Introduction
Desmoplastic malignant pleural mesothelioma (DMM) is
a rare histological variant of malignant pleural mesothelioma
(MPM), accounting for 5–10% of all cases of MPM. DMM is categorized
as a subtype of sarcomatoid tumor (1) and characterized by dense,
collagenized tissue (>50%) separated by atypical cells arranged
in a storiform or ‘patternless pattern’ in the tumor specimen
(2). The pathological diagnosis of
DMM is difficult, with fibrous pleuritis and reactive mesothelial
hyperplasia as potential differential diagnoses (3). The role of immunohistochemistry is
important for the diagnosis of DMM and broad-spectrum staining of
cytokeratins is crucial to diagnose DMM correctly. In addition, p16
deletion is useful to distinguish DMM from benign pleuritic
(2). The characteristic computed
tomography (CT) findings include unilateral pleural effusion,
thickening of the mediastinal pleura, circumferential and nodular
pleural thickening of >1 cm and interlobar fissure thickening
(4–6). MPM is refractory to chemotherapy and
radiation therapy, and the combination of pemetrexed and cisplatin
has become the standard first-line chemotherapy regimen for MPM
based on the results of a randomized phase III trial (7). However, this regimen has only
improved median survival from 9.3 months for treatment with
cisplatin alone to 12.1 months (7)
and it lacks the impact on survival that second-line therapy is
expected to have (8). Thus,
further investigations for novel therapeutics have long been
desired for MPM and DMM (9).
Patient-derived cancer cells that retain the genetic
and phenotypic profiles of their original tumor tissue are crucial
for the elucidation of molecular mechanisms underlying the
malignant features of tumors and for the development of novel
therapies (10). In particular,
patient-derived cell lines provide a model for screening numerous
anti-cancer agents and investigate their modes of action in a
high-throughput manner. Furthermore, cell lines have a utility in
the development of predictive biomarkers and identification of
therapeutic targets and previous studies have included numerous
cell lines of various cancer types (11–13).
Of note, the availability of cell lines is crucial for drawing the
benefits of modern oncology technology. However, according to the
largest cell line database, Cellosaurus (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5945021/),
only two patient-derived cell lines of DMM, namely TCPH-MM02
(14) and TCPH-MM11 (15), have been reported in the literature
and they are not available from any cell banks. The database of
patient-derived xenografts (PDXs), PDXFinder (16), reveals no PDXs of DMM available
from public biobanks. The paucity of adequate patient-derived
cancer models is one factor hindering the development of novel
therapies for DMM and the establishment of patient-derived DMM
models is an urgent requirement.
In the present study, a novel cell line, designated
NCC-DMM1-C1, was established from tumor tissue of DMM and
characterized. Its utility in high-throughput screening of
anti-cancer agents for anti-proliferative effects was then
evaluated. To the best of our knowledge, the present study is the
third report describing the establishment of a patient-derived cell
line of DMM.
Materials and methods
Patient history
The patient was a 73-year-old male who visited the
National Cancer Center Hospital (Tokyo, Japan) with a major symptom
of progressive dyspnea. The patient had been exposed to asbestos
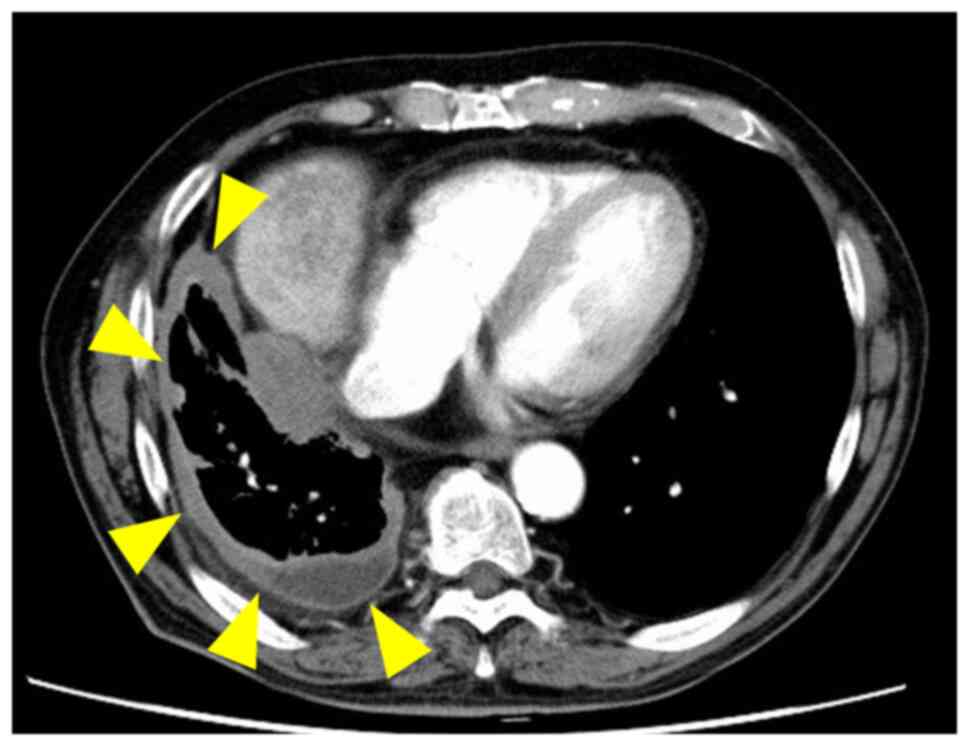
from the age of 25 to 60 years. Enhanced CT detected pleural
thickening and pleural effusion on the right side (Fig. 1A). Pathological diagnosis using
pleural biopsy suggested DMM. Pleurectomy/decortication was
performed and the definitive diagnosis from surgically resected
tumor tissues was DMM (pT2 pN1 cM0 pStage II). The tumor tissue
obtained at the time of surgery was used to establish the cell
line. After seven months, multiple liver metastases occurred and
the patient developed jaundice and died from pneumonia and
respiratory failure due to DMM recurrence.
Cell culture procedure
Cell culture was performed according to a previous
study by our group (17). Informed
consent was obtained from the patient. In brief, the resected tumor
tissue was mechanically dissected with scissors in tissue culture
plates (Thermo Fisher Scientific, Inc.). The cells were maintained
in Dulbecco's modified Eagle's medium (DMEM)/F12 (Sigma-Aldrich;
Merck KGaA) supplemented with 10% heat-inactivated fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U
penicillin G and 100 µg/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.). After reaching sub-confluence, the cells were
treated with 0.25% trypsin-EDTA (Gibco; Thermo Fisher Scientific,
Inc.) and transferred to a new tissue culture plate.
