Introduction
Lung cancer is one of the most aggressive
malignancies and the leading cause of cancer-associated mortality
worldwide (1,2). Lung cancer can be classified into two
major histological groups: Small cell lung cancer and non-small
cell lung cancer. With the development of medical technologies in
the past decades, therapeutic strategies, such as surgery,
radiation therapy, chemotherapy and targeted therapy, have been
greatly improved along with improvements of the outcome of patients
with lung cancer (3). However, the
prognosis of patients with lung cancer remains unsatisfactory,
especially that in patients with metastasis, recurrence and drug
resistance (4–6). Therefore, an improved understanding
of lung cancer might be beneficial for the improvement of
diagnostic and therapeutic strategies.
Mucin 13 (MUC13) was first identified as a member of
the transmembrane glycoprotein mucins and is expressed by
epithelial and hematopoietic cells (7). MUC13 could protect against intestinal
inflammation, and a deficiency in MUC13 might aggravate the
development of severe acute colitis upon treatment with dextran
sodium sulfate (8). Increasing
evidence has demonstrated that dysregulated MUC13 expression is
observed in various tumors, including gastric, ovarian, pancreatic
and colon cancer (9–12). In colorectal cancer, MUC13 could
activate NF-κB signaling and protect colorectal cancer cells from
apoptosis (13). Additionally,
MUC13 has been reported to promote the development of intrahepatic
cholangiocarcinoma via activation of the EGFR/PI3K/AKT signaling
pathway (14). Studies have
revealed that MUC13 expression could be post-transcriptionally
regulated by microRNAs (miRs), such as miR-145 and miR132-3p
(15,16). Serum MUC13 expression has been
reported to be elevated in certain carcinomas, such as ovarian
cancer and gastric cancer (9,17),
and could be developed as a novel biomarker (18). However, to the best of our
knowledge, the expression and function of MUC13 in lung cancer
remain unknown.
Therefore, to elucidate the function and mechanism
of MUC13 in lung cancer, the expression pattern of MUC13 was first
investigated in lung cancer tissues and cell lines. Subsequently,
MUC13 was silenced in lung cancer cell lines, followed by
proliferation, apoptosis, migration and invasion analyses to
determine the regulatory role of MUC13 in lung cancer. Furthermore,
a xenograft tumor model was applied to confirm the findings at the
cell level. Based on these investigations, the present study aimed
to provide novel insights for the understanding of lung cancer, as
well as underlying therapeutic targets for the treatment of lung
cancer.
Materials and methods
Patient specimens
Lung adenocarcinoma cancer tissues and adjacent
normal tissues (1 cm from tumor margin) sections were collected
from 20 patients (14 male and 6 female patients) aged 46.50±10.30
years (range, 28–64 years) who underwent surgical resection at
Gansu Provincial Hospital (Lanzhou, China) between May 2018 and
April 2019. Patients were enrolled in the present study if they met
the following criteria: i) Aged 18 years old; ii) pathology
confirmed as lung adenocarcinoma; iii) presented with clear TNM
stage (19) and detailed clinical
information; and iv) had informed consent to participate this
research. Patients were excluded if they met any of the following
criteria: i) Complicated with other cancer; ii) congenital lung
hypoplasia; iii) had infectious disease; or iv) other lung
diseases. The clinical characteristics of the patients are
summarized in Table I. The patient
specimens were snap-frozen and stored in liquid nitrogen (−196°C)
until reverse transcription-quantitative PCR (RT-qPCR) and western
blotting were performed. All patients provided written informed
consent. The study was approved by the Institutional Ethical Review
Board of Gansu Provincial Hospital (Lanzhou, China; approval no.
GSRMYYLL-2019-95).
 | Table I.Clinicopathological characteristics of
patients (n=20). |
Table I.
Clinicopathological characteristics of
patients (n=20).
| Characteristics | No. of patients |
|---|
| Age, years |
|
|
<60 | 7 |
| ≥60 | 13 |
| Sex |
|
| Male | 14 |
|
Female | 6 |
| TNM stage |
|
| T |
|
| T1-2 | 8 |
| T3 | 9 |
| T4 | 3 |
| N |
|
| N0 | 10 |
| N1 | 2 |
| N2 | 4 |
| N3 | 4 |
| M |
|
| M0 | 18 |
|
M1a | 2 |
In silico analysis of MUC13
expression
In the present study, MUC13 expression in lung
adenocarcinoma was analyzed using the starBase online tool (version
3.1; http://starbase.sysu.edu.cn/panGeneDiffExp.php) based
on 526 cancer samples and 59 normal samples, which were downloaded
from The Cancer Genome Atlas project via the Genomic Data Commons
Data Portal (http://portal.gdc.cancer.gov/), and the expression
values of genes from RNA-seq data were scaled with
log2(FPKM+0.01).
Cell culture
A total of four lung cancer cell lines (NCI-H460,
squamous carcinoma epithelial cells; H1703, squamous carcinoma
epithelial cells; A549, adenocarcinoma epithelial cancer cells; and
NCI-H1650, adenocarcinoma epithelial cells) were obtained from The
Cell Bank of Type Culture Collection of The Chinese Academy of
Sciences. A control cell line [normal lung epithelial cells
(NLECs), derived from normal human primary lobar epithelial cells]
was purchased from the Shanghai Baiye Biotechnology Center, and
human bronchial epithelial (HBE) cells were obtained from Procell
Life Science & Technology Co., Ltd. Cells were cultured in DMEM
(HyClone; Cytiva) supplemented with 10% fetal bovine serum
(HyClone; Cytiva) and 1% penicillin-streptomycin (Thermo Fisher
Scientific, Inc.). Cells were maintained in a cell incubator at
37°C with 5% CO2.
Transfection
The MUC13 short hairpin RNAs (shRNAs) and scramble
shRNA plasmids cloned into pLKO.1 were purchased from Shanghai
GenePharma Co., Ltd. Transfection (2 µg/well of a 6-well plate) was
performed in A549 or NCI-H1650 cells using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Cells
were incubated at 37°C for 48 h. At 48 h post-transfection, cells
were harvested for the following experiments.
