Introduction
Lung cancer is a prevalent type of cancer worldwide,
accounting for the highest number of new cases (11.4% of all cases)
and deaths (18.0% of all deaths) in men in 2020 (1). Despite recent advances in treatment,
and although numerous molecular signatures for diagnostic and
prognostic purposes have been proposed, the incidence and mortality
of lung cancer remain high. Increasing evidence has suggested that
circulating microRNAs (miRNAs/miRs) and proteins in the blood may
be dysregulated, and associated with cancer development and
progression (2,3).
miRNAs, which are small single-stranded non-coding
RNA molecules, serve important roles in various biological
processes, such as cell proliferation, apoptosis, migration and
invasion (4). miRNAs suppress gene
expression at the post-transcriptional level by binding to the
3′-untranslated regions of mRNA (4). Among the miRNAs, miR-145 has been
suggested to have a role in various types of cancer (5,6),
including non-small cell lung cancer (NSCLC) (7,8).
miR-145 exerts its functions by targeting specific genes, one of
which is vascular endothelial growth factor (VEGF), a key regulator
of angiogenesis (9). Zou et
al (10) reported that miR-145
served an inhibitory role, both in vitro and in vivo,
in the angiogenesis of breast cancer cells via VEGF-A regulation.
Based on bioinformatics analysis in our previous study, VEGF was
identified to be a downstream target gene of miR-145 (11). Therefore, the evaluation of both
miR-145 and VEGF levels may provide further insight into their
clinical use in NSCLC.
Previous studies have shown that serum miR-145 and
VEGF levels may be associated with diagnosis and prognosis in
glioblastoma (12), ovarian cancer
(13,14) and NSCLC (15,16).
Although there are an increasing number of studies that have
supported the possibility of using serum miR-145 and VEGF as
biomarkers for various purposes in lung cancer, the reports have
been unclear regarding the prognostic role of these two markers
(12,15–18).
Furthermore, most of the studies have been conducted using healthy
individuals as the control group, which could have led to an
overestimation of the diagnostic accuracy (19).
The present study aimed to investigate the
diagnostic performance of serum miR-145 and VEGF levels in patients
with NSCLC and included patients with other lung diseases (OLDs) as
the control group. The prognostic role of the two markers and the
biological function of miR-145 in NSCLC cells were also
evaluated.
Materials and methods
Study participants and blood
samples
Clinical information on the study participants, as
well as remaining samples, were obtained from our two previous
studies following ethics approval from the Human Research Ethics
Committee of the Faculty of Medicine, Prince of Songkla University
(Songkhla, Thailand; approval nos. 59-011-05-1 and 60-350-04-2)
(11,20). The participants were enrolled at
Songklanagarind Hospital (Songkhla, Thailand) between January 2016
and December 2018. From the original 230 samples (117 NSCLC and 113
OLDs) of the previous study, 215 samples (106 NSCLC and 109 OLDs)
were adequately obtained for the analysis of miR-145 expression and
183 samples (92 NSCLC and 91 OLDs) for VEGF determination. Patients
with suspected lung cancer were included if they presented with a
chronic cough for ≥8 weeks or hemoptysis more than once. In
addition, information on smoking and drinking status, as well as
the family history of cancer, for each participant were obtained by
interviewing the patients. The demographic and clinical
characteristics of the patients were retrieved from the hospital
registry. Patients with a previous history of cancer or
chemotherapy were excluded. During or after data collection, no
personal information that could be used to identify individuals was
available.
Confirmation of cancer was obtained via a
pathological diagnosis. The histological type of NSCLC was
diagnosed according to the 2015 World Health Organization
classification of lung and pleural tumors (21). The clinical staging was based on
the Tumor Node Metastasis staging system of the Cancer Staging
Manual (7th Edition) of the American Joint Committee on Cancer
(22). Patients with OLDs were
diagnosed via routine procedures, including laboratory tests, chest
X-rays and tissue biopsies. The mortality data were obtained from
the civil registry.
Peripheral blood (5 ml) was collected at the time of
diagnosis after the patients had provided their written informed
consent, and samples were processed as previously described
(11,20). Briefly, blood samples were
coagulated for 30 min at room temperature and centrifuged at 3,400
× g for 10 min at room temperature. Isolated serum was filtered
through a polyvinylidene difluoride syringe filter with a pore size
of 0.22-mm (cat. no. SLGS033SB; MilliporeSigma) and prepared for
miRNA isolation.
Cell culture
Human NSCLC cell lines, A549 and H1792 (both lung
adenocarcinoma), were purchased from the American Type Culture
Collection. Cells were cultured in RPMI-1640 medium (cat. no.
R6504; Sigma-Aldrich; Merck KGaA) supplemented with 10% (v/v) FBS
(cat. no. 10091148; Gibco; Thermo Fisher Scientific, Inc.), and
maintained at 37°C in a 5% CO2 atmosphere.
miRNA transfection
miR-145 mimic (50 nM, 5′-GUCCAGUUUUCCCAGGAAUCCCU-3′)
and mimic negative control (NC) (50 nM,
5′-UCACAACCUCCUAGAAAGAGUAGA-3′) were purchased from GE Healthcare
Dharmacon, Inc. Transfection was conducted using
Lipofectamine® 2000 (cat. no. 11668019; Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. Briefly, A549 (2×105) and H1792
(4×105) cells were suspended in medium containing 10%
FBS and seeded into a T25 cell culture flask (Corning, Inc.) for 24
h before miRNA transfection. miR-145 mimic and mimic NC were
diluted in Opti-MEM (cat. no. 31985088; Gibco; Thermo Fisher
Scientific, Inc.), mixed with Lipofectamine 2000, and incubated for
30 min at room temperature. Subsequently, cells were incubated with
the transfection mixture at 37°C in a 5% CO2 atmosphere.
Culture medium was replaced after 10 h, and the cells were further
incubated at 37°C in a 5% CO2 atmosphere for 4 days.
After transfection, the cells were harvested and subjected to
subsequent experiments. The efficiency of miRNA transfection was
confirmed via reverse transcription-quantitative PCR (RT-qPCR).
Untreated cells (cell group), cells treated with Opti-MEM and
Lipofectamine 2000 (mock group) and cells transfected with mimic NC
(mimic NC groups) were assigned as the control groups.
Detection of miR-145 expression using
RT-qPCR
Total RNA was extracted from cells using
TRIzol® reagent (cat. no. 15596018; Invitrogen; Thermo
Fisher Scientific, Inc.) and from serum (250 µl) using a miRNeasy
serum/plasma kit (cat. no. 217184; Qiagen GmbH), according to the
manufacturers' protocols. miR-145 expression levels were measured
using RT-qPCR, as previously described (11). Briefly, total RNA (50 ng) was
reverse transcribed into cDNA using the miScript II RT kit (cat.
no. 218161; Qiagen GmbH) at 37°C for 60 min and 95°C for 5 min
using a thermal cycler (Bio-Rad Laboratories, Inc.). After dilution
in 200 µl RNase-free water, miRNA amplification was performed using
a miScript SYBR® Green PCR kit (cat. no. 218073; Qiagen,
Inc.) on a CFX96 Touch Real-time qPCR Detection system (Bio-Rad
Laboratories, Inc.). The miScript primer assay used for miR-145 and
U6 small nuclear RNA 2 (RNU6-2, an internal control) was purchased
from Qiagen, Inc. Mature miRNA sequences were used for designing
the forward primers: miR-145, 5′-GUCCAGUUUUCCCAGGAAUCCCU-3′ (cat.
no. MS00003528; Qiagen GmbH) and RNU6-2,
5′-CGCTTCGGCAGCACATATACTA-3′ (cat. no. MS00033740; Qiagen GmbH).