Authentication and quality control of
the established cell line
The cell line was authenticated by examining the
short tandem repeats (STRs) in 10 loci using the GenePrint 10
System (Promega Corporation), as previously described (17). Genomic DNA was extracted from the
cells and the tumor tissue using a DNeasy Blood & Tissue kit
(Qiagen GmbH) and quantified using a NanoDrop 8000
Spectrophotometer (Thermo Fisher Scientific, Inc.). STRs were
amplified and sequenced using a 3500×L Genetic Analyzer (Applied
Biosystems; Thermo Fisher Scientific, Inc.). STR profiling was
performed using the GeneMapper Software (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The similarity of STRs among the reported
cell lines was examined using the CLASTR function of Cellosaurus
(18). Based on the Tanabe
algorithm (19), the STR match
ratio between the established cell lines and the corresponding
original tumor was calculated. The score met the standard match
threshold of 80% (19) based on
Tanabe algorithms and non-empty marker mode that computed the score
to handle cases in which allele data is missing for the query or
the reference. Mycoplasma DNA was assessed in the tissue
culture medium using an e-Myco Mycoplasma PCR Detection Kit (Intron
Biotechnology).
Single nucleotide polymorphism (SNP)
array
Copy number alterations were examined using an
Infinium OmniExpressExome-8 v.1.4 BeadChip (Illumina, Inc.)
following the manufacturer's protocol. Detailed descriptions of
these procedures are provided in previous reports (20). Abnormal copy number regions were
detected using the circular binary segmentation algorithm (21,22)
using the R package DNAcopy from Bioconductor (23). Amplifications were defined as
regions for which the copy number was >3. Deletions were defined
as regions of which <1 copy was present in the tumor cells.
Among identified genes with copy number alterations, a search for
‘cancer-related genes’ was performed in the Cancer Gene Census in
the Catalogue Of Somatic Mutations In Cancer database (GRCh 37
v.91) (24).
Histological evaluation
All specimens for H&E staining were cut into
4-µm slices and placed on the slides. After deparaffinization, the
nuclei were immersed in hematoxylin staining solution (Muto
Chemical) for 15 min, followed by washing out the solution with tap
water. Subsequently, cytoplasm and stromal matrix were stained with
eosin (Muto Chemical). The slides were washed in water and mounted.
Immunohistochemical staining was performed on formalin-fixed,
paraffin-embedded specimens obtained through surgery and on
NCC-DMM1-C1 cells. NCC-DMM1-C1 cells were detached by treatment
with trypsin and cell suspensions were solidified using iPGell
(Genostaff) according to the manufacturer's protocol. Cell masses
were fixed with 10% formalin and embedded in paraffin. Cell blocks
were cut into sections that were processed for immunolabeling and
H&E staining. Sections were incubated with antibodies against
various proteins. The expression of cytokeratin, D2-40, HEG1 and
WT1 was evaluated using the following primary antibodies:
Cytokeratin AE1/3 (AE1/AE3; 1:200 dilution; cat. no. sc-81714;
Santa Cruz Biotechnology, Inc.), D2-40 (D2-40; 1:200 dilution; cat.
no. PDM558; Diagnostic Biosystem), sialylated heart development
protein with EGF-like domains 1 (SKM9-2; prediluted; cat. no.
418231; Nichirei Biosciences, Inc.) and WT1 (6F-H2; M3561, 1:50
dilution; Agilent Technologies, Inc.). Antigen detection was
performed using a Dako Autostainer and EnVision Detection System
(Dako; Agilent Technologies, Inc.) according to the manufacturer's
protocol. The slides were counterstained with hematoxylin.
Cell proliferation assay
The cells were seeded into 24-well culture plates at
a density of 2.5×104 cells/well as described previously
(17). The cell viability was
monitored using Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc.). The absorbance of each well at 450 nm was
recorded at multiple time-points using a microplate reader (Bio-Rad
Laboratories, Inc.). Growth curves were constructed by plotting the
absorbance as a function of culture time and these were used to
estimate the population doubling time. The cells were maintained in
a humidified atmosphere of 5% CO2 at 37°C.
Spheroid formation assay
Spheroid formation was performed by placing
1×104 cells in a 96-well Clear Flat Bottom Ultra-Low
Attachment Microplate (Corning, Inc.) in DMEM/F12 medium containing
10% FBS, as described previously (17). After three days of plating,
spheroid formation was monitored using a BZ-9000 fluorescence
microscope (Keyence). Spheroid formation assays were performed in
duplicate.
Transwell cell invasion assay
The invasive capability of NCC-DMM1-C1 cells and
MG63 osteosarcoma cells (The Japanese Collection of Research
Bioresources) (25) was measured
as previously described (17).
Cells (1×105 and 2×105) were seeded in the
upper chamber. Following incubation, the cells in the three
separate areas were counted under a microscope (Keyence) at ×100
magnification.
Tyrosine kinase activity assay
Tyrosine kinase activity was examined for multiple
kinases using the PamChip TK peptide microarray system (PamGene
International B.V.) as previously described (26). The protein lysate (5 µg) was
extracted from the tissue cultured cells and tumor tissue and
hybridized to the membrane array, which included 144 peptides. The
average signal intensity of the 144 hybridized peptides based on
the end levels of the phosphorylation curve was used for the
activity analysis. All data analyses were performed using
BioNavigator v.6.3.67.0 (PamGene International B.V.). Active
kinases were predicted using PhosphoSitePlus (27), the UniProt database (28) and the Human Protein Reference
Database (29).
Screening for anti-proliferative
effects of anti-cancer agents
The anti-proliferative effects of 213 anti-cancer
agents (Table SI) were examined
using the CCK-8 assay, as described previously (17). The cells were seeded at
5×103 cells/well in DMEM/F12 medium supplemented with
10% FBS using the Bravo Automated Liquid Handling Platform (Agilent
Technologies, Inc.). The following day, anti-cancer agent compounds
(10 µM; Selleck Chemicals) and 0.1% DMSO were added using the Bravo
Automated Liquid Handler. After 72 h, survival rates were assessed
using the CCK-8 reagent according to the manufacturer's protocol.
The response rate was calculated relative to that of the DMSO
control. Dose-response experiments were performed to validate the
candidate anti-cancer agents that were identified during the pilot
screening. The compounds were dispensed into 384-well plates with
serial dilution at 10 different concentrations, ranging from 0.1 to
100,000 nM, using an Echo 555 Acoustic Liquid Handler (Labcyte
Inc.) and IC50 values were determined. CCK-8 absorbance
was measured using an Epoch multimode multiplate reader (BioTek
Instruments). Absorbance values were plotted as a function of the
compound concentrations to obtain IC50 values using
GraphPad Prism 8.1.0 software (GraphPad Inc.). This screening was
performed in duplicate.
