Introduction
Breast cancer is a disease characterised by
molecular heterogeneity (1,2).
Gene expression profiling has identified molecular subtypes of
breast cancer with differences in prognosis and therapeutic
options. In particular, the HER2-enriched subtype is characterised
by upregulation of the HER2 gene (3), while the HER2 gene is amplified in
15–20% of all breast carcinomas (4).
The HER2/neu protein is a component of a four-member
family of closely related growth factor receptors, including EGFR
or HER1 (ERBB1), HER2 (ERBB2), HER3 (ERBB3) and HER4 (ERBB4). HER
receptors exist as monodimers, and they can form homodimers or
heterodimers when they bind to a ligand. Numerous ligands are
associated with HER1, HER3 and HER4, while HER2 is characterised as
an ‘orphan’ receptor, since there is no known ligand that can
promote homodimers of HER2. In terms of the HER ligands, epidermal
growth factor (EGF), transforming growth factor α, the heparin
binding EGF-like growth factor (HBEGF), betacellulin (BTC),
amphiregulin (AREG) and epiregulin (EREG) bind to the HER1 receptor
(5). Neuregulin 1 (NRG1) and
neuregulin 2 bind to HER3, while HBEGF, BTC, EREG and neuregulins
1, 2, 3 and 4 bind to HER4 (6,7).
The importance of HER2 as a prognostic or predictive
marker in invasive breast cancer is well recognised and, therefore,
HER2 status should be determined in all cases of invasive (early
stage or recurrent) breast cancer (8,9).
HER2 testing includes the evaluation of HER2 protein upregulation
assessed by immunohistochemistry (IHC) and HER2 gene amplification
assessed by in situ hybridisation, fluorescence in
situ hybridisation (FISH), chromogenic in situ
hybridisation, silver-enhanced in situ hybridisation or
quantitative PCR (10). Although
mRNA expression profiling has been used to classify breast tumours
into molecular subtypes, assessment of oestrogen receptors
(ER)/progesterone receptors (PgR)/HER2 status via IHC is currently
the standard in clinical practice for the selection of patients
that are more likely to respond to hormone or anti-HER2 treatments,
according to international guidelines (11–13).
Previous work has demonstrated that ERBB/HER
ligands, including AREG, BTC, EREG, EGF, HBEGF, NRG1 and
transforming growth factor α (TGFA), are co-expressed at the mRNA
level in breast cancer in various combinations alongside their
receptors, whereas associations have also been established among
the mRNA levels of the aforementioned ligands and the four HER
receptors (14,15). Furthermore, the mRNA expression
patterns of EGF, AREG, TGFA and HBEGF have been linked to
clinicopathological parameters, including tumour size and
histoprognostic grading (14).
However, in the case of EGF, an association with improved prognosis
has also been observed for overall survival and relapse-free
survival, at least in univariate analyses (14). Specific ERBB/HER ligands, have been
linked to treatment response with anti-HER2 therapeutic agents,
including trastuzumab, as shown by preclinical or clinical studies
(16–18).
Although most HER2-positive patients derive benefit
from trastuzumab and other approved anti-HER2 targeted therapies,
resistance will eventually develop and cause disease progression
(19). Furthermore, there is a
subset of HER2-positive patients that will not respond to
trastuzumab due to primary resistance (18). Therefore, it is crucial to identify
biomarkers that will allow for the categorisation of patients most
likely to either respond to or develop primary and secondary
resistance to trastuzumab.
Apart from the HER ligands, the mRNA levels of other
molecules, including insulin-like growth factor binding protein 4
(IGFBP4), a member of the family of proteins binding to
insulin-like growth factors, transforming growth factor β1 (TGFB1),
a TGFβ signalling component, the thyroid hormone receptor α (THRA),
and the retinoic acid receptor α (RARA), have been studied in
breast cancer for their prognostic significance (20–25). The
present study retrospectively examined the mRNA expression of
several HER ligands and other potential prognostic biomarkers of
interest, including IGFBP4, TGFB1, THRA and RARA, in patients with
metastatic breast cancer (MBC) who were treated with trastuzumab.
Their relationship with other tumour and pathological
characteristics, as well as ER/PgR/HER2 status, assessed centrally
by IHC, was evaluated and their prognostic role in terms of
progression and survival in this subset of patients was
explored.
Materials and methods
Patients and tissue processing
The present study was conducted on a previously
reported group of patients (26)
that was enriched with additional cases to achieve a total study
population of 145 cases (all female). The eligibility criteria were
as follows: i) Treatment with trastuzumab for histologically
confirmed MBC; ii) adequate clinical data on patient history,
demographics, tumour characteristics, treatment details (i.e., drug
dosages, schedule of administration and serious toxicities) and
clinical outcome; and iii) adequate tumour tissue available for
biological marker evaluation.
Formalin-fixed paraffin-embedded tumour tissue
samples were retrospectively collected from patients treated with
trastuzumab-based regimens in the metastatic setting, as previously
described in detail (26–28). All tumours were characterised by
local pathologists as HER2-positive based on American Society of
Clinical Oncology/College of American Pathologists (ASCO/CAP)
criteria current at that time (29). Consequently, all patients received
trastuzumab as part of their treatment. All tumour samples were
centrally re-evaluated by IHC for oestrogen receptors (ER),
progesterone receptors (PgR), HER2 and the expression of the
proliferation marker Ki67. Additionally, HER2 amplification status
was assessed using the FISH method, as later described.
The translational research protocol was approved by
the Bioethics Committee of the Aristotle University of Thessaloniki
School of Medicine (Protocol #4283; January 14, 2008; Thessaloniki,
Greece) under the general title ‘Investigation of major mechanisms
of resistance to treatment with trastuzumab in patients with
metastatic breast cancer’. All patients included in the study in
2005 and later provided written informed consent for the provision
of biological material for future research studies before receiving
any treatment. A waiver of consent was obtained from the Bioethics
Committee for patients included in the study before 2005.
Tissue microarrays (TMAs)
Representative haematoxylin-eosin-stained 2-µm
sections from the tissue blocks were reviewed by a pathologist. A
total of 17 TMA blocks were constructed from the 145 eligible cases
using a manual tissue microarrayer (Beecher Instruments), as
previously described (26). For
the construction of the TMA blocks, two core samples (1.5 mm in
diameter) were obtained from representative regions of each tumour
in the donor blocks. All IHC and FISH markers were assessed on the
TMA sections. The cases not represented, which had damaged or
inadequate cores on the TMA sections, were re-cut from the original
blocks if still available. These sections were used for protein and
gene analyses as previously described (26–28,30). Further method
details are provided in Data
S1.
Immunohistochemistry (IHC)-tumor
infiltrating lymphocytes (TILs)
Serial 2.5-µm-thick TMA sections or whole tissue
sections were stained for ER (clone 6F11; cat. no. NCL-L-ER-6F11;
Leica Microsystems, Ltd.), PgR (clone 1A6; cat. no. NCL-L-PGR;
Leica Microsystems, Ltd.), HER2 (polyclonal Ab; cat. no. A0485;
Dako; Agilent Technologies, Inc.), Ki67 (clone MIB-1; cat. no.
M7240; Dako; Agilent Technologies, Inc.), phosphorylated mTOR at
serine 2448 (p-mTOR; clone 49F9; cat. no. 2976; Cell Signaling
Technology, Inc.; dilution, 1:30; 20 min) and stPTEN (clone 6H2.1;
cat. no. M3627; Dako; Cell Signaling Technology, Inc.; dilution,
1:200; 30 min), using the Bond Max™ autostainer (Leica
Microsystems, Ltd.) as previously described in detail (31) (Table
SI). All sections were stained in one run for each antibody and
were evaluated by pathologists experienced in breast cancer and
blinded to the patient's clinical and survival data. Positive
controls were used for all antibodies from known positive breast
cancer cases, while negative controls were obtained by omitting the
primary antibody as previously described (26–28,30).
All IHC stains were evaluated according to the
formerly outlined interpretation (32). Stromal TIL density was assessed on
whole H&E sections according to the guidelines from the
International TILs Working Group (33) and was analysed as a continuous
variable. HER2 protein expression was scored according to the 2007
ASCO/CAP guidelines (29) (scores
between 0 and 3+). A positive for HER2 protein expression (IHC 3+)
was defined as uniform intense membrane staining of >30% of
invasive tumour cells. Further method details are provided in
Data S1.
Fluorescence in situ hybridisation
(FISH)
TMA sections or whole tissue sections (5-µm-thick)
were used for FISH analysis using the ZytoLight® SPEC
HER2/TOP2A/CEN17 triple colour probe kit for HER2 (code Z-2093;
ZytoVision GmbH). FISH was performed according to the
manufacturer's protocol with minor modifications (pepsin solution
was applied to the specimens and incubated for 11–12 min at 37°C in
a humidified chamber) in all cases (i.e., not only the HER2 IHC 2+
cases). Digital images were constructed using software specifically
developed for cytogenetics (XCyto-Gen, ver 1.2.11; Alphelys) and
evaluated as previously described (26,31).