The reverse primer was specifically designed for use with and
contained in the miScript SYBR Green PCR kits (cat. no. 218073;
Qiagen, Inc.). The qPCR amplification conditions were as follows:
95°C for 15 min for polymerase activation, followed by 50 cycles of
denaturation at 94°C for 15 sec, amplification at 55°C for 30 sec
and extension at 70°C for 30 sec. The difference between the cycle
quantification (Cq) value of miR-145 and RNU6-2 (∆Cq) was
calculated and the relative expression levels were quantified using
the 2−∆∆Cq method (23).
Detection of mRNA expression using
RT-qPCR
Total RNA was extracted from the cells using TRIzol
reagent, according to the manufacturer's protocol. To synthesize
cDNA, 50 ng total RNA was reverse transcribed using iScript™
Reverse Transcription SuperMix (cat. no. 1708840; Bio-Rad
Laboratories, Inc.) at 46°C for 20 min and 95°C for 1 min using a
thermal cycler. qPCR was subsequently performed using a
Luna® Universal qPCR Master Mix (cat. no. M3003L; New
England BioLabs, Inc.). The following thermocycling conditions were
used for the amplification of CDK4, cyclin A, CDK1 and GAPDH: 95°C
for 5 min for polymerase activation, followed by 40 cycles of
denaturation at 95°C for 10 sec, amplification at 60°C for 30 sec
and extension at 72°C for 30 sec. A CFX96 Touch Real-time qPCR
Detection system was used for the analysis and the relative
expression levels of the genes were calculated using the
2−∆∆Cq method (24),
using GAPDH as the endogenous control. The following primer
sequences were used for qPCR: CDK4, forward
5′-GGAGGCCTTTGAACATCCCA-3′ and reverse 5′-ACTGGCGCATCAGATCCTTA-3′;
cyclin A, forward 5′-TAGACACCGGCAACTCAAG-3′ and reverse
5′-TCTTCAGACTGGGAGAGGAGA-3′; CDK1, forward
5′-TGGAGAAGGTACCTATGGAGTTG-3′ and reverse
5′-AGGAACCCCTTCCTCTTCAC-3′; and GAPDH, forward,
5′-GACTTCAACAGCGACACCCACTCC-3′ and reverse
5′-AGGTCCACCACCCTGTTGCTGTAG-3′.
ELISA
Serum VEGF levels were measured using a sandwich
human ELISA kit (cat. no. 900-K10; PeproTech, Inc.) according to
the manufacturer's instructions. Briefly, an ELISA plate was coated
with 100 µl capture antibody and incubated overnight at room
temperature. After blocking with 1% BSA (cat. no. A9418;
Sigma-Aldrich; Merck KGaA) for 1 h at room temperature, 100 µl
serum sample was added to each well, followed by 100 µl
HRP-conjugated detection antibody, after which the reaction was
incubated for 1 h at room temperature. After washing, colorimetric
detection was performed using an avidin-HRP conjugate (1:2,000)
using ABTS liquid substrate solution (cat. no. 002024; Thermo
Fisher Scientific, Inc.) for 10 min. Color development was measured
at a wavelength of 405 nm, with a reference wavelength of 650 nm,
using a microplate reader (Molecular Devices, LLC). VEGF levels
were quantified using the constructed standard curve.
Cell viability assay
Cell suspension aliquots were mixed with an equal
amount of 0.4% (w/v) trypan blue solution (cat. no. 15250061;
Gibco; Thermo Fisher Scientific, Inc.) and the viable cells were
counted under light microscopy using a hemocytometer. The number of
cells in the cell group was considered as 100%, whereas the number
of cells in the other groups was expressed as a percentage of that
compared with the cell group.
In addition, a cell suspension was prepared at a
concentration of 1×106 cells/ml in culture medium and a
50-µl cell suspension was mixed with 450 µl Muse® Count
& Viability reagent (cat. no. 637365; MilliporeSigma). The
mixed cells were incubated at room temperature in the dark for 5
min, then the live and dead cells were counted using a Muse cell
analyzer (MilliporeSigma). The number of total cells in the cell
group was considered as 100%, whereas the number of total cells in
the other groups was expressed as a percentage of that compared
with the cell group. The percentage of live and dead cells in a
population was determined.
Cell cycle analysis
After transfection, cells were trypsinized and
centrifuged at 300 × g for 5 min at room temperature. Subsequently,
1×106 cells were fixed with ice-cold 70% methanol (cat.
no. 67-56-1; RCI Labscan Ltd.) overnight at −20°C. Fixed cells were
centrifuged at 300 × g for 5 min and washed with 500 µl cold PBS.
The cells were subsequently resuspended in 1 ml 1X binding buffer
and a 100-µl cell suspension (1×105 cells) was mixed
with 5 µl PI (cat. no. 556463; BD Biosciences) and 1 µl RNase A,
DNase and protease-free (10 mg/ml; cat. no. EN0531; Invitrogen;
Thermo Fisher Scientific, Inc.). After incubation for 15 min at
room temperature in the dark, 400 µl 1X binding buffer was added to
the cells, and the cell cycle distribution was analyzed using an
Amnis® ImageStream®X Mk II and IDEAS
software, version 6.0 (both Luminex Corporation). The percentage of
cells distributed in each phase of the cell cycle was
determined.
Statistical analysis
Statistical analysis was performed using R
statistical software, version 3.4.4 (RStudio, Inc.) and GraphPad
Prism 5.0 software (GraphPad Software, Inc.). The experiments were
performed in triplicate. Clinical characteristics of the patients
were presented as numbers and percentages. The continuous data of
the cell viability assay and cell cycle analysis, which were
obtained from three independent experiments, were presented as the
mean ± SD. Serum miR-145 and VEGF levels were presented as the
median and interquartile range (IQR). The association between
clinical variables and patient status (NSCLC and OLDs) was analyzed
using a χ2 test. The statistical differences between
multiple groups were determined using one-way ANOVA followed by
Tukey's post hoc test. Logistic regression was used to evaluate the
variables associated with the cancer diagnosis. The diagnostic
performance was assessed using a receiver operating characteristic
(ROC) curve and area under the curve (AUC) analysis, which was
reported with 95% CIs. The cut-off values for miR-145 and VEGF were
obtained from the best coordinate in the ROC to provide a maximum
sum of sensitivity and specificity. For the survival analysis, a
Kaplan-Meier curve was constructed and the statistical differences
in overall survival (OS) among the various categories of variables
were determined using a log-rank test. A multivariate Cox
proportional hazards model was used to identify independent
prognostic variables. P<0.05 was considered to indicate a
statistically significant difference.
Results
Demographic and clinical characteristics
of the patients
The present study included 215 patients, 106 with
NSCLC and 109 with OLDs. The diagnoses for the patients with OLDs
were tuberculosis, bronchiectasis, interstitial lung disease,
pneumonia or chronic obstructive pulmonary disease. Table I lists the clinicopathological
characteristics of the patients, and shows the significant
differences identified in sex (P=0.030), age (P<0.001) and
smoking status (P<0.001) between the NSCLC and control groups.