Results
Authentication and quality control of
the established cell line
NCC-DMM1-C1 was maintained for >16 months and
passaged >30 times under tissue culture conditions. The
characterization of NCC-DMM1-C1 cells was performed by STR
profiling at the 20th passage. The same patterns of the peaks in
all loci composed of D5S818, D13S317, D7S820, D16S539, vWA, TH01,
TP0X, CSF1P0, and the sex chromosomal marker amelogenin were
observed in both the normal tissues and the NCC-DMM1-C1 cells. The
normal tissues were composed of the adjacent tissue to the tumor
tissue which did not include the tumor cells near the chest wall.
The authentication indicated that the normal tissues and the
NCC-DMM1-C1 cells were derived from the same origin (Table I, Fig. S1). Calculated based on the Tanabe
algorithm (19), the STR match
ratio between the NCC-DMM1-C1 cells and the corresponding original
tumor was 100%. The score met the standard match threshold of 80%
(19). According to Cellosaurus,
the profile did not match that of cell lines deposited in public
cell banks. It was therefore indicated that NCC-DMM1-C1 is a novel
cell line. The DNA sequence of Mycoplasma was not detected
in the tissue culture medium (data not shown) and it was concluded
that NCC-DMM1-C1 was not contaminated with Mycoplasma.
 | Table I.Results of short tandem repeat
analysis. |
Table I.
Results of short tandem repeat
analysis.
|
