Introduction
Globally, breast cancer is the second most common
cancer and the most common cancer in women. In Taiwan, the
age-standardized incidence rate (ASIR) of female breast cancer was
70.7 per 100,000 individuals in 2014, and the ASIR gradually
increased at a rate of 3.5 per 100,000 person-years (1,2). The
age of breast cancer onset is currently 10 years younger than that
in Western countries (3). Thus,
early detection and early treatment are important objectives for
patients with breast cancer (4).
At present, diagnosis and disease monitoring primarily involve
tissue biopsy and imaging. Tissue biopsy is an invasive method
limited to specific regions (5).
Mammography is the gold standard for breast cancer detection due to
its high sensitivity and specificity; however, numerous women tend
to avoid this due the exposure to radiation (6). A novel minimally invasive biomarker
for breast cancer is needed.
Liquid biopsy from bodily fluid, such as circulating
cell-free DNA (cfDNA), circulating tumor cells and exosomes, is
minimally invasive and has proven both convenient and effective in
cancer diagnosis (7,8). The quantity of cfDNA in the blood can
be used to characterize tumorigenesis, inflammatory disease and
stroke (9). The Epi-proColon blood
test for circulating methylated DNA has been approved for the
detection of colorectal cancer (10,11).
It has been revealed that genomic alterations, such as human
epidermal growth factor receptor 2 (HER2) amplification, can serve
as a predictive biomarker for breast cancer (12); however, the identification of valid
diagnostic biomarkers for breast cancer from liquid biopsy is
needed.
DNA methylation, occurring at CpG dinucleotides, is
a hallmark of cancer (13). As
this epigenetic change occurs early in tumorigenesis, it can be
used as a biomarker for early detection, disease prognosis and
monitoring (14). Several studies
have reported the presence of methylated DNA in serum samples from
patients with cancer of the gastrointestinal tract, lung, head and
neck, liver and breast (15–17).
In cases of breast cancer, DNA methylation signature is related to
the clinicopathological characteristics of the tumor, such as tumor
stage and grade (18,19). It is possible that DNA
methylation-mediated epigenetic silencing of tumor suppressors
could affect the progression and prognosis of breast cancer
(20–22). One previous study demonstrated that
hypermethylation of seven biomarkers, including protocadherin β15
(PCDHB15), could differentiate patients with breast cancer
into the high- and low-risk groups (23).
PCDHB15 is a member of the cadherin
superfamily and calcium-dependent cell-cell adhesion molecules,
which encodes for PCDHB15 protein in humans (24,25).
Several cell adhesion molecules, such as CDH1 (also known as
E-cadherin), act as epithelial-mesenchymal transition suppressors
(26). In this regard, the
epigenetic silencing of CDH1 has frequently been observed in
cases of human cancer, including breast cancer (27–29).
Nonetheless, the role played by the epigenetic silencing of
PCDHB15 in cases of breast cancer remains unclear. In the
present study, PCDHB15 was identified as a potential tumor
suppressor gene in breast cancer, based on the observation that
PCDHB15 expression is positively correlated with the
likelihood of relapse-free survival. The detection of
PCDHB15 methylation in serum samples of patients with breast
cancer could be a novel minimally invasive biomarker for the
diagnosis and prognosis of breast cancer.
Materials and methods
Cell culture
MCF7 and MDA-MB-231 human breast cancer cell lines
were maintained in DMEM (Gibco; Thermo Fisher Scientific, Inc.),
supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo
Fisher Scientific, Inc.) and 50 U/ml penicillin/streptomycin
(Invitrogen; Thermo Fisher Scientific, Inc.). The cells were
incubated under 5% CO2 at 37°C. DNA demethylation
involved treating the cells with 0.1 µM DNA methyltransferase
(DNMT) inhibitor 5′-aza-2′-deoxycytidine, (5aza; Merck KGaA) or
DMSO (as control) at 37°C for 72 h. Culture media and drugs were
replenished every 24 h. Following treatment, the cells were
harvested for RNA analysis.