For the assessment of HER2 status, the 2007 ASCO/CAP guidelines
(29) were used with the addition
of the ≥6-HER2-copies criterion (34), as patients had locally received
trastuzumab according to this classification. HER2 status was
considered positive if HER2 was amplified by FISH and/or a HER2
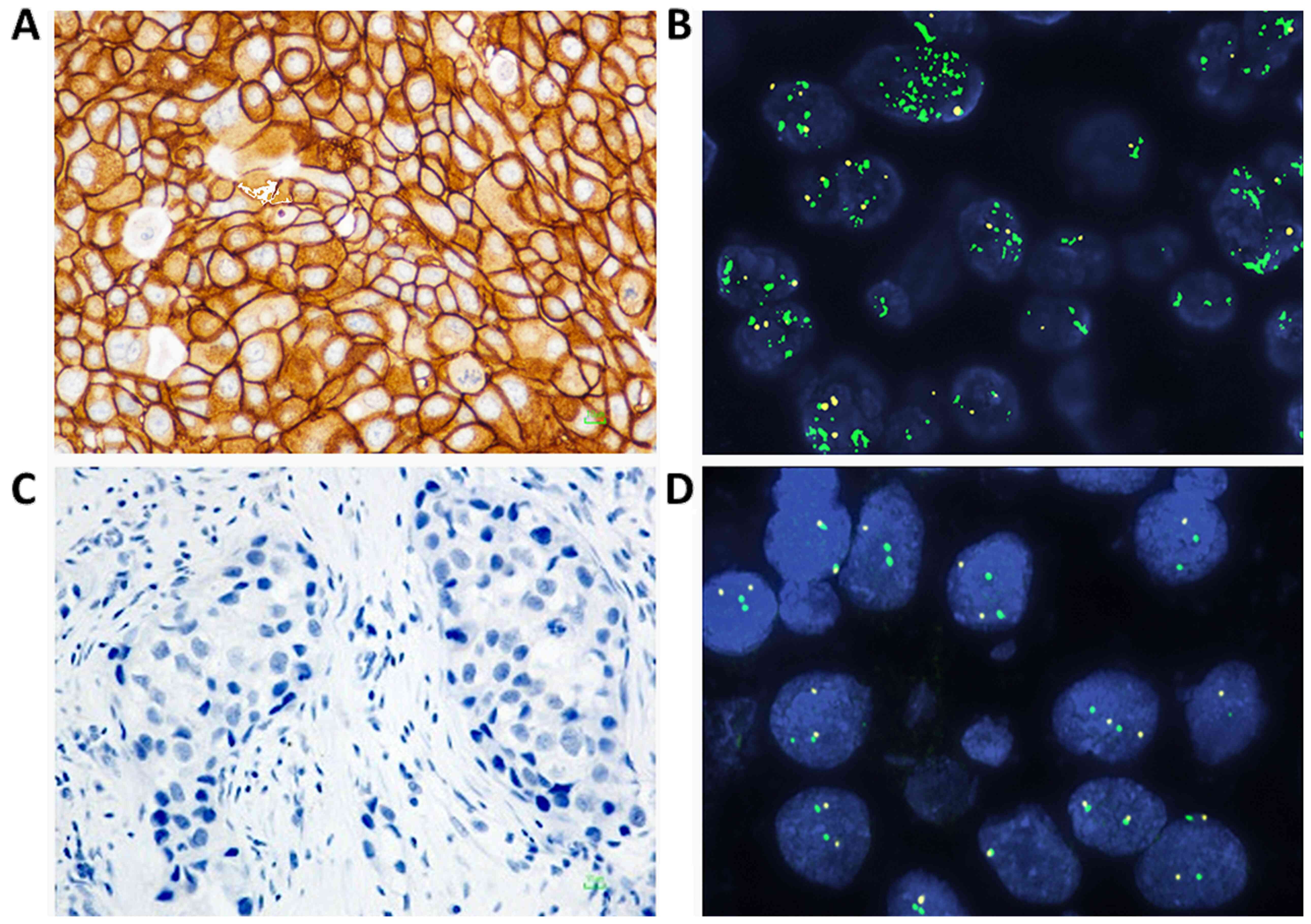
score of 3+ was obtained by IHC. Representative IHC and FISH images
from invasive breast carcinoma cases are presented in Fig. 1. Further method details are
provided in Data S1.
Dual nucleic acid extraction and mRNA
expression analysis
Simultaneous isolation of DNA and RNA from whole or
macrodissected 10-µm paraffin sections, the latter in cases with
<50% tumour cell content, was performed for the 145 examined
tumours with iron oxide beads coated with a nanolayer of silica
using the VERSANT Tissue Preparation Reagents kit (Siemens
Healthcare Diagnostics) as previously described (35). The nucleic acid extract of each
sample was divided into two aliquots. DNase I was then added to one
aliquot in order to remove DNA and ensure the presence of pure RNA
for downstream mRNA analyses. cDNA synthesis was performed with
random primers and SuperScript III Reverse Transcriptase (catalogue
number 48190011 and 18080044; Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. cDNAs were
assessed in duplicate with quantitative PCR using an ABI7900HT
system under default thermal cycling conditions as follows: 50°C
for 2 min, 95°C for 10 min, followed by 45 cycles at 95°C for 15
sec and 60°C for 1 min, respectively.
mRNA expression analysis was performed with pre-made
exon-spanning TaqMan-MGB assays (Applied Biosystems; Thermo Fisher
Scientific, Inc.) targeting the following gene transcripts (data in
parentheses refer to assay ID; exon boundary; amplicon length):
AREG Hs00950669_m1; 3–4; 66 bp), BTC Hs01101204_m1; 4–5, 5–6; 139
bp), EGF Hs01099999_m1; 19–20, 20–21: 70 bp), EREG ID
Hs00914313_m1; 3–4; 65 bp), HBEGF Hs00181813_m1; 3–4; 78 bp),
IGFBP4 Hs00181767_m1; 1–2: 91 bp), NRG1 Hs00247620_m1; 2–3: 93 bp),
RARA Hs00940446_m1; 4–5, 5–6, 6–7: 68 bp), TGFA Hs00608187_m1; 4–5:
70 bp), TGFB1 Hs00171257_m1; 1–2: 63 bp), THRA Hs00268470_m1; 4–5:
89 bp) (Table I). A TaqMan-MGB
expression assay targeting the glucuronidase β (GUSB) gene
(Hs99999908_m1; 9–10, 10–11, 11–12; 81 bp) was used as the
endogenous reference for relative quantification. The commercially
available TaqMan Control Total RNA (cat. no. 4307281; Applied
Biosystems; Thermo Fisher Scientific, Inc.) was applied as a
positive control for interrun evaluation of the PCR assay
efficiency, together with no-template controls. To obtain linear
relative quantification (RQ) values, relative expression was
assessed as 40-∆Cq, whereby ∆Cq was calculated as (average target
Cq) - (average GUSB Cq) from all eligible measurements, as
previously described (35). Only
samples with average GUSB Cq values <36 and a ∆RQ value for each
duplicate sample pair (intrarun variation) of <1 were considered
eligible for further analysis in the present study.
 | Table I.Premade TaqMan-MGB assays for mRNA
expression analysis. |
Table I.
Premade TaqMan-MGB assays for mRNA
expression analysis.
| Gene symbol | Assay ID, Hs | Size, bp | Exons | Gene name | Genbank ref |
|---|
| AREG | Hs00950669_m1 | 66 | 3-4 | Amphiregulin | NM_001657.3 |
| BTC | Hs01101204_m1 | 139 | 4-5, 5–6 | Betacellulin | NM_001316963.1,
NM_001729.3 |
| EGF | Hs01099999_m1 | 70 | 19-20, 19–20,
20–21 | Epidermal growth
factor | NM_001178130.2,
NM_001178131.2, NM_001963.5 |
| EREG | Hs00914313_m1 | 65 | 3-4 | Epiregulin | NM_001432.2 |
| HBEGF | Hs00181813_m1 | 78 | 3-4 | Heparin binding EGF
like growth factor | NM_001945.2 |
| IGFBP4 | Hs00181767_m1 | 91 | 1-2 | Insulin like growth
factor binding protein 4 | NM_001552.2 |
| NRG1 | Hs00247620_m1 | 93 | 2-3 | Neuregulin 1 | NM_001159995.2,
NM_001159999.2, NM_001160001.2, NM_001160002.1, NM_001160004.2,
NM_001160005.1, NM_001160007.1, NM_001160008.1, NM_013956.4,
NM_013957.4, NM_013958.3, NM_013960.4, NM_013962.2, NM_013964.4,
NM_004495.3 |
| RARA | Hs00940446_m1 | 68 | 4-5, 5–6, 6–7,
6–7 | Retinoic acid
receptor α | NM_001145302.2,
NM_001024809.3, NM_001145301.2, NM_000964.3 |
| TGFA | Hs00608187_m1 | 70 | 4-5 | Transforming growth
factor α | NM_001099691.1,
NM_001308158.1, NM_001308159.1 |
| TGFB1 | Hs00171257_m1 | 63 | 1-2 | Transforming growth
factor β1 | NM_000660.5 |
| THRA | Hs00268470_m1 | 89 | 4-5 | Thyroid hormone
receptor α | NM_001190918.1,
NM_001190919.1, NM_003250.5, NM_199334.3 |
| GUSB | Hs99999908_m1 | 81 | 9-10, 9–10, 10–11,
11–12 | Glucuronidase
β | NM_001284290.1,
NM_001293105.1, NM_001293104.1, NM_000181.3 |
Statistical analysis
Frequencies with the corresponding percentages and
medians with range were used to summarize categorical and
continuous variables, respectively. Comparisons of categorical data
were performed using the χ2 or Fisher's exact test (if
more appropriate), while the non-parametric Wilcoxon rank-sum test
was applied for the comparison of categorical with continuous
variables. Associations of the markers of interest were assessed
using Spearman's correlations.