| Allele number |
|---|
|
|
|
|---|
| STR locus
(chromosome) | NCC-DMM1-C1 | Normal tissue |
|---|
| Amelogenin (X
Y) | X, Y | X, Y |
| TH01 (3) | 9 | 9 |
| D21S11 (21) | 29, 32.2 | 29, 32.2 |
| D5S818 (5) | 9, 11 | 9, 11 |
| D13S317 (13) | 12 | 12 |
| D7S820 (7) | 8 | 8 |
| D16S539 (16) | 9, 12 | 9, 12 |
| CSF1PO (5) | 12 | 12 |
| vWA (12) | 18, 19 | 18, 19 |
| TPOX (2) | 8, 11 | 8, 11 |
Cell line characteristics
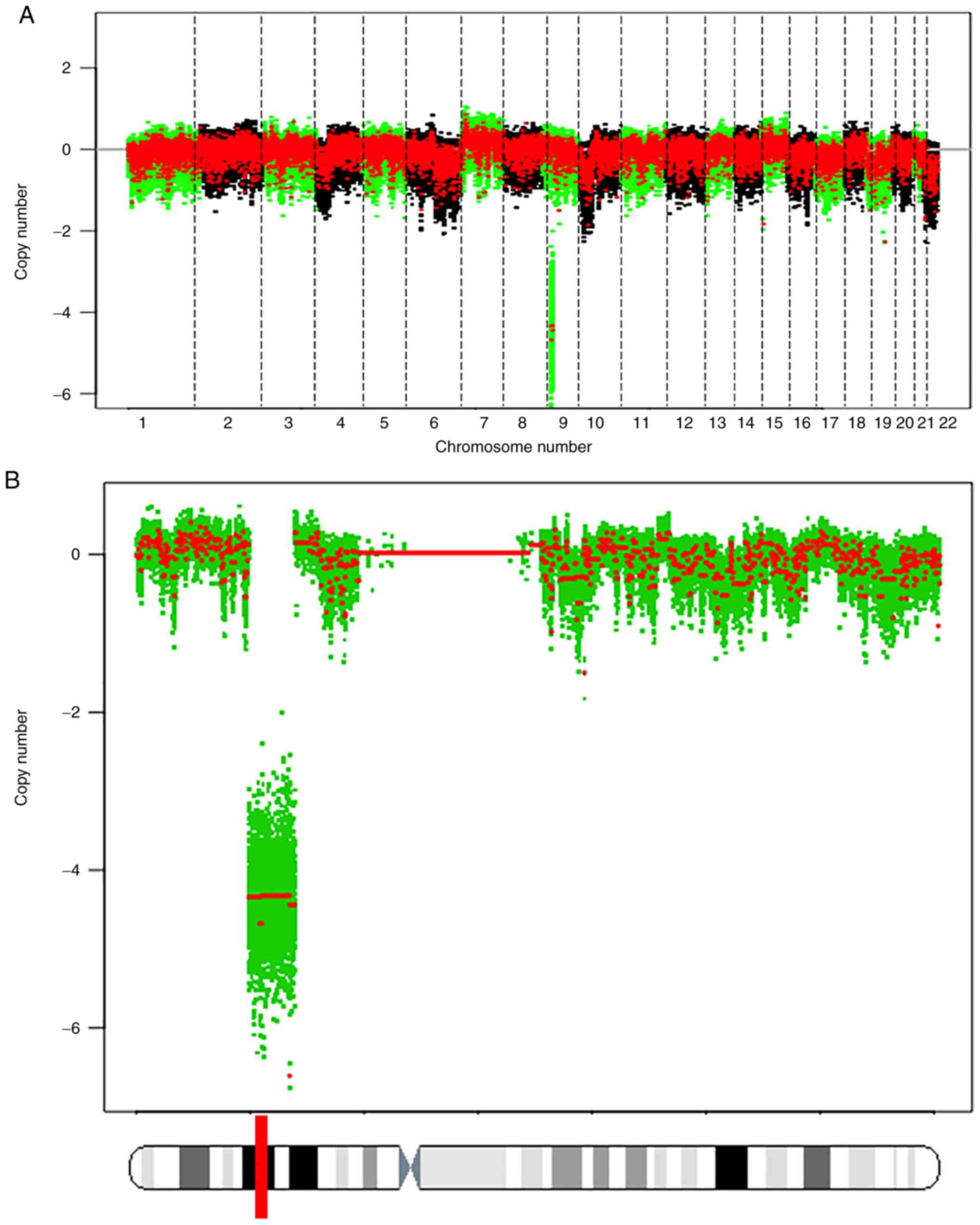
SNP array experiments revealed the presence of copy
number variants mostly involving partial deletions of chromosomal
arms (9p) (Fig. 2A) and
cyclin-dependent kinase inhibitor 2A was located in the loci of
deletions (Fig. 2B). NCC-DMM1-C1
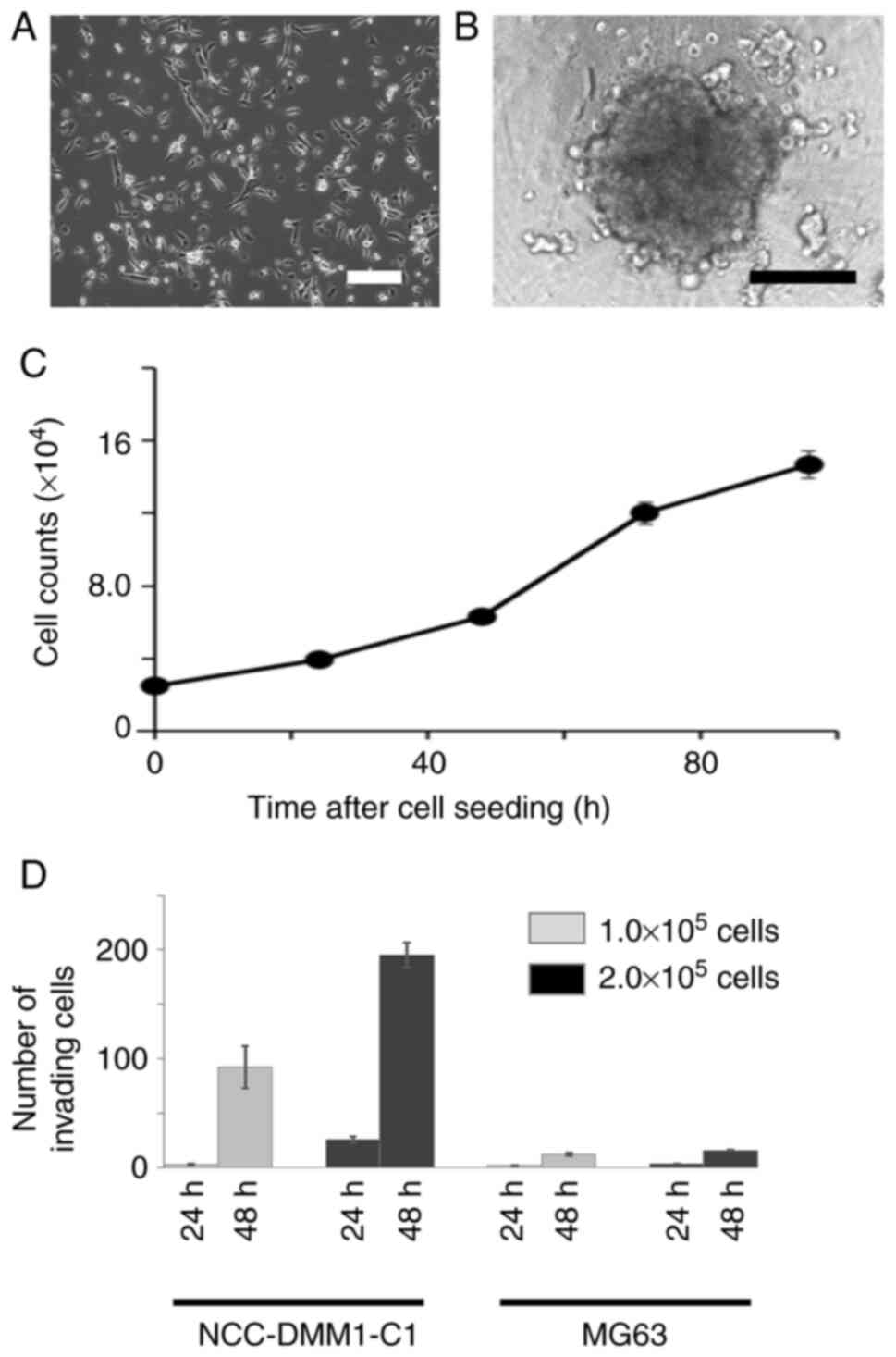
cells exhibited a polygonal or spindle-shaped cytoplasm, arranged
in a partially cohesive sheet-like structure, with enlarged and
variably sized nuclei (Fig. 3A).
The NCC-DMM1-C1 cells formed spheroids in low-attachment substrates
(Fig. 3B) and exhibited constant
growth (Fig. 3C). The population
doubling time based on the growth curve was 25.9 h (Fig. 3C). The invasion capability of
NCC-DMM1-C1 cells was higher than that of MG63 osteosarcoma cells
(Figs. 3D and S2).
Histological evaluation
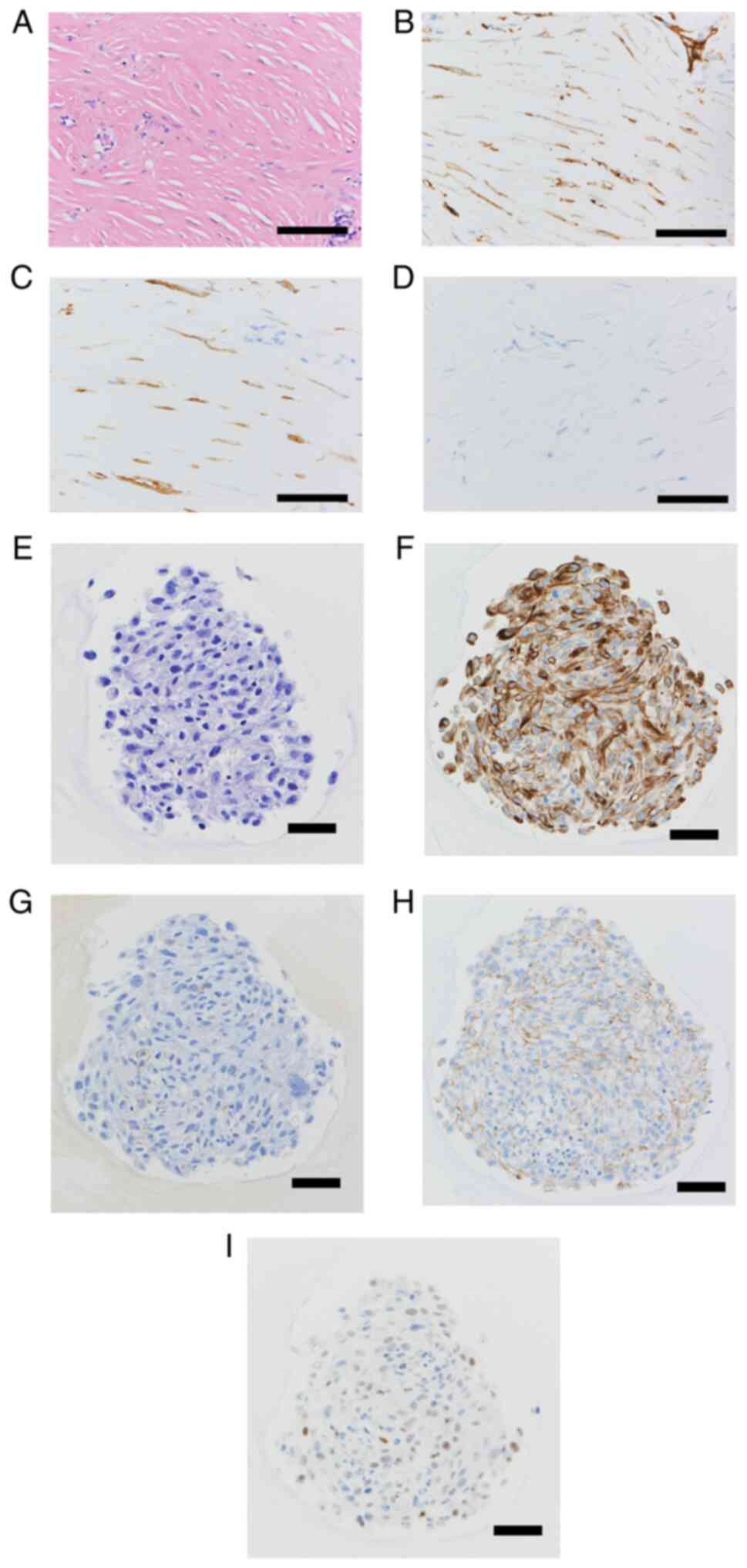
H&E staining of the sectioned tissue indicated
that the spindle-shaped cells had a storiform pattern and
proliferated densely (Fig. 4A).