Patient samples
All patient samples were collected from the Biobank
of the Ditmanson Medical Foundation Chiayi Christian Hospital,
Chiayi, Taiwan (Table I). The
cancer group (age range, 30–78 years) was comprised of patients
with confirmed breast cancer, whereas the control group (age range,
20–53 years) was comprised of patients diagnosed with benign
tumors. The inclusion criteria were patients >20 years old and
who were undergoing biopsy or mastectomy; exclusion criteria were
patients who could not undergo any surgery or blood sampling. Serum
samples obtained from patients with breast cancer (n=49) and
patients with benign tumors (n=49) were used for quantitative
methylation-specific PCR (qMSP) analysis. Briefly, blood samples
were drawn into a 10-ml K2-EDTA blood tube (BD
Biosciences) and centrifuged at 1,358 × g at room temperature for
10 min, whereupon the serum was collected and stored at −80°C. The
present study was approved (approval no. IRB2019006 on 2019/3/5) by
the Institutional Review Board of the Ditmanson Medical Foundation
Chiayi Christian Hospital (Chiayi, Taiwan) and performed in strict
accordance with approved guidelines. Written informed consent was
obtained from all participants.
 | Table I.Summary of cliniopathological data of
plasma samples. |
Table I.
Summary of cliniopathological data of
plasma samples.
|
| Plasma samples |
|---|
|
|
|
|---|
| Clinicopathological
characteristic | Cancer (n=49) | Benign (n=49) |
|---|
| Age, years |
55.12±10.59a | 34.96±9.92 |
| Histological
gradeb |
|
|
| Low
grade | 4 |
|
| High
grade | 37 |
|
|
Unknown | 8 |
|
| Estrogen
receptor |
|
|
| - | 18 |
|
| + | 30 |
|
|
Unknown | 1 |
|
| Progesterone
receptor |
|
|
| - | 20 |
|
| + | 28 |
|
|
Unknown | 1 |
|
| Human epidermal
growth factor receptor 2 |
|
|
| - | 19 |
|
| + | 28 |
|
|
Unknown | 2 |
|
Plasmid transfection and colony
formation
The full-length human PCDHB15 expression
plasmid was a gift from Professor Jun Yu (Chinese University of
Hong Kong, Hong Kong). PCDHB15-expressing or empty vectors
(5 µg; pCMV6-XL5) were transfected into MDA-MB-231 cells (a
triple-negative breast cancer cell line with lower expression of
PCDHB15) using TransIT-LT1 Transfection Reagent (Mirus Bio
LLC) in accordance with the manufacturer's protocol. After 72 h of
incubation at 37°C, the transfection reagents were removed and
replaced with fresh medium. Transfected cells were cultured in
fresh medium at 37°C prior to further experiments.
For the colony-formation analysis, a total of
1×104 transfected cells per well were seeded in three
6-cm dishes with complete culture medium. Cells were cultured in
fresh culture medium at 37°C for 5–7 days, and the culture medium
was replaced at intervals of 3 days. Surviving colonies were
stained with 0.4% crystal violet (solubilized in 50% methanol) at
room temperature for 30 min. The number of colonies (as defined by
the size of the colonies >5 pixel2) was then
calculated by using ImageJ 1.53e software (National Institutes of
Health).
RNA extraction and reverse
transcription-quantitative PCR
Total RNA was extracted from the MCF7 and MDA-MB-231
cells using TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.) in accordance with the manufacturer's protocol.
Briefly, 1 µg of total RNA was treated with DNase I (Amplification
grade; Invitrogen; Thermo Fisher Scientific, Inc.), before it
underwent reverse transcription. First-strand cDNA synthesis was
performed using MMLV Reverse Transcriptase (Epicentre; Illumina,
Inc.) with oligo dT primers. Briefly, RNA was denatured and the
oligo dT primers were annealed at 65°C for 2 min, then chilled on
ice for 1 min. The mixture was gently mixed with dNTP, DTT, RNase
inhibitor, X10 RT reaction buffer and MMLV Reverse Transcriptase.