There were no significant differences identified between the two
groups for alcohol consumption and family history of cancer.
 | Table I.Demographic and clinical
characteristics, and miR-145 and VEGF levels among the study
participants. |
Table I.
Demographic and clinical
characteristics, and miR-145 and VEGF levels among the study
participants.
|
| Number of patients
(%) |
|
|---|
|
|
|
|
|---|
| Variable | OLD (n=109) | NSCLC (n=106) | P-value |
|---|
| Sex |
|
| 0.030 |
|
Male | 55 (50.5) | 69 (65.1) |
|
|
Female | 54 (49.5) | 37 (34.9) |
|
| Age, years |
|
| <0.001 |
|
<60 | 57 (52.3) | 31 (29.2) |
|
|
≥60 | 52 (47.7) | 75 (70.8) |
|
| Smoking status |
|
| <0.001 |
|
Non-smoker | 69 (63.3) | 41 (38.7) |
|
|
Smoker | 40 (36.7) | 65 (61.3) |
|
| Alcohol
drinking |
|
| 0.760 |
|
Non-drinker | 67 (61.5) | 63 (59.4) |
|
|
Drinker | 42 (38.5) | 43 (40.6) |
|
| Family history of
cancer |
|
| 0.649 |
| No | 74 (67.9) | 75 (70.8) |
|
|
Yes | 35 (32.1) | 31 (29.2) |
|
| Histology |
|
|
|
|
ADE | - | 77 (72.6) |
|
|
SCC | - | 22 (20.8) |
|
|
Uncertain NSCLC | - | 7 (6.6) |
|
| Clinical stage |
|
|
|
| I | - | 9 (8.5) |
|
| II | - | 5 (4.7) |
|
|
III | - | 12 (11.3) |
|
| IV | - | 80 (75.5) |
|
| miR-145
expression |
|
| 0.002 |
|
Low | 20 (18.3) | 39 (36.8) |
|
|
High | 89 (81.7) | 67 (63.2) |
|
| VEGF
levela |
|
| <0.001 |
|
Low | 66 (72.5) | 41 (44.6) |
|
|
High | 25 (27.5) | 51 (55.4) |
|
Serum miR-145 and VEGF levels
The median serum miR-145 level was lower in the
NSCLC group (17.15; IQR, 1.50-136.72) compared with that in the
control group (25.46; IQR, 4.56-109.90). The median VEGF level was
higher in the NSCLC group (0.04; IQR, 0.02-0.05) compared with that
in the control group (0.02; IQR, 0.00-0.03). The cut-off values for
miR-145 and VEGF levels were set as follows: ≤3.434 for miR-145 and
≤0.032 for VEGF for the lower value and >0.434 for miR-145 and
>0.032 for VEGF for the upper value. A larger proportion of the
NSCLC group had lower miR-145 levels compared with those in the
control group (36.8 vs. 18.3%; P=0.002; Table I). By contrast, a larger proportion
of the NSCLC group had higher VEGF levels compared with those in
the control group (55.4 vs. 27.5%; P<0.001).
Diagnostic performance of serum miR-145
and VEGF
The logistic regression analysis revealed that
smoking status, and miR-145 and VEGF levels were significant
predictors for diagnosing NSCLC (Table II). Age also exhibited a moderate
trend as a predictor (P=0.023). Low miR-145 expression levels
[adjusted odds ratio (aOR), 0.26; 95% CI, 0.12-0.57] and high VEGF
expression levels (aOR, 3.19; 95% CI, 1.64-6.20) were associated
with a NSCLC diagnosis.
 | Table II.Logistic regression analysis of
predictors of the diagnosis of non-small cell lung carcinoma. |
Table II.
Logistic regression analysis of
predictors of the diagnosis of non-small cell lung carcinoma.
| Variable | Crude OR (95%
CI) | Adjusted OR (95%
CI) | P-value |
|---|
| Sex |
|
|
|
| Male
vs. female | 0.57
(0.32-1.03) |
|
|
| Age |
|
|
|
| ≥60 vs.
<60 years | 2.43
(1.33-4.43) | 2.16
(1.11-4.21) | 0.023 |
| Smoking status |
|
|
|
| Smoker
vs. non-smoker | 2.38
(1.31-4.30) | 2.92
(1.48-5.76) | 0.002 |
| Alcohol
drinking |
|
|
|
| Drinker
vs. non-drinker | 1.13
(0.62-2.04) |
|
|
| Family history of
cancer |
|
|
|
| Yes vs.
no | 0.80
(0.43-1.50) |
|
|
| miR-145
expression |
|
|
|
| High
vs. low | 0.31
(0.15-0.61) | 0.26
(0.12-0.57) | <0.001 |
| VEGF level |
|
|
|
| High
vs. low | 3.28
(1.77-6.09) | 3.19
(1.64-6.20) | <0.001 |
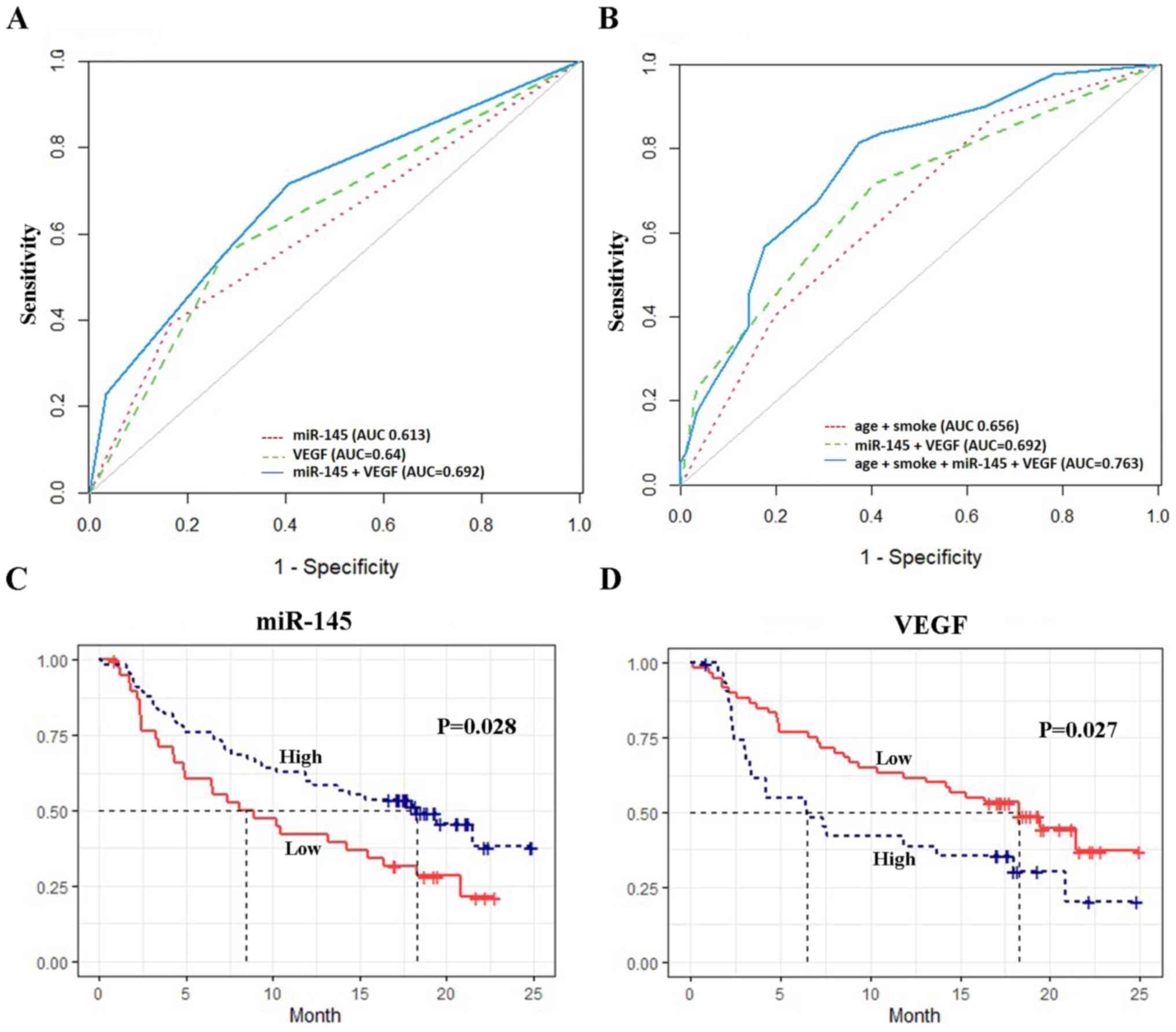
The diagnostic power of miR-145 and VEGF, with or
without other risk factors, was evaluated using ROC curve analysis.
