Introduction
Liver cancer is the fourth leading cause of
cancer-related mortality and is ranked sixth in terms of cancer
incidence; it is estimated that over one million individuals will
succumb to the disease by 2030 (1). In the US, the 5-year survival rate
for patients with liver cancer is 18.1% (2), while in China, it is only 12.1%
(3). The majority of primary liver
cancers are pathologically diagnosed as hepatocellular carcinoma,
accounting for 70-85% of cases (4). Previous studies have reported that
the majority of liver cancers occur in patients with chronic liver
diseases, including hepatitis B virus or hepatitis C virus
infection (5), non-alcoholic fatty
liver disease (6) and alcohol
abuse (7). To date, surgical
excision, radiofrequency ablation, interventional embolization,
liver transplantation, as well as targeted and systemic therapies
have been applied in the treatment of liver cancer, which have
improved the prognosis of those patients to a certain extent
(8). Thus, further elucidation of
the molecular mechanisms involved in the occurrence and development
of liver cancer is warranted, as this may contribute to the
development of novel preventive, diagnostic and therapeutic
strategies for liver cancer.
Previous studies have indicated that zinc, an
important nutrient (9), has a key
role in organisms, including growth, development, reproduction,
enzyme function, immune function, DNA repair, gene expression and
endocrine function, as well as cancer biology (9–12).
Under normal conditions, 98% of zinc is localized in the
intracellular compartment (11).
The balance of intracellular and extracellular zinc is regulated by
the Zrt- and Irt-like proteins [ZIP/solute carrier (SLC)39 family,
SLC39A] and the SLC30 family (SLC30A/ZnT) (13,14).
The ZnT family member functions to move zinc from the inside of the
cell to the outside (13), while
the ZIP family member (SLC39A1-14) serves to pass zinc into the
cytoplasm (14). An imbalance of
zinc homeostasis may be associated with the development of a number
of diseases. It has been demonstrated that zinc levels in prostate
cancer cells are significantly reduced and are associated with
metabolic reprogramming (15).
However, increased zinc levels may also be associated with
tumorigenesis and progression; for instance, SLC39A8 expression has
been determined to be upregulated in early renal clear cell
carcinoma (16), and SLC39A7 has
been reported to have a key role in the growth and survival of
breast cancer cells (17). To the
best of our knowledge, the role of the SLC39A family in liver
cancer has remained elusive. The present study performed an
integrated bioinformatics analysis of several independent studies
to explore the role of the SLC39A family in the development and
progression of liver cancer. In addition, the function of SLC39A6,
a key gene of the SLC39A family, in liver cancer was also
investigated.
Materials and methods
UALCAN database
UALCAN is a public web portal that provides
comprehensive tumor transcriptome data from The Cancer Genome Atlas
(TCGA) (http://ualcan.path.uab.edu/). The
relative mRNA expression of SLC39A in liver cancer and
corresponding normal tissues was detected using the UALCAN
database.
Gene expression profiling interactive
analysis (GEPIA) database
The mRNA expression data of SLC39A family genes in
liver cancer tissues, as well as the clinicopathological and
survival data of patients with liver cancer were downloaded from
the University of California Santa Cruz (UCSC) Xena browser
(https://xenabrowser.net/). To determine the
potential prognostic values of SLC39A family genes in patients with
liver cancer, the association between SLC39A mRNA expression and
the overall survival (OS) of patients with liver cancer was
analyzed using the GEPIA database (http://gepia.cancer-pku.cn/) or by GraphPad Prism 6.0
(GraphPad Software, Inc.). The association of SLC39A family genes
with disease-free survival (DFS) was also analyzed. Furthermore,
the prognostic value of SLC39A6 mRNA expression in patients with
different disease grades of liver cancer was also analyzed.
Oncomine database
Oncomine is a cancer microarray database to
facilitate discovery from genome-wide expression analysis data
(https://www.oncomine.org/resource/main.html). The
relative expression of SLC39A6 in the Mas (18), Chen (19) and Roessler (20) liver cancer and normal samples were
downloaded from the Oncomine database.
Gene set enrichment analysis
(GSEA)
To identify the hallmark effect gene sets associated
with SLC39A6 mRNA expression in the TCGA-Liver Hepatocellular
Carcinoma dataset, GSEA was performed using GSEA software (v.
4.1.0; http://www.gsea-msigdb.org/gsea/login.jsp). For
enriched genomes, pathways with false discovery rate values
<0.25 and P<0.05 were considered after 1,000 permutations as
significantly enriched pathways.
Tissue samples
The study population included 12 patients who had
been diagnosed with liver cancer (T3/T4) composed of 6 males and 6
females with a median age of 63.93 years (range, 56–78 years)
between March 2021 and June 2021 at the Department of General
Surgery, The First Affiliated Hospital of Nanchang University
(Nanchang, China). Patients or their families who refused to
provide liver cancer specimens were excluded. The present study was
approved by the Human Research Ethics Committee of Nanchang
University (no. 2021-027) and all the patients were fully informed
of the study procedures orally and signed an informed consent form.
The tumor tissues and normal tissues (~5 cm away from tumor tissue)
were collected and then either immediately frozen in liquid
nitrogen or fixed in formalin after the surgery.
Histological analysis
Three pairs of tumors and healthy tissue samples
were fixed in 4% paraformaldehyde and embedded in paraffin. The
sections were immunohistochemically (IHC) stained with SLC39A6
rabbit polyclonal antibody (cat. no. 14236-1-AP; 1:200 dilution;
Proteintech Group, Inc.) and observed using microscopy (IX71;
Olympus Corporation). IHC scores (21) were used to evaluate the expression
level of SLC39A6. SLC39A6 staining intensity was scored as 0-3
points (0, negative; 1, weak; 2, mild; 3, strong). The percentage
score of SLC39A6-positive cells was as follows: 0, unstained; 1,
1–25; 2, 26–50; 3, 51–75; 4, 76–100% stained cells. The staining
intensity score was then multiplied with the positive cell
proportion score to obtain the final score.
Cells and cell culture
The human HepG2 and Hep3B liver cancer cell lines
were obtained from Procell Life Science & Technology Co., Ltd.,
and cultured in high-glucose DMEM (cat. no. SH30022.01) containing
10% FBS (cat. no. SH30084.03) and 1% penicillin-streptomycin (cat.
no. SV30010; all from HyClone; Cytiva) in an incubator with 5%
CO2 at 37°C. The authenticity of the cell lines was
confirmed using short tandem repeat methods.
Cell transfection
The human SLC39A6 small interfering (si)RNA
expression vector (si-SLC39A6) and corresponding negative control
siRNA (si-NC) were synthesized and provided by Vigene Biosciences,
Inc. The HepG2 and Hep3B cells were transfected with si-SLC39A6 and
si-NC using Lipofectamine 2000® according to the
manufacturer's protocol. The cells were collected at 48 h following
transfection for use in subsequent experiments. Short hairpin
(sh)RNA for SLC39A6 (Shanghai Genechem Co., Ltd.) and the
corresponding control vector (Shanghai Genechem Co., Ltd.) were
transfected into the HepG2 cells at a multiplicity of infection of
20 using enhanced infection solution (Shanghai Genechem Co., Ltd.)
according to the manufacturer's protocol. The plasmid backbone
(GV493) for shRNA-containing plasmid was provided by Shanghai
Genechem Co., Ltd. Protein lysates and total RNA were collected 96
h following transfection to verify the transfection efficiency and
for use in subsequent animal experiments. The sequences were as
follows: si-SLC39A6, 5′-UUCCAUUGCUGGUUCUUCAUGGCUA-3′ and si-NC,
5′-CGCTTCCGCGGCCCGTTCAA-3′; shRNA for SLC39A6,
5′-UUCCAUUGCUGGUUCUUCAUGGCUA-3′ and the respective NC-shRNA,
5′-CGCTTCCGCGGCCCGTTCAA-3′.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from 12 pairs of tumor and
normal tissue samples from patients with liver cancer and liver
cancer cell lines (HepG2 and Hep3B) using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and
reverse-transcribed into cDNA according to the manufacturer's
protocol using an RT kit (Vazyme Biotech Co., Ltd.) at 95°C for 5
min and 65°C for 60 min, and then kept at 4°C for storage. qPCR was
performed using SYBR-Green qPCR Master Mix (Vazyme Biotech Co.,
Ltd.). The qPCR conditions were used as follows: Initial
denaturation at 95°C for 5 min, followed by 40 cycles of
denaturation at 95°C for 30 sec, annealing at 52°C for 30 sec and
extension at 72°C for 30 sec. Each sample was repeated in
triplicate in qPCR step. The relative mRNA levels were normalized
to β-actin. The primer sequences were as follows: SLC39A6 forward,
5′-GCCGCGGAGACTGTTTCAAT-3′ and reverse, 3′-TGTTGCATTCAGCGGAACCT-5′;
and β-actin forward, 5′-GGCTCTTTTCCAGCCTTCCT-3′ and reverse,
3′-AATGCCAGGGTACATGGTGG-5′. The relative gene expression level was
analyzed by the 2−ΔΔCq method (22).