The median value of the mRNA expression of the
examined markers was used as the optimal cut-off to classify
tumours into high- and low-expression groups and assess their
prognostic significance and the associations with several
clinicopathological characteristics (including age, menopausal
status, histological grade, TIL density, PTEN expression and bone
metastasis). Additionally, we evaluated the upper and lower
quartiles of the mRNA distribution as potential thresholds. Time to
progression (TTP) was defined as the time interval between the
initiation of trastuzumab-based treatment for advanced disease
(with or without parallel administration of chemotherapy or
hormonal therapy) and the first documented disease progression.
Non-progressors were censored at the date of last follow-up.
Survival was also estimated from the initiation of
trastuzumab-based therapy until death (from any cause) or last
follow-up. Time-to-event distributions were estimated with the
Kaplan-Meier method and comparisons of groups were performed with
the log-rank test. Cox proportional hazard regression models
(univariate and multivariate) were applied to estimate the effect
of the examined markers on TTP and survival.
The TTP and survival analyses were conducted
separately in the subgroup of HER2-positive and HER2-negative
patients (as defined by HER2 central assessment), while the
interactions with the ER/PgR status among patients with
HER2-positive tumours and with the disease presentation status in
the entire study population were also assessed for all examined
markers with respect to TTP and survival to detect whether the
effect of the marker expression on patients' outcome varied between
the subgroups defined by ER/PgR (positive vs. negative) and disease
presentation status [relapsed MBC (R-MBC) vs. de novo
MBC].
Model choice was performed in multivariate analyses
using the following variables in the first step: Menopausal status,
performance status, mTOR protein expression, PTEN protein
expression and each one of the markers that showed significance in
univariate analysis. The final model was selected using backward
selection criteria with P<0.10. All tests were two-sided and
significance was set at 5%. The SAS software (version 9.3; SAS
Institute, Inc.) was used for the statistical analyses.
Results
Patient characteristics and
trastuzumab exposure
A total of 145 patients with at least one
measurement in the markers of interest, including HER family
receptor ligands and other potential prognostic biomarkers, were
included in the present study. Among them, 109 (75.2%) had
available mRNA data for all 11 markers. Although all patients were
found to be HER2-positive when assessed at the local institutions
and, therefore, were treated with trastuzumab, only 98 patients
(67.6%) were classified as HER2-positive by the central
re-evaluation of HER2 status. Therefore, 47 patients (32.4%) were
treated with trastuzumab despite being HER2-negative (Fig. 2).
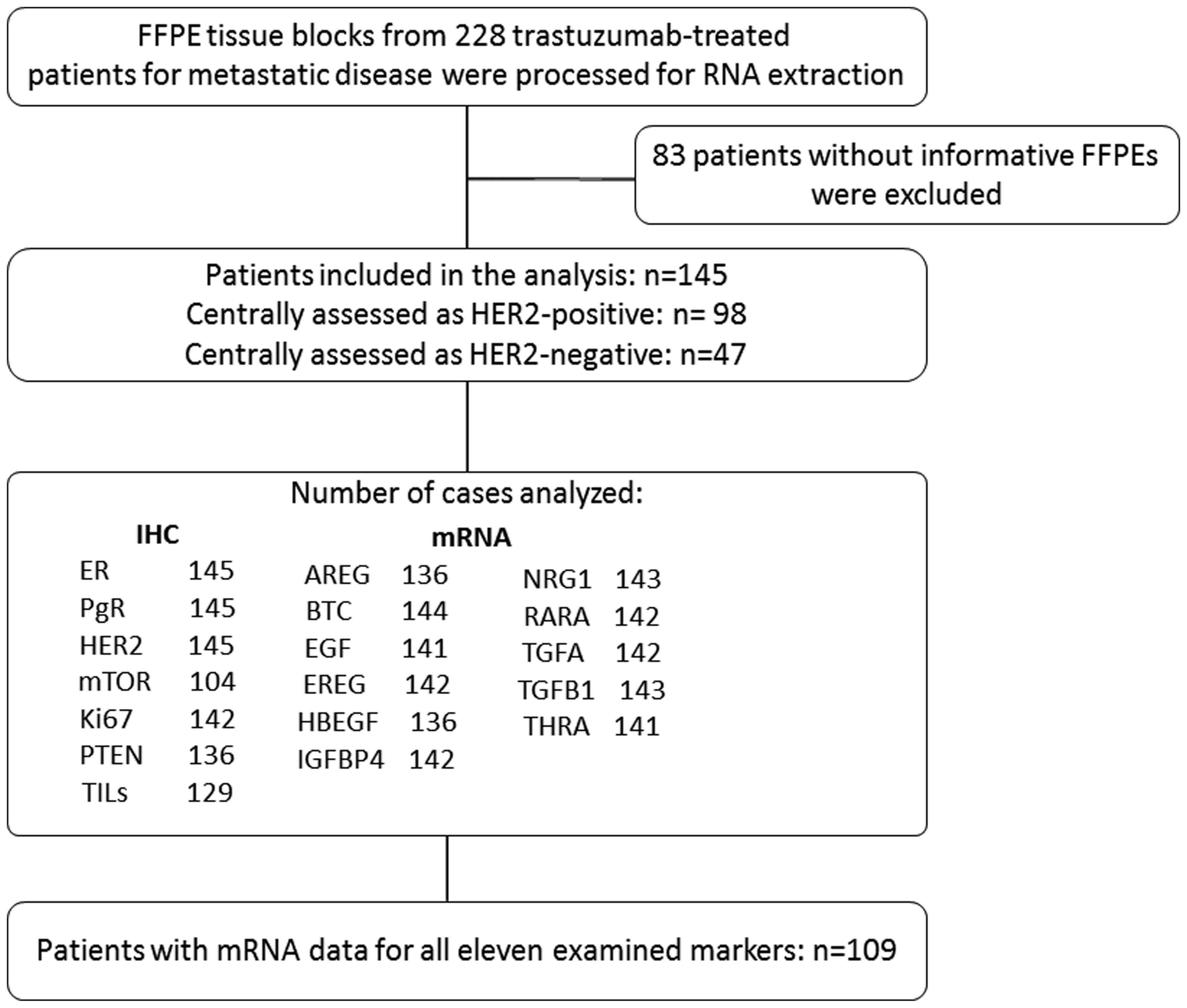 | Figure 2.Reporting recommendations for tumour
marker prognostic studies (REMARK) diagram. AREG, amphiregulin;
BTC, betacellulin; EGF, epidermal growth factor; ER, oestrogen
receptors; EREG, epiregulin; FFPE, formalin-fixed
paraffin-embedded; HBEGF, heparin Binding EGF like growth factor;
IGFBP4, insulin-like growth factor binding protein 4; IHC,
immunohistochemistry; NRG1, neuregulin 1; PgR, progesterone
receptors; RARA, retinoic acid receptor α; TGFA, transforming
growth factor α; TGFB1, transforming growth factor β1; THRA,
thyroid hormone receptor α; TILs, tumour-infiltrating
lymphocytes. |
Selected patient and tumour characteristics for the
entire study population and by HER2 status, as defined by the
central assessment, are shown in Table II. A total of 69 patients (47.6%)
had stage IV disease at the time of diagnosis (de novo MBC),
while 52.4% had R-MBC. Most women were of postmenopausal status at
the time of trastuzumab initiation (72.4%), while the majority of
the patients with R-MBC had received adjuvant chemotherapy (85.5%).
The median age at the initiation of trastuzumab-based therapy was
56 years (range, 29–86 years).
 | Table II.Selected patient and tumour
characteristics in groups of patients divided by HER2 status. |
Table II.