Immunohistochemistry revealed diffusely positive immunoreactivity
for cytokeratin AE1/AE3 (Fig. 4B)
and D2-40 (Fig. 4C) and negative
immunoreactivity for WT1 (Fig.
4D). H&E staining of the NCC-DMM1-C1 cells indicated the
same pattern as that of the tissue section with a storiform pattern
and proliferated density and the composition of one type of cells,
i.e. tumor cells with no other cells (Fig. 4E). Immunohistochemistry revealed
diffusely positive staining for cytokeratin AE1/AE3 (Fig. 4F), negativity for D2-40 (Fig. 4G), weakly but diffusely positive
staining for HEG1 (Fig. 4H) and
mostly positive staining for WT1 (Fig.
4I).
Kinase activity assay
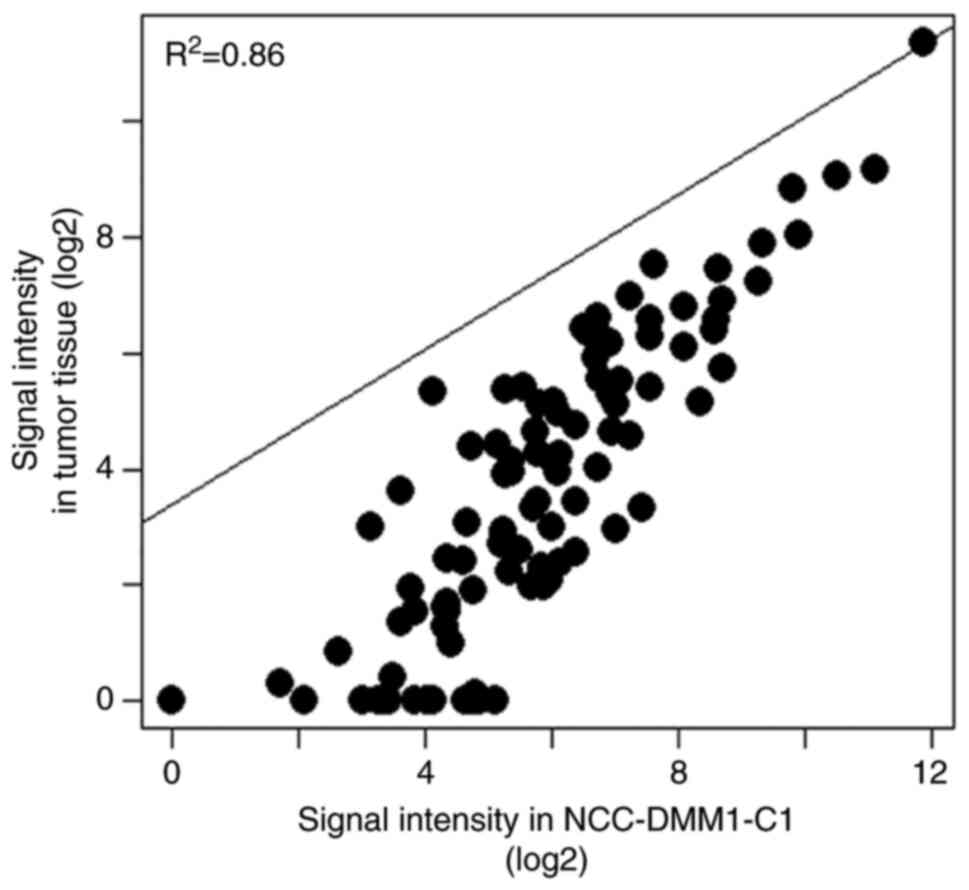
Pearson's correlation scatter plot revealed high
similarity of kinase activity between NCC-DMM1-C1 cells and tumor
tissue (r2=0.86; Fig.
4, Table SII). The 25 most
highly phosphorylated peptides were identified in both NCC-DMM1-C1
cells and tumor tissue (Table
SIII). These peptides included FES, Wee1, platelet-derived
growth factor receptor (PDGFR)-β and Src (Table SIII).
Sensitivity to anti-cancer agents
The IC50 values of 13 of the anti-cancer
agents applied were calculated, including four anti-cancer agents
used for DMM and nine drugs that had relatively high inhibitory
effects on cancer cell proliferation at 10 µM (Fig. S3, Table SIV). The calculated
IC50 values are summarized in Table II and the representative growth
curves of five anti-cancer agents with IC50 values
<100 nM (bortezomib, gemcitabine, homoharringtonine,
fostamatinib and vinorelbine) are displayed in Fig. 5.
 | Table II.Summary of IC50
values. |
Table II.
Summary of IC50
values.
| CAS no. | Name of drug | IC50
(µM) |
|---|
| 15663-27-1 | Cisplatin | 5.38 |
| 95058-81-4 | Gemcitabine | 0.053 |
| 901119-35-5 | Fostamatinib | 0.097 |
| 443913-73-3 | Vandetanib | 13.75 |
| 366017-09-6 | Mubritinib | 0.43 |
| 179324-69-7 | Bortezomib | 0.055 |
| 26833-87-4 |
Homoharringtonine | 0.072 |
| 65271-80-9 | Mitoxantrone | 0.26 |
| 267243-28-7 | Canertinib | 0.76 |
| 357166-30-4 | Pemetrexed disodium
hydrate | 0.49 |
| 149647-78-9 | Vorinostat | 1.26 |
| 71486-22-1 | Vinorelbine | 0.04 |
| 252916-29-3 | Orantinib | 30.06 |
Discussion
DMM is an aggressive neoplasm with poor prognosis.