The AUC was 0.61 (95% CI, 0.55-0.68) for miR-145 alone and 0.64
(95% CI, 0.57-0.71) for VEGF alone. When combined, miR-145 and VEGF
improved the discrimination, with an AUC of 0.69 (95% CI,
0.62-0.76) (Fig. 1A). The
combination of age, smoking status, miR-145 and VEGF expression
proved to be the best model for differentiating patients with NSCLC
from those with OLDs (AUC, 0.76; 95% CI, 0.69-0.83), with an
optimal sensitivity and specificity of 80.4 and 65.9%, respectively
(Table III; Fig. 1B).
 | Table III.Discriminative performance of
miR-145, VEGF and clinical factors for the diagnosis of non-small
cell lung carcinoma. |
Table III.
Discriminative performance of
miR-145, VEGF and clinical factors for the diagnosis of non-small
cell lung carcinoma.
|
| Diagnostic
performance |
|---|
| Variable | AUC (95% CI) | Sensitivity (95%
CI) | Specificity (95%
CI) |
|---|
| miR-145 | 0.61
(0.55-0.68) | 0.39
(0.29-0.49) | 0.84
(0.76-0.91) |
| VEGF | 0.64
(0.57-0.71) | 0.55
(0.45-0.65) | 0.73
(0.63-0.81) |
| miR-145 + VEGF | 0.69
(0.62-0.76) | 0.70
(0.27-0.80) | 0.64
(0.52-0.97) |
| Age + Smoking | 0.76
(0.69-0.83) | 0.80
(0.55-0.89) | 0.66
(0.54-0.87) |
| + miR-145 +
VEGF |
|
|
|
Association between serum miR-145 and
VEGF levels and OS
The patients had a median survival time of 14.24
months. The Kaplan-Meier curve showed that low miR-145 levels were
associated with a significantly shorter OS (P=0.028), whereas low
VEGF levels were associated with a longer OS (P=0.027) (Fig. 1C and D). Table IV shows the results of the
multivariate Cox regression analysis. High miR-145 expression was
independently associated with a higher OS [hazard ratio (HR), 0.48;
95% CI, 0.27-0.85]. By contrast, high VEGF levels were associated
with a lower OS (HR, 1.47; 95% CI, 0.81-2.68); however, the
association was not statistically significant. Stage and
histological types were also significantly associated with OS,
whereas age, smoking status, drinking status and family history of
cancer were not.
 | Table IV.Cox regression analysis for overall
survival in non-small cell lung carcinoma (n=106). |
Table IV.
Cox regression analysis for overall
survival in non-small cell lung carcinoma (n=106).
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
Male | Reference |
|
|
|
|
|
|
Female | 0.67 | 0.39-1.16 | 0.152 |
|
|
|
| Age |
|
|
|
|
|
|
| <60
years | Reference |
|
|
|
|
|
| ≥60
years | 0.89 | 0.52-1.55 | 0.696 |
|
|
|