Western blot analysis
Proteins were extracted from tissues from patients
with liver cancer and cell lines (HepG2 and Hep3B) using
radioimmunoprecipitation assay buffer containing protease
inhibitor. The protein concentration was determined using a
bicinchoninic acid assay. For western blot analysis, 50 µg of
protein per lane was separated by 15% SDS-PAGE and transferred onto
nitrocellulose membranes (cat. no. FFN08; Beyotime Institute of
Biotechnology). The membranes were blocked with 5% non-fat milk for
1 h at room temperature and incubated with polyclonal primary
antibodies [SLC39A6 rabbit polyclonal antibody (cat no. 14236-1-AP;
1:1,000 diluted; Proteintech Group, Inc.) and β-actin mouse
monoclonal antibody (cat. no. 66009-1-Ig; 1:5,000 diluted;
Proteintech Group, Inc.)] in Tris-buffered saline containing
Tween-20 (TBST) buffer overnight at 4°C. The membranes were washed
with TBST solution three times and incubated with secondary
antibody [HRP-conjugated Affinipure goat anti-rabbit IgG(H+L); cat.
no. SA00001-2; 1:5,000 diluted; Proteintech Group, Inc.] for 1 h at
room temperature. The target protein was visualized using enhanced
chemiluminescence reagent (cat. no. 21050; Thermo Fisher
Scientific, Inc.).
Colony formation assay
The HepG2 and Hep3B cells transfected with
si-SLC39A6 or si-NC were seeded in 6-well plates at a concentration
of 1,000 cells per well in 3 ml complete medium. Following
incubation at 37°C for 9 days, the cell colonies were fixed with 4%
paraformaldehyde for 15 min, stained with 0.04% crystal violet for
20 min at room temperature and images were acquired.
Cell proliferation assay
Cell proliferation was assessed using the Cell
Counting Kit-8 (CCK-8) assay (cat. no. HY-K030; MedChemExpress).
HepG2 and Hep3B cells transfected with si-SLC39A6 or si-NC were
seeded in 96-well plates at a concentration of 1,000 cells per well
in 100 µl complete medium. After culturing the cells for 1–4 days,
the medium was removed and 10 µl CCK-8 stain was added per well.
The absorbance at a wavelength of 450 nm was measured following
incubation for 1 h using an automated microplate reader (Infinite
F50; Tecan Group).
In vitro migration and invasion
assays
In vitro migration and invasion assays were
performed using 24-well Transwell chambers with 8-µm pore size
membranes. For the migration assay, the HepG2 and Hep3B cells
transfected with si-SLC39A6 and si-NC were seeded in the upper
chamber at a concentration of 2×104 cells per well in
200 µl FBS-free medium, while 500 µl DMEM with 10% FBS was added to
the lower chamber. Following incubation at 37°C for 36 h, the cells
on the upper surface of the filters were removed and the cells that
had migrated to the lower surface were fixed in 4%
paraformaldehyde, subsequently stained with 0.04% crystal violet
for 20 min at room temperature and counted under a microscope
(IX71; Olympus Corporation). For the invasion assay, the procedure
was repeated as described above, but the membrane was initially
coated with Matrigel® at 4°C.
Animal experiment
A total of 2×106 HepG2 cells transfected
with shRNA for SLC39A6 or the corresponding control vector were
subcutaneously injected into the right infra-axillary dermis of
8-week-old female nude mice (n=5 per group; the total number of the
animals was 10 and the weight was 20.23±1.13 g). All mice were
provided by GemPharmatech Co., Ltd., and housed in a specific
pathogen-free facility (temperature, 18–29°C; relative humidity,
50–80%). Tumor volumes were measured using a digital caliper every
3 days to access tumor growth and the tumor weight was measured
when the mice were sacrificed on day 30 following cell
implantation. The mice were anesthetized with 1% pentobarbital
sodium at 50 mg/kg body weight by intraperitoneal injection and
sacrificed by cervical dislocation. Tumor sizes were measured every
3 days, commencing at 6 days after the cells were implanted, for a
total of nine times. According to previous authoritative studies
(23–26), when the tumor volumes reached 2,000
mm3, the mice underwent euthanasia as the humane
end-point of the study. The mean diameter did not exceed 1.5 cm. In
addition, if body weight loss exceeded 20%, the animals became
moribund or the subcutaneous tumor reached a volume of 2,000
mm3 or became necrotic, humane endpoints were considered
to have been reached in the present study (25–28).
All the animal experiments were performed in accordance with the
guidelines provided by the UK Animals (Scientific Procedures) Act,
1986, the EU Directive 2010/63/EU and approved by the Ethics
Committee for Animal Experiments of Nanchang Royo Biotech Co. Ltd.,
(no. RYE2021041402).
Statistical analysis
Statistical analysis was performed using SPSS 21.0
software (IBM Corp.) and GraphPad Prism 6.0 (GraphPad Software,
Inc.). Values are expressed as the mean ± standard deviation. All
comparisons between the two groups were performed using unpaired
Student's t-tests. For the two paired-group comparisons, the paired
Student's t-test was used. The survival curves were compared by the
log-rank test, while two-stage log-rank analysis (29) was used for those survival plots
where late-stage crossover between the groups was present. Based on
literature reports (29), P1
represents the P-value prior to curve crossing, P2 represents the
P-value after curve crossing and P represents the P-value for the
whole. Univariate and multivariate analyses of OS and DFS according
to prognostic factors were analyzed using Cox regression analysis.
A value of P<0.05 was considered to indicate a statistically
significant difference.
Results
Relative mRNA expression of SLC39A
family genes in liver cancer
To examine the expression levels of SLC39A family
genes in liver cancer, the expression data of 14 SLC39A genes in
patients with liver cancer from the TCGA database were analyzed
using the UALCAN database. As presented in Fig. 1A-N, the expression of SLC39A1,
SLC39A3, SLC39A4, SLC39A6, SLC39A7, SLC39A10 and SLC39A13 in the
cancer tissues was significantly higher than that in healthy
tissues; however, SLC39A5, SLC39A9, SLC39A11 and SLC39A14 were
expressed at markedly lower levels in the tumor tissues compared
with the normal tissues. The expression of SLC39A2, SLC39A8 and
SLC39A12 exhibited no significant difference between cancer and
normal tissues.
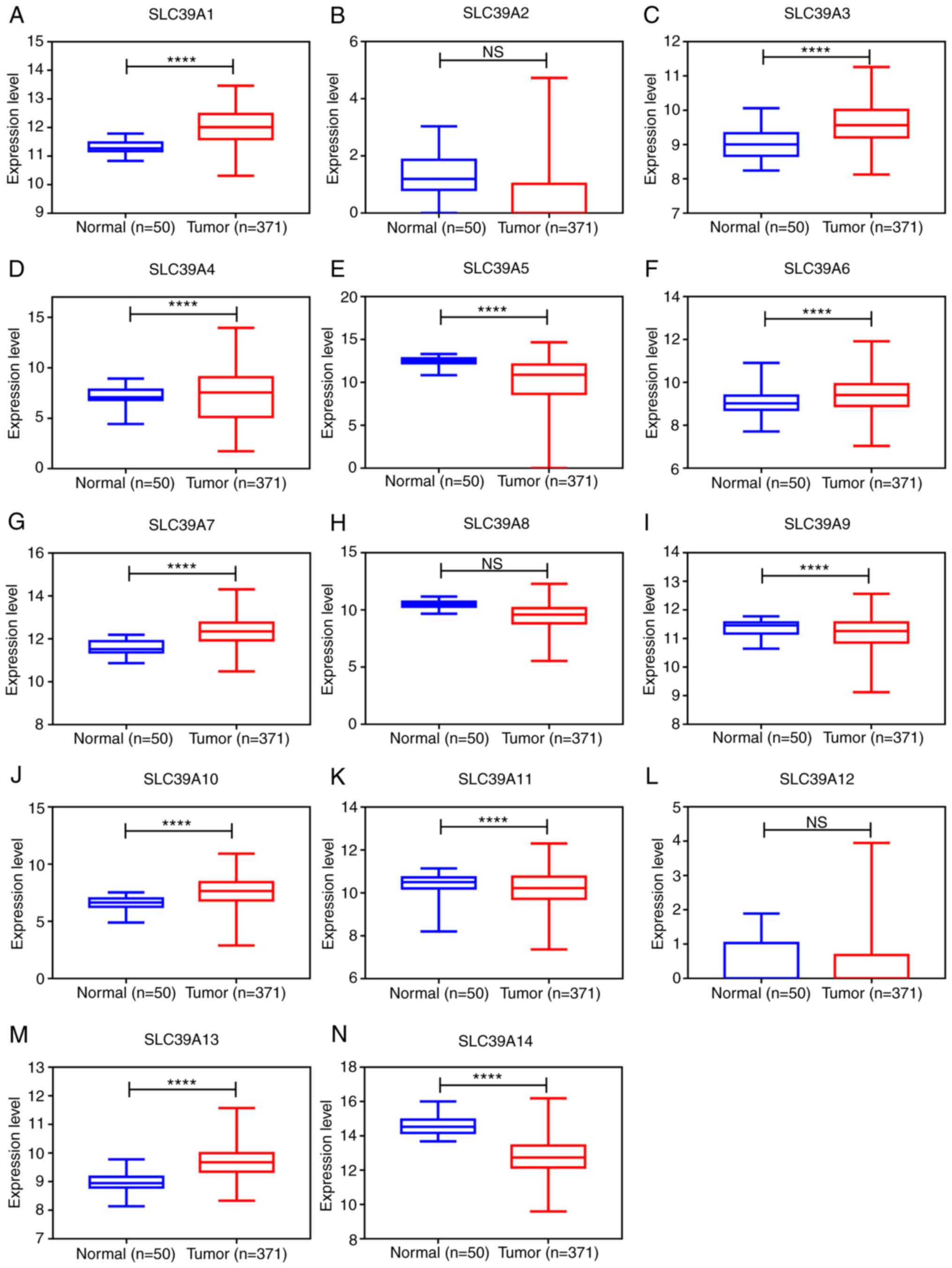 | Figure 1.mRNA expression of SLC39A family
genes in primary tumor and corresponding normal tissues in patients
with liver cancer using the dataset from the UALCAN database. (A)
SLC39A1, (B) SLC39A2, (C) SLC39A3, (D) SLC39A4, (E) SLC39A5, (F)
SLC39A6, (G) SLC39A7, (H) SLC39A8, (I) SLC39A9, (J) SLC39A10, (K)
SLC39A11, (L) SLC39A12, (M) SLC39A13 and (N) SLC39A14.