The effects of surgery, chemotherapy and radiotherapy have remained
to be fully defined. Thus, it is necessary to improve the current
knowledge regarding the molecular mechanisms underlying disease
progression of DMM and develop assay systems to evaluate novel
anti-cancer agents. However, due to the rarity of the disease, only
a small number of DMM cell lines have been reported and cell lines
and xenografts for DMM are not readily available from public cell
banks. The present study reported on the establishment a novel DMM
cell line, designated as NCC-DMM1-C1 and determined its
characteristics. Furthermore, anti-cancer drugs were screened using
NCC-DMM1-C1 cells to evaluate whether the cell line may be used to
screen drug candidates or not.
NCC-DMM1-C1 cells exhibited a spindle cell
morphology and demonstrated constant growth and aggressive
invasion, which reflect the characteristics of the original tumor.
The spheroid formation capability contributes to the understanding
of the behavior of DDM in a 3D environment. While NCC-DMM1-C1 cells
had these features reflecting the original tumor, the
immunohistochemical staining for D2-40 and WT1 was different
between the tumor tissue and NCC-DMM1-C1 cells. NCC-DMM1-C1 cells
stained negative for D2-40 expression and the tumor tissue
demonstrated positive staining for D2-40 expression. Furthermore,
NCC-DMM1-C1 cells demonstrated mostly positive WT1 expression and
the tumor tissue demonstrated negative WT1 expression. Tumor tissue
of DMM is composed of collagen tissue occupying >50% of the
tumor and tumor cells (2) and
NCC-DMM1-C1 was composed of tumor cells only. In addition, the
expression of D2-40 and WT1 in NCC-DMM1-C1 cells was different from
that of the tumor tissue due to heterogeneity. The cell components
and the heterogeneity may have led to the distinct differences in
D2-40 and WT1 expression.
The drug screening assay identified bortezomib,
gemcitabine, homoharringtonine, fostamatinib and vinorelbine as
anti-cancer agents with low IC50 values in NCC-DMM1-C1.
Gemcitabine is a DNA-damaging agent and has been investigated as a
first-line single treatment in chemotherapy-naive patients or as a
second- or third-line combination therapy in patients with MPM
(8,30,31).
Vinorelbine is a microtubule-damaging agent and its efficacy has
been reported in patients with progressive disease after treatment
with pemetrexed-platinum chemotherapy (8,31,32).
Bortezomib is a proteasome inhibitor and its cytotoxic effects were
revealed in a preclinical study using six cell lines of MPM
(33). However, two subsequent
Phase II studies did not reach the desired endpoint in MPM
(34,35). Further clinical trials are
necessary to clarify the clinical utility of these three
anti-cancer drugs for DMM. Homoharringtonine, a natural plant
alkaloid extracted from Cephalotaxus harringtonia used in
Chinese Traditional Medicine (36), has efficient inhibitory activity
against myelocytic leukemia (37,38).
Fostamatinib is a pro-drug inhibitor of spleen tyrosine kinase,
which is a key mediator of Fc and B-cell receptor signaling in
inflammatory cells and has efficacy in non-Hodgkin lymphoma and
chronic lymphocytic leukemia (39). The treatment utility of
homoharringtonine and fostamatinib in MPM has not been previously
reported and it is worth elucidating their effects on MPM using
NCC-DMM1-C1 cells.
Comprehensive kinase activity assays revealed that
kinase activity was similar between NCC-DMM1-C1 cells and the
original tumor tissue, suggesting that the NCC-DMM1-C1 cell line
may be useful for examining the effects of kinase inhibitors in
vitro. It was also determined that the kinases FES, Wee1,
PDGFR-β and Src were highly activated in both NCC-DMM1-C1 cells and
the tumor tissue. Tsao et al (40) demonstrated that Src was expressed
and activated in MPM cell lines and 46 MPM tumor specimens.
Additionally, the MPM cell lines were sensitive to an Src
inhibitor, as determined using in vitro cytotoxicity assays.
Wee1 is a tyrosine kinase that phosphorylates and inactivates
cyclin-dependent kinase 1 and is involved in G2 checkpoint
signaling (41). Xu et al
(41) reported that a kinome-wide
CRISPR/Cas9 knockout screen identified Wee1, the loss of function
of which sensitizes cells to standard combination cisplatin and
pemetrexed chemotherapy in MPM cell lines and PDXs. Several groups
have reported that the PDGF/PDGFR pathway is involved in
mesothelioma carcinogenesis (42,43).
Melaiu et al (44) reported
that, in MPM cells, PDGFRB silencing causes a decrease in the
proliferation rate and reduces the colony formation capacity, which
highlights the utility of PDGFR-β as a drug target. In the present
study, high FES activity was observed, which has not been
previously reported in MPM, to the best of our knowledge.
Experiments to evaluate kinase activity cannot be perfect and to
compensate for the drawbacks of each experiment, it is required to
use different types of experiments prior to clinical trials.
Therefore, kinase activities identified in the present study need
to be validated using other methods such as western blot analysis.
The present findings confirm that kinase activity may be used as a
drug target and biomarker to assess the utility of kinase
inhibitors.
In conclusion, in the present study, a novel cell
line was established from tumor tissue of DMM, designated as
NCC-DMM1-C1, and its characteristics and utility in high-throughput
drug screening were demonstrated. However, the results of the
present study were obtained from only a single cell line; further
cell lines from different patients are required to generate
reliable results. In addition, the present results regarding the
candidate anti-cancer drugs should be validated in other
patient-derived cancer models, including organoids and xenografts,
prior to clinical trials. In addition, the candidate anti-cancer
drugs were identified using the drug screening based on a single
agent. Reflecting the practical clinical situation, drug screening
based on several agents of the combination treatment will be
performed in the future. Furthermore, evaluation of highly
phosphorylated kinases and the relevant molecular pathway should be
investigated by other proteomics techniques such as mass
spectrometry. The present findings, along with future validations,
will provide a deeper understanding of DMM, suggesting the possible
utility of NCC-DMM1-C1 cells in drug development.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
The authors appreciate the technical support
provided by Miss Yu Kuwata (Division of Rare Cancer Research,
National Cancer Center, Tokyo, Japan).
Funding
This research was supported by the Japan Agency for Medical
Research and Development (grant no. 20ck0106537h0001).