Selected patient and tumour
characteristics in groups of patients divided by HER2 status.
|
| HER2 status (by
central assessment) |
|---|
|
|
|
|---|
|
Characteristics | Total (n=145) | Negative
(n=47) | Positive
(n=98) |
|---|
| Median age, years
(range)a | 56 (29–86) | 58 (33–76) | 54 (29–86) |
| De novo MBC,
n (%) | 69 (47.6) | 18 (38.3) | 51 (52.0) |
| R-MBC, n (%) | 76 (52.4) | 29 (61.7) | 47 (48.0) |
| History of adjuvant
CT, n (%)b | 65 (85.5) | 26 (89.7) | 39 (83.0) |
|
Anthracycline-based adjuvant
CT, n (%)b | 52 (68.4) | 20 (69.0) | 32 (68.1) |
|
Taxane-containing CT, n
(%)b | 33 (43.4) | 9 (31.0) | 24 (51.1) |
|
CMF-based CT, n
(%)b | 32 (42.1) | 15 (51.7) | 17 (36.2) |
| History of adjuvant
HT, n (%)b | 46 (60.5) | 20 (69.0) | 26 (55.3) |
| History of adjuvant
RT, n (%)b | 43 (56.6) | 16 (55.2) | 27 (57.4) |
| Histological grade,
n (%) |
|
I–II | 50 (34.5) | 19 (40.4) | 31 (31.6) |
|
III | 88 (60.7) | 25 (53.2) | 63 (64.3) |
|
Unknown | 7 (4.8) | 3 (6.4) | 4 (4.1) |
| Menopausal status,
n (%)a |
|
Premenopausal | 39 (26.9) | 13 (27.7) | 26 (26.5) |
|
Postmenopausal | 105 (72.4) | 34 (72.3) | 71 (72.4) |
|
Unknown | 1 (0.7) | 0 (0.0) | 1 (1.0) |
| N of trastuzumab
lines, n (%) |
| 1 | 59 (40.7) | 22 (46.8) | 37(37.8) |
| 2 | 32 (22.1) | 10 (21.3) | 22 (22.4) |
| 3 | 24 (16.6) | 6 (12.8) | 18 (18.4) |
| ≥4 | 30 (20.7) | 9 (19.1) | 21 (21.4) |
| Performance status,
n (%)a |
|
0-1 | 137 (94.5) | 41 (87.2) | 96 (98.0) |
|
2-3 | 5 (3.4) | 4 (8.5) | 1 (1.0) |
|
Unknown | 3 (2.1) | 2 (4.3) | 1 (1.0) |
| Subtype
classification, n (%) |
| Luminal
A | 7 (4.8) | 7 (14.9) | 0 (0.0) |
| Luminal
B | 30 (20.7) | 30 (63.8) | 0 (0.0) |
|
Luminal-HER2 | 56 (38.6) | 0 (0.0) | 56 (57.1) |
|
HER2-enriched | 42 (29.0) | 0 (0.0) | 42 (42.9) |
|
TNBC | 8 (5.5) | 8 (17.0) | 0 (0.0) |
|
Unknown | 2 (1.4) | 2 (4.3) | 0 (0.0) |
| N of metastatic
sites, n (%)a |
|
1-3 | 126 (86.9) | 37 (78.7) | 89 (90.8) |
| ≥4 | 16 (11.0) | 8 (17.0) | 8 (8.2) |
|
Unknown | 3 (2.1) | 2 (4.3) | 1 (1.0) |
| Sites of
metastasis, n (%)a |
|
Locoregional | 44 (30.3) | 17 (36.2) | 27 (27.6) |
|
Distant | 127 (87.6) | 41 (87.2) | 86 (87.8) |
| Only
locoregional | 8 (5.5) | 3 (6.4) | 5 (5.1) |
| Only
distant | 90 (62.1) | 27 (27.6) | 63 (64.3) |
|
Bones | 60 (41.4) | 21 (44.7) | 39 (39.8) |
|
Nodes | 28 (19.3) | 10 (21.3) | 18 (18.4) |
|
Visceral metastases | 98 (67.6) | 29 (61.7) | 69 (70.4) |
In total, 132 patients (91.0%) received trastuzumab
as a first-line treatment, while 9 patients (6.2%) were treated
with trastuzumab as a second-line treatment. The remaining patients
received trastuzumab as third- to seventh-line therapy. In 130
patients (89.7%), trastuzumab was administered with chemotherapy,
whereas 13 patients received hormonal therapy at the time of
trastuzumab initiation, and 2 patients received trastuzumab as
monotherapy. In addition, 6 patients (4.1%) had been previously
treated with a trastuzumab regimen in the adjuvant and/or
neoadjuvant setting.
Marker distribution by HER2
status
The distribution of the markers of interest based on
the normalised expression of mRNA-encoding genes is presented in
Fig. S1. The distribution of all
examined markers by HER2 status (based on central assessment) is
presented in Table SII, while
Table SIII presents the
distribution of markers by ER/PgR status among HER2-positive
patients. Table SIV shows the
distribution of markers by disease presentation status for the
entire study population. HER2-positive patients presented with
higher HBEGF, TGFB1 and THRA mRNA expression compared with patients
with HER2-negative tumours (P=0.026, P<0.001 and P<0.001,
respectively; Table SII), while
IGFBP4 mRNA expression was higher in HER2-positive patients with
positive ER/PgR status compared with HER2-positive patients with
ER/PgR-negative tumours (P=0.004; Table SII). No significant differences
were observed in the distribution of the markers of interest
between patients with de novo MBC and R-MBC (Table SIV).
Correlations among HER family receptor
ligands
IGFBP4 was positively strongly correlated with RARA
(rho=0.45; P<0.001), TGFB1 (rho=0.60; P<0.001) and THRA
(rho=0.45; P<0.001). In addition, RARA was strongly and
positively correlated with THRA (rho=0.52; P<0.001). A strong
correlation was also detected between TGFB1 and THRA (rho=0.51;
P<0.001) and between BTC and EREG (rho=0.47; P<0.001;
Fig. 3).
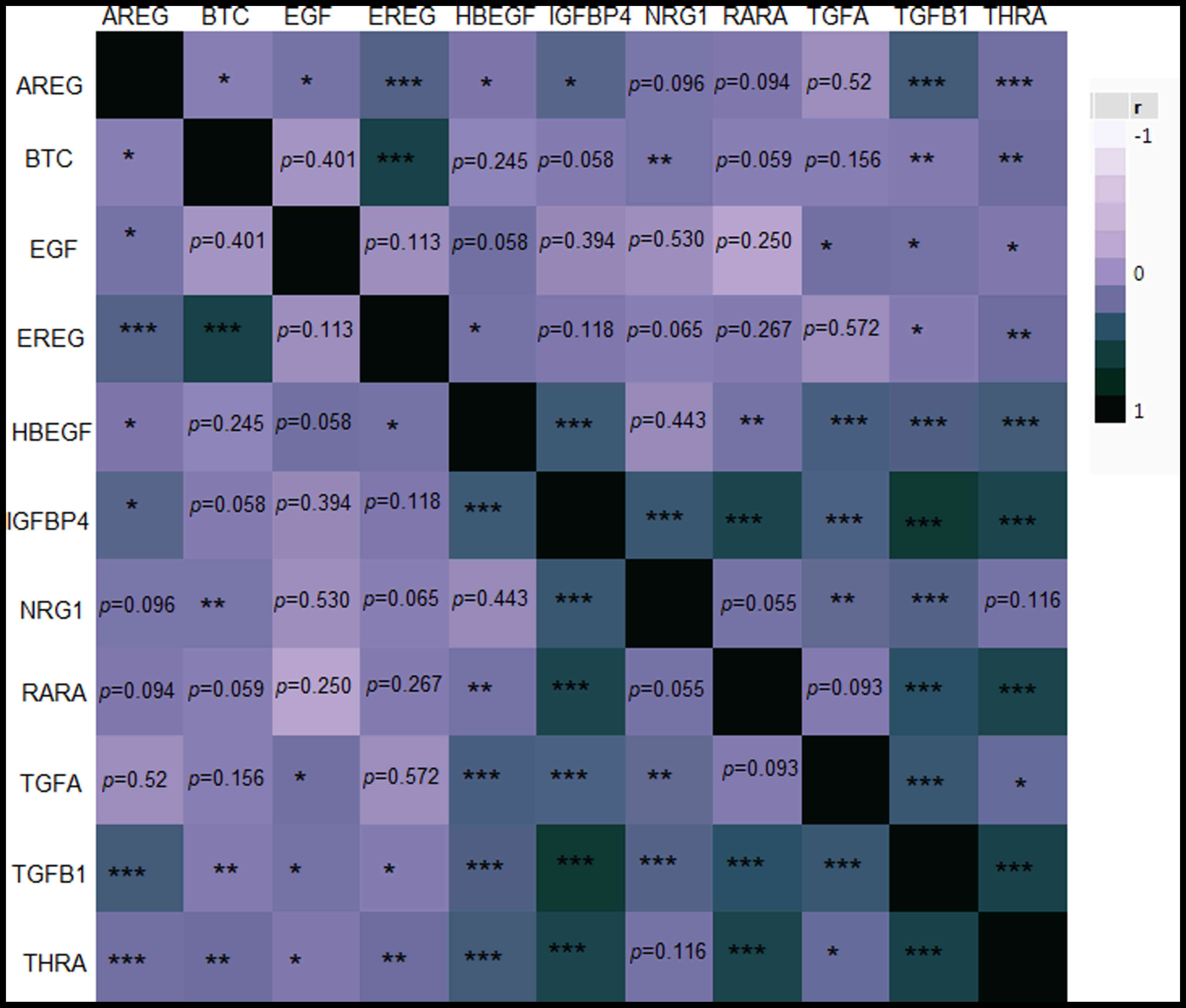 | Figure 3.Heatmap matrix plot showing
Spearman's correlation coefficients (rho) among the examined
markers in the entire cohort. The value of r ranges between −1
(light purple) and 1 (dark green) as explained in the legend
corresponding to negative or positive correlations between the
markers. *P<0.050, **P<0.010 and ***P<0.001, respectively.