****P<0.0001. SLC, solute carrier; NS, no significance. |
SLC39A genes have a high prognostic
value in patients with liver cancer in the GEPIA dataset
The potential prognostic values of SLC39A family
genes in liver cancer were investigated using the GEPIA database.
As presented in Fig. 2A-N,
positive associations were observed between high expression of
several genes and a significantly unfavorable OS in patients with
liver cancer, including SLC39A1, SLC39A6, SLC39A10 and SLC39A13.
However, no positive associations were observed between high
expression of genes and a significantly poorer DFS (Fig. S1A-N). Furthermore, univariate and
multivariate analyses of OS and DFS were performed to identify key
genes of the SLC39A family in patients with liver cancer. The
results suggested that SLC39A1, SLC39A6 or SLC39A13 overexpression
was an influencing factor for poor prognosis of patients with liver
cancer and may thus be a potential prognostic factor for liver
cancer (Tables I and II). In this study, SLC39A6 was selected
for follow-up experiments.
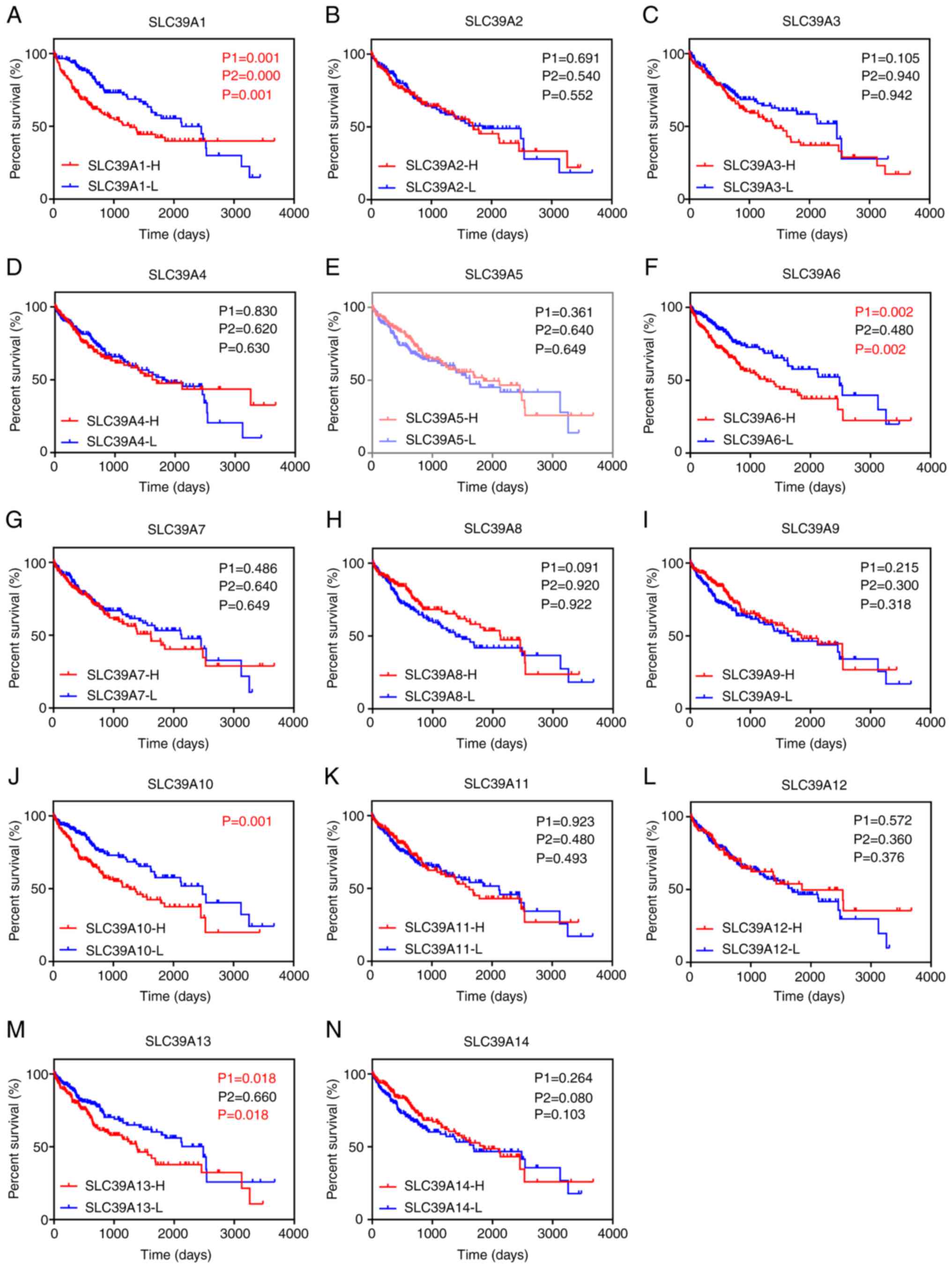 | Figure 2.Association of SLC39A family genes
with overall survival of patients with liver cancer based on data
from the Gene Expression Profiling Interactive Analysis database.
(A) SLC39A1, (B) SLC39A2, (C) SLC39A3, (D) SLC39A4, (E) SLC39A5,
(F) SLC39A6, (G) SLC39A7, (H) SLC39A8, (I) SLC39A9, (J) SLC39A10,
(K) SLC39A11, (L) SLC39A12, (M) SLC39A13 and (N) SLC39A14. P1 and
P2 represent the P-value prior to and after curve crossing,
respectively, and P represents the P-value for the whole graphs.
P<0.05 was considered to indicate statistical significance. SLC,
solute carrier; H, high; L, low. |
 | Table I.Univariate and multivariate analyses
of the influence of SLC39A mRNA levels and other factors on patient
overall survival. |
Table I.
Univariate and multivariate analyses
of the influence of SLC39A mRNA levels and other factors on patient
overall survival.
|
| Univariate
analysis | Multivariate
analysis |
|---|
| Variable | HR | 95%CI | P-value | HR | 95%CI | P-value |
|---|
| SLC39A1 | 1.799 | 1.266-2.556 | 0.001 | 1.462 | 1.011-2.112 | 0.043 |
| SLC39A2 | 1.072 | 0.760-1.514 | 0.691 |
|
|
|
| SLC39A3 | 1.334 | 0.940-1.893 | 0.107 |
|
|
|
| SLC39A4 | 1.039 | 0.735-1.468 | 0.830 |
|
|
|
| SLC39A5 | 0.852 | 0.603-1.202 | 0.361 |
|
|
|
| SLC39A6 | 1.736 | 1.222-2.465 | 0.002 | 1.505 | 1.029-2.203 | 0.035 |
| SLC39A7 | 1.130 | 0.801-1.595 | 0.487 |
|
|
|
| SLC39A8 | 0.742 | 0.525-1.050 | 0.092 |
|
|
|
| SLC39A9 | 0.216 | 1.568-1.136 | 0.216 |
|
|
|
| SLC39A10 | 1.801 | 1.265-2.565 | 0.001 | 1.341 | 0.908-1.983 | 0.141 |
| SLC39A11 | 0.983 | 0.697-1.388 | 0.923 |
|
|
|
| SLC39A12 | 0.899 | 0.621-1.301 | 0.572 |
|
|
|
| SLC39A13 | 1.518 | 1.072-2.149 | 0.019 | 1.429 | 1.000-2.041 | 0.050 |
| SLC39A14 | 0.822 | 0.582-1.161 | 0.265 |
|
|
|
| Age | 1.186 | 0.836-1.683 | 0.339 |
|
|
|
| Sex | 0.800 | 0.562-1.141 | 0.218 |
|
|
|
| T stage | 1.682 | 1.403-2.017 | <0.001 | 0.558 | 0.064-4.870 | 0.597 |
| N stage | 2.012 | 0.493-8.212 | 0.330 |
|
|
|
| M stage | 1.055 | 1.274-12.906 | 0.018 | 1.475 | 0.281-7.740 | 0.646 |
| TNM stage | 1.670 | 1.361-2.048 | <0.001 | 0.245 | 0.032-1.845 | 0.172 |
| Grade | 1.114 | 0.881-1.408 | 0.368 |
|
|
|
| Recurrence | 1.610 | 1.099-2.357 | 0.014 | 0.640 | 0.392-1.043 | 0.073 |
 | Table II.Univariate and multivariate analyses
of the influence of SLC39A mRNA levels and other factors on patient
progression-free interval. |
Table II.