Availability of data and materials
The cell line of the current study is available from
the corresponding author on reasonable request. The datasets
generated during the current study are available from the Gene
Expression Omnibus (GEO) repository (45). The present data of the peptide
microarray are accessible through GEO (series accession no.
GSE185107; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185107)
and the data of the SNP array are accessible through GEO (series
accession no. GSE185549; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185549).
Authors' contributions
RN and YYoshimatsu established the cell line. NM and
YYa performed the pathological diagnoses and immunohistochemical
staining. SW, YYoshida and AS prepared tumor tissues and obtained
clinical information of the donor patient. RN, YYoshimatsu and TO
performed all experiments, including authentication, drug screening
assay and kinase assay. TK was responsible for study conception and
design. RN, YYoshimatsu and TK prepared the manuscript, including
the figures and tables. RN, YYoshimatsu and TK edited the
manuscript. All authors read and approved the final manuscript
prior to submission. RN and TK checked and confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
The use of clinical materials for this study was
approved by the ethics committee of the National Cancer Center
(approval no. 2015-108) and written informed consent was obtained
from the patient. The informed consent was for collection of
tissues from the patient for the present study and publication of
data obtained using the tissues. All procedures followed were in
accordance with the ethical standards of the responsible committee
on human experimentation (institutional and national) and with the
Helsinki Declaration.
Patient consent for publication
Although detailed clinical data were not included in
the present study, consent for publication was obtained from the
patient included in the present study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gibbs AR and Thunnissen FB: Histological
typing of lung and pleural tumours: Third edition. J Clin Pathol.
54:498–499. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Galateau-Salle F, Churg A, Roggli V and
Travis WD; World Health Organization Committee for Tumors of the
Pleura, : The 2015 World Health Organization classification of
tumors of the pleura: Advances since the 2004 classification. J
Thorac Oncol. 11:142–154. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hashimoto K, Okuma Y, Hosomi Y and Hishima
T: Malignant mesothelioma of the pleura with desmoplastic
histology: A case series and literature review. BMC Cancer.
16:7182016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Leung AN, Müller NL and Miller RR: CT in
differential diagnosis of diffuse pleural disease. AJR Am J
Roentgenol. 154:487–492. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kawashima A and Libshitz HI: Malignant
pleural mesothelioma: CT manifestations in 50 cases. AJR Am J
Roentgenol. 155:965–969. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang ZJ, Reddy GP, Gotway MB, Higgins CB,
Jablons DM, Ramaswamy M, Hawkins RA and Webb WR: Malignant pleural
mesothelioma: Evaluation with CT, MR imaging, and PET.
Radiographics. 24:105–119. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vogelzang NJ, Rusthoven JJ, Symanowski J,
Denham C, Kaukel E, Ruffie P, Gatzemeier U, Boyer M, Emri S,
Manegold C, et al: Phase III study of pemetrexed in combination
with cisplatin versus cisplatin alone in patients with malignant
pleural mesothelioma. J Clin Oncol. 21:2636–2644. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zauderer MG, Kass SL, Woo K, Sima CS,
Ginsberg MS and Krug LM: Vinorelbine and gemcitabine as second- or
third-line therapy for malignant pleural mesothelioma. Lung Cancer.
84:271–274. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Christoph DC and Eberhardt WE: Systemic
treatment of malignant pleural mesothelioma: New agents in clinical
trials raise hope of relevant improvements. Curr Opin Oncol.
26:171–181. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sharma SV, Haber DA and Settleman J: Cell
line-based platforms to evaluate the therapeutic efficacy of
candidate anticancer agents. Nat Rev Cancer. 10:241–253. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Barretina J, Caponigro G, Stransky N,
Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV,
Sonkin D, et al: The cancer cell line encyclopedia enables
predictive modelling of anticancer drug sensitivity. Nature.
483:603–607. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iorio F, Knijnenburg TA, Vis DJ, Bignell
GR, Menden MP, Schubert M, Aben N, Gonçalves E, Barthorpe S,
Lightfoot H, et al: A landscape of pharmacogenomic interactions in
cancer. Cell. 166:740–754. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Teicher BA, Polley E, Kunkel M, Evans D,
Silvers T, Delosh R, Laudeman J, Ogle C, Reinhart R, Selby M, et
al: Sarcoma cell line screen of oncology drugs and investigational
agents identifies patterns associated with gene and microRNA
expression. Mol Cancer Ther. 14:2452–2462. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Oey H, Daniels M, Relan V, Chee TM,
Davidson MR, Yang IA, Ellis JJ, Fong KM, Krause L and Bowman RV:
Whole-genome sequencing of human malignant mesothelioma tumours and
cell lines. Carcinogenesis. 40:724–734. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Relan V, Morrison L, Parsonson K, Clarke
BE, Duhig EE, Windsor MN, Matar KS, Naidoo R, Passmore L, McCaul E,
et al: Phenotypes and karyotypes of human malignant mesothelioma
cell lines. PLoS One. 8:e581322013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Conte N, Mason JC, Halmagyi C, Neuhauser
S, Mosaku A, Yordanova G, Chatzipli A, Begley DA, Krupke DM,
Parkinson H, et al: PDX Finder: A portal for patient-derived tumor
xenograft model discovery. Nucleic Acids Res. 47(D1): D1073–D1079.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yoshimatsu Y, Noguchi R, Tsuchiya R, Kito
F, Sei A, Sugaya J, Nakagawa M, Yoshida A, Iwata S, Kawai A and
Kondo T: Establishment and characterization of NCC-CDS2-C1: A novel
patient-derived cell line of CIC-DUX4 sarcoma. Hum Cell.