The final 20-µl mixture was incubated at 37°C for 60 min followed
by 85°C for 5 min. qPCR was performed using an ABI Step-One
real-time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) with specific primers and Power SYBR Green Master Mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: Initial denaturation at
95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, 60°C for
30 sec and 72°C for 30 sec. The primer sequences were as follows:
PCDHB15 forward, 5′-agcctttcaggagaaattcgactaat-3′ and
reverse, 5′-gcaccttaacagagacagagcatttt-3′; and GAPDH
forward, 5′-ccccttcattgacctcaactacat-3′ and reverse,
5′-cgctcctggaagatggtga-3′. Relative gene expression was calculated
by comparing the quantification cycle (Cq) value of PCDHB15
gene against the Cq value of GAPDH in a given sample (i.e.,
2−ΔΔCq) (30).
Extraction and bisulphite conversion
of DNA
DNA was extracted from serum samples using the
QIAamp Circulating Nucleic Acid kit (Qiagen GmbH) in accordance
with the manufacturer's protocol. Extracted DNA was
bisulphite-modified using the EZ DNA methylation kit (ZYMO Research
Corp.) in accordance with the manufacturer's protocol, as
previously described (31).
MSP and qMSP
PCDHB15 methylation in serum samples was
detected by subjecting the bisulphite-modified DNA to MSP and qMSP,
as previously described (32).
Briefly, 4 µl bisulphite-converted DNA was subjected to MSP within
the specific promoter PCDHB15 region. The thermocycling
conditions were as follows: Initial denaturation at 95°C for 10
min, followed by 40 cycles of 95°C for 15 sec, 60°C for 30 sec and
72°C for 30 sec. The PCR products were analyzed by electrophoresis
in 10% polyacrylamide gel and subsequent the gel was stained with
ethidium bromide. For qMSP analysis, 4 µl bisulphite-converted DNA
was subjected to qMSP within the specific promoter PCDHB15
region using an ABI Step One real time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and Power SYBR Green
Master Mix. The thermocycling conditions were as follows: Initial
denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for
15 sec, 60°C for 30 sec and 72°C for 30 sec. The primer sequences
for PCDHB15 were as follows: Forward,
5′-acgttttttttaaggaatcg-3′ and reverse, 5′-acgaaccaatatctccga-3′
(130 bp). The presence of cfDNA in serum samples was detected via
collagen type II α1 chain (COL2A1) MSP using the forward primer
5′-tct aac aat tat aaa ctc caa cca cca a-3′ and the reverse primer
5′-gggaagatgggatagaagggaatat-3′. The quantity of methylated DNA was
determined in terms of Cq value against a standard curve generated
using an in vitro methylated DNA-MSP cloned fragment, as
previously described (33).
TCGA data analysis
The Cancer Genome Atlas (TCGA) breast cancer (BRCA)
Methylation450K dataset was downloaded from UCSC Xena (http://xena.ucsc.edu). The methylation level of CpG
sites in PCDHB15 between solid normal tissues and primary
tumor tissues were compared and analyzed. Associations between the
expression and methylation (cg17023770) of PCDHB15 were
analyzed.
Statistical analysis
All statistical analysis was performed using
GraphPad Prism Version 5.0 software packages for Windows (GraphPad
Software, Inc.). Differences between two groups were analyzed using
an unpaired Student's t-test or the Mann-Whitney U test. Pearson's
correlation analysis was used to analyze correlation between gene
expression, methylation status of a gene or protein expression.
Locoregional relapse-free survival was assessed by Kaplan-Meier
analysis, and differences between groups were estimated by the
log-rank test. P<0.05 was considered to indicate a statistically
significant difference.
Results
PCDHB15 is hypermethylated in breast
tumor samples compared with that in normal samples
Breast cancer data from TCGA were first used to
analyze DNA methylation profiles for PCDHB15. The present
study included 785 primary tumors and 98 solid tissue normal
samples. The methylation of PCDHB15 was higher in tumor
samples than that in normal solid tissue (P<0.001; Fig. 1A and B). As expected, a negative
association between the methylation of a particular CG site
(cg17023770) in the promoter region and the expression of
PCDHB15 was observed in this TCGA cohort (Fig. 1C; r=−0.2, P<0.0001).
Kaplan-Meier Plotter (34)
revealed that patients with lower PCDHB15 expression were
associated with shorter relapse-free survival times (Fig. 1D; HR, 0.74; P=0.00011), but not at
all with overall survival times (data not shown).