Univariate and multivariate analyses
of the influence of SLC39A mRNA levels and other factors on patient
progression-free interval.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | 95%CI | P-value | HR | 95%CI | P-value |
|---|
| SLC39A1 | 1.096 | 0.819-1.466 | 0.539 |
|
|
|
| SLC39A2 | 0.868 | 0.647-1.165 | 0.347 |
|
|
|
| SLC39A3 | 0.911 | 0.680-1.219 | 0.530 |
|
|
|
| SLC39A4 | 0.828 | 0.618-1.110 | 0.207 |
|
|
|
| SLC39A5 | 0.932 | 0.697-1.247 | 0.637 |
|
|
|
| SLC39A6 | 1.279 | 0.956-1.712 | 0.098 |
|
|
|
| SLC39A7 | 0.941 | 0.703-1.260 | 0.685 |
|
|
|
| SLC39A8 | 0.918 | 0.686-1.229 | 0.567 |
|
|
|
| SLC39A9 | 1.020 | 0.762-1.366 | 0.895 |
|
|
|
| SLC39A10 | 1.347 | 1.006-1.803 | 0.046 | 1.295 | 0.907-1.850 | 0.155 |
| SLC39A11 | 0.859 | 0.641-1.151 | 0.309 |
|
|
|
| SLC39A12 | 0.837 | 0.611-1.145 | 0.264 |
|
|
|
| SLC39A13 | 1.083 | 0.809-1.449 | 0.593 |
|
|
|
| SLC39A14 | 0.875 | 0.654-1.170 | 0.368 |
|
|
|
| Age | 1.011 | 0.755-1.354 | 0.941 |
|
|
|
| Sex | 0.979 | 0.718-1.334 | 0.893 |
|
|
|
| T stage | 1.622 | 1.391-1.892 | <0.001 | 1.284 | 0.588-2.805 | 0.530 |
| N stage | 1.379 | 0.340-5.586 | 0.653 |
|
|
|
| M stage | 3.460 | 1.086-11.025 | 0.036 | 1.094 | 0.315-3.795 | 0.888 |
| TNM stage | 1.660 | 1.397-1.973 | <0.001 | 1.100 | 0.485-2.497 | 0.820 |
| Grade | 1.103 | 0.908-1.340 | 0.322 |
|
|
|
| Recurrence | 23.799 | 12.902-43.900 | <0.001 | 40.509 | 16.469-99.640 | <0.001 |
Association between the expression of
SLC39A6 and the prognosis of patients with liver cancer
The Oncomine database and UCSC Xena browser were
used to analyze the sequencing data of SLC39A6 in liver cancer. As
presented in Fig. 3A-D, the mRNA
expression levels of SLC39A6 were significantly higher in the tumor
vs. normal tissues in the datasets by Mas et al (18), Chen et al (19) and Roessler et al (20). The expression levels of SLC39A6 in
patients with different stages of liver cancer were also evaluated
(Fig. 3E and F). The results
revealed that increased expression of SLC39A6 mainly occurred in
the late stages of liver cancer, while its expression level
remained unaltered in patients with early-stage liver cancer.
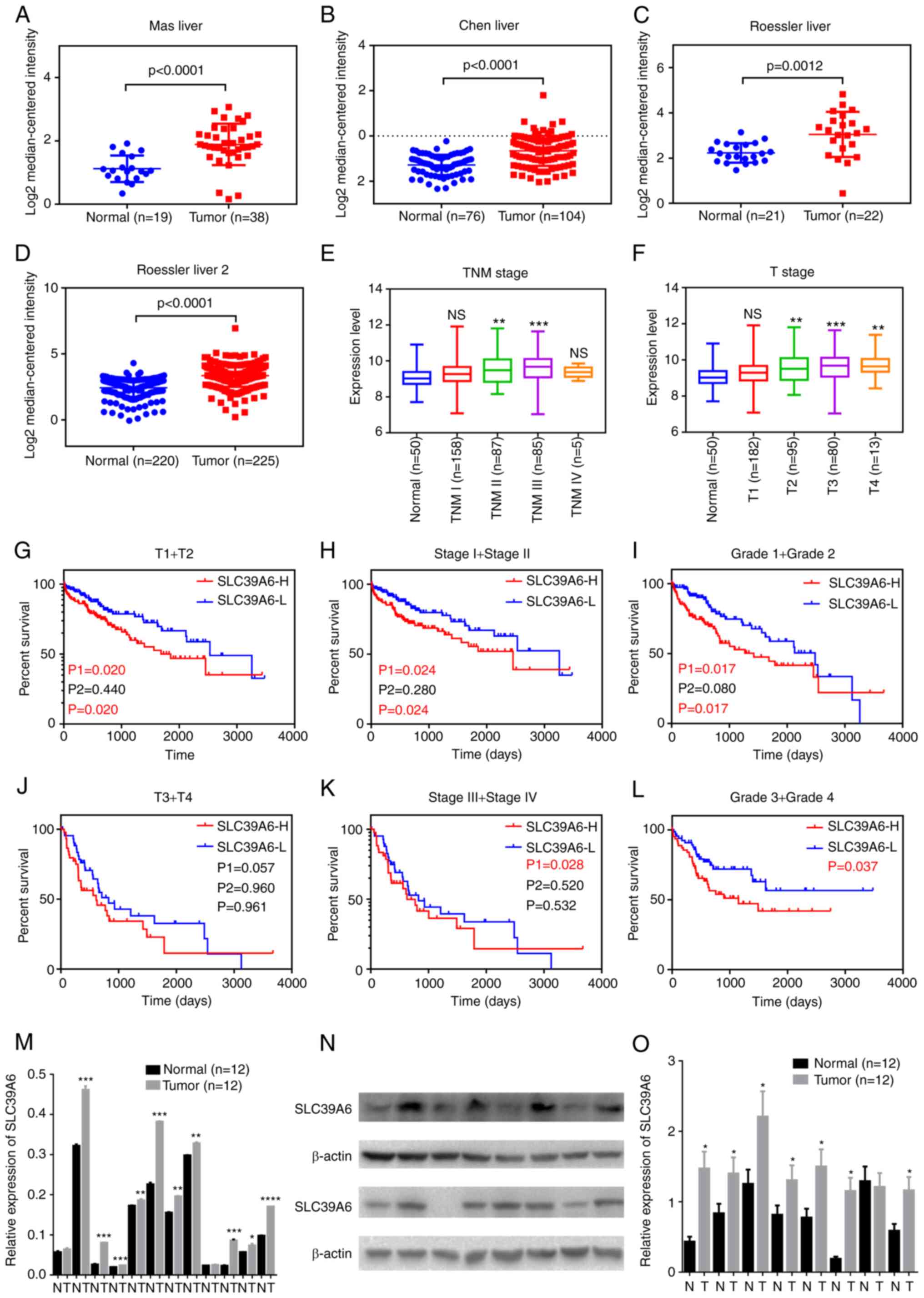 | Figure 3.SLC39A6 expression is upregulated in
patients with liver cancer and associated with poor prognosis.
(A-D) Expression of SLC39A6 in patients with liver cancer based on
the Oncomine database, including (A) Mas, (B) Chen and (C) Roessler
Liver 1 and (D) Roessler Liver 2. (E and F) mRNA expression of
SLC39A family genes in patients with liver cancer of different
stages: (E) TNM stage and (F) T stage. (G-L) Association of SLC39A6
with overall survival of patients with liver cancer with different
stages according to The Cancer Genome Atlas dataset. (G) T1 + T2;
(H) stage I and II; (I) grade 1 and 2; (J) T3 + T4; (K) stage III
and IV; (L) grade 3 and 4. P1 and P2 represent the P-value prior to
and after curve crossing, respectively, and P represents the
P-value for the whole graphs. (M) mRNA expression of SLC39A6 in
liver cancer tissues and the corresponding noncancerous tissues. (N
and O) Protein expression of SLC39A6 in liver cancer tissues and
the corresponding noncancerous tissues: (N) Representative western
blot and (O) quantified expression values. *P<0.05, **P<0.01,
***P<0.001 vs. normal. SLC, solute carrier; H, high; L, low; NS,
no significance; T, tumor; N, normal tissues. |
Furthermore, survival curve analysis indicated that
high expression of SLC39A6 was associated with markedly shorter OS
and DFS of patients with different stages of liver cancer (Fig. 3G-L). These results suggested that
the expression levels of SLC39A6 had a good diagnostic value for
patients with liver cancer.