RT-qPCR
Total RNA was isolated from tissues samples or
cultured NLECs, HBE, NCI-H460, H1703, A549 and NCI-1650 cells using
an AccuRef RNA isolation kit (AccuRef Scientific) and
reverse-transcribed into cDNA using the QuantiTect Reverse
Transcription kit (Qiagen GmbH) according to the manufacturers'
instructions. qPCR analysis was performed using SYBR master mix
(Takara Bio, Inc.) on an ABI 7500 instrument with the following
conditions: 94°C for 5 min, followed by 40 cycles of 94°C for 10
sec, 58°C for 30 sec, and 72°C for 10 sec. The relative gene
expression was analyzed using the 2−ΔΔCq method
(20) with β-actin as the internal
control. The primer sequences used were: hsa-MUC13 forward,
5′-CTGCGGATGACTGCCTCAATGG-3′ and hsa-MUC13 reverse,
5′-ATTGCTTGTGCTGTGCGTTGC-3′; and hsa-β-actin forward,
5′-CCTGTGGCATCCACGAAACT-3′, and hsa-β-actin reverse,
5′-GAAGCATTTGCGGTGGACGAT-3′.
Western blotting
Total protein was extracted from tissues samples or
cultured A549 and NCI-1650 cells using RIPA lysis buffer (AccuRef
Scientific) and quantified using the BCA method (Thermo Fisher
Scientific, Inc.). After boiling with an equal volume of loading
buffer, a total of 25 µg/lane for each sample was subjected to 12%
SDS-PAGE separation. Protein was transferred to a PVDF membrane
(MilliporeSigma) and the membrane was blocked with 5% non-fat milk
(non-phosphorylation-specific) or 5% BSA (phosphorylation-specific;
Sangon Biotech Co., Ltd.) at room temperature for 1 h.
Subsequently, the membranes were incubated with primary antibodies
overnight at 4°C, followed by incubation with an HRP-conjugated
goat anti-rabbit secondary antibody (dilution, 1:10,000; cat. no.
ab6721; Abcam) at room temperature for 1 h. The protein bands were
visualized using an enhanced chemiluminescence kit (Pierce; Thermo
Fisher Scientific, Inc.). Using β-actin as the internal control,
protein bands were semi-quantified using ImageJ software (version
1.48; National Institutes of Health) and the relative expression of
protein was determined and compared with other groups. The primary
antibodies used in the present study were: MUC13 rabbit monoclonal
antibody (mAb) (dilution, 1:1,000; cat. no. ab235450; Abcam), ERK
rabbit mAb (dilution, 1:5,000; cat. no. ab184699; Abcam),
phosphorylated (p- ERK (ERK1 phospho T202+ERK2 phospho T185) rabbit
mAb (dilution, 1:100; cat. no. ab214036; Abcam), JNK rabbit mAb
(dilution, 1:1,000; cat. no. ab179461; Abcam), p-JNK (phospho
T183+T183+T221) rabbit mAb (dilution, 1:1,000; cat. no. ab124956;
Abcam), p38 rabbit mAb (dilution, 1:2,000; cat. no. ab170099;
Abcam) and p-p38 rabbit mAb (dilution, 1:1,000; cat. no. ab178867;
Abcam).
Cell Counting Kit-8 (CCK-8) assay
For the CCK-8 assay, A549 cells or NCI-H1650 cells
were seeded into 96-well plates (2,000 cells/well). Subsequently,
10 µl CCK-8 reagent (Dojindo Molecular Technologies, Inc.) was
added to each well at 0, 24, 48 and 72 h according to the
manufacturer's protocol, and the cells were incubated for 2 h at
37°C. Next, the absorbance of each well was measured at 450 nm
using an ELX800 microplate reader (BioTek Instruments, Inc.) to
evaluate the cell viability.
Colony formation assay
For the colony formation assay, A549 cells or
NCI-1650 cells were seeded into 6-well plates at a density of 500
cells/well and cultured for 14 days. Cell colonies were fixed with
100% methanol at room temperature for 15 min and stained with 1%
crystal violet in 20% methanol at room temperature for 30 min.
Subsequently, the staining solution was removed and cells were
washed with slow running water, followed by natural drying and
counting under a light microscope (Nicon Eclipase E600; Nikon
Corporation).
Apoptosis assay
A549 cells or NCI-1650 cells were washed with
ice-cold PBS in triplicate and suspended in 200 µl binding buffer
at a concentration of 1.0×106 cells/ml. Subsequently,
cells were stained with FITC-Annexin V/PI using a cell apoptosis
detection kit (BD Biosciences) for 30 min in the dark according to
the manufacturer's protocol. Subsequently, 330 µl binding buffer
was added to each sample and apoptosis of cells was analyzed using
a flow cytometer (CytoFlex; Beckman Coulter, Inc.). The apoptotic
cells were defined as Annexin V-positive and PI-negative and data
were analyzed using Kaluza Analysis Software v2 (Beckman Coulter,
Inc.).
Wound healing assay
A549 cells or NCI-1650 cells were seeded into 6-well
plates and cultured until they reached 100% confluency. The
monolayer of cells was scratched using a 200-µl sterile pipette
tip, and the culture was continued for another 48 h in serum-free
medium. Wound closure was imaged at 0 and 48 h using a light
microscope (Nicon Eclipase E600; Nikon Corporation). The width of
the wound was measured using ImageJ (version 1.48u; National
Institutes of Health), and the relative wound width was calculated
to indicate cell migration.
Transwell assay
A Transwell assay was performed to evaluate the
migration and invasion of A549 cells or NCI-1650 cells using a
Transwell chamber with or without pre-coated Matrigel (8-µm pore
size; Corning, Inc.). For pre-coating, Matrigel was moved from
−20°C to 4°C overnight and diluted with serum-free DMEM (1:5) on
ice with a pre-cooled pipette. Subsequently, an equal volume of
diluted Matrigel was added to the upper transwell chamber for
coverage. After incubation at 37°C for 1 h, the coated chamber was
rinsed with serum-free DMEM to remove the uncombined Matrigel,
followed by addition of 50 µl serum-free medium with 10 g/l BSA at
37°C. A total of 5×104 cells were suspended in
serum-free DMEM and seeded into the upper chamber. Subsequently,
500 µl DMEM supplemented with 10% FBS was added to the lower
chamber. After incubation at 37°C for 48 h, the migrated or invaded
cells were fixed with 4% formaldehyde at room temperature for 20
min, stained with 1% crystal violet at room temperature for 15 min,
and counted under a light microscope (Nicon Eclipase E600; Nikon
Corporation).