Non-significant P-values are stated explicitly. AREG, amphiregulin;
BTC, betacellulin; EGF, epidermal growth factor; EREG, epiregulin;
HBEGF, heparin Binding EGF like growth factor; IGFBP4, insulin-like
growth factor binding protein 4; NRG1, neuregulin 1; r, Spearman's
correlation coefficient; RARA, retinoic acid receptor α; TGFA,
transforming growth factor α; TGFB1, transforming growth factor β1;
THRA, thyroid hormone receptor α. |
Association of HER family ligand
receptors with clinicopathological characteristics
Patients carrying tumours with high AREG mRNA
expression (using the median value as a cut-off point) were of
younger age at the time of trastuzumab initiation and were more
frequently premenopausal compared with those with low AREG mRNA
expression (P=0.002 and P=0.002, respectively). Younger age was
also associated with high NRG1 mRNA expression (P=0.030), while
lower histological grade was associated with high IGFBP4 mRNA
expression (P=0.008). Patients with high mRNA expression levels of
RARA, as compared with those with low expression (using the median
value as a cut-off point), more frequently exhibited bone
metastasis (P=0.012). Higher TIL density was observed in tumours
with high TGFA mRNA expression (using the median value as a cut-off
point), while PTEN loss was associated with low mRNA expression
levels of TGFA and THRA, using the median values as cut-off points
(P=0.043, P=0.045 and P=0.003, respectively; Table SV).
Association of markers with patient
outcome
The median follow-up time for HER2-positive and
HER2-negative patients was 120.3 and 114.2 months, respectively.
During this time, 108 patients (74.5%) died (72 HER2-positive,
73.5%; 36 HER2-negative, 76.6%), while 113 (77.9%) experienced
disease progression (76 HER2-positive, 77.6%; 37 HER2-negative,
78.7%). The median TTP was 15.1 months (95% CI, 12.6-19.6) and 11.6
months (95% CI, 7.1-17.6) for HER2-positive and HER2-negative
patients, respectively. The median survival was 48.5 months (95%
CI, 39.2-59.8) for HER2-positive patients and 38.1 months (95% CI,
25.8-49.1) for HER2-negative patients, while no significant
differences were observed between HER2-positive and HER2-negative
patients in terms of TTP or survival (P=0.33 and P=0.26,
respectively).
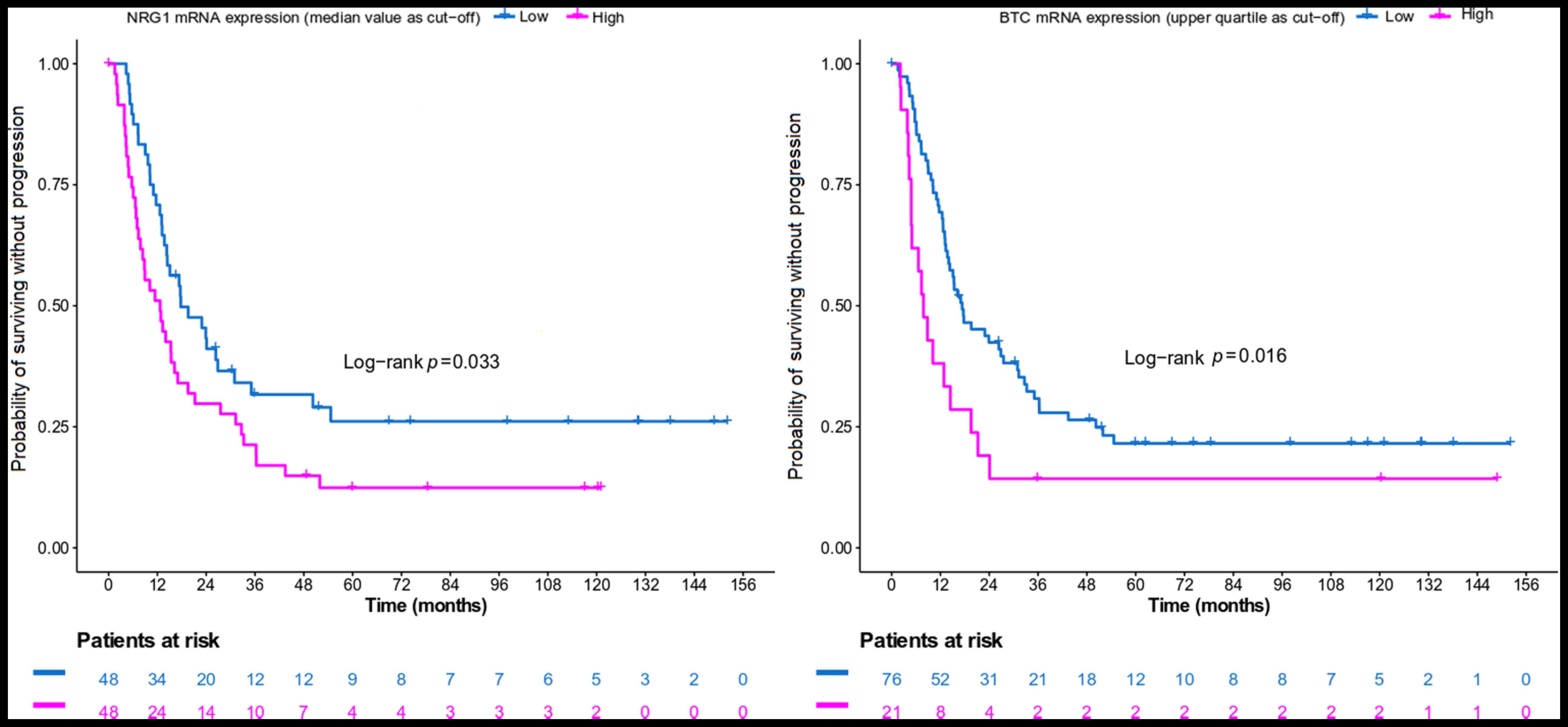
High NRG1 mRNA expression (using the median value as
a cut-off) and high BTC mRNA expression (using the upper quartile
as a cut-off) was associated with an increased risk of progression
in the HER2-positive population (Table III; Fig. 4). In the HER2-negative population,
high EREG mRNA expression (using the median value as a cut-off) was
univariately associated with a decreased risk of progression
(hazard ratio = 0.45; 95% CI, 0.23-0.90; P=0.025). However, this
was not retained upon the adjustment for clinicopathological
parameters (P=0.12).
 | Table III.Hazard ratios and 95% confidence
intervals estimated by univariate and multivariate Cox regression
analyses for TTP and survival. |
Table III.
Hazard ratios and 95% confidence
intervals estimated by univariate and multivariate Cox regression
analyses for TTP and survival.
|
| Univariate |
Multivariatea |
|---|
|
|
|
|
|---|
| Parameter,
endpoint | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| TTP |
|
|
|
|
|
|
|
HER2-positive patientsb |
|
|
|
|
|
|
|
NRG1 mRNA
expression (median as high vs. low) | 1.63 | 1.03-2.57 | 0.035 | 1.78 | 1.03-3.09 | 0.040 |
|
BTC mRNA
expression (upper quartile as cut-off value; high vs. low) | 1.90 | 1.12-3.24 | 0.018 | 2.00 | 1.02-3.93 | 0.043 |
|
Patients with de novo
MBC |
|
|
|
|
|
|
|
EGF mRNA
expression (median value as cut-off; high vs. low) | 0.55 | 0.32-0.94 | 0.029 | 0.52 | 0.25-1.08 | 0.080 |
|
Survival |
|
|
|
|
|
|
|
Patients with
de novo MBC |
|
|
|
|
|
|
|
EGF mRNA
expression (lower quartile as cut-off; high vs. low) | 0.46 | 0.24-0.87 | 0.017 | 0.40 | 0.19-0.87 | 0.020 |
A significant interaction was observed between the
disease presentation status and EGF mRNA expression (using the
median value as a cut-off) for TTP (interaction P=0.037). High EGF
mRNA expression was associated with a decreased risk of progression
among patients with de novo MBC (Table III), while the hazard ratio was
of the opposite direction in the subgroup of R-MBC women, even
though significance was not reached (P=0.43). After adjustment for
clinicopathological parameters, a non-significant trend towards
improved TTP was observed for patients with de novo MBC
carrying tumours with high EGF mRNA expression compared with those
with low expression.
In terms of survival, a significant interaction was
identified between disease presentation status and EGF expression
(using the lower quartile as a cut-off; interaction P=0.045). In
the subgroup of patients with de novo MBC, high EGF
expression was associated with a decreased risk of death (Table III). No significance was reached
among patients with R-MBC (P=0.82).