Protein expression and biological
function of SLC39A6 in patients with liver cancer
To investigate the role of SLC39A6 in the
progression of liver cancer, the mRNA and protein expression of
SLC39A6 in T3 liver cancer tissues was analyzed using RT-qPCR,
western blot analysis and IHC. The results revealed that SLC39A6
mRNA and protein expression in the tumor tissues was markedly
higher than that in the corresponding non-cancerous tissues,
indicating that SLC39A6 is an important target for liver cancer
(Figs. 3M-O, 4A and S2A). GSEA was performed to evaluate the
Kyoto Encyclopedia of Genes and Genomes and Gene Ontology gene sets
in patients with liver cancer (Fig.
4B). The results revealed that the group with high expression
of SLC39A6 was significantly enriched in gene sets associated with
liver cancer, tumor adhesion and the tumor metastasis pathway.
Thus, these results demonstrated that the expression of SLC39A6 was
significantly upregulated in liver cancer tumor tissues and that it
was associated with the tumor metastasis pathway.
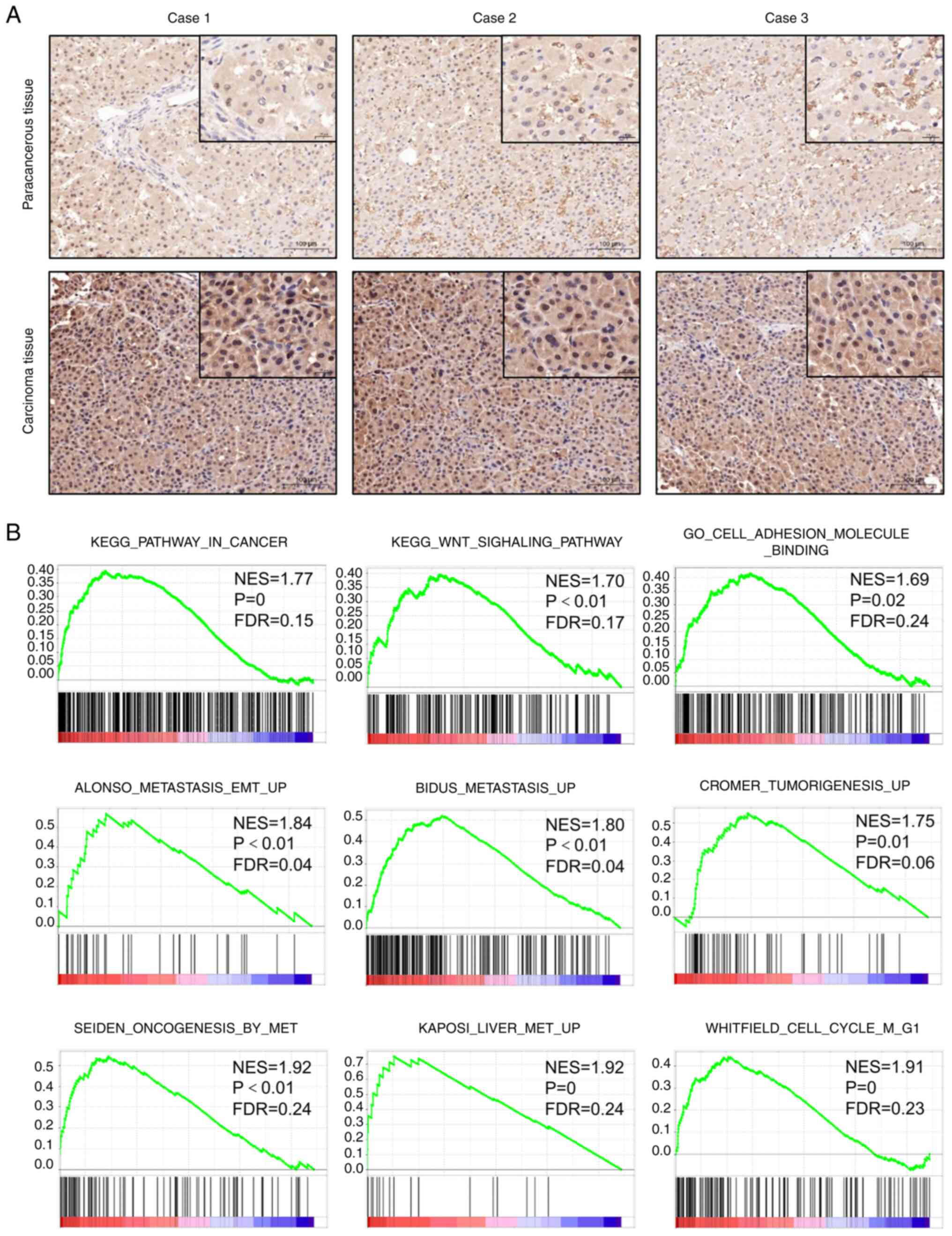 | Figure 4.Expression and biological function of
SLC39A6 in patients with liver cancer. (A) Typical
immunohistochemical images of SLC39A6 expression in liver cancer
tissues and the corresponding noncancerous tissues in three
representative cases (magnification and scale bar, ×100 and 100 µm,
or ×400 and 20 µm in the magnified windows, respectively). (B) The
liver cancer, tumor adhesion and tumor metastasis pathway were
highly enriched in patients with liver cancer with SLC39A6 high
expression according to gene set enrichment analysis of The Cancer
Genome Atlas Liver Hepatocellular Carcinoma dataset. SLC, solute
carrier; FDR, false discovery rate; KEGG, Kyoto Encyclopedia of
Genes and Genomes; GO, Gene Ontology; EMT,
epithelial-to-mesenchymal transition; NES, normalized enrichment
score. |
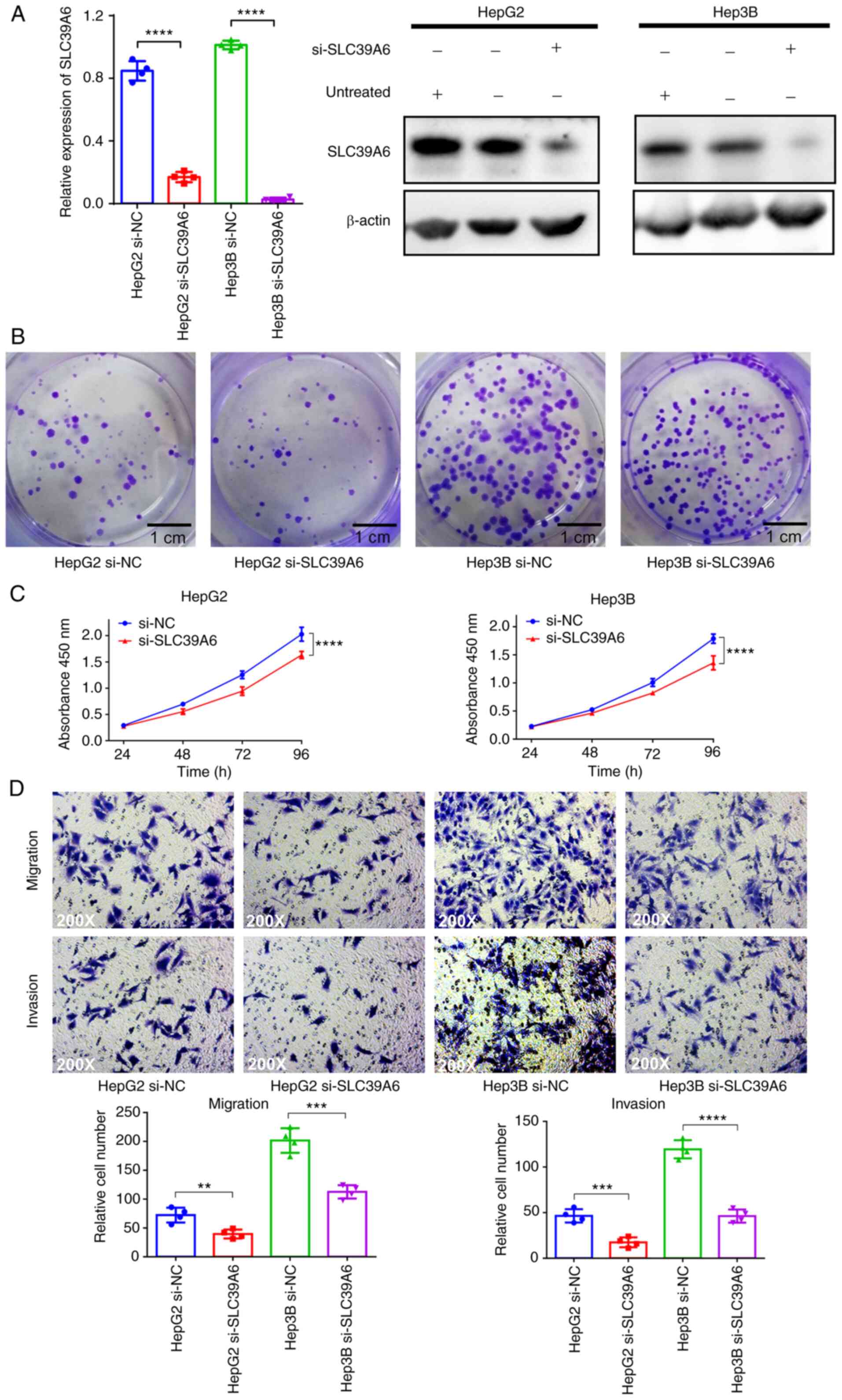
Downregulation of SLC39A6 suppresses
liver cancer cell growth, migration and invasion
To analyze the function of SLC39A6 in the biological
behavior of liver cancer cells, HepG2 and Hep3B cells were
transfected with si-SLC39A6 to downregulate the expression of
SLC39A6. HepG2 and Hep3B were the most commonly used cell lines to
study liver cancer (30–34), so these cell lines were selected
for the present study. In addition, the Cancer Cell Line
Encyclopedia database (35–38)
was checked and the results indicated that HepG2 and Hep3B were the
liver cancer cell lines that expressed high SLC39A6. Thus, it was
considered appropriate to select these two cell lines to study
SLC39A6. As presented in Figs. 5A
and S2B, the mRNA and protein
expression of SLC39A6 was significantly decreased in the
si-SLC39A6-transfected HepG2 and Hep3B cells. To determine whether
SLC39A6 was essential for liver cancer cell proliferation, colony
formation and CCK-8 assays were performed. In the colony formation
assay, the colonies of the si-SLC39A6-transfected HepG2 and Hep3B
cells were much smaller than those of the control cells (Figs. 5B and S2C). The results of the CCK-8 assay also
demonstrated that the growth of the si-SLC39A6-transfected HepG2
and Hep3B cells was significantly inhibited compared with that of
the control cells (Fig. 5C).