Xenograft tumor model
A total of 10 BALB/c male nude mice (age, 6 weeks;
weight, 18–20 g), were purchased from Shanghai SLAC Laboratory
Animal Co., Ltd. and housed in a specific pathogen-free room with a
controlled humidity of 40–60%, a temperature of 24–26°C, a 12/12 h
light/dark cycle, and free access to food and water. After
adaptation for 1 week, the xenograft tumor model was established by
subcutaneously inoculating A549 cells (5×106) stably
transfected with sh-MUC13 or scrambled shRNA in PBS into the right
flank of the mice (age, 7 weeks). Tumor growth was measured every 5
days. On day 25, the mice were euthanatized with CO2
exposure at a flow rate of 30% volume per minute, and death was
confirmed by continued CO2 exposure for at least 15 min
after breathing stopped. The xenograft tumors were dissected,
weighed, and fixed with 4% formaldehyde at 4°C for >24 h or
stored at −80°C. All animal experiments were performed according to
the guidelines of the Chinese Experimental Animal Administration
Legislation and approved by the Institutional Animal Care and Use
Committee of Gansu Provincial Hospital (Lanzhou, China; approval
no. RMYYLAC202033-2).
Immunohistochemistry (IHC)
staining
Xenograft tumor tissues or patient tissue samples
were washed twice with PBS and cut into pieces of the appropriate
size (5 mm3). Subsequently, the tissues were fixed with
4% formaldehyde at 4°C for 2 days and dehydrated using gradient
ethanol. Next, the tissues were paraffin-embedded at 60°C overnight
and cut into 5-µm thick sections. The sections were then heated at
60°C for 1 h, de-waxed using xylene, and rehydrated with gradient
alcohol solution (100, 95, 80 and 70% for 2 min each at room
temperature) followed by antigen retrieval in a microwave oven in
boiled 0.01 M sodium citrate buffer (pH 6.0) three times for 3 min
each with 5 min intervals. This was followed by the addition of 3%
H2O2 to the sections for 10 min to block
endogenous peroxidase, after which the sections were washed three
times with PBS. Next, the sections were blocked with 5% goat serum
(Beyotime Institute of Biotechnology) at room temperature for 1 h,
followed by incubation overnight at 4°C with anti-Ki-67 antibodies
(dilution, 1:200; cat. no. ab15580; Abcam). After washing with PBS
three times, sections were then incubated with donkey anti-rabbit
secondary antibody (1:500; cat. no. ab207999; Abcam) the next day
at room temperature for 45 min. Biotin-labeled HRP was added to the
tissue sections and the staining was visualized using a DAB
Substrate Kit (cat. no. ab64238; Abcam). Following this, sections
were counterstained with hematoxylin for nuclei staining at room
temperature for 5–10 min. Then, slices were washed with running
water, dehydrated with ethanol, hyalinized by xylene, mounted with
neutral resins (cat. no. E675007; Sangon Biotech Co., Ltd.) and
analyzed under a light microscope (Nicon Eclipase E600; Nikon
Corporation). To evaluate MUC13 as a transmembrane glycoprotein,
the MUC13-positive area was calculated for three different fields
and its expression was recorded as a percentage of the total area
of the field using ImageJ software (version 1.48u; National
Institutes of Health). To detect Ki-67 in the nucleus, the number
of Ki-67-positive cells was calculated for three different fields
and the number of positive cells per field was recorded for
analysis.
Statistical analysis
Results are presented as the mean ± SD of at least
three experiments. The statistical analysis was conducted using
GraphPad Prism (V6; GraphPad Software, Inc.). Student's paired
t-test was used for comparisons between clinical samples and
unpaired Student's t-test was performed for comparisons between two