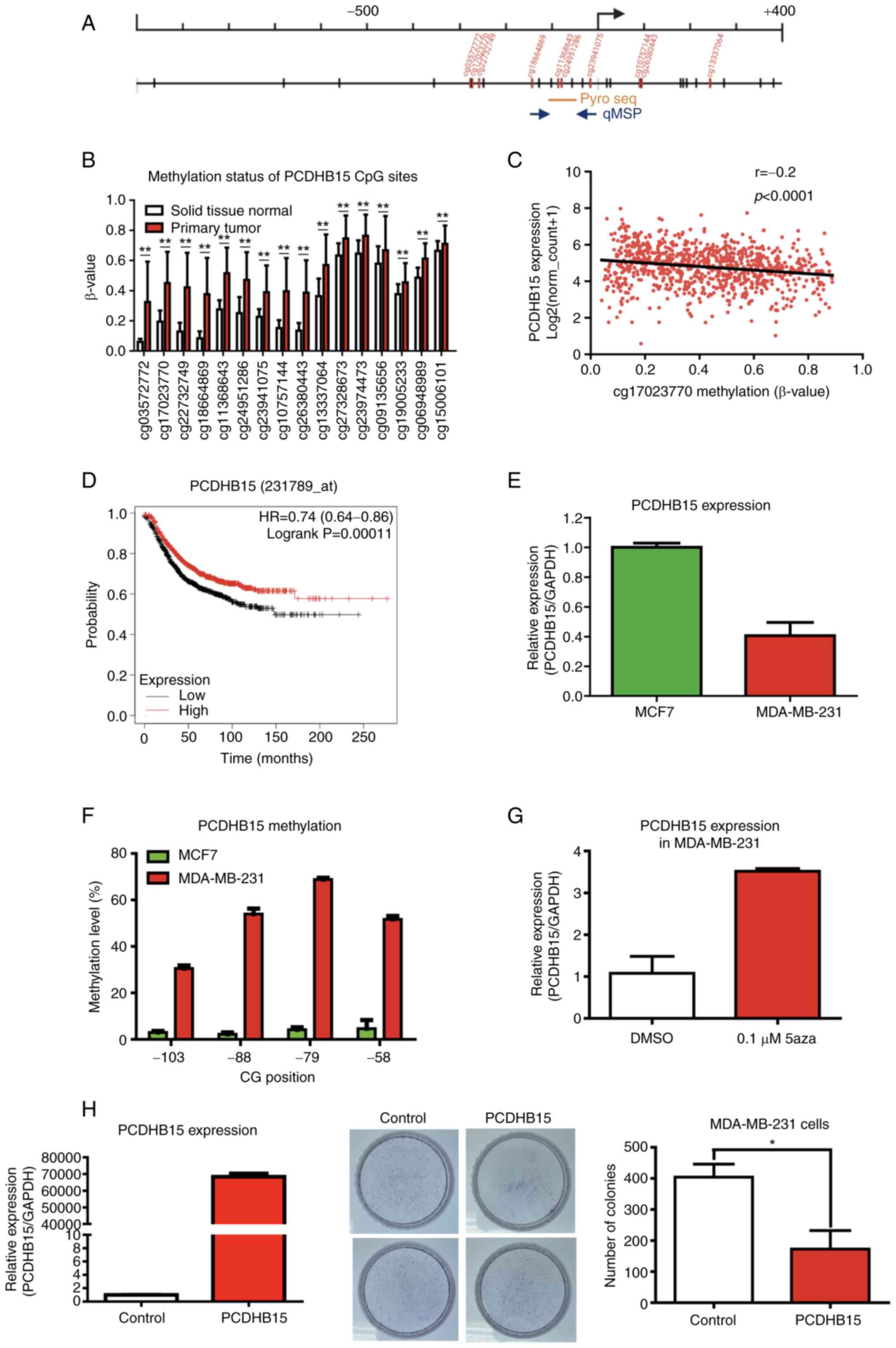 | Figure 1.PCDHB15 may be a tumor
suppressor gene that is epigenetically silenced in in breast
cancer. (A) Schematic diagram depicting the genomic structure and
position of the CG sites (vertical dashes) in the PCDHB15
promoter region (from-1,000 to +400 with respect to the
transcriptional start site). The location of the microarray probes
(red vertical dashes), bisulphite pyrosequencing (yellow horizontal
line) and qMSP primers (blue solid arrows) are indicated. (B) DNA
methylation level (β-value from Illumina Infinium 450 K microarray)
of the PCDHB15 CpG island from-278 (cg03572772) to +2252
(cg15006101) in solid tissue normal (white) vs. primary tumor (red)
in TCGA breast cancer dataset. Primary tumor tissues (n=785) had
higher methylation levels than solid tissue normal tissues (n=98).