Furthermore, the effects of SLC39A6 on the migration
and invasion of liver cancer cells were investigated. As presented
in Fig. 5D, the migratory and
invasive ability of the si-SLC39A6-transfected HepG2 and Hep3B
cells was significantly decreased compared with that of the control
cells. These results revealed that knockdown of SLC39A6 inhibited
the proliferation, migration and invasion of liver cancer
cells.
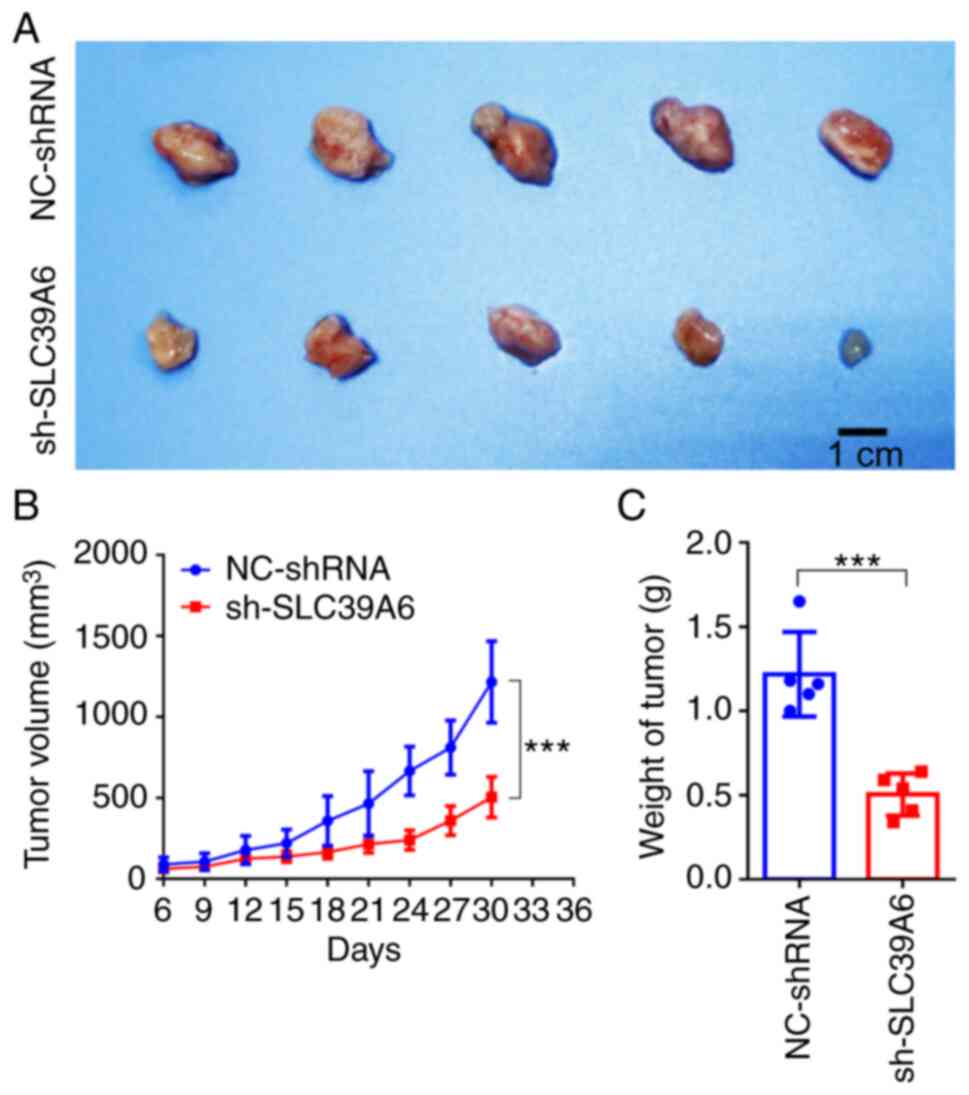
Knockdown of SLC39A6 significantly
suppresses liver cancer progression in vivo
In order to further verify the aforementioned
observations, the role of SLC39A6 in the occurrence and development
of liver cancer was investigated in vivo. As presented in
Fig. S2D, the prognosis of
patients with liver cancer was not significantly associated with
sex. Furthermore, for the animal experiment, only female mice were
available. HepG2 cells transfected with shRNA targeting SLC39A6 or
the corresponding control vector were implanted subcutaneously into
nude mice. The tumor size was strictly controlled to remain
<2,000 mm3 and the maximum percentage of body weight
loss was 8.65% (Figs. 6A and
S2E), fully complying with the
ethical animal standards. The maximum tumor size observed was
1,648.70 mm3. The present results demonstrated that
knockdown of SLC39A6 significantly reduced both the volume and
weight of the tumors in vivo (Fig. 6A-C).
Discussion
Liver cancer is one of the most common malignant
solid tumor types worldwide, with >42,000 new cases and 30,000
associated mortalities in 2020 (39). Among the primary cancers of the
liver, hepatocellular carcinoma is the main histological subtype,
accounting for 70–85% of the total cases (40). Although a variety of molecular
changes have been detected over the past years (41–44),
to the best of our knowledge, no sensitive and specific biomarkers
and no accurate indicators are yet available for the early
diagnosis or for predicting the prognosis of patients with liver
cancer. Therefore, the early detection of liver cancer is of utmost
importance in order to improve the prognosis of patients with liver
cancer.
Zinc usually has an important role in tumor events
and low levels of zinc have been detected in serum and tumor
tissues of patients with cancer (45,46).
SLC39A6 is a subfamily member of zinc transporters, which is
involved in maintaining the intracellular homeostasis of zinc
(47–49). Previous studies have demonstrated
that SLC39A6 is overexpressed in breast (50,51),
pancreatic and cervical cancer (52,53),
as well as in esophageal squamous cell carcinoma (54,55).
In liver cancer, a previous study indicated that miR-192 was a
prognostic indicator, reducing liver cancer metastasis through the
SLC39A6/SNAIL pathway (56).
However, whether SLC39A6 expression is an independent prognostic
factor for patients with liver cancer warrants further
exploration.
In the present study, the mRNA expression of SLC39A
family genes in liver cancer tissues and their association with OS
of patients with liver cancer was analyzed. The results revealed
that the expression levels of SLC39A1, SLC39A3, SLC39A4, SLC39A6,
SLC39A7, SLC39A10 and SLC39A13 were significantly higher in the
tumor compared with healthy tissues, while SLC39A5, SLC39A9,
SLC39A11 and SLC39A14 were expressed at higher levels in healthy
tissues. The results demonstrated that high expression of SLC39A1,
SLC39A6, SLC39A10 and SLC39A12 was associated with unfavorable OS
of patients with liver cancer. The expression of SLC39A members was
associated with the prognosis of specific patients with liver
cancer by subgroup analysis. In addition, Cox regression analysis
confirmed that SLC39A6 was a key gene influencing the survival of
patients with liver cancer. All of these results indicated that
SLC39A6 may be a biomarker for the early detection and prognosis of
liver cancer.