Discussion
The present study examined the expression of the
ERBB family receptor (ERBB1, ERBB3 and ERBB4) ligands and of other
putative prognostic biomarkers, including THRA, RARA, TGFB1 and
IGFBP4, and their association with clinicopathological parameters
and clinical outcomes in patients treated with trastuzumab-based
therapy for MBC. Tissue samples from the primary tumours were
examined for all patients in the study. In addition, a cohort of
metastatic HER2-negative patients treated with trastuzumab was
assessed. In the present study, 47 patients (32.4%) were found to
be HER2-positive in the local institution and HER2 negative in the
central re-assessment and were treated with anti-HER2 therapy.
In the literature, discordance between local and
central laboratories in HER2 results, using either IHC or FISH, has
been reported (9). Repeat testing
is recommended if results appear contradictory to other
histopathologic findings (8). In
an N9831 study, the concordance for central HercepTest and central
FISH assays was 92%, while a National Surgical Adjuvant Breast and
Bowel Project (NSABP-B) prospective adjuvant trastuzumab study
revealed an 18% discordance rate between local and central
laboratories in HER2 testing (36–38). A number of factors may
contribute to this discordance, such as tumour heterogeneity,
borderline HER2-positive samples, difficulty in evaluating tumours
with chromosome 17 polysomy, methodologic factors, such as antibody
sensitivity, antigen retrieval, tissue processing, lack of
concordance between IHC and FISH, and lack of experience of
pathologists, especially in the early years of trastuzumab use, and
low-volume testing laboratories (36,39,40).
In the present study, additional factors, such as the limited
experience of local pathologists with the IHC HER2 assessment and
the adoption of a 4-point scale of HER2 status for the
administration of trastuzumab, especially during the early years of
its registration, may have contributed to the observed
discordance.
This discordance highlights the main advantage of
the central assessment of HER2 testing. Testing must be performed
in central laboratories, which are able to show high concordance
with a validated HER2 test on a large set of specimens. Expression
of HER2 is a predictive factor of response to anti-HER2 therapies
and therefore, accurate testing of HER2 is of great importance.
According to the NSABP B47 study, HER2-low tumours do not derive
any benefit from the addition of 1 year of trastuzumab to standard
chemotherapy (41). In the present
study, no significant difference was found between HER2-positive
and HER2-negative patients in terms of TTP or survival, a finding
that can be explained by the improvement of prognosis of
HER2-positive tumours due to trastuzumab. It is clear that the use
of trastuzumab improved the outcomes of the HER2-positive breast
tumours and transformed those tumours into less aggressive ones
(42). The question that remains,
however, is whether treatment with trastuzumab equalised the
prognosis between HER2-positive and HER2-negative MBC.
The amplification or upregulation of the HER2
oncogene identifies patients for whom HER2-directed therapy is
appropriate. There is a subset of HER2-positive patients that will
not respond to trastuzumab or other approved anti-HER2 targeted
therapies due to primary or secondary resistance (19). A number of mechanisms of resistance
to trastuzumab have been described, and one of these is increased
signalling from other ERBB family receptors (18,43).
The initial signal is generated by the extracellular ligands of the
ERBB family receptors, which leads to the dimerisation of two HER
family receptors and transphosphorylation of their intracellular
regions, with subsequent activation of a number of downstream
signalling pathways (44).
The present study examined the mRNA expression of
the specific HER family receptor ligands and their association with
patient outcomes. It was demonstrated that NRG1 had a negative
prognostic value in the trastuzumab-treated HER2-positive MBC
population, as high NRG1 mRNA expression was associated with an
increased risk for disease progression in both univariate and
multivariate analyses. In breast cancer, NRG1 is known as a ligand
for the HER3 receptor, which has no intrinsic tyrosine kinase
activity. When activated by NRG1 binding, the HER3 receptor forms a
heterodimer with other HER family receptors and mediates downstream
signalling pathways, leading to multiple effects, including growth,
proliferation, decreased apoptosis, cellular migration and
angiogenesis (45).
In the literature, NRG expression is associated with
poor outcomes and high-risk features of breast carcinoma, as this
gene promotes metastatic dissemination of breast cancer cells
(46). Similarly, a previous study
has revealed that NRG1/HER3 activation is one of the key factors
inducing primary resistance to trastuzumab in HER2-overexpressing
breast cancer cells and that a HER3 antibody may reverse primary
trastuzumab resistance by inhibiting the activation of
NRG1-dependent HER3 (47). The
present results are consistent with the literature and suggest
upregulation of NRG1 as a potential mechanism of resistance to
trastuzumab that leads to an increased risk of disease progression.
This finding is important because it could lead to the development
of novel drugs to overcome the resistance to trastuzumab and other
anti-HER2 treatments.
Another study demonstrated that upregulation of the
NRG1-HER3 axis is a mechanism of resistance in HER2-positive breast
cancer cell lines and xenografts treated with anti-HER2 therapy,
and that multitargeted antibody mixtures, such as Pan-HER, inhibit
the growth of drug-sensitive and drug-resistant HER2+ cancers
(48). In addition, in our cohort
of HER2-positive patients, BTC mRNA expression was found to have a
negative prognostic value for TTP. This was a novel finding
regarding the effect of BTC on the clinical outcome of patients
with HER2-positive disease that should be taken into consideration
and interpreted with caution until it can be further validated in
larger cohorts.
In HER2-negative patients, high EREG mRNA expression
was univariately associated with a decreased risk of progression,
but this was not retained in the multivariate analysis. At the same
time, a significant interaction was detected between EGF mRNA
expression and disease presentation status in all patients with
respect to TTP. More specifically, the univariate analysis revealed
that EGF mRNA might represent a positive prognostic factor for TTP
in de novo metastatic patients. In the multivariate
analysis, a trend associated with a lower risk of progression was
observed for high EGF mRNA, while longer survival was confirmed for
patients with tumours with high EGF mRNA expression in the same
subgroup of patients.
EGF expression in breast cancer is associated with
poor outcomes and aggressive phenotypes, such as low hormone
receptor levels, high proliferation index and HER2 upregulation
(49). In a review article by Ross
and Fletcher (50), EGFR
expression was associated with a higher risk of relapse in patients
with breast cancer. It has been hypothesized that high EGF mRNA is
a potential mechanism of trastuzumab resistance, as the growth
inhibition by trastuzumab in HER2-positive breast cancer cell lines
is blocked by increased levels of the HER family ligands, such as
heregulin and EGF (43,51). Furthermore, there is evidence that
EGF stimulates the synthesis of its own receptor (EGFR) in a human
breast cancer cell line (52).
Additionally, in the North Central Cancer Treatment Group N9831
(Alliance) trial, patients with high expression levels of EGFR
derived a decreased benefit from adjuvant trastuzumab administered
concurrently with chemotherapy (53).
Based on all the aforementioned results, one can
reasonably assume that high EGF mRNA and subsequent activation of
EGFR-mediated signalling pathways act as a potential mechanism of
resistance to trastuzumab and, therefore, should be related to a
higher risk of progression and death in trastuzumab-treated
HER2-positive patients. The results of the present study, however,
are not consistent with the literature, since in the present study,
high EGF mRNA expression was associated with a decreased risk of
progression among patients with de novo MBC. One explanation
could be that de novo (vs. recurrent) metastatic patients
were not pre-treated with trastuzumab and had not developed
resistance to trastuzumab, although this assumption cannot explain
the positive prognostic value of high EGF in this subgroup of
patients. In addition, only a small subset (4.1%) of patients in
the present study received trastuzumab in the adjuvant or
neoadjuvant setting.
Another explanation could be based on the ‘EGFR
paradox’ in primary vs. MBC (54).
According to this paradox, there are fundamental changes in EGFR
signalling in metastases compared with primary breast tumours, such
as EGF-induced apoptosis and EGF-induced growth arrest. This means
that EGFR acts as a tumour driver in primary breast cancer and as a
tumour suppressor in metastatic disease (55). In the present study, in recurrent
metastatic patients, tissue from the primary tumour and not from
the metastases was examined, which could explain the positive
prognostic value of high EGF mRNA only in the de novo
metastatic patients.
The present study is an exploratory study consisting
mainly of hypotheses generated with limited samples. The results
should be further validated in a larger cohort. Among the strengths
of the present study are the long follow-up of patients and the
central assessment of HER2 status, which precludes any
false-positive cases. Furthermore, this is the first study to
collectively evaluate ligands of all three HER2 receptors in
patients with MBC.
In conclusion, the present results provided evidence
suggesting that there may be an association between HER family
ligand expression and clinical outcome in patients with
HER2-positive and HER2-negative MBC who received trastuzumab-based
therapy. High NGR1 mRNA expression was a negative prognostic factor
for progression among patients with HER2-positive MBC, while high
EGF mRNA expression was a positive prognostic factor for
progression only in patients with de novo MBC. In addition,
it was observed that the HER2-negative trastuzumab-treated subgroup
of patients had similar TTP and survival compared with the
HER2-positive group.
MBC is a heterogeneous disease with poor prognosis.
Although survival in HER2-positive breast cancer has been improved
due to anti-HER2 directed therapies, further studies are required
to improve the prognostic outcomes of patients with this
disease.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Ms. Emily Daskalaki
(Department of Pathology, Aristotle University of Thessaloniki,
School of Health Sciences, Faculty of Medicine, Thessaloniki,
Greece) for providing technical assistance with molecular methods,
including mRNA expression analysis by qPCR, Ms. Eneida Jaupaj for
tissue sample collection and Ms. Maria Moschoni for data
coordination (both Hellenic Cooperative Oncology Group, Data
Office, Athens, Greece).