groups of cell samples. One-way ANOVA followed by Tukey's post hoc
test was used for comparisons among multiple groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
MUC13 is highly expressed in lung
cancer
Previous studies have demonstrated that MUC13 is
abnormally expressed in multiple malignancies (9–12).
In the present study, the expression levels of MUC13 were first
analyzed in lung tumor tissues and adjacent normal tissues. RT-qPCR
demonstrated that the expression levels of MUC13 in lung tumor
tissues were significantly higher than those in the adjacent normal
tissues (Fig. 1A). Western
blotting also demonstrated that MUC13 protein expression was
increased in lung tumor tissues compared with that in adjacent
normal tissues (Fig. 1B).
Furthermore, IHC analysis revealed that MUC13 expression was
upregulated in lung tumor tissues compared with that in adjacent
normal tissues (Fig. 1C). MUC13
expression was also analyzed and compared between 526 cancer
samples and 59 normal samples derived from patients with lung
adenocarcinoma using StarBase datasets. Consistently, the results
demonstrated that MUC13 expression was significantly upregulated in
lung cancer samples (Fig. 1D).
Furthermore, MUC13 expression was markedly higher in lung cancer
cell lines than in control NLECs; however, but no significant
difference was identified between NLECs and HBE cells (Fig. 1E). These findings suggested that
the expression levels of MUC13 were abnormally upregulated in lung
cancer.
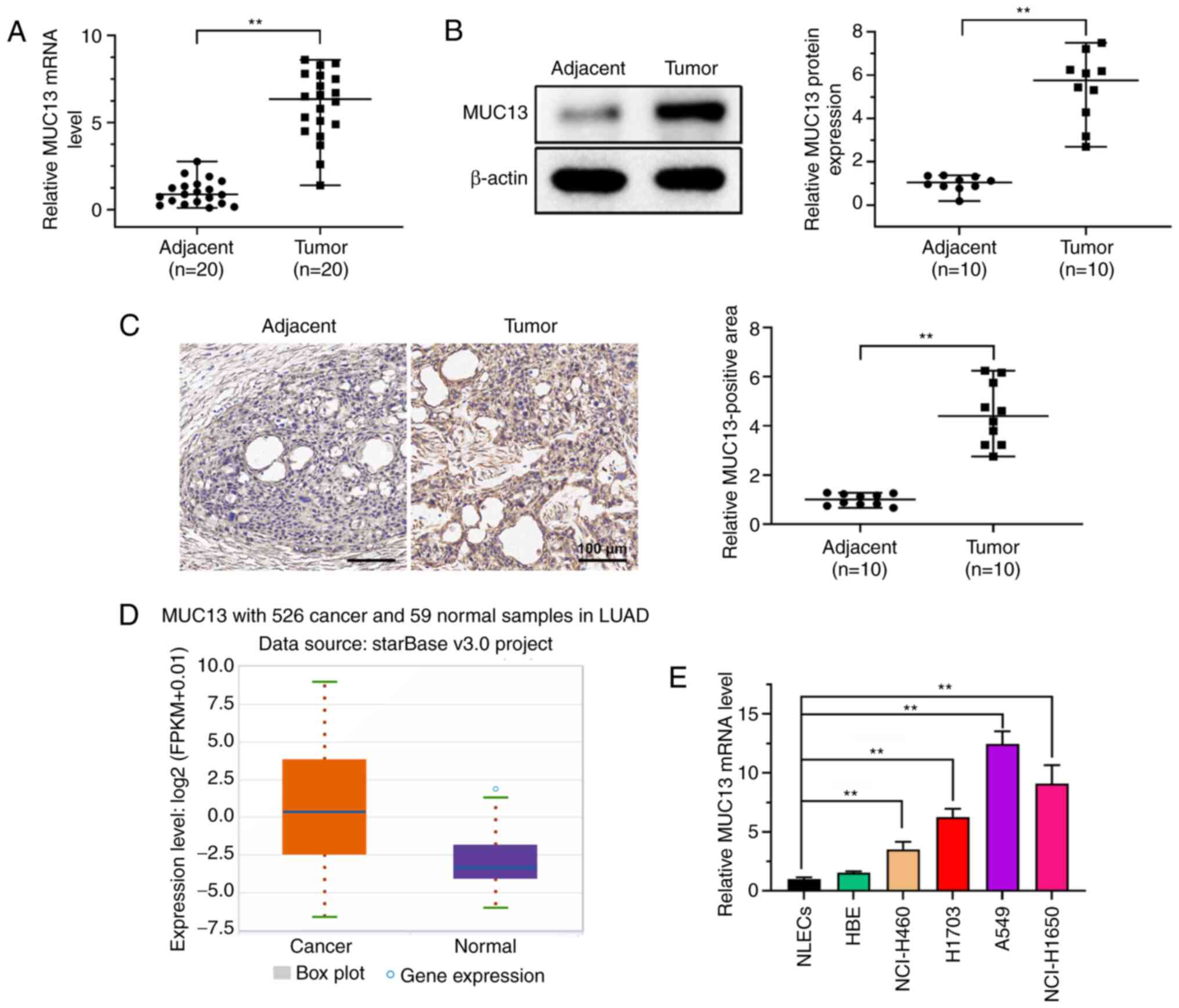 | Figure 1.MUC13 expression is increased in lung
cancer. (A) mRNA expression levels of MUC13 in lung tumor tissues
and adjacent normal tissues were analyzed by RT-qPCR. (B) Paired
lung cancer and normal control tissues were analyzed by western
blotting and a representative example is presented in this figure.
(C) Immunohistochemical staining was performed to examine MUC13
expression in lung tumor and adjacent normal control tissues. Scale
bar, 100 µm. (D) Expression levels of MUC13 in 526 cancer and 59
normal samples in lung adenocarcinoma were analyzed using StarBase
datasets. (E) mRNA expression levels of MUC13 in lung cancer cell
lines (NCI-H460, H1703, A549 and NCI-H1650), control cells (NLECs)
and HBEs were analyzed by RT-qPCR (n=3). **P<0.01. FPKM,
fragments per kilo base per million mapped reads; HBEs, human
bronchial epithelial cells; LUAD, lung adenocarcinoma; MUC13, mucin
13; NLECs, normal lung epithelial cells; RT-qPCR, reverse
transcription-quantitative PCR. |
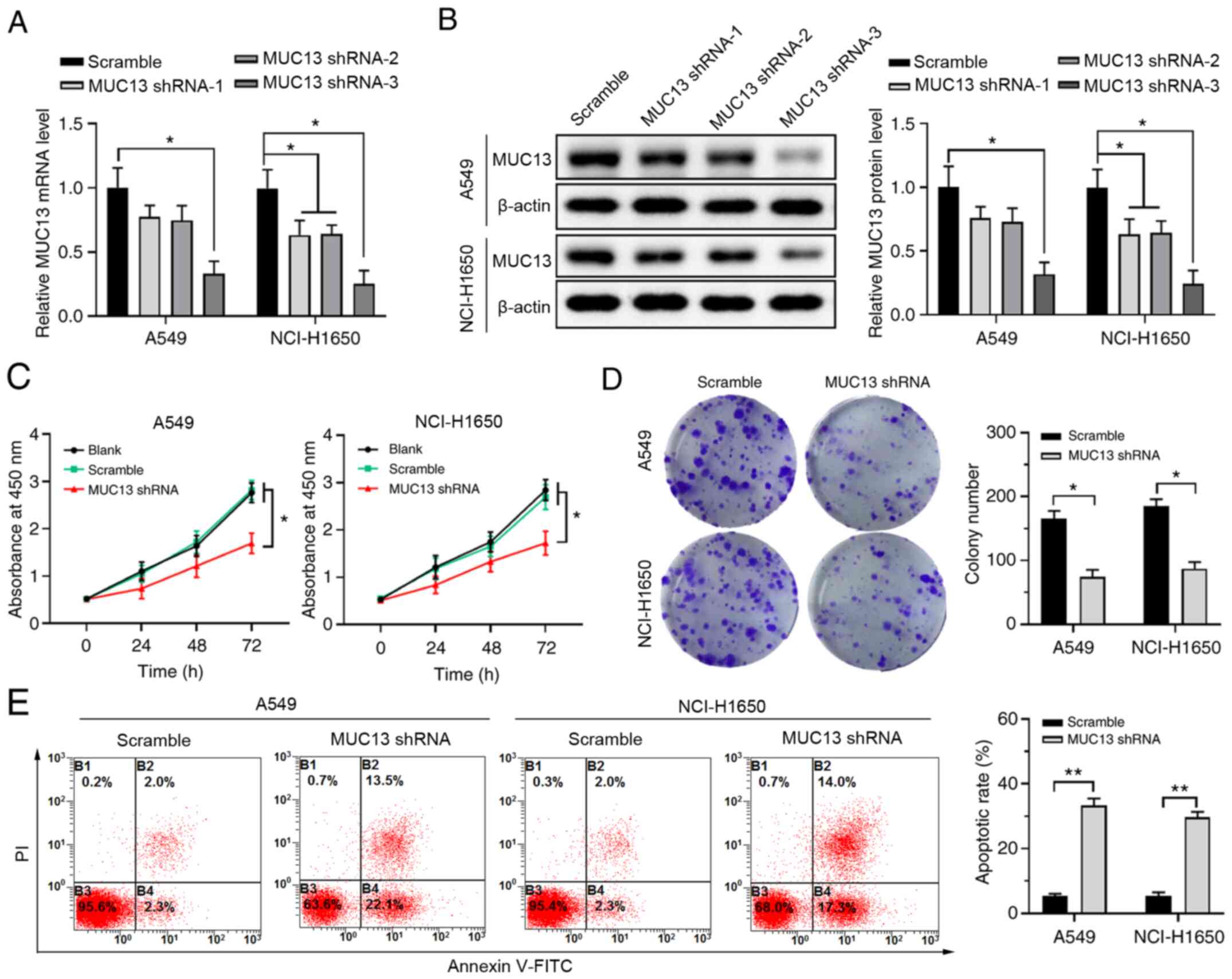
Knockdown of MUC13 inhibits
proliferation and promotes apoptosis of lung cancer cells
To investigate the function of MUC13 in lung cancer
development, MUC13 expression was silenced in A549 or NCI-H1650
cells using the shRNA method. Both RT-qPCR and western blotting
demonstrated that MUC13 shRNA-1 and shRNA-2 could only slightly
suppress MUC13 expression in NCI-1650 cells but exerted no obvious
influence on MUC13 expression in A549 cells, whereas MUC13 shRNA-3
could significantly suppress MUC13 expression compared with that in
the scramble group in both A549 and NCI-H1650 cells (Fig. 2A and B). Therefore, the most
efficient shRNA (MUC13 shRNA-3) was used for subsequent
experiments. The results of CCK-8 and colony formation assays
demonstrated that knockdown of MUC13 inhibited the proliferation
and colony formation of A549 and NCI-H1650 cells (Fig. 2C and D). Furthermore, Annexin V/PI
double-staining revealed that silencing of MUC13 expression could
significantly increase the apoptosis of A549 and NCI-H1650 cells
(Fig. 2E). These results
demonstrated that inhibiting MUC13 expression could significantly
suppress the proliferation and promote the apoptosis of lung cancer
cells.