As metastasis is the main cause of liver
cancer-related mortality, the identification of new genes involved
in this process is crucial in order to elucidate the molecular
mechanism of liver cancer metastasis (57,58).
The present study observed the upregulation of SLC39A6 expression
in liver cancer tumor tissues and cell lines. GSEA also revealed
that SLC39A6 was associated with liver cancer tumor proliferation
and metastasis.
In recent years, studies have indicated that SLC39A6
has an important role in tumor progression, metastasis and
invasion. With regard to breast cancer, the expression of SLC39A6
has been indicated to be significantly increased compared with that
in normal breast tissue and the expression of the adhesion protein,
E-cadherin, has been reported to be significantly decreased
following overexpression of SLC39A6. It was thus revealed that
SLC39A6 may promote epithelial-mesenchymal transition (EMT) in
breast cancer (59). Furthermore,
it was reported that SLC39A6 is activated by STAT3 in breast tumor
cells, triggering the influx of zinc ions and promoting Snail to
remain in the nucleus as an E-cadherin transcription inhibitor,
thus promoting the migration process. Therefore, SLC39A6 has been
proposed as a novel target for breast cancer (60). In addition, the expression of
SLC39A6 has been indicated to be markedly higher in pancreatic
tumors than in normal pancreatic tissues. Furthermore, the
proliferative and migratory ability of pancreatic cancer cells has
been indicated to decrease significantly following SLC39A6
knockdown (61). Another study
demonstrated that SLC39A6 enhanced the invasive phenotype by
inducing the EMT of pancreatic cancer cells (52). With regard to non-small cell lung
cancer, knockdown of SLC39A6 has been reported to significantly
inhibit the proliferation, migration and invasion of lung cancer
cells and to induce cell cycle arrest in G1 phase (62). These results indicate that SLC39A6
promotes the development of multiple tumor types, which is
consistent with the findings of the present study on liver
cancer.
In the present study, in order to verify whether
SLC39A6 promotes the proliferation, migration and invasion of liver
cancer cells, the expression of SLC39A6 in HepG2 and Hep3B cells
was knocked down. The results of a CCK-8 assay indicated that
knockdown of SLC39A6 significantly decreased the viability and
proliferation of HepG2 and Hep3B cells. Furthermore, in the
Transwell assays, it was observed that knockdown of SLC39A6
significantly reduced the number of migrating and invading HepG2
and Hep3B cells. These results demonstrated that SLC39A6
significantly regulated the proliferation, migration and invasion
of liver cancer cells, thus regulating the malignant progression of
liver cancer. Of note, in vivo, it was demonstrated that the
knockdown of SLC39A6 significantly inhibited the progression of
liver cancer.
In conclusion, in the present study, SLC39A6 was
identified as a key gene of the SLC39A family in liver cancer. The
results demonstrated that SLC39A6 was upregulated in liver cancer,
indicating that it may be used as a potential biomarker for the
diagnosis of liver cancer. Furthermore, knockdown of SLC39A6
expression significantly reduced the proliferation, migration and
invasion of HepG2 and Hep3B cells in vitro and also
significantly suppressed liver cancer growth in vivo.
Collectively, the results of the present study demonstrated that
SLC39A6 may have a crucial tumor-promoting role in liver cancer and
may exhibit the potential for use as a therapeutic target for liver
cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the National Nature
Science Foundation of China (grant no. 81760137) and the Nature
Science Foundation of Jiangxi Province (grant no.
20202BAB206016).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZW participated in the conception and design of the
study. ZW and XW performed the experiments, data analysis and
interpretation. ZW provided the resources. ZW and XW wrote the
manuscript. ZW and XW confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Human Research
Ethics Committee of Nanchang University (Nanchang, China; approval
no. 2021-027). All of the patients were fully informed of the study
procedures orally and signed an informed consent form.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Villanueva A: Hepatocellular carcinoma. N
Engl J Med. 380:1450–1462. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Ward EM, Johnson CJ, Cronin KA,
Ma J, Ryerson B, Mariotto A, Lake AJ, Wilson R, Sherman RL, et al:
Annual report to the nation on the status of cancer, 1975-2014,
featuring survival. J Natl Cancer Inst. 109:djx0302017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zheng R, Qu C, Zhang S, Zeng H, Sun K, Gu
X, Xia C, Yang Z, Li H, Wei W, et al: Liver cancer incidence and
mortality in China: Temporal trends and projections to 2030. Chin J
Cancer Res. 30:571–579. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sia D, Villanueva A, Friedman SL and
Llovet JM: Liver cancer cell of origin, molecular class, and
effects on patient prognosis. Gastroenterology. 152:745–761. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Idilman R, De Maria N, Colantoni A and Van
Thiel DH: Pathogenesis of hepatitis B and C-induced hepatocellular
carcinoma. J Viral Hepat. 5:285–299. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kanwal F, Kramer JR, Duan Z, Yu X, White D
and El-Serag HB: Trends in the burden of nonalcoholic fatty liver
disease in a united states cohort of veterans. Clin Gastroenterol
Hepatol. 14:301–8.e1-e2. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Morgan TR, Mandayam S and Jamal MM:
Alcohol and hepatocellular carcinoma. Gastroenterology. 127((5
Suppl 1)): S87–S96. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pan Z, Choi S, Ouadid-Ahidouch H, Yang JM,
Beattie JH and Korichneva I: Zinc transporters and dysregulated
channels in cancers. Front Biosci (Landmark Ed). 22:623–643. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ho E and Ames BN: Low intracellular zinc
induces oxidative DNA damage, disrupts p53, NFkappa B, and AP1 DNA
binding, and affects DNA repair in a rat glioma cell line. Proc
Natl Acad Sci USA. 99:16770–16775. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vallee BL and Falchuk KH: The biochemical
basis of zinc physiology. Physiol Rev. 73:79–118. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Andreini C, Banci L, Bertini I and Rosato
A: Counting the zinc-proteins encoded in the human genome. J
Proteome Res. 5:196–201. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang L and Tepaamorndech S: The SLC30
family of zinc transporters-a review of current understanding of
their biological and pathophysiological roles. Mol Aspects Med.
34:548–560. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jeong J and Eide DJ: The SLC39 family of
zinc transporters. Mol Aspects Med. 34:612–619. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Costello LC and Franklin RB: The clinical
relevance of the metabolism of prostate cancer; zinc and tumor
suppression: Connecting the dots. Mol Cancer. 5:172006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu L, Hou Y, Hu J, Zhou L, Chen K, Yang X
and Song Z: SLC39A8/Zinc suppresses the progression of clear cell
renal cell carcinoma. Front Oncol. 11:6519212021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu L, Yang J and Wang C: Analysis of the
prognostic significance of solute carrier (SLC) family 39 genes in
breast cancer. Biosci Rep. 40:BSR202007642020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mas VR, Maluf DG, Archer KJ, Yanek K, Kong
X, Kulik L, Freise CE, Olthoff KM, Ghobrial RM, McIver P and Fisher
R: Genes involved in viral carcinogenesis and tumor initiation in
hepatitis C virus-induced hepatocellular carcinoma. Mol Med.
15:85–94. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen X, Cheung ST, So S, Fan ST, Barry C,
Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, et al: Gene expression
patterns in human liver cancers. Mol Biol Cell. 13:1929–1939. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Roessler S, Jia HL, Budhu A, Forgues M, Ye
QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX and Wang XW: A
unique metastasis gene signature enables prediction of tumor
relapse in early-stage hepatocellular carcinoma patients. Cancer
Res. 70:10202–10212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen P, Yao Y, Yang N, Gong L, Kong Y and
Wu A: Circular RNA circCTNNA1 promotes colorectal cancer
progression by sponging miR-149-5p and regulating FOXM1 expression.
Cell Death Dis. 11:5572020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Workman P, Aboagye EO, Balkwill F, Balmain
A, Bruder G, Chaplin DJ, Double JA, Everitt J, Farningham DA,
Glennie MJ, et al: Guidelines for the welfare and use of animals in
cancer research. Br J Cancer. 102:1555–1577. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kang JC, Sun W, Khare P, Karimi M, Wang X,
Shen Y, Ober RJ and Ward ES: Engineering a HER2-specific
antibody-drug conjugate to increase lysosomal delivery and
therapeutic efficacy. Nat Biotechnol. 37:523–526. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao H, Guo Y, Li B, Li X, Wang Y, Han S,
Cheng D and Shuai X: M2-like tumor-associated macrophage-targeted
codelivery of STAT6 inhibitor and IKKβ siRNA induces M2-to-M1
repolarization for cancer immunotherapy with low immune side
effects. ACS Cent Sci. 6:1208–1222. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang X, Wilhelm J, Li W, Li S, Wang Z,
Huang G, Wang J, Tang H, Khorsandi S, Sun Z, et al:
Polycarbonate-based ultra-pH sensitive nanoparticles improve
therapeutic window. Nat Commun. 11:58282020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chongsathidkiet P, Jackson C, Koyama S,
Loebel F, Cui X, Farber SH, Woroniecka K, Elsamadicy AA, Dechant
CA, Kemeny HR, et al: Sequestration of T cells in bone marrow in
the setting of glioblastoma and other intracranial tumors. Nat Med.