| Smoking status |
|
|
|
|
|
|
|
Non-smoker | Reference |
|
|
|
|
|
|
Smoker | 1.56 | 0.92-2.64 | 0.097 |
|
|
|
| Drinking
status |
|
|
|
|
|
|
|
Non-drinker | Reference |
|
|
|
|
|
|
Drinker | 1.58 | 0.96-2.6 | 0.069 |
|
|
|
| Family history of
cancer |
|
|
|
|
|
|
| No | Reference |
|
|
|
|
|
|
Yes | 0.70 | 0.39-1.26 | 0.237 |
|
|
|
| Staging |
|
|
|
|
|
|
|
I–II | Reference |
|
|
|
|
|
|
III–IV | 3.63 | 1.31-10.06 | 0.013 | 3.13 | 1.10-8.90 | 0.013 |
| Histologic
type |
|
|
|
|
|
|
|
ADC | Reference |
|
|
|
|
|
|
SCC | 1.13 | 0.62-2.07 | 0.694 | 1.13 | 0.62-2.07 | 0.693 |
|
Uncertain NSCLC | 3.10 | 1.31-7.34 | 0.010 | 3.09 | 1.31-7.34 | 0.010 |
| miR-145
expression |
|
|
|
|
|
|
|
Low | Reference |
|
|
|
|
|
|
High | 0.58 | 0.35-0.95 | 0.030 | 0.48 | 0.27-0.85 | 0.012 |
| VEGF level |
|
|
|
|
|
|
|
Low | Reference |
|
|
|
|
|
|
High | 1.84 | 1.07-3.18 | 0.029 | 1.47 | 0.81-2.68 | 0.203 |
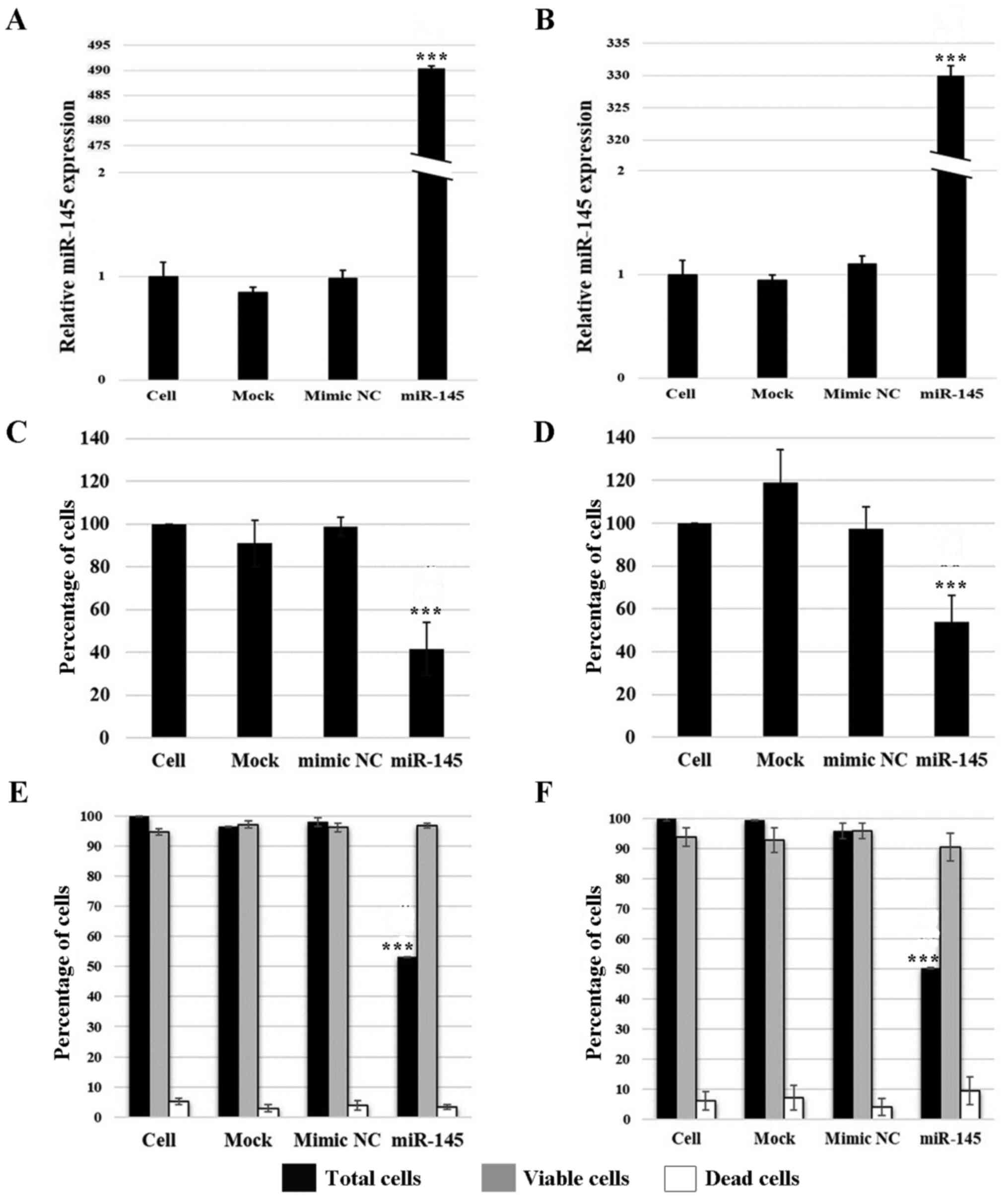
Effects of upregulated miR-145 expression
on cell viability
The functional role of miR-145 in A549 and H1792
adenocarcinoma cells was further explored following its
overexpression. Transfection success was confirmed by measuring
miR-145 expression levels using RT-qPCR. The results revealed that
miR-145 expression levels were upregulated by 494- and 330-fold in
transfected A549 and H1792 cells, respectively, compared with the
cell group (P≤0.001; Fig. 2A and
B). The expression of miR-145 was not significantly different
among the control groups.
As determined using the trypan blue assay, miR-145
overexpression resulted in a 58.5% reduction in total A549 cell
numbers and a 42.9% reduction in H1792 cell numbers compared with
the cell group (P≤0.001; Fig. 2C and
D). Similarly, the results of the cell viability assay using
the Muse cell analyzer revealed that the total cell numbers were
significantly decreased following the overexpression of miR-145, by
~45.0% for A549 and 49.4% for H1792 cells compared with those in
the cell group (P≤0.001; Fig. 2E and
F). However, miR-145 overexpression had no effect on the number
of viable and dead cells compared with in the control cells.
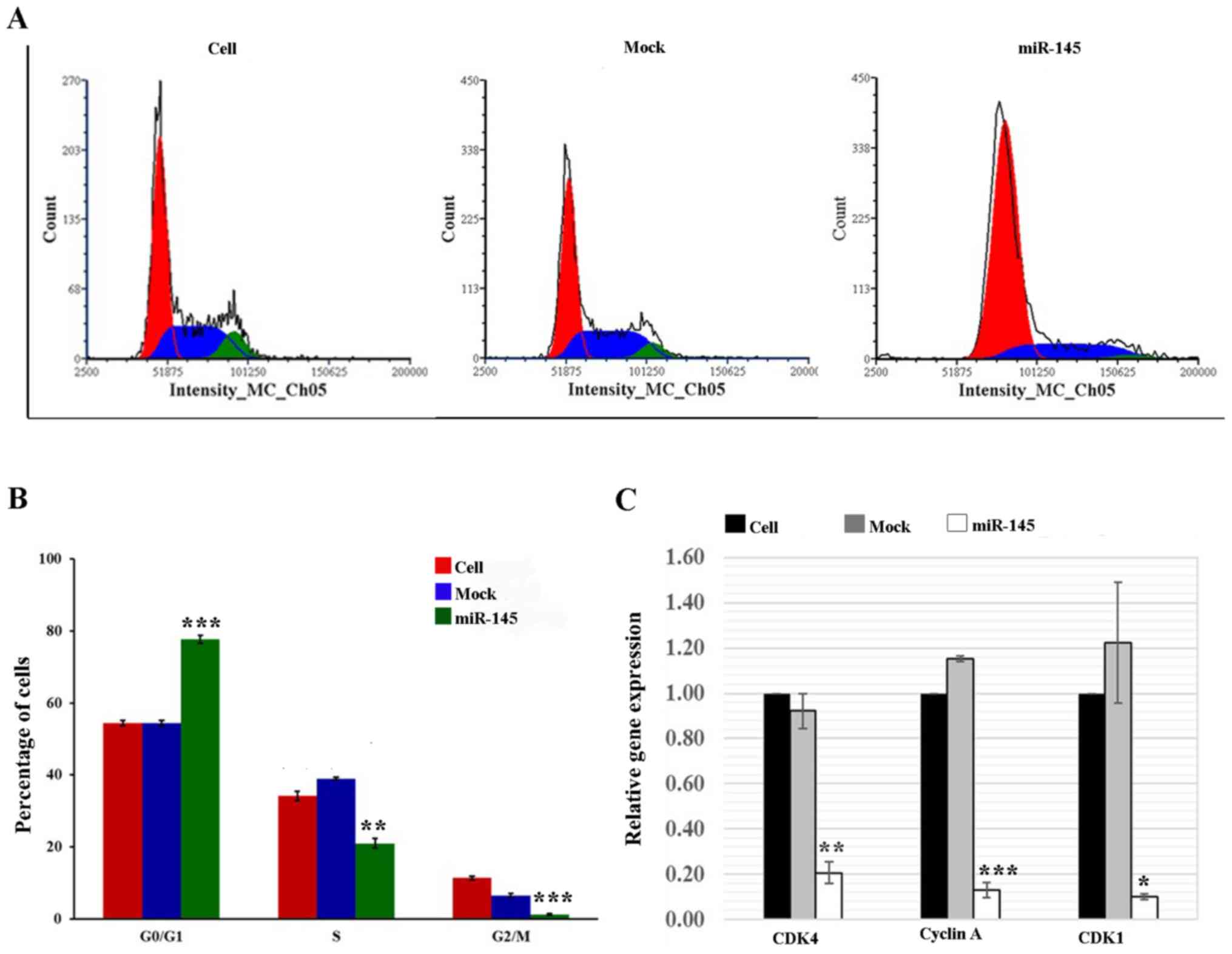
Effects of upregulated miR-145 expression
on the cell cycle distribution
To further explore whether a reduction in total cell
numbers following miR-145 overexpression was caused by cell cycle
arrest, cell cycle analysis was performed using flow cytometry.