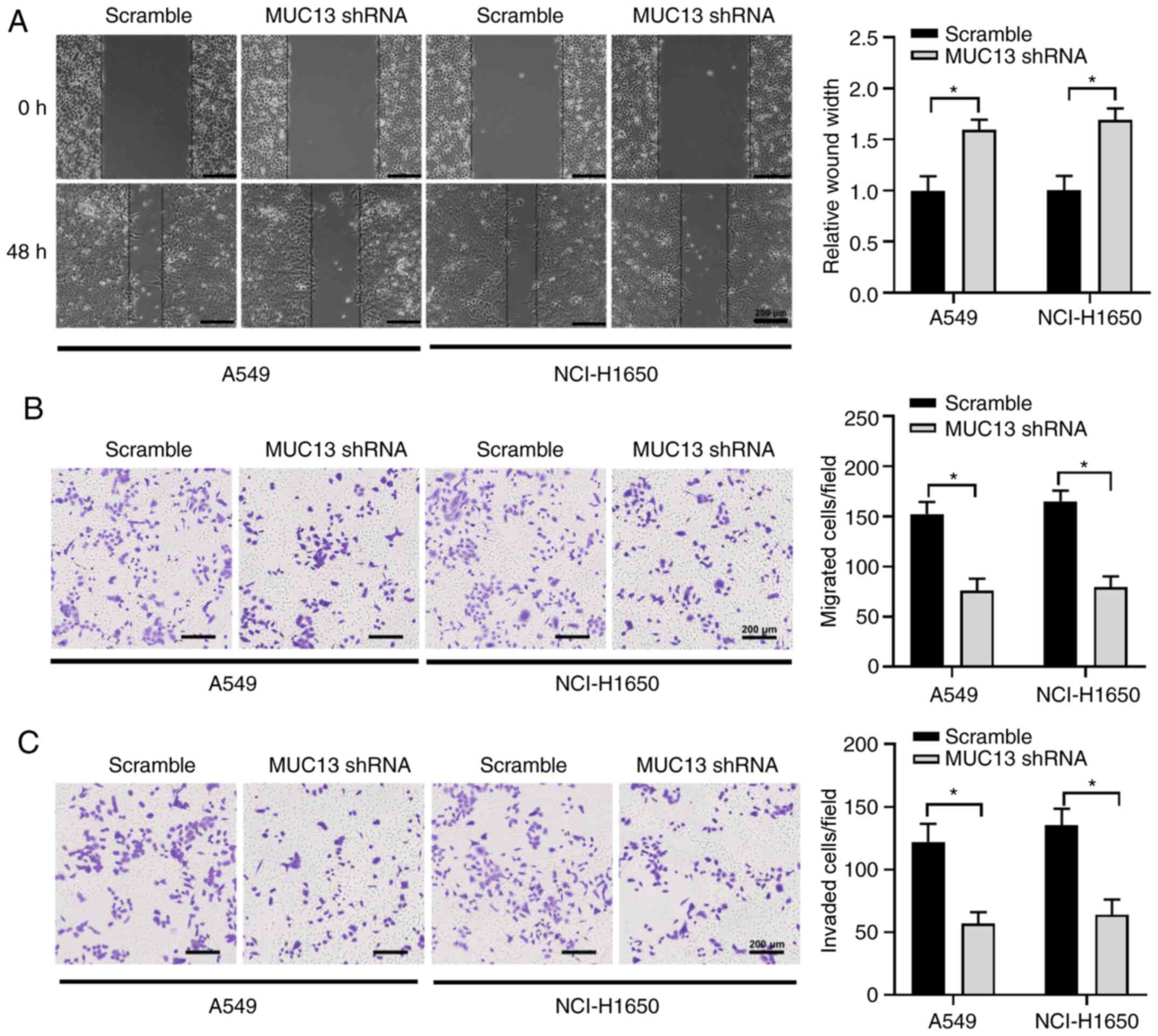
Knockdown of MUC13 suppresses the
migration and invasion of lung cancer cells
The effect of MUC13 on the migration and invasion of
lung cancer cells was also determined. The results of a
wound-healing assay revealed that knockdown of MUC13 significantly
suppressed the wound width reduction of A549 and NCI-H1650 cells
compared with the scramble group at 48 h (Fig. 3A). Consistently, the Transwell
assays also revealed that knockdown of MUC13 decreased the
migration and invasion of lung cancer cells (Fig. 3B and C). This suggested that
silencing MUC13 could markedly inhibit the migration and invasion
of lung cancer cells.