The x-axis indicates the name of the probe on the microarray. (C)
Scatter plot showing correlation between PCDHB15 promoter
methylation (cg17023770; x-axis) and expression (y-axis) in TCGA
breast cancer dataset (n=873). A negative association between
promoter methylation and expression was observed. (D) Kaplan-Meier
analysis of PCDHB15 mRNA expression in tumor tissues for
relapse-free survival of patients with breast cancer. Patients with
breast cancer with lower PCDHB15 expression demonstrated
shorter relapse-free survival times than patients with higher
PCDHB15 expression (log-rank test, P=0.00011). (E) Relative
expression level of PCDHB15 mRNA in MCF7 and MDA-MB-231
breast cancer cells. (F) Methylation analysis of PCDHB15
promoter in breast cancer cell lines using bisulphite
pyrosequencing. (G) Relative expression level of PCDHB15 in
0.1 µM 5aza-treated MDA-MB-231 breast cancer cells, compared with
DMSO control. (H) Ectopic expression of PCDHB15 inhibited
tumor proliferation by colony formation assay. MDA-MB-231 breast
cancer cells were transfected with empty (control) or
PCDHB15 expression vector. Left panel, reverse
transcription-quantitative PCR confirmed overexpression of
PCDHB15 in MDA-MB-231 cells transiently transfected with
PCDHB15 expression vector. Medium panel, MDA-MB-231 breast cancer
cells overexpressing PCDHB15 had significantly fewer
colonies than the control. Right panel, quantitative analysis of
the colony formation assay. Colony formation assay were performed
in duplicate and in two independent experiments (mean ± SD).
*P<0.05 and **P<0.001. PCDHB15, protocadherin β15; TCGA, The
Cancer Genome Atlas; 5aza, 5′-aza-2′-deoxycytidine; qMSP,
quantitative methylation-specific PCR. |
The association between promoter methylation and the
expression of PCDHB15 in breast cancer cell lines was then
investigated. It was determined that the expression of
PCDHB15 was downregulated in MDA-MB-231 cells compared with
that in MCF7 cells (Fig. 1E).
Concomitantly, bisulphite pyrosequencing revealed that promoter
methylation was higher in MDA-MB-231 cells than that in MCF7 cells
at CpG sites located in the upstream promoter region of
PCDHB15 (Fig. 1F). Notably,
the expression of PCDHB15 in MDA-MB-231 cells was restored
upon treatment using DNMT inhibitor (0.1 µM 5aza; Fig. 1G).
To examine the function of PCDHB15, PCDHB15
was overexpressed in MDA-MB-231 breast cancer cells, showing a
lower PCDHB15 expression, as compared with MCF7 cells. The
induced overexpression of PCDHB15 in MDA-MB-231 cells led to
a significant decrease in the number of colonies, compared with the
control vector (P<0.05; Fig.
1H). Taken together, these results suggested that PCDHB15 may
be a tumor suppressor subject to epigenetic silencing via promoter
hypermethylation in breast cancer.