Funding
The study was supported by a research grant from F. Hoffmann-La
Roche and by an internal HeCOG research grant (HE TRANS_BR).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available at https://files.hecog.gr/RQ_mRNA_data.xls.
Authors' contributions
VR, VK and GF were responsible for the
conceptualization of the study. Formal analysis was performed by
GAK. Experiments and data collection were completed by KP, MB, KC,
SC and VK. VR, EM, IB, GP, DB, CC, IN, MS, CM, AKou, PP, AKot, ERa,
AP, DT, DP, ERe, AA and GF were involved in patient provision, data
acquisition and analysis, and critical revision of the manuscript.
EM, VK and GF supervised the study. VR, EM, GAK, KP, SC, VK and GF
were responsible for writing the original draft. KP and GF confirm
the authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The translational research protocol was approved by
the Bioethics Committee of the Aristotle University of Thessaloniki
School of Medicine (Protocol #4283; January 14, 2008; Thessaloniki,
Greece) under the general title ‘Investigation of major mechanisms
of resistance to treatment with trastuzumab in patients with
metastatic breast cancer’. All patients included in the study from
2005 onwards provided written informed consent for the provision of
biological material for future research studies before receiving
any treatment. A waiver of consent was obtained from the Bioethics
Committee for patients included in the study before 2005.
Patient consent for publication
Not applicable.
Competing interests
EM Advisory boards: Roche; GP advisory role: Roche;
research funding: Roche. CC Advisory role: Roche; honoraria: Roche;
advisory role: Roche. PP Advisory role: Roche; honoraria: Roche. AK
Consulting or advisory role: Roche. ER Travel: Roche. AP
Consultation Fees: Roche; honoraria: Roche. DP Advisory role:
Roche; honoraria: Roche. GF Advisory Board: Roche.
Glossary
Abbreviations
Abbreviations:
|
ER
|
oestrogen receptors
|
|
FISH
|
fluorescence in situ hybridization
|
|
IHC
|
immunohistochemistry
|
|
MBC
|
metastatic breast cancer
|
|
PgR
|
progesterone receptors
|
|
R-MBC
|
relapsed metastatic breast cancer
|
|
RARA
|
retinoic acid receptor α
|
|
THRA
|
thyroid hormone receptor α
|
|
TTP
|
time to progression
|
References
|
1
|
Hu Z, Fan C, Oh DS, Marron JS, He X,
Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, et al: The
molecular portraits of breast tumors are conserved across
microarray platforms. BMC Genomics. 7:962006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
O'Brien KM, Cole SR, Tse CK, Perou CM,
Carey LA, Foulkes WD, Dressler LG, Geradts J and Millikan RC:
Intrinsic breast tumor subtypes, race, and long-term survival in
the Carolina Breast Cancer Study. Clin Cancer Res. 16:6100–6110.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rilke F, Colnaghi MI, Cascinelli N,
Andreola S, Baldini MT, Bufalino R, Della Porta G, Ménard S,
Pierotti MA and Testori A: Prognostic significance of HER-2/neu
expression in breast cancer and its relationship to other
prognostic factors. Int J Cancer. 49:44–49. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cronin KA, Harlan LC, Dodd KW, Abrams JS
and Ballard-Barbash R: Population-based estimate of the prevalence
of HER-2 positive breast cancer tumors for early stage patients in
the US. Cancer Invest. 28:963–968. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alroy I and Yarden Y: The ErbB signaling
network in embryogenesis and oncogenesis: Signal diversification
through combinatorial ligand-receptor interactions. FEBS Lett.
410:83–86. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rubin I and Yarden Y: The basic biology of
HER2. Ann Oncol. 12 (Suppl 1):S3–S8. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ferreira PMP and Pessoa C: Molecular
biology of human epidermal receptors, signaling pathways and
targeted therapy against cancers: new evidences and old challenges.
Braz J Pharm Sci. 53:1–17. 2017. View Article : Google Scholar
|
|
8
|
Harris L, Fritsche H, Mennel R, Norton L,
Ravdin P, Taube S, Somerfield MR, Hayes DF and Bast RC Jr; American
Society of Clinical Oncology, : American Society of Clinical
Oncology 2007 update of recommendations for the use of tumor
markers in breast cancer. J Clin Oncol. 25:5287–5312. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wolff AC, Hammond ME, Hicks DG, Dowsett M,
McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M,
Fitzgibbons P, et al American Society of Clinical Oncology; College
of American Pathologists, : Recommendations for human epidermal
growth factor receptor 2 testing in breast cancer: American Society
of Clinical Oncology/College of American Pathologists clinical
practice guideline update. J Clin Oncol. 31:3997–4013. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Noone AM, Cronin KA, Altekruse SF,
Howlader N, Lewis DR, Petkov VI and Penberthy L: Cancer Incidence
and Survival Trends by Subtype Using Data from the Surveillance
Epidemiology and End Results Program, 1992-2013. Cancer Epidemiol
Biomarkers Prev. 26:632–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wolff AC, Hammond MEH, Allison KH, Harvey
BE, Mangu PB, Bartlett JMS, Bilous M, Ellis IO, Fitzgibbons P,
Hanna W, et al: Human Epidermal Growth Factor Receptor 2 Testing in
Breast Cancer: American Society of Clinical Oncology/College of
American Pathologists Clinical Practice Guideline Focused Update. J
Clin Oncol. 36:2105–2122. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Allison KH, Hammond MEH, Dowsett M,
McKernin SE, Carey LA, Fitzgibbons PL, Hayes DF, Lakhani SR,
Chavez-MacGregor M, Perlmutter J, et al: Estrogen and Progesterone
Receptor Testing in Breast Cancer: American Society of Clinical
Oncology/College of American Pathologists Guideline Update. Arch
Pathol Lab Med. 144:545–563. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Filipits M, Rudas M, Singer CF, Fitzal F,
Bago-Horvath Z, Greil R, Balic M, Lax SF, Halper S, Hulla W, et al:
ESR1, PGR, ERBB2, and MKi67 mRNA expression in postmenopausal women
with hormone receptor-positive early breast cancer: Results from
ABCSG Trial 6. ESMO Open. 6:1002282021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Révillion F, Lhotellier V, Hornez L,
Bonneterre J and Peyrat JP: ErbB/HER ligands in human breast
cancer, and relationships with their receptors, the
bio-pathological features and prognosis. Ann Oncol. 19:73–80. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gajria D and Chandarlapaty S:
HER2-amplified breast cancer: Mechanisms of trastuzumab resistance
and novel targeted therapies. Expert Rev Anticancer Ther.
11:263–275. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smith BL, Chin D, Maltzman W, Crosby K,
Hortobagyi GN and Bacus SS: The efficacy of Herceptin therapies is
influenced by the expression of other erbB receptors, their ligands
and the activation of downstream signalling proteins. Br J Cancer.
91:1190–1194. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Motoyama AB, Hynes NE and Lane HA: The
efficacy of ErbB receptor-targeted anticancer therapeutics is
influenced by the availability of epidermal growth factor-related
peptides. Cancer Res. 62:3151–3158. 2002.PubMed/NCBI
|
|
18
|
Vernieri C, Milano M, Brambilla M,
Mennitto A, Maggi C, Cona MS, Prisciandaro M, Fabbroni C, Celio L,
Mariani G, et al: Resistance mechanisms to anti-HER2 therapies in
HER2-positive breast cancer: Current knowledge, new research
directions and therapeutic perspectives. Crit Rev Oncol Hematol.
139:53–66. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wong ALA and Lee SC: Mechanisms of
Resistance to Trastuzumab and Novel Therapeutic Strategies in
HER2-Positive Breast Cancer. Int J Breast Cancer. 2012:4151702012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang J, Luo XX, Tang YL, Xu JX and Zeng
ZG: The prognostic values of insulin-like growth factor binding
protein in breast cancer. Medicine (Baltimore). 98:e155612019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen C, Zhao KN, Masci PP, Lakhani SR,
Antonsson A, Simpson PT and Vitetta L: TGFβ isoforms and receptors
mRNA expression in breast tumours: Prognostic value and clinical
implications. BMC Cancer. 15:10102015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Heublein S, Mayr D, Meindl A, Angele M,
Gallwas J, Jeschke U and Ditsch N: Thyroid Hormone Receptors
Predict Prognosis in BRCA1 Associated Breast Cancer in Opposing
Ways. PLoS One. 10:e01270722015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ditsch N, Toth B, Himsl I, Lenhard M,
Ochsenkühn R, Friese K, Mayr D and Jeschke U: Thyroid hormone
receptor (TR)alpha and TRbeta expression in breast cancer. Histol
Histopathol. 28:227–237. 2013.PubMed/NCBI
|
|
24
|
Hua S, Kittler R and White KP: Genomic
antagonism between retinoic acid and estrogen signaling in breast
cancer. Cell. 137:1259–1271. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ross-Innes CS, Stark R, Holmes KA, Schmidt
D, Spyrou C, Russell R, Massie CE, Vowler SL, Eldridge M and
Carroll JS: Cooperative interaction between retinoic acid
receptor-alpha and estrogen receptor in breast cancer. Genes Dev.