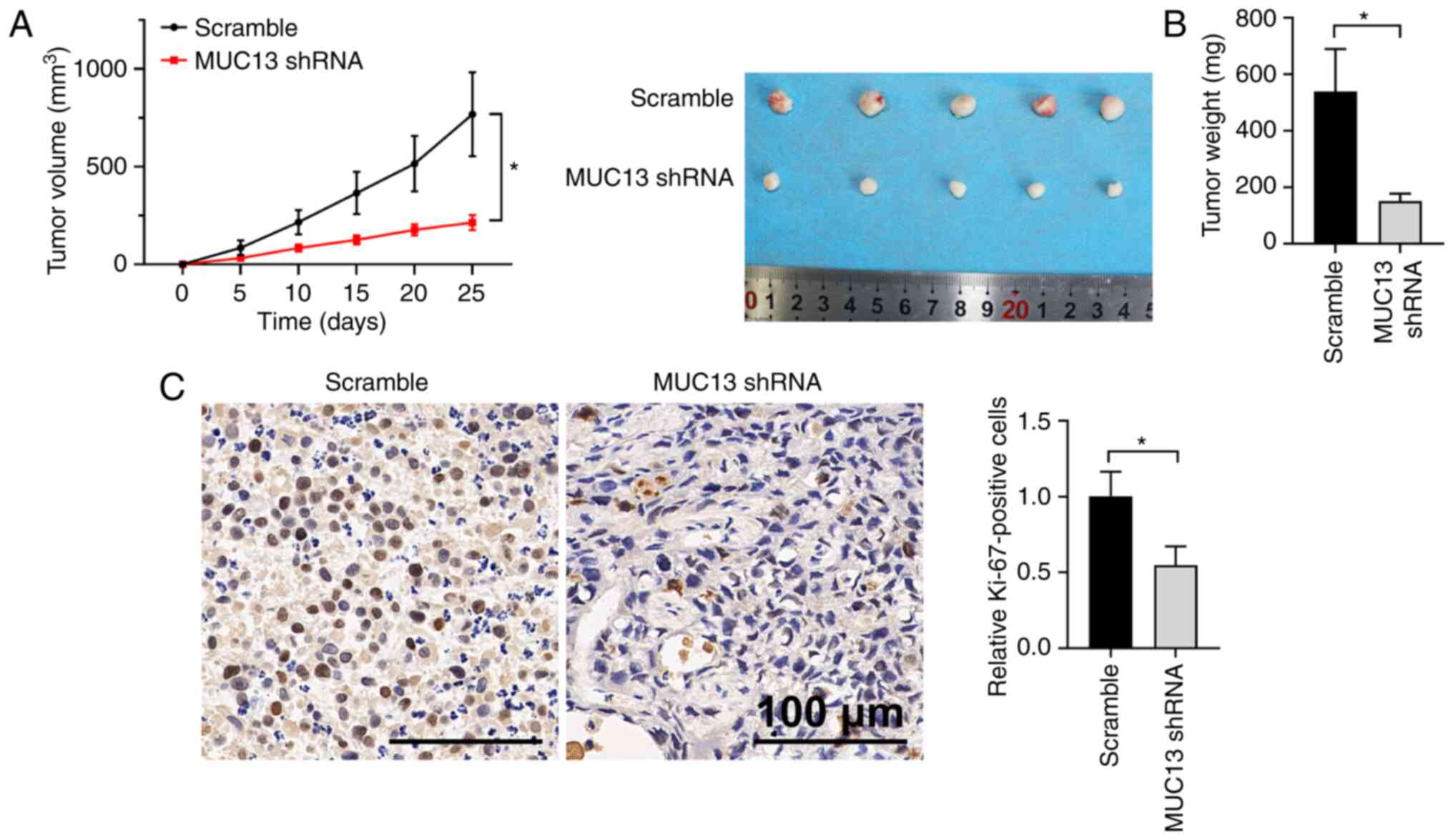
Knockdown of MUC13 inhibits xenograft
lung tumor development in vivo
To further explore the function of MUC13 in lung
cancer development, a xenograft tumor model was established by
subcutaneous injection of A549 cells with stable knockdown of MUC13
or injection of control cells. Silencing of MUC13 expression
significantly delayed the increase of tumor volume in vivo
compared with the scramble group (Fig.
4A). The weights of tumors collected from the MUC13-knockdown
group were lower than those from the control group (Fig. 4B). Furthermore, IHC staining of the
proliferation marker Ki-67 demonstrated that knockdown of MUC13
inhibited the proliferation of lung cancer cells in vivo, as
indicated by lower Ki-67 expression (Fig. 4C). Overall, these results indicated
that knockdown of MUC13 could markedly inhibit the growth of
xenograft lung tumors.
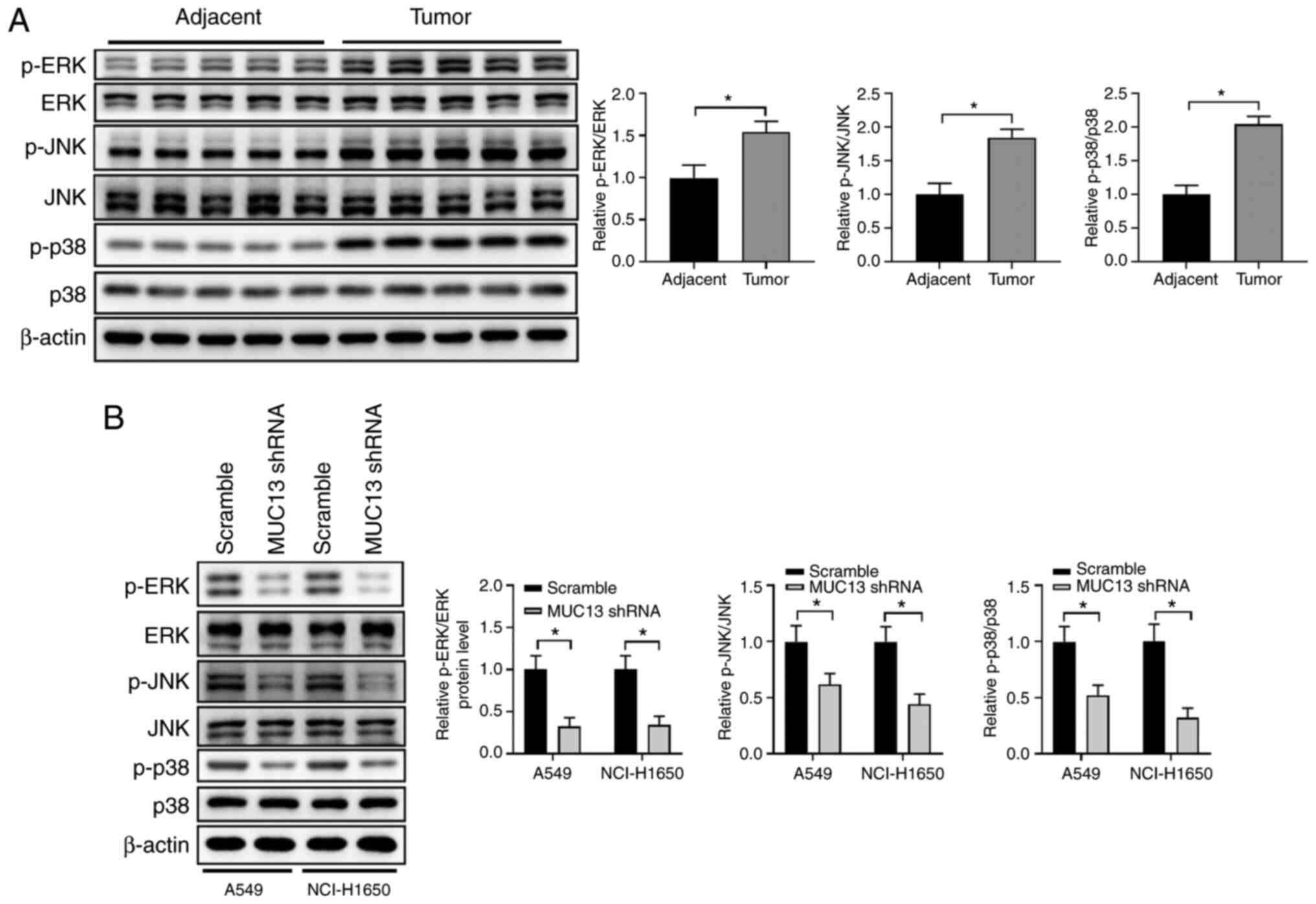
MUC13 promotes lung cancer development
via regulation of ERK signaling
To further examine the mechanism of MUC13 regulation
in lung cancer, ERK, an underlying signaling pathway of MUC13
(14), was evaluated in the
present study. Compared with those in the adjacent control tissues,
the phosphorylation levels of ERK, JNK and p38 in the lung cancer
tissues were significantly increased (Fig. 5A). By contrast, knockdown of MUC13
in A549 and NCI-H1650 cells markedly decreased the levels of p-ERK,
p-JNK and p-p38 (Fig. 5B).