24:1459–1468. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu YL, Ding CL, Qian CL, Qi ZT and Wang W:
Retinoid acid induced 16 deficiency aggravates colitis and
colitis-associated tumorigenesis in mice. Cell Death Dis.
10:9582019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Qiu P and Sheng J: A two-stage procedure
for comparing hazard rate functions. J R Statist Soc B. 70:191–208.
2008.
|
|
30
|
Moh MC, Lee LH, Yang X and Shen S: HEPN1,
a novel gene that is frequently down-regulated in hepatocellular
carcinoma, suppresses cell growth and induces apoptosis in HepG2
cells. J Hepatol. 39:580–586. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Notas G, Kolios G, Mastrodimou N, Kampa M,
Vasilaki A, Xidakis C, Castanas E, Thermos K and Kouroumalis E:
Cortistatin production by HepG2 human hepatocellular carcinoma cell
line and distribution of somatostatin receptors. J Hepatol.
40:792–798. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu JM, Xu Y, Skill NJ, Sheng H, Zhao Z, Yu
M, Saxena R and Maluccio MA: Autotaxin expression and its
connection with the TNF-alpha-NF-kappaB axis in human
hepatocellular carcinoma. Mol Cancer. 9:712010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chang Y, Yan W, He X, Zhang L, Li C, Huang
H, Nace G, Geller DA, Lin J and Tsung A: miR-375 inhibits autophagy
and reduces viability of hepatocellular carcinoma cells under
hypoxic conditions. Gastroenterology. 143:177–187.e8. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Prieto-Domínguez N, Ordóñez R, Fernández
A, Méndez-Blanco C, Baulies A, Garcia-Ruiz C, Fernández-Checa JC,
Mauriz JL and González-Gallego J: Melatonin-induced increase in
sensitivity of human hepatocellular carcinoma cells to sorafenib is
associated with reactive oxygen species production and mitophagy. J
Pineal Res. 61:396–407. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jaffe JD, Wang Y, Chan HM, Zhang J,
Huether R, Kryukov GV, Bhang HE, Taylor JE, Hu M, Englund NP, et
al: Global chromatin profiling reveals NSD2 mutations in pediatric
acute lymphoblastic leukemia. Nat Genet. 45:1386–1391. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ghandi M, Huang FW, Jané-Valbuena J,
Kryukov GV, Lo CC, McDonald ER III, Barretina J, Gelfand ET,
Bielski CM, Li H, et al: Next-generation characterization of the
cancer cell line encyclopedia. Nature. 569:503–508. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li H, Ning S, Ghandi M, Kryukov GV, Gopal
S, Deik A, Souza A, Pierce K, Keskula P, Hernandez D, et al: The
landscape of cancer cell line metabolism. Nat Med. 25:850–860.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nusinow DP, Szpyt J, Ghandi M, Rose CM,
McDonald ER III, Kalocsay M, Jané-Valbuena J, Gelfand E, Schweppe
DK, Jedrychowski M, et al: Quantitative proteomics of the cancer
cell line encyclopedia. Cell. 180:387–402.e16. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Perz JF, Armstrong GL, Farrington LA,
Hutin YJF and Bell BP: The contributions of hepatitis B virus and
hepatitis C virus infections to cirrhosis and primary liver cancer
worldwide. J Hepatol. 45:529–538. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hartke J, Johnson M and Ghabril M: The
diagnosis and treatment of hepatocellular carcinoma. Semin Diagn
Pathol. 34:153–159. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ayuso C, Rimola J, Vilana R, Burrel M,
Darnell A, García-Criado Á, Bianchi L, Belmonte E, Caparroz C,
Barrufet M, et al: Diagnosis and staging of hepatocellular
carcinoma (HCC): Current guidelines. Eur J Radiol. 101:72–81. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Piñero F, Dirchwolf M and Pessôa MG:
Biomarkers in hepatocellular carcinoma: Diagnosis, prognosis and
treatment response assessment. Cells. 9:13702020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chowdhury MMH, Salazar CJJ and Nurunnabi
M: Recent advances in bionanomaterials for liver cancer diagnosis
and treatment. Biomater Sci. 9:4821–4842. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fukada T, Yamasaki S, Nishida K, Murakami
M and Hirano T: Zinc homeostasis and signaling in health and
diseases: Zinc signaling. J Biol Inorg Chem. 16:1123–1134. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kolenko V, Teper E, Kutikov A and Uzzo R:
Zinc and zinc transporters in prostate carcinogenesis. Nat Rev
Urol. 10:219–226. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bafaro E, Liu Y, Xu Y and Dempski RE: The
emerging role of zinc transporters in cellular homeostasis and
cancer. Signal Transduct Target Ther. 2:170292017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Eide DJ: The SLC39 family of metal ion
transporters. Pflugers Arch. 447:796–800. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Schweigel-Röntgen M: The families of zinc
(SLC30 and SLC39) and copper (SLC31) transporters. Curr Top Membr.
73:321–355. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Grattan BJ and Freake HC: Zinc and cancer:
Implications for LIV-1 in breast cancer. Nutrients. 4:648–675.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Taylor KM, Morgan HE, Smart K, Zahari NM,
Pumford S, Ellis IO, Robertson JF and Nicholson RI: The emerging
role of the LIV-1 subfamily of zinc transporters in breast cancer.
Mol Med. 13:396–406. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Unno J, Satoh K, Hirota M, Kanno A, Hamada
S, Ito H, Masamune A, Tsukamoto N, Motoi F, Egawa S, et al: LIV-1
enhances the aggressive phenotype through the induction of
epithelial to mesenchymal transition in human pancreatic carcinoma
cells. Int J Oncol. 35:813–821. 2009.PubMed/NCBI
|
|
53
|
Zhao L, Chen W, Taylor KM, Cai B and Li X:
LIV-1 suppression inhibits HeLa cell invasion by targeting
ERK1/2-Snail/Slug pathway. Biochem Biophys Res Commun. 363:82–88.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wu C, Li D, Jia W, Hu Z, Zhou Y, Yu D,
Tong T, Wang M, Lin D, Qiao Y, et al: Genome-wide association study
identifies common variants in SLC39A6 associated with length of
survival in esophageal squamous-cell carcinoma. Nat Genet.
45:632–638. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Cui XB, Shen YY, Jin TT, Li S, Li TT,
Zhang SM, Peng H, Liu CX, Li SG, Yang L, et al: SLC39A6: A
potential target for diagnosis and therapy of esophageal carcinoma.
J Transl Med. 13:3212015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lian J, Jing Y, Dong Q, Huan L, Chen D,
Bao C, Wang Q, Zhao F, Li J, Yao M, et al: miR-192, a prognostic
indicator, targets the SLC39A6/SNAIL pathway to reduce tumor
metastasis in human hepatocellular carcinoma. Oncotarget.
7:2672–2683. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ge Y, Mu W, Ba Q, Li J, Jiang Y, Xia Q and
Wang H: Hepatocellular carcinoma-derived exosomes in organotropic
metastasis, recurrence and early diagnosis application. Cancer
Lett. 477:41–48. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Subbotin VM: Privileged portal metastasis
of hepatocellular carcinoma in light of the coevolution of a
visceral portal system and liver in the chordate lineage: A search
for therapeutic targets. Drug Discov Today. 23:548–564. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chandler P, Kochupurakkal BS, Alam S,
Richardson AL, Soybel DI and Kelleher SL: Subtype-specific
accumulation of intracellular zinc pools is associated with the
malignant phenotype in breast cancer. Mol Cancer. 15:22016.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Hogstrand C, Kille P, Ackland ML, Hiscox S
and Taylor KM: A mechanism for epithelial-mesenchymal transition
and anoikis resistance in breast cancer triggered by zinc channel
ZIP6 and STAT3 (signal transducer and activator of transcription
3). Biochem J. 455:229–237. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Unno J, Masamune A, Hamada S and
Shimosegawa T: The zinc transporter LIV-1 is a novel regulator of
stemness in pancreatic cancer cells. Scand J Gastroenterol.
49:215–221. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wan X, Kong Z, Chu K, Yi C, Hu J, Qin R,
Zhao C, Fu F, Wu H, Li Y and Huang Y: Co-expression analysis
revealed PTCH1-3′UTR promoted cell migration and invasion by
activating miR-101-3p/SLC39A6 axis in non-small cell lung cancer:
Implicating the novel function of PTCH1. Oncotarget. 9:4798–4813.
2018. View Article : Google Scholar : PubMed/NCBI
|