24:171–182. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Razis E, Bobos M, Kotoula V, Eleftheraki
AG, Kalofonos HP, Pavlakis K, Papakostas P, Aravantinos G, Rigakos
G, Efstratiou I, et al: Evaluation of the association of PIK3CA
mutations and PTEN loss with efficacy of trastuzumab therapy in
metastatic breast cancer. Breast Cancer Res Treat. 128:447–456.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fountzilas G, Christodoulou C, Bobos M,
Kotoula V, Eleftheraki AG, Xanthakis I, Batistatou A,
Pentheroudakis G, Xiros N, Papaspirou I, et al: Topoisomerase II
alpha gene amplification is a favorable prognostic factor in
patients with HER2-positive metastatic breast cancer treated with
trastuzumab. J Transl Med. 10:2122012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Koumarianou A, Karayannopoulou G,
Gourgioti G, Batistatou A, Bobos M, Efstratiou I, Miliaras D,
Galani E, Pentheroudakis G, Pectasides D, et al: PAI-1 and HER2
interaction in advanced breast cancer disease: Evidence for added
benefit from trastuzumab in HER2-negative patients. Cancer
Chemother Pharmacol. 75:1289–1301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wolff AC, Hammond ME, Schwartz JN, Hagerty
KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer
A, et al American Society of Clinical Oncology; College of American
Pathologists, : American Society of Clinical Oncology/College of
American Pathologists guideline recommendations for human epidermal
growth factor receptor 2 testing in breast cancer. J Clin Oncol.
25:118–145. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pavlakis K, Bobos M, Batistatou A, Kotoula
V, Eleftheraki AG, Stofas A, Timotheadou E, Pentheroudakis G,
Psyrri A, Koutras A, et al: p85 protein expression is associated
with poor survival in HER2-positive patients with advanced breast
cancer treated with trastuzumab. Pathol Oncol Res. 21:273–282.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gogas H, Kotoula V, Alexopoulou Z,
Christodoulou C, Kostopoulos I, Bobos M, Raptou G, Charalambous E,
Tsolaki E, Xanthakis I, et al: MYC copy gain, chromosomal
instability and PI3K activation as potential markers of
unfavourable outcome in trastuzumab-treated patients with
metastatic breast cancer. J Transl Med. 14:1362016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Christodoulou C, Oikonomopoulos G, Koliou
GA, Kostopoulos I, Kotoula V, Bobos M, Pentheroudakis G, Lazaridis
G, Skondra M, Chrisafi S, et al: Evaluation of the Insulin-like
Growth Factor Receptor Pathway in Patients with Advanced Breast
Cancer Treated with Trastuzumab. Cancer Genomics Proteomics.
15:461–471. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Salgado R, Denkert C, Demaria S, Sirtaine
N, Klauschen F, Pruneri G, Wienert S, Van den Eynden G, Baehner FL,
Penault-Llorca F, et al International TILs Working Group 2014, :
The evaluation of tumor-infiltrating lymphocytes (TILs) in breast
cancer: Recommendations by an International TILs Working Group
2014. Ann Oncol. 26:259–271. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sauter G, Lee J, Bartlett JM, Slamon DJ
and Press MF: Guidelines for human epidermal growth factor receptor
2 testing: Biologic and methodologic considerations. J Clin Oncol.
27:1323–1333. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Economopoulou P, Kotoula V, Koliou GA,
Papadopoulou K, Christodoulou C, Pentheroudakis G, Lazaridis G,
Arapantoni-Dadioti P, Koutras A, Bafaloukos D, et al: Prognostic
Impact of Src, CDKN1B, and JAK2 Expression in Metastatic Breast
Cancer Patients Treated with Trastuzumab. Transl Oncol. 12:739–748.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Perez EA, Suman VJ, Davidson NE, Martino
S, Kaufman PA, Lingle WL, Flynn PJ, Ingle JN, Visscher D and
Jenkins RB: HER2 testing by local, central, and reference
laboratories in specimens from the North Central Cancer Treatment
Group N9831 intergroup adjuvant trial. J Clin Oncol. 24:3032–3038.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Roche PC, Suman VJ, Jenkins RB, Davidson
NE, Martino S, Kaufman PA, Addo FK, Murphy B, Ingle JN and Perez
EA: Concordance between local and central laboratory HER2 testing
in the breast intergroup trial N9831. J Natl Cancer Inst.
94:855–857. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Paik S, Bryant J, Tan-Chiu E, Romond E,
Hiller W, Park K, Brown A, Yothers G, Anderson S, Smith R, et al:
Real-world performance of HER2 testing - National Surgical Adjuvant
Breast and Bowel Project experience. J Natl Cancer Inst.
94:852–854. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kaufman PA, Bloom KJ, Burris H, Gralow JR,
Mayer M, Pegram M, Rugo HS, Swain SM, Yardley DA, Chau M, et al:
Assessing the discordance rate between local and central HER2
testing in women with locally determined HER2-negative breast
cancer. Cancer. 120:2657–2664. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Perez EA, Cortés J, Gonzalez-Angulo AM and
Bartlett JM: HER2 testing: Current status and future directions.
Cancer Treat Rev. 40:276–284. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fehrenbacher L, Cecchini RS, Geyer CE Jr,
Rastogi P, Costantino JP, Atkins JN, Crown JP, Polikoff J, Boileau
JF, Provencher L, et al: NSABP B-47/NRG Oncology Phase III
Randomized Trial Comparing Adjuvant Chemotherapy With or Without
Trastuzumab in High-Risk Invasive Breast Cancer Negative for HER2
by FISH and With IHC 1+ or 2. J Clin Oncol. 38:444–453. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Exman P and Tolaney SM: HER2-positive
metastatic breast cancer: A comprehensive review. Clin Adv Hematol
Oncol. 19:40–50. 2021.PubMed/NCBI
|
|
43
|
Fiszman GL and Jasnis MA: Molecular
Mechanisms of Trastuzumab Resistance in HER2 Overexpressing Breast
Cancer. Int J Breast Cancer. 2011:3521822011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sergina NV and Moasser MM: The HER family
and cancer: Emerging molecular mechanisms and therapeutic targets.
Trends Mol Med. 13:527–534. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Britsch S: The neuregulin-I/ErbB signaling
system in development and disease. Adv Anat Embryol Cell Biol.
190:1–65. 2007.PubMed/NCBI
|
|
46
|
Seoane S, Montero JC, Ocaña A and
Pandiella A: Breast cancer dissemination promoted by a
neuregulin-collagenase 3 signalling node. Oncogene. 35:2756–2765.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang L, Li Y, Shen E, Cao F, Li L, Li X,
Wang X, Kariminia S, Chang B, Li H, et al: NRG1-dependent
activation of HER3 induces primary resistance to trastuzumab in
HER2-overexpressing breast cancer cells. Int J Oncol. 51:1553–1562.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Schwarz LJ, Hutchinson KE, Rexer BN,
Estrada MV, Gonzalez Ericsson PI, Sanders ME, Dugger TC, Formisano
L, Guerrero-Zotano A, Red-Brewer M, et al: An ERBB1-3 Neutralizing
Antibody Mixture With High Activity Against Drug-Resistant HER2+
Breast Cancers With ERBB Ligand Overexpression. J Natl Cancer Inst.
109:djx0652017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Rimawi MF: EGFR Expression in Breast
Cancer Association with biologic phenotype and clinical outcomes.
Cancer. 116:1234–1242. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ross JS and Fletcher JA: The HER-2/neu
Oncogene in Breast Cancer: Prognostic Factor, Predictive Factor,
and Target for Therapy. Oncologist. 3:237–252. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Robinson AG, Turbin D, Thomson T, Yorida
E, Ellard S, Bajdik C, Huntsman D and Gelmon K: Molecular
predictive factors in patients receiving trastuzumab-based
chemotherapy for metastatic disease. Clin Breast Cancer. 7:254–261.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kudlow JE, Cheung CY and Bjorge JD:
Epidermal growth factor stimulates the synthesis of its own
receptor in a human breast cancer cell line. J Biol Chem.
261:4134–4138. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cheng H, Ballman K, Vassilakopoulou M,
Dueck AC, Reinholz MM, Tenner K, Gralow J, Hudis C, Davidson NE,
Fountzilas G, et al: EGFR expression is associated with decreased
benefit from trastuzumab in the NCCTG N9831 (Alliance) trial. Br J
Cancer. 111:1065–1071. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ali R and Wendt MK: The paradoxical
functions of EGFR during breast cancer progression. Signal
Transduct Target Ther. 2:160422017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wendt MK, Williams WK, Pascuzzi PE,
Balanis NG, Schiemann BJ, Carlin CR and Schiemann WP: The
Antitumorigenic Function of EGFR in Metastatic Breast Cancer is
Regulated by Expression of Mig6. Neoplasia. 17:124–133. 2015.
View Article : Google Scholar : PubMed/NCBI
|