Measuring PCDHB15 methylation in
clinical human serum specimens
The present study also sought to determine whether
PCDHB15 methylation could be used as a serum biomarker for
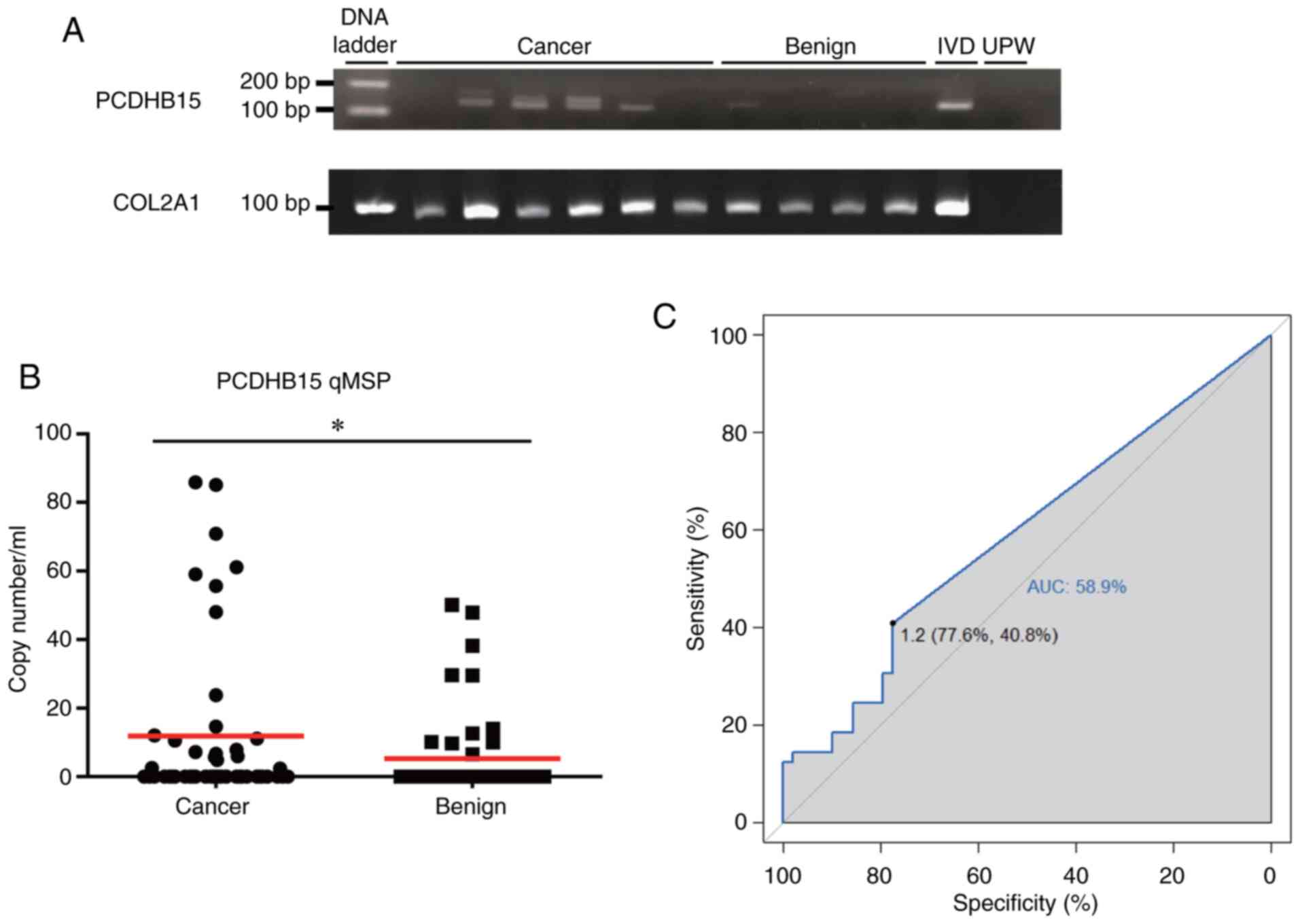
breast cancer. MSP revealed PCDHB15 methylation in 66% (4/6)
of cfDNA extracted from serum samples of patients with breast
cancer, but only in 25% of the samples (1/4) from patients with
benign tumors (Figs. 2A and
S1). The presence of COL2A1 MSP
products suggested the presence the cfDNA in serum samples that
were devoid of PCDHB15 methylation (Fig. 2A). The PCDHB15 methylation
was further examined using qMSP in serum samples from 49 patients
with cancer and 49 patients with benign tumors (Table I). The quantity of methylated
PCDHB15 was higher in patients with breast cancer than that
in samples from patients with benign tumors (P<0.05; Fig. 2B). Based on the cutoff value
generated by the area under the receiver operating characteristic
curve (0.589; a cutoff value of 1.2 copy number/ml), PCDHB15
methylation in serum samples provided a sensitivity of 40.8% and
specificity of 77.6% in breast cancer detection (Fig. 2C; Table II). These results demonstrated the
feasibility of using PCDHB15 methylation in the cfDNA of
serum samples as a minimally invasive biomarker for breast
cancer.
 | Table II.Summary of protocadherin β15
quantitative methylation-specific PCR analysis. |
Table II.