33:427–436. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bairoch A: The cellosaurus, a cell-line
knowledge resource. J Biomol Tech. 29:25–38. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Capes-Davis A, Reid YA, Kline MC, Storts
DR, Strauss E, Dirks WG, Drexler HG, MacLeod RA, Sykes G, Kohara A,
et al: Match criteria for human cell line authentication: Where do
we draw the line? Int J Cancer. 132:2510–2519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Noguchi R, Yoshimatsu Y, Ono T, Sei A,
Hirabayashi K, Ozawa I, Kikuta K and Kondo T: Establishment and
characterization of NCC-PLPS1-C1, a novel patient-derived cell line
of pleomorphic liposarcoma. Hum Cell. 34:688–697. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Olshen AB, Venkatraman ES, Lucito R and
Wigler M: Circular binary segmentation for the analysis of
array-based DNA copy number data. Biostatistics. 5:557–572. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Venkatraman ES and Olshen AB: A faster
circular binary segmentation algorithm for the analysis of array
CGH data. Bioinformatics. 23:657–663. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Willenbrock H and Fridlyand J: A
comparison study: Applying segmentation to array CGH data for
downstream analyses. Bioinformatics. 21:4084–4091. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Forbes SA, Tang G, Bindal N, Bamford S,
Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, et al: COSMIC
(the catalogue of somatic mutations in cancer): A resource to
investigate acquired mutations in human cancer. Nucleic Acids Res
38 (Database Issue). D652–D657. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Billiau A, Edy VG, Heremans H, Van Damme
J, Desmyter J, Georgiades JA and De Somer P: Human interferon: Mass
production in a newly established cell line, MG-63. Antimicrob
Agents Chemother. 12:11–15. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sikkema AH, Diks SH, den Dunnen WF, ter
Elst A, Scherpen FJ, Hoving EW, Ruijtenbeek R, Boender PJ, de Wijn
R, Kamps WA, et al: Kinome profiling in pediatric brain tumors as a
new approach for target discovery. Cancer Res. 69:5987–5995. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hornbeck PV, Zhang B, Murray B, Kornhauser
JM, Latham V and Skrzypek E: PhosphoSitePlus, 2014: Mutations, PTMs
and recalibrations. Nucleic Acids Res 43 (Database Issue).
D512–D520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
UniProt Consortium: UniProt: A worldwide
hub of protein knowledge. Nucleic Acids Res. 47(D1): D506–D515.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Keshava Prasad TS, Goel R, Kandasamy K,
Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R,
Shafreen B, Venugopal A, et al: Human protein reference
database-2009 update. Nucleic Acids Res 37 (Database Issue).
D767–D772. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
van Meerbeeck JP, Baas P, Debruyne C,
Groen HJ, Manegold C, Ardizzoni A, Gridelli C, van Marck EA, Lentz
M and Giaccone G: A Phase II study of gemcitabine in patients with
malignant pleural mesothelioma. European organization for research
and treatment of cancer lung cancer cooperative group. Cancer.
85:2577–2582. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zucali PA, Ceresoli GL, Garassino I, De
Vincenzo F, Cavina R, Campagnoli E, Cappuzzo F, Salamina S, Soto
Parra HJ and Santoro A: Gemcitabine and vinorelbine in
pemetrexed-pretreated patients with malignant pleural mesothelioma.
Cancer. 112:1555–1561. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zucali PA, Perrino M, Lorenzi E, Ceresoli
GL, De Vincenzo F, Simonelli M, Gianoncelli L, De Sanctis R,
Giordano L and Santoro A: Vinorelbine in pemetrexed-pretreated
patients with malignant pleural mesothelioma. Lung Cancer.
84:265–270. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Szulkin A, Nilsonne G, Mundt F, Wasik AM,
Souri P, Hjerpe A and Dobra K: Variation in drug sensitivity of
malignant mesothelioma cell lines with substantial effects of
selenite and bortezomib, highlights need for individualized
therapy. PLoS One. 8:e659032013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
O'Brien ME, Gaafar RM, Popat S, Grossi F,
Price A, Talbot DC, Cufer T, Ottensmeier C, Danson S, Pallis A, et
al: Phase II study of first-line bortezomib and cisplatin in
malignant pleural mesothelioma and prospective validation of
progression free survival rate as a primary end-point for
mesothelioma clinical trials (European organisation for research
and treatment of cancer 08052). Eur J Cancer. 49:2815–2822. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fennell DA, McDowell C, Busacca S, Webb G,
Moulton B, Cakana A, O'Byrne KJ, Meerbeeck JV, Donnellan P,
McCaffrey J and Baas P: Phase II clinical trial of first or
second-line treatment with bortezomib in patients with malignant
pleural mesothelioma. J Thorac Oncol. 7:1466–1470. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang CC, Han CS, Yue XF, Shen CM, Wang
SW, Wu FG and Xu B: Cytotoxicity and sister chromatid exchanges
induced in vitro by six anticancer drugs developed in the People's
Republic of China. J Natl Cancer Inst. 71:841–847. 1983.PubMed/NCBI
|
|
37
|
Kantarjian HM, Keating MJ, Walters RS,
Koller CA, McCredie KB and Freireich EJ: Phase II study of low-dose
continuous infusion homoharringtonine in refractory acute
myelogenous leukemia. Cancer. 63:813–817. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Quintás-Cardama A, Kantarjian H,
Garcia-Manero G, O'Brien S, Faderl S, Estrov Z, Giles F, Murgo A,
Ladie N, Verstovsek S and Cortes J: Phase I/II study of
subcutaneous homoharringtonine in patients with chronic myeloid
leukemia who have failed prior therapy. Cancer. 109:248–255. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Friedberg JW, Sharman J, Sweetenham J,
Johnston PB, Vose JM, Lacasce A, Schaefer-Cutillo J, De Vos S,
Sinha R, Leonard JP, et al: Inhibition of Syk with fostamatinib
disodium has significant clinical activity in non-Hodgkin lymphoma
and chronic lymphocytic leukemia. Blood. 115:2578–2585. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tsao AS, He D, Saigal B, Liu S, Lee JJ,
Bakkannagari S, Ordonez NG, Hong WK, Wistuba I and Johnson FM:
Inhibition of c-Src expression and activation in malignant pleural
mesothelioma tissues leads to apoptosis, cell cycle arrest, and
decreased migration and invasion. Mol Cancer Ther. 6:1962–1972.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu D, Liang SQ, Yang H, Bruggmann R,
Berezowska S, Yang Z, Marti TM, Hall SRR, Gao Y, Kocher GJ, et al:
CRISPR screening identifies WEE1 as a combination target for
standard chemotherapy in malignant pleural mesothelioma. Mol Cancer
Ther. 19:661–672. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dorai T, Kobayashi H, Holland JF and
Ohnuma T: Modulation of platelet-derived growth factor-beta mRNA
expression and cell growth in a human mesothelioma cell line by a
hammerhead ribozyme. Mol Pharmacol. 46:437–444. 1994.PubMed/NCBI
|
|
43
|
Langerak AW, Dirks RP and Versnel MA:
Splicing of the platelet-derived-growth-factor A-chain mRNA in
human malignant mesothelioma cell lines and regulation of its
expression. Eur J Biochem. 208:589–596. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Melaiu O, Catalano C, De Santi C,
Cipollini M, Figlioli G, Pellè L, Barone E, Evangelista M,
Guazzelli A, Boldrini L, et al: Inhibition of the platelet-derived
growth factor receptor beta (PDGFRB) using gene silencing,
crenolanib besylate, or imatinib mesylate hampers the malignant
phenotype of mesothelioma cell lines. Genes Cancer. 8:438–452.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|