Overall, these findings suggested that MUC13 might promote lung
cancer development via regulation of ERK signaling.
Discussion
The oncogenic role of MUC13 in regulating cancer
cell proliferation, apoptosis, migration and invasion has been
reported in various malignancies, including pancreatic cancer and
intraductal papillary mucinous neoplasms (21,22).
However, its role in lung cancer remains unknown. The results of
the present study demonstrated that MUC13 expression was
upregulated in lung cancer cells and that knockdown of MUC13
suppressed lung cancer cell proliferation, migration and invasion,
which may further help elucidate the mechanism of lung cancer
development.
Upregulation of MUC13 expression has been identified
in pancreatic cancer, and MUC13 could enhance cell motility and
proliferation via activation of p21-activated kinase 1 and ERK
signaling (11). In colon cancer
cells, upregulated MUC13 expression has been reported to enhance
the tumorigenic features of colon cancer cells and to increase the
phosphorylation of HER2 and ERK (12). Similarly, the present study
revealed that lung cancer cells exhibited enhanced phosphorylation
of ERK, JNK and p38, whereas knockdown of MUC13 significantly
suppressed the phosphorylation of ERK, JNK and p38 in lung cancer
cells. Additionally, Sheng et al (23) demonstrated that MUC13 promotes
renal cell carcinoma progression and drug resistance by inducing
the cell cycle regulator cyclin D1, and that it inhibits cell
apoptosis by inducing Bcl-XL and survivin expression. Whether MUC13
regulates the cell cycle in lung cancer cells requires further
investigation.
Since MUC13 functions as an oncogenic glycoprotein
in multiple malignancies, including hepatocellular carcinoma,
colorectal cancer and esophageal squamous cell carcinoma (24–26),
it is of significant interest to explore the regulation of MUC13 in
tumors. A chromatin immunoprecipitation assay has demonstrated that
the transcription factor upstream transcription factor 1 could bind
to the promoter region of MUC13 and enhance MUC13 expression in
glioblastoma (27). Additionally,
the expression levels of MUC13 are regulated post-transcriptionally
by miR-145 in pancreatic cancer and colorectal cancer (15). In another study, the inhibition of
miR-132-3p led to elevated expression levels of MUC13 and enhanced
gastric cancer cell proliferation (16). However, the mechanism by which
MUC13 expression is regulated in lung cancer development requires
further investigation. A previous study demonstrated that patients
with high MUC13 expression in the cytoplasm and nucleus presented
with a larger colon tumor size and poorly differentiated grade
compared with the patients with only membrane localization
(28). In addition, MUC13 is
upregulated and co-localized with parasites during hepatic
infection, and could be a hallmark of Plasmodium
exoerythrocytic infection (29).
Considering these aforementioned results (24–28),
it is of significance to detect the subcellular location and
downstream target of MUC13 in subsequent investigations to further
clarify the mechanism of MUC13 in lung cancer.
In conclusion, MUC13 functions as an oncogenic
glycoprotein mucin in the development of lung cancer via activation
of downstream ERK/JNK/p38 signaling. The results of the present
study suggested that MUC13 served a vital role in the progression
of lung cancer. It is important to further elucidate the function
and mechanism of MUC13 in lung cancer, and this may provide novel
insights for improving the understanding and treatment of the
disease.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study available from the corresponding author on reasonable
request.
Authors' contributions
YP, YZ and ZJZ participated in the conception and
design of the study. HYZ and WHW performed the research. GJ and JWL
performed the statistical analyses and evaluated the results. YZ
drafted the paper. YP and ZJZ contributed to the enrollment of
patients and analysis of data. YP, YZ and ZJZ confirmed the
authenticity of all raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients and the study protocol was approved by the Ethics
Committee of Gansu Provincial Hospital (Lanzhou, China). Animal
care and experiments were approved by the Institutional Animal Care
and Use Committee of Gansu Provincial Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bade BC and Dela Cruz CS: Lung Cancer
2020: Epidemiology, Etiology, and Prevention. Clin Chest Med.
41:1–24. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang D, Liu Y, Bai C, Wang X and Powell
CA: Epidemiology of lung cancer and lung cancer screening programs
in China and the United States. Cancer Lett. 468:82–87. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ Jr, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu WJ, Du Y, Wen R, Yang M and Xu J: Drug
resistance to targeted therapeutic strategies in non-small cell
lung cancer. Pharmacol Ther. 206:1074382020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yan X, Jiao SC, Zhang GQ, Guan Y and Wang
JL: Tumor-associated immune factors are associated with recurrence
and metastasis in non-small cell lung cancer. Cancer Gene Ther.
