Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Invest Urol. 17:16–23. 1979.PubMed/NCBI
|
|
8
|
Korenchuk S, Lehr JE, MClean L, Lee YG, Whitney S, Vessella R, Lin DL and Pienta KJ: VCaP, a cell-based model system of human prostate cancer. In vivo. 15:163–168. 2001.PubMed/NCBI
|
|
9
|
Navone NM, Olive M, Ozen M, Davis R, Troncoso P, Tu SM, Johnston D, Pollack A, Pathak S, von Eschenbach AC and Logothetis CJ: Establishment of two human prostate cancer cell lines derived from a single bone metastasis. Clin Cancer Res. 3:2493–2500. 1997.PubMed/NCBI
|
|
10
|
Sramkoski RM, Pretlow TG II, Giaconia JM, Pretlow TP, Schwartz S, Sy MS, Marengo SR, Rhim JS, Zhang D and Jacobberger JW: A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell Dev Biol Anim. 35:403–409. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mertz KD, Setlur SR, Dhanasekaran SM, Demichelis F, Perner S, Tomlins S, Tchinda J, Laxman B, Vessella RL, Beroukhim R, et al: Molecular characterization of TMPRSS2-ERG gene fusion in the NCI-H660 prostate cancer cell line: A new perspective for an old model. Neoplasia. 9:200–206. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schwank G, Andersson-Rolf A, Koo BK, Sasaki N and Clevers H: Generation of BAC transgenic epithelial organoids. PLoS One. 8:e768712013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Drost J, Karthaus WR, Gao D, Driehuis E, Sawyers CL, Chen Y and Clevers H: Organoid culture systems for prostate epithelial tissue and prostate cancer tissue. Nat Protoc. 11:347–358. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ and Clevers H: Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459:262–265. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Koo BK, Stange DE, Sato T, Karthaus W, Farin HF, Huch M, van Es JH and Clevers H: Controlled gene expression in primary Lgr5 organoid cultures. Nat Methods. 9:81–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bartucci M, Ferrari AC, Kim IY, Ploss A, Yarmush M and Sabaawy HE: Personalized medicine approaches in prostate cancer employing patient derived 3D organoids and humanized mice. Front Cell Dev Biol. 4:642016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M, et al: Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell stem cell. 6:25–36. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stange DE, Koo BK, Huch M, Sibbel G, Basak O, Lyubimova A, Kujala P, Bartfeld S, Koster J, Geahlen JH, et al: Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 155:357–368. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sachs N and Clevers H: Organoid cultures for the analysis of cancer phenotypes. Curr Opin Genet Dev. 24:68–73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jung P, Sato T, Merlos-Suarez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco MA, Sancho E, et al: Isolation and in vitro expansion of human colonic stem cells. Nat Med. 17:1225–1227. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Taguchi A, Kaku Y, Ohmori T, Sharmin S, Ogawa M, Sasaki H and Nishinakamura R: Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 14:53–67. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Takasato M, Er PX, Becroft M, Vanslambrouck JM, Stanley EG, Elefanty AG and Little MH: Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat Cell Biol. 16:118–126. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Antonica F, Kasprzyk DF, Opitz R, Iacovino M, Liao XH, Dumitrescu AM, Refetoff S, Peremans K, Manto M, Kyba M and Costagliola S: Generation of functional thyroid from embryonic stem cells. Nature. 491:66–71. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Koehler KR, Mikosz AM, Molosh AI, Patel D and Hashino E: Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature. 500:217–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T and Sasai Y: Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 472:51–56. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP and Knoblich JA: Cerebral organoids model human brain development and microcephaly. Nature. 501:373–379. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Agarwal S, Hynes PG, Tillman HS, Lake R, Abou-Kheir WG, Fang L, Casey OM, Ameri AH, Martin PL, Yin JJ, et al: Identification of different classes of luminal progenitor cells within prostate tumors. Cell Rep. 13:2147–2158. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim J, Koo BK and Knoblich JA: Human organoids: Model systems for human biology and medicine. Nat Rev Mol Cell Biol. 21:571–584. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, Paulsen B, Nguyen L, Adiconis X and Regev A: Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 570:523–527. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel H, Sachs N, et al: Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 159:163–175. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK, et al: Organoid cultures derived from patients with advanced prostate cancer. Cell. 159:176–187. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vela I and Chen Y: Prostate cancer organoids: A potential new tool for testing drug sensitivity. Expert Rev Anticancer Ther. 15:261–263. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Puca L, Bareja R, Prandi D, Shaw R, Benelli M, Karthaus WR, Hess J, Sigouros M, Donoghue A, Kossai M, et al: Patient derived organoids to model rare prostate cancer phenotypes. Nat Commun. 9:24042018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Richards Z, McCray T, Marsili J, Zenner ML, Manlucu JT, Garcia J, Kajdacsy-Balla A, Murray M, Voisine C, Murphy AB, et al: Prostate stroma increases the viability and maintains the branching phenotype of human prostate organoids. iScience. 12:304–317. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Drost J and Clevers H: Organoids in cancer research. Nat Rev Cancer. 18:407–418. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Elbadawy M, Abugomaa A, Yamawaki H, Usui T and Sasaki K: Development of prostate cancer organoid culture models in basic medicine and translational research. Cancers (Basel). 12:7772020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gleave AM, Ci X, Lin D and Wang Y: A synopsis of prostate organoid methodologies, applications, and limitations. Prostate. 80:518–526. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Beshiri ML, Tice CM, Tran C, Nguyen HM, Sowalsky AG, Agarwal S, Jansson KH, Yang Q, McGowen KM, Yin J, et al: A PDX/organoid biobank of advanced prostate cancers captures genomic and phenotypic heterogeneity for disease modeling and therapeutic screening. Clin Cancer Res. 24:4332–4345. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Njoroge RN, Unno K, Zhao JC, Naseem AF, Anker JF, McGee WA, Nonn L and Abdulkadir SA: Organoids model distinct vitamin E effects at different stages of prostate cancer evolution. Sci Rep. 7:162852017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Santamaria PG, Moreno-Bueno G, Portillo F and Cano A: EMT: Present and future in clinical oncology. Mol Oncol. 11:718–738. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Park JW, Lee JK, Phillips JW, Huang P, Cheng D, Huang J and Witte ON: Prostate epithelial cell of origin determines cancer differentiation state in an organoid transformation assay. Proc Natl Acad Sci USA. 113:4482–4487. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al: The genomic complexity of primary human prostate cancer. Nature. 470:214–220. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fontugne J, Davis K, Palanisamy N, Udager A, Mehra R, McDaniel AS, Siddiqui J, Rubin MA, Mosquera JM and Tomlins SA: Clonal evaluation of prostate cancer foci in biopsies with discontinuous tumor involvement by dual ERG/SPINK1 immunohistochemistry. Mod Pathol. 29:157–165. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Barbieri CE, Bangma CH, Bjartell A, Catto JW, Culig Z, Grönberg H, Luo J, Visakorpi T and Rubin MA: The mutational landscape of prostate cancer. Eur Urol. 64:567–576. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Haffner MC, Mosbruger T, Esopi DM, Fedor H, Heaphy CM, Walker DA, Adejola N, Gürel M, Hicks J, Meeker AK, et al: Tracking the clonal origin of lethal prostate cancer. J Clin Invest. 123:4918–4922. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, Brewer DS, Kallio HM, Högnäs G, Annala M, et al: The evolutionary history of lethal metastatic prostate cancer. Nature. 520:353–357. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cheaito K, Bahmad HF, Jalloul H, Hadadeh O, Msheik H, El-Hajj A, Mukherji D, Al-Sayegh M and Abou-Kheir W: Epidermal growth factor is essential for the maintenance of novel prostate epithelial cells isolated from patient-derived organoids. Front Cell Dev Biol. 8:5716772020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bahmad HF, Cheaito K, Chalhoub RM, Hadadeh O, Monzer A, Ballout F, El-Hajj A, Mukherji D, Liu YN, Daoud G and Abou-Kheir W: Sphere-formation assay: Three-dimensional in vitro culturing of prostate cancer stem/progenitor sphere-forming cells. Front Oncol. 8:3472018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Love MI, Huber W and Anders S: Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nelson JW, Sklenar J, Barnes AP and Minnier J: The START app: A web-based RNAseq analysis and visualization resource. Bioinformatics. 33:447–449. 2017.PubMed/NCBI
|
|
51
|
Edgar R, Domrachev M and Lash AE: Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A, Wadi L, Meyer M, Wong J, Xu C, et al: Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, cytoscape and EnrichmentMap. Nat Protoc. 14:482–517. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T and Buetow KH: PID: The pathway interaction database. Nucleic Acids Res. 37:D674–D679. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kandasamy K, Mohan SS, Raju R, Keerthikumar S, Kumar GS, Venugopal AK, Telikicherla D, Navarro JD, Mathivanan S, Pecquet C, et al: NetPath: A public resource of curated signal transduction pathways. Genome Biol. 11:R32010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Romero P, Wagg J, Green ML, Kaiser D, Krummenacker M and Karp PD: Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 6:R22005. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al: Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 39:D691–697. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al: Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al: The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 33:D284–288. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Martin AM, Nirschl TR, Nirschl CJ, Francica BJ, Kochel CM, van Bokhoven A, Meeker AK, Lucia MS, Anders RA, DeMarzo AM and Drake CG: Paucity of PD-L1 expression in prostate cancer: Innate and adaptive immune resistance. Prostate Cancer Prostatic Dis. 18:325–332. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhu YP, Wan FN, Shen YJ, Wang HK, Zhang GM and Ye DW: Reactive stroma component COL6A1 is upregulated in castration-resistant prostate cancer and promotes tumor growth. Oncotarget. 6:14488–14496. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Rasool RU, Natesan R, Deng Q, Aras S, Lal P, Effron SS, Mitchell-Velasquez E, Posimo JM, Carskadon S, Baca SC, et al: CDK7 inhibition suppresses castration-resistant prostate cancer through MED1 inactivation. Cancer Discov. 9:1538–1555. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Velardi E, Tsai JJ, Radtke S, Cooper K, Argyropoulos KV, Jae-Hung S, Young LF, Lazrak A, Smith OM, Lieberman S, et al: Suppression of luteinizing hormone enhances HSC recovery after hematopoietic injury. Nat Med. 24:239–246. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
O'Neill AJ, Prencipe M, Dowling C, Fan Y, Mulrane L, Gallagher WM, O'Connor D, O'Connor R, Devery A, Corcoran C, et al: Characterisation and manipulation of docetaxel resistant prostate cancer cell lines. Mol Cancer. 10:1262011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Shen MM and Abate-Shen C: Molecular genetics of prostate cancer: New prospects for old challenges. Genes Dev. 24:1967–2000. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Peehl DM: Primary cell cultures as models of prostate cancer development. Endocr Relat Cancer. 12:19–47. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Rosenbluth JM, Schackmann RCJ, Gray GK, Selfors LM, Li CM, Boedicker M, Kuiken HJ, Richardson A, Brock J, Garber J, et al: Organoid cultures from normal and cancer-prone human breast tissues preserve complex epithelial lineages. Nat Commun. 11:17112020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Donjacour AA, Thomson AA and Cunha GR: FGF-10 plays an essential role in the growth of the fetal prostate. Dev Biol. 261:39–54. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yamamoto H, Masters JR, Dasgupta P, Chandra A, Popert R, Freeman A and Ahmed A: CD49f is an efficient marker of monolayer- and spheroid colony-forming cells of the benign and malignant human prostate. PLoS One. 7:e469792012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Guo C, Liu H, Zhang BH, Cadaneanu RM, Mayle AM and Garraway IP: Epcam, CD44, and CD49f distinguish sphere-forming human prostate basal cells from a subpopulation with predominant tubule initiation capability. PLoS One. 7:e342192012. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Fatehullah A, Tan SH and Barker N: Organoids as an in vitro model of human development and disease. Nat Cell Biol. 18:246–254. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, Clevers H and Lutolf MP: Designer matrices for intestinal stem cell and organoid culture. Nature. 539:560–564. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Huang Y, Hamana T, Liu J, Wang C, An L, You P, Chang JY, Xu J, Jin C, Zhang Z, et al: Type 2 fibroblast growth factor receptor signaling preserves stemness and prevents differentiation of prostate stem cells from the basal compartment. J Biol Chem. 290:17753–17761. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Cook C, Vezina CM, Allgeier SH, Shaw A, Yu M, Peterson RE and Bushman W: Noggin is required for normal lobe patterning and ductal budding in the mouse prostate. Dev Biol. 312:217–230. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Bjerke GA, Yang CS, Frierson HF, Paschal BM and Wotton D: Activation of Akt signaling in prostate induces a TGFβ-mediated restraint on cancer progression and metastasis. Oncogene. 33:3660–3667. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Qin J, Wu SP, Creighton CJ, Dai F, Xie X, Cheng CM, Frolov A, Ayala G, Lin X, Feng XH, et al: COUP-TFII inhibits TGF-β-induced growth barrier to promote prostate tumorigenesis. Nature. 493:236–240. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R, et al: SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 470:269–273. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Montanari M, Rossetti S, Cavaliere C, D'Aniello C, Malzone MG, Vanacore D, Franco RD, Mantia EL, Iovane G, Piscitelli R, et al: Epithelial-mesenchymal transition in prostate cancer: An overview. Oncotarget. 8:35376–35389. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Lu X and Kang Y: Epidermal growth factor signalling and bone metastasis. Br J Cancer. 102:457–461. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Wang S, Gao D and Chen Y: The potential of organoids in urological cancer research. Nat Rev Urol. 14:401–414. 2017. View Article : Google Scholar : PubMed/NCBI
|




