Since there were no significant differences among the three control
groups (data not shown), untreated cells and mock cells were used
as the controls for subsequent experiments on the effects of
miR-145 overexpression on cell cycle distribution and the
expression of cell cycle regulatory genes. In A549 cells, the
results revealed that the percentage of cells in
G0/G1 phase was significantly increased by
42.7% (P≤0.001) in the miR-145 mimic-transfected cells compared
with the cell group. Significant reductions in the number of cells
in the S-phase (38.5%; P≤0.01) and G2/M-phase (88.9%; P≤0.001) were
also observed compared with the cell group (Fig. 3A and B). By contrast, the miR-145
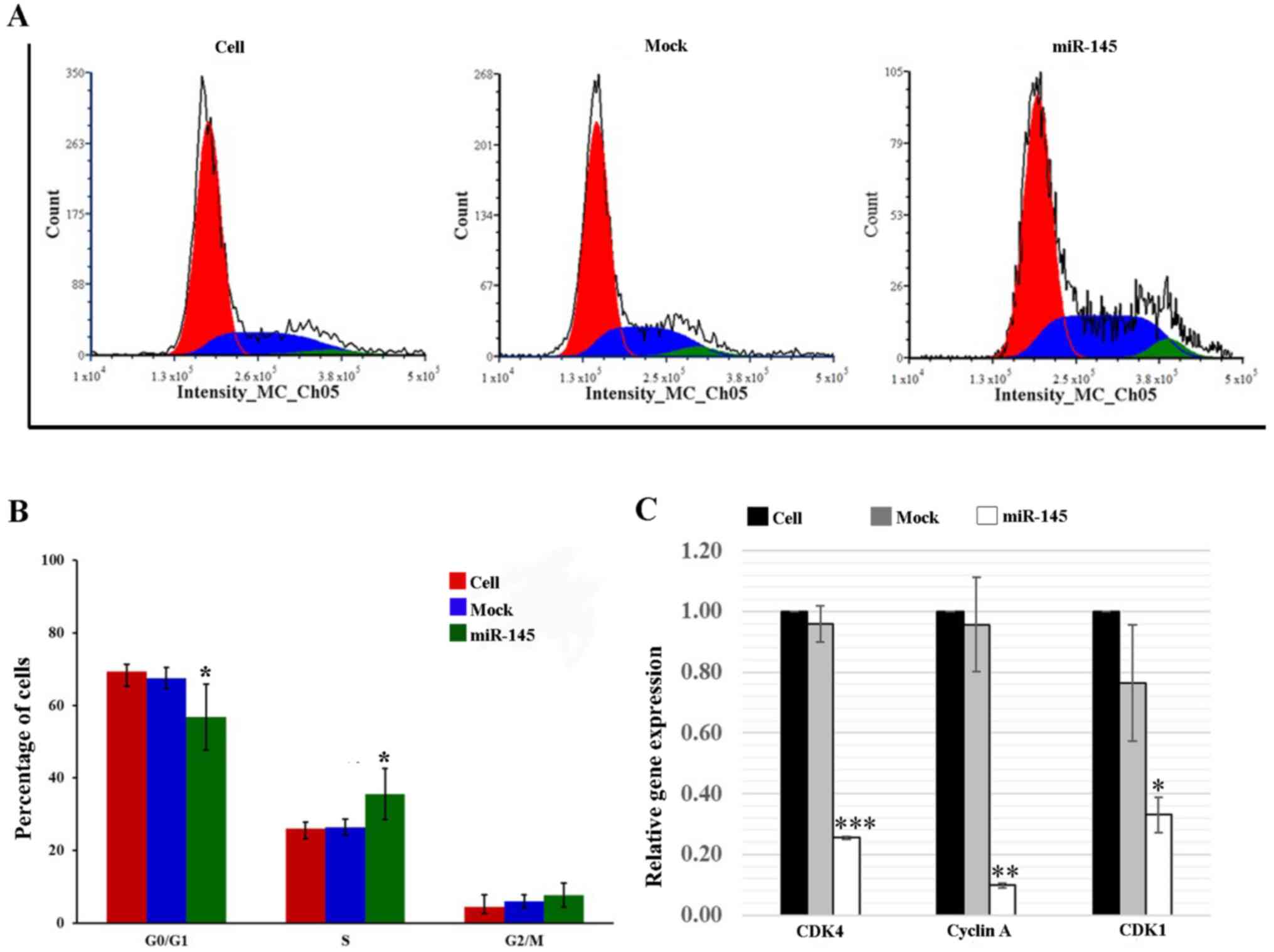
mimic-transfected H1792 cells exhibited an 18.1% decrease in the
percentage of cells in G0/G1 phase compared
with that in the cell group (P≤0.05), and a 36.4% increase in the
percentage of cells in the S phase compared with in the cell group
(P≤0.05; Fig. 4A and B).
Effects of upregulated miR-145 expression
on cell cycle regulatory genes
In A549 cells, the expression levels of CDK4, cyclin
A and CDK1 were significantly downregulated in the miR-145
mimic-transfected cells by 81.0 (P≤0.01), 88.6 (P≤0.001) and 91.5%
(P≤0.05), respectively, compared with those in the cell group
(Fig. 3C). Similar to the A549
cells, there was a significant reduction in CDK4 (76.1%; P≤0.001),
cyclin A (91.7%; P≤0.01) and CDK1 (68.5%; P≤0.05) expression in the
miR-145 mimic-transfected H1792 cells when compared with the cell
group (Fig. 4C).
Discussion
The results of the present study demonstrated that
miR-145 and VEGF may be useful in terms of their clinical use as
diagnostic and prognostic biomarkers in NSCLC. The data revealed
that combining the evaluation of patient clinical factors,
including age and smoking status, with the measurement of serum
miR-145 and VEGF levels was the best model for differentiating
patients with NSCLC from those with OLDs, yielding an AUC of 0.76
with 80.4% sensitivity and 65.9% specificity. The results also
revealed that downregulated miR-145 and upregulated VEGF levels
were significantly associated with poor survival, and that miR-145
expression was an independent prognostic biomarker for NSCLC. The
in vitro experimental results also revealed that miR-145
overexpression reduced cell viability via the induction of cell
cycle arrest.
Serum miR-145 expression was significantly decreased
in patients with NSCLC compared with that in the control group.
miR-145 expression has also been shown to be reduced in the serum
of patients with glioblastoma (12) and ovarian cancer (13) when compared with healthy controls.
However, the results of the present study differ from those of Wang
et al (15), which reported
higher serum miR-145 levels in patients with NSCLC (n=70) compared
with those in healthy controls (n=70). The discrepancy may be due
to the difference in histological ratio. The study by Wang et
al (15) included 48 patients
with adenocarcinoma and 20 with squamous cell carcinoma, whereas
the present study included 77 patients with adenocarcinoma and 22
patients with squamous cell carcinoma. Similar to serum miR-145
levels, the trend in serum VEGF levels in the current study was
similar to those of other studies. Numerous studies have reported
higher serum VEGF levels in patients with cancer compared with in
healthy individuals, including within breast (25), gastrointestinal (26), ovarian (14) and lung (27) cancer. Similarly, Zhang et al
(27) revealed that VEGF levels
were upregulated in patients with lung cancer compared with those
with tuberculosis.