24:57–63. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hung JJ, Jeng WJ, Hsu WH, Chou TY, Huang
BS and Wu YC: Predictors of death, local recurrence, and distant
metastasis in completely resected pathological stage-I
non-small-cell lung cancer. J Thorac Oncol. 7:1115–1123. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Williams SJ, Wreschner DH, Tran M, Eyre
HJ, Sutherland GR and McGuckin MA: Muc13, a novel human cell
surface mucin expressed by epithelial and hemopoietic cells. J Biol
Chem. 276:18327–18336. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sheng YH, Lourie R, Lindén SK, Jeffery PL,
Roche D, Tran TV, Png CW, Waterhouse N, Sutton P, Florin TH, et al:
The MUC13 cell-surface mucin protects against intestinal
inflammation by inhibiting epithelial cell apoptosis. Gut.
60:1661–1670. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shimamura T, Ito H, Shibahara J, Watanabe
A, Hippo Y, Taniguchi H, Chen Y, Kashima T, Ohtomo T, Tanioka F, et
al: Overexpression of MUC13 is associated with intestinal-type
gastric cancer. Cancer Sci. 96:265–273. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chauhan SC, Vannatta K, Ebeling MC,
Vinayek N, Watanabe A, Pandey KK, Bell MC, Koch MD, Aburatani H,
Lio Y, et al: Expression and functions of transmembrane mucin MUC13
in ovarian cancer. Cancer Res. 69:765–774. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chauhan SC, Ebeling MC, Maher DM, Koch MD,
Watanabe A, Aburatani H, Lio Y and Jaggi M: MUC13 mucin augments
pancreatic tumorigenesis. Mol Cancer Ther. 11:24–33. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gupta BK, Maher DM, Ebeling MC, Stephenson
PD, Puumala SE, Koch MR, Aburatani H, Jaggi M and Chauhan SC:
Functions and regulation of MUC13 mucin in colon cancer cells. J
Gastroenterol. 49:1378–1391. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sheng YH, He Y, Hasnain SZ, Wang R, Tong
H, Clarke DT, Lourie R, Oancea I, Wong KY, Lumley JW, et al: MUC13
protects colorectal cancer cells from death by activating the NF-κB
pathway and is a potential therapeutic target. Oncogene.
36:700–713. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tiemin P, Fanzheng M, Peng X, Jihua H,
Ruipeng S, Yaliang L, Yan W, Junlin X, Qingfu L, Zhefeng H, et al:
MUC13 promotes intrahepatic cholangiocarcinoma progression via
EGFR/PI3K/AKT pathways. J Hepatol. 72:761–773. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Khan S, Ebeling MC, Zaman MS, Sikander M,
Yallapu MM, Chauhan N, Yacoubian AM, Behrman SW, Zafar N, Kumar D,
et al: MicroRNA-145 targets MUC13 and suppresses growth and
invasion of pancreatic cancer. Oncotarget. 5:7599–7609. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
He L, Qu L, Wei L, Chen Y and Suo J:
Reduction of miR 132 3p contributes to gastric cancer proliferation
by targeting MUC13. Mol Med Rep. 15:3055–3061. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Moniaux N, Escande F, Porchet N, Aubert JP
and Batra SK: Structural organization and classification of the
human mucin genes. Front Biosci. 6:D1192–D1206. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Filippou PS, Ren AH, Korbakis D,
Dimitrakopoulos L, Soosaipillai A, Barak V, Frenkel S, Pe'er J,
Lotem M, Merims S, et al: Exploring the potential of mucin 13
(MUC13) as a biomarker for carcinomas and other diseases. Clin Chem
Lab Med. 56:1945–1953. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lim W, Ridge CA, Nicholson AG and
Mirsadraee S: The 8th lung cancer TNM classification and clinical
stageing system: Review of the changes and clinical implications.
Quant Imaging Med Surg. 8:709–718. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Δ Δ C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kumari S, Khan S, Gupta SC, Kashyap VK,
Yallapu MM, Chauhan SC and Jaggi M: MUC13 contributes to rewiring
of glucose metabolism in pancreatic cancer. Oncogenesis. 7:192018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stiles ZE, Khan S, Patton KT, Jaggi M,
Behrman SW and Chauhan SC: Transmembrane mucin MUC13 distinguishes
intraductal papillary mucinous neoplasms from non-mucinous cysts
and is associated with high-risk lesions. HPB (Oxford). 21:87–95.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sheng Y, Ng CP, Lourie R, Shah ET, He Y,
Wong KY, Seim I, Oancea I, Morais C, Jeffery PL, et al: MUC13
overexpression in renal cell carcinoma plays a central role in
tumor progression and drug resistance. Int J Cancer. 140:2351–2363.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dai Y, Liu L, Zeng T, Liang JZ, Song Y,
Chen K, Li Y, Chen L, Zhu YH, Li J, et al: Overexpression of MUC13,
a poor prognostic predictor, promotes cell growth by activating Wnt
signaling in hepatocellular carcinoma. Am J Pathol. 188:378–391.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Doxtater K, Sekhri R, Mishra U, Jaggi M,
Tripathi M and Chauhan S: MUC13 enhances anchorage independent
survival and cooperates with YAP1 and β-catenin towards colorectal
cancer metastasis. Cancer Res. 80 (Suppl):49162020.
|
|
26
|
Wang H, Shen L, Lin Y, Shi Q, Yang Y and
Chen K: The expression and prognostic significance of Mucin 13 and
Mucin 20 in esophageal squamous cell carcinoma. J Cancer Res Ther.
11 (Suppl 1):C74–C79. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li P, Wang H, Hou M, Li D and Bai H:
Upstream stimulating factor1 (USF1) enhances the proliferation of
glioblastoma stem cells mainly by activating the transcription of
mucin13 (MUC13). Pharmazie. 72:98–102. 2017.PubMed/NCBI
|
|
28
|
Gupta BK, Maher DM, Ebeling MC, Sundram V,
Koch MD, Lynch DW, Bohlmeyer T, Watanabe A, Aburatani H, Puumala
SE, et al: Increased expression and aberrant localization of mucin
13 in metastatic colon cancer. J Histochem Cytochem. 60:822–831.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
LaMonte GM, Orjuela-Sanchez P, Calla J,
Wang LT, Li S, Swann J, Cowell AN, Zou BY, Abdel-Haleem Mohamed AM,
Villa Galarce ZH, et al: Dual RNA-seq identifies human mucosal
immunity protein Mucin-13 as a hallmark of Plasmodium
exoerythrocytic infection. Nat Commun. 10:4882019. View Article : Google Scholar : PubMed/NCBI
|