Summary of protocadherin β15
quantitative methylation-specific PCR analysis.
| Diagnosis | Valid specimens
(n) | Positive specimens
(n) | Negative specimens
(n) | Positive rate
(%) |
|---|
| Breast cancer | 49 | 20 | 29 | 40.8 |
| Low
grade (G1) | 4 | 1 | 3 | 25.0 |
| High
grade (≥G2) | 37 | 13 | 24 | 35.1 |
|
Unknown | 8 | 6 | 2 | 75.0 |
| Benign | 49 | 11 | 38 | 22.4 |
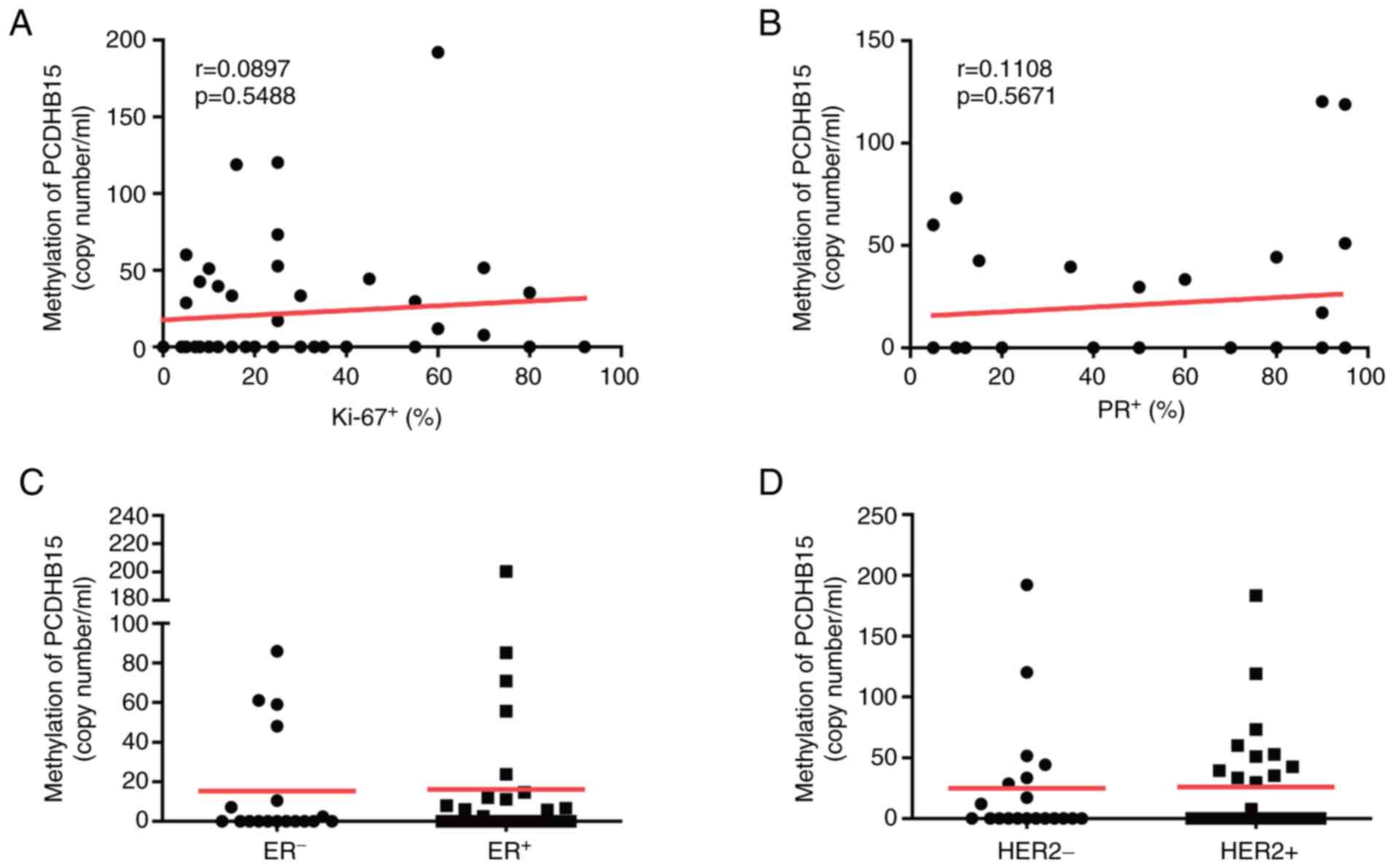
PCDHB15 methylation is not correlated
with other clinicopathological features
Several proteins, including HER2, Ki-67, estrogen
receptor and progesterone receptor, are important parameters in
subtyping breast cancer and pathogenesis characterization (6). Nonetheless, our analysis did not
reveal any correlation between PCDHB15 methylation and any
of those clinical parameters (Fig.