For diagnostic evaluations, the present study
results demonstrated that the AUC of serum miR-145 was 0.61. By
contrast, a study by Wang et al (15) indicated that serum miR-145 was a
good biomarker for differentiating patients with NSCLC from healthy
controls, with an AUC of 0.84 (15). The AUC for VEGF in the present
study was 0.64, which is similar to that measured in a study by
Chakra et al (16), which
reported an AUC value of 0.66 (16). The conflicting findings between the
present results and those of the study by Wang et al
(15) may be due to the use of a
different control; the present study enrolled patients with OLDs as
the control group, whereas the study by Wang et al included
healthy individuals as the control group.
In the present study, clinical risk factors (age and
smoking status) were measured alongside miR-145 and VEGF, and an
increased AUC value was observed for the predictive model (AUC,
0.76), which is consistent with the findings of other studies
(20,28). For example, in a previous study,
the combination of 24 miRNAs with major lung cancer risk factors,
such as sex, age and smoking status, was discovered to improve the
AUC to 0.94 compared with using the risk factors in isolation (AUC,
0.72) (28). Similar to the
present study, our previous study showed that the AUC value of the
combined clinical factors and miR-339-3p was increased to 0.71
compared with the AUC of miR-339-3p alone, which was 0.62 (20). This result suggested that a
combination of biomarkers and risk factors may provide valuable
insights for predictive accuracy.
In terms of the prognostic role, the findings of the
present study revealed that low serum miR-145 and high VEGF levels
were associated with poor survival in NSCLC. Zhang et al
(12) revealed that patients with
glioblastoma with low serum miR-145 levels had a shorter survival
compared with those with high miR-145 levels. Similarly, Liang
et al (13) demonstrated
that low miR-145 expression was associated with the prognosis of
ovarian cancer. These results indicated that miR-145 may be
involved in lung tumorigenesis and may serve as an invasive
prognostic biomarker for NSCLC. For VEGF, the findings of the
present study identified no significant role as an independent
prognostic biomarker for NSCLC. Previous studies have indicated
that high serum VEGF levels may be an independent prognostic factor
in ovarian (14) and
gastrointestinal cancer (26). In
lung cancer, two studies have indicated that high VEGF levels were
useful for predicting a shorter OS (16,29).
By contrast, a study by Akın Kabalak et al (18) showed that serum VEGF levels did not
significantly predict the prognosis of lung cancer, either in NSCLC
or small cell lung cancer (18).
Similarly, Zhang et al (27) found no differences in OS between
patients with high and low serum VEGF levels in lung cancer. The
results of the current study are consistent with those of the two
latter studies, suggesting that high VEGF levels were not an
independent prognostic biomarker. The small number of patients with
NSCLC investigated in the present study may be one of the reasons
for this lack of significance.
In the present study, the biological function of
miR-145 in NSCLC was also examined. The results revealed that
miR-145 led to a decrease in cell number, with no effect on cell
death. Furthermore, miR-145 overexpression induced cell cycle
arrest at the G0/G1-phase for A549 cells and at the S-phase for
H1792 cells. This may indicate that miR-145 affected the cell cycle
at different phases depending on the cell type. In addition, the
present results revealed that miR-145 inhibited the cell cycle
progression by modulating the expression of cell cycle regulators,
including CDK1, CDK4 and cyclin A, in both A549 and H1792 cells.
These results are consistent with previous studies from other
groups, which reported that miR-145 prevented cell proliferation in
gastric cancer (30),
hepatocellular carcinoma (31) and
NSCLC (32–34). In addition, previous studies have
shown that increased miR-145 expression inhibited invasion and
metastasis in numerous types of cancer, including osteosarcoma
(35), hepatocellular carcinoma
(31), nasopharyngeal carcinoma
(36) and lung cancer (8). Taken together, the findings of the
present study suggested that miR-145 may serve a crucial role in
NSCLC as a tumor suppressor miRNA.
Nevertheless, there are several limitations to the
current study that should be addressed in future research. Firstly,
transcript levels only are not sufficient to predict protein
levels; however, due to budget restrictions, only the alterations
in gene expression levels were detected. Cell cycle analysis was
performed and gene expression of related cell cycle regulators was
determined, and the results suggested a potential role of miR-145
in cell cycle regulation. Regarding the clinical implications of
miR-145 and VEGF, the ideal cancer diagnostic biomarker would be
most beneficial if the cancer could be detected as early as
possible. This type of study should include a substantial
proportion of early-stage disease; however, in the present study,
>80% of the patients had late-stage disease, which may lead to
an overestimated diagnostic power. However, OLDs were included as
control subjects rather than healthy individuals, as this may have
caused an overestimation of the diagnostic performance. Therefore,
using the current study design, the resultant diagnostic power
estimation, to a certain extent, was balanced.
In conclusion, the findings of the present study
demonstrated that patients with NSCLC had lower serum miR-145
expression levels and higher serum VEGF levels compared with those
in patients with OLDs. Serum miR-145 expression was identified to
be a useful diagnostic and prognostic marker in NSCLC, whereas
serum VEGF was only discovered to have potential as a diagnostic
biomarker. The present study, which was based on findings obtained
from clinical samples and in vitro experiments, suggested a
role for miR-145 as a tumor suppressor in NSCLC. In addition, to
the best of our knowledge, the present study led to the innovative
finding that combined evaluation of miR-145 and VEGF enhanced the
diagnostic performance of lung cancer diagnosis. However, further
studies regarding the molecular target and specific biological
pathway of miR-145 are warranted.
Acknowledgements
Not applicable.
Funding
The present study was funded by grants from the Faculty of
Medicine, Prince of Songkla University (grant no. REC6131242) and
the Prince of Songkla University, Songkhla, Thailand (grant no.
MED61020002S).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PR, PT and SLG conceived and designed the study. PR,
PC and KT developed and designed the methodology. SLG and WK
acquired the clinical data and managed the patients. PR, SB and PT
performed the statistical analysis and interpreted the data. PR and
PT supervised the study. PR and PT confirmed the authenticity of
all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The experiments were approved by the Human Research
Ethics Committee of the Faculty of Medicine, Prince of Songkla
University (approval nos. 59-011-05-1 and 60-350-04-2). Patients
provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
miRNA
|
microRNA
|
|
VEGF
|
vascular endothelial growth factor
|
|
OLDs
|
other lung diseases
|
|
AUC
|
area under the curve
|
|
ROC
|
receiver operating characteristic
|
|
OS
|
overall survival
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cui M, Wang H, Yao X, Zhang D, Xie Y, Cui
R and Zhang X: Circulating MicroRNAs in Cancer: Potential and
Challenge. Front Genet. 10:6262019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hirales Casillas CE, Flores Fernández JM,
Padilla Camberos E, Herrera López EJ, Leal Pacheco G and Martínez
Velázquez M: Current status of circulating protein biomarkers to
aid the early detection of lung cancer. Future Oncol. 10:1501–1513.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu W, Chang J, Du X and Hou J: Long
non-coding RNA PCAT-1 contributes to tumorigenesis by regulating
FSCN1 via miR-145-5p in prostate cancer. Biomed Pharmacother.
95:1112–1118. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Wen X, Hu XL, Cheng LZ, Yu JY and
Wei ZB: Downregulation of miR-145-5p correlates with poor prognosis
in gastric cancer. Eur Rev Med Pharmacol Sci. 20:3026–3030.
2016.PubMed/NCBI
|
|
7
|
Chang Y, Yan W, Sun C, Liu Q, Wang J and
Wang M: miR-145-5p inhibits epithelial-mesenchymal transition via
the JNK signaling pathway by targeting MAP3K1 in non-small cell
lung cancer cells. Oncol Lett. 14:6923–6928. 2017.PubMed/NCBI
|
|
8
|
Wang M, Wang J, Deng J, Li X, Long W and
Chang Y: MiR-145 acts as a metastasis suppressor by targeting
metadherin in lung cancer. Med Oncol. 32:3442015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kieran MW, Kalluri R and Cho YJ: The VEGF
pathway in cancer and disease: Responses, resistance, and the path
forward. Cold Spring Harb Perspect Med. 2:a0065932012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zou C, Xu Q, Mao F, Li D, Bian C, Liu LZ,
Jiang Y, Chen X, Qi Y, Zhang X, et al: MiR-145 inhibits tumor
angiogenesis and growth by N-RAS and VEGF. Cell Cycle.
11:2137–2145. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chaniad P, Trakunran K, Geater SL,
Keeratichananont W, Thongsuksai P and Raungrut P: Serum miRNAs
associated with tumor-promoting cytokines in non-small cell lung
cancer. PLoS One. 15:e02415932020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Ta WW, Sun PF, Meng YF and Zhao
CZ: Diagnostic and prognostic significance of serum miR-145-5p
expression in glioblastoma. Int J Clin Exp Pathol. 12:2536–2543.
2019.PubMed/NCBI
|
|
13
|
Liang H, Jiang Z, Xie G and Lu Y: Serum
microRNA-145 as a novel biomarker in human ovarian cancer. Tumour
Biol. 36:5305–5313. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Komatsu H, Oishi T, Itamochi H, Shimada M,
Sato S, Chikumi J, Sato S, Nonaka M, Sawada M, Wakahara M, et al:
Serum vascular endothelial growth factor-A as a prognostic
biomarker for epithelial ovarian cancer. Int J Gynecol Cancer.
27:1325–1332. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang RJ, Zheng YH, Wang P and Zhang JZ:
Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in
non-small cell lung cancer. Int J Clin Exp Pathol. 8:765–771.
2015.PubMed/NCBI
|
|
16
|
Chakra M, Pujol JL, Lamy PJ, Bozonnat MC,
Quantin X, Jacot W and Daurès JP: Circulating serum vascular
endothelial growth factor is not a prognostic factor of non-small
cell lung cancer. J Thorac Oncol. 3:1119–1126. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shen H, Shen J, Wang L, Shi Z, Wang M,
Jiang BH and Shu Y: Low miR-145 expression level is associated with
poor pathological differentiation and poor prognosis in non-small
cell lung cancer. Biomed Pharmacother. 69:301–305. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Akın Kabalak P, Çiledağ A, Demir N, Çelik
G, Yüksel C, Köycü G, Gökmen Öztuna D, Taner A, Kaya A, Kutlay H,
et al: Prognostic significance of serum vascular endothelial growth
factor and Angiopoietin-2 in patients with lung cancer. Tuberk
Toraks. 63:71–77. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bossuyt PM: Clinical validity: Defining
biomarker performance. Scand J Clin Lab Invest Suppl. 242:46–52.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Trakunram K, Chaniad P, Geater SL,
Keeratichananont W, Chittithavorn V, Uttayamakul S, Buya S,
Raungrut P and Thongsuksai P: Serum miR-339-3p as a potential
diagnostic marker for non-small cell lung cancer. Cancer Biol Med.
17:652–663. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Travis WD, Brambilla E, Nicholson AG,
Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E,
Flieder DB, et al WHO Panel, : The 2015 World Health Organization
Classification of Lung Tumors: Impact of genetic, clinical and
radiologic advances since the 2004 classification. J Thorac Oncol.
10:1243–1260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: the 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rao X, Huang X, Zhou Z and Lin X: An
improvement of the 2ˆ(−delta delta CT) method for quantitative
real-time polymerase chain reaction data analysis. Biostat
Bioinforma Biomath. 3:71–85. 2013.PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li S, Wang L, Meng Y, Chang Y, Xu J and
Zhang Q: Increased levels of LAPTM4B, VEGF and survivin are
correlated with tumor progression and poor prognosis in breast
cancer patients. Oncotarget. 8:41282–41293. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang L, Hochwald S, Deng S, Chen Y, Tan
C, Zhong Q and Huang H: Diagnostic and prognostic performance of
serum vascular endothelial growth factor, vascular endothelial
growth factor receptor 2, and osteopontin for gastrointestinal
cancers. Clin Lab. 65:652019. View Article : Google Scholar
|
|
27
|
Zhang Y, Yu LK, Lu GJ, Xia N, Xie HY, Hu
W, Hao KK, Xu CH and Qian Q: Prognostic values of VEGF and
endostatin with malignant pleural effusions in patients with lung
cancer. Asian Pac J Cancer Prev. 15:8435–8440. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wozniak MB, Scelo G, Muller DC, Mukeria A,
Zaridze D and Brennan P: Circulating microRNAs as non-invasive
biomarkers for early detection of non-small-cell lung cancer. PLoS
One. 10:e01250262015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shimanuki Y, Takahashi K, Cui R, Hori S,
Takahashi F, Miyamoto H and Fukurchi Y: Role of serum vascular
endothelial growth factor in the prediction of angiogenesis and
prognosis for non-small cell lung cancer. Lung. 183:29–42. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zeinali T, Karimi L, Hosseinahli N,
Shanehbandi D, Mansoori B, Mohammadi A, Hajiasgharzadeh K, Babaloo
Z, Majidi-Zolbanin J and Baradaran B: Overexpression of miRNA-145
induces apoptosis and prevents proliferation and migration of
MKN-45 gastric cancer cells. EXCLI J. 19:1446–1458. 2020.PubMed/NCBI
|
|
31
|
Ding W, Tan H, Zhao C, Li X, Li Z, Jiang
C, Zhang Y and Wang L: MiR-145 suppresses cell proliferation and
motility by inhibiting ROCK1 in hepatocellular carcinoma. Tumour
Biol. 37:6255–6260. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pan Y, Ye C, Tian Q, Yan S, Zeng X, Xiao
C, Wang L and Wang H: miR-145 suppresses the proliferation,
invasion and migration of NSCLC cells by regulating the BAX/BCL-2
ratio and the caspase-3 cascade. Oncol Lett. 15:4337–4343.
2018.PubMed/NCBI
|
|
33
|
Ye Z, Shen N, Weng Y, Li K, Hu L, Liao H,
An J, Liu L, Lao S and Cai S: Low miR-145 silenced by DNA
methylation promotes NSCLC cell proliferation, migration and
invasion by targeting mucin 1. Cancer Biol Ther. 16:1071–1079.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen Z, Zeng H, Guo Y, Liu P, Pan H, Deng
A and Hu J: miRNA-145 inhibits non-small cell lung cancer cell
proliferation by targeting c-Myc. J Exp Clin Cancer Res.
29:1512010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fan L, Wu Q, Xing X, Wei Y and Shao Z:
MicroRNA-145 targets vascular endothelial growth factor and
inhibits invasion and metastasis of osteosarcoma cells. Acta
Biochim Biophys Sin (Shanghai). 44:407–414. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li YQ, He QM, Ren XY, Tang XR, Xu YF, Wen
X, Yang XJ, Ma J and Liu N: MiR-145 inhibits metastasis by
targeting fascin actin-bundling protein 1 in nasopharyngeal
carcinoma. PLoS One. 10:e01222282015. View Article : Google Scholar : PubMed/NCBI
|