3A-D).
Discussion
PCDHB15 of the protocadherin superfamily is involved
in calcium-dependent cell-cell adhesion. The epigenetic silencing
of other cadherins (e.g., E-cadherin) has previously been
demonstrated; however, the role of PCDHB15 in breast cancer has yet
to be fully understood. In the current study, based on the TCGA
database and in-house samples, it was determined that
PCDHB15 methylation was more pronounced in patients with
breast cancer compared with that in patients with benign tumors or
in normal controls. Overall, the higher expression level of
PCDHB15 was positively associated with relapse-free
survival. Further analysis on specific cell lines revealed that
PCDHB15 may be a tumor suppressor downregulated via promoter
hypermethylation. It is also noteworthy to point out that the
correlation between PCDHB15 methylation and relapse-free
survival could not be determined, as DNA methylation data was not
available from the Kaplan-Meier Plotter. The prognostic
significance of PCDHB15 methylation requires further
investigation. It was recently reported that PCDHB15, acting
as a tumor suppressor through the inhibition of WNT/β-catenin
signaling, was epigenetically silenced in KRAS-mutated colorectal
cancer (35). This phenomenon can
perhaps be attributed to the overexpression of mitochondria
glutamate transporter, SLC25A22, in KRAS-mutated colorectal cancer.
Nonetheless, determining whether the overexpression of SLC25A22 is
responsible for PCDHB15 methylation in breast cancer is
worthy of further investigation.
In the present study, PCDHB15 methylation was
higher in cfDNA from serum samples of patients with breast cancer
than that from patients with benign tumors. Nonetheless,
PCDHB15 methylation was not associated with any clinical
parameters, thereby suggesting that PCDHB15 methylation may
occur early in the carcinogenesis. It also suggested that
PCDHB15 methylation is common to all molecular subtypes of
breast cancer. The fact that 22.4% (11 out of 49) of the benign
tumor samples tested positive for PCDHB15 methylation
supports these hypotheses; however, further clinical analysis using
a larger sample size and different molecular subtypes will be
required to demonstrate whether PCDHB15 methylation is
involved in the field defect of breast cancer (36).
The sensitivity of PCDHB15 methylation
(40.8%) in breast cancer detection in serum would not allow its use
as a sole biomarker; however it could potentially serve as one
epigenetic biomarker in a ‘methylation signature panel’ for the
diagnosis and/or prognosis of breast cancer (37–39).
Previous studies have identified distinct methylation biomarkers
indicating the signaling-mediated epigenetic silencing of tumor
suppressors, regardless of different subtypes of breast (40–42).
Multiple research groups are currently evaluating the methylation
signature panel of GSTP1, RASSF1 and RARB for the
detection of breast cancer (29).
The PITX2 methylation assay, which has been already
certified for in vitro diagnosis, has proven effective as a
prognostic and predictive biomarker for breast cancer (43). It is possible that methylation of
PCDHB15 could be used in conjunction with these markers to
form a novel epigenetic panel by which to characterize the
progression of breast cancer.
In conclusion, PCDHB15 is a potential tumor
suppressor subject to epigenetic silencing via promoter methylation
in breast cancer. PCDHB15 methylation in serum cfDNA may
provide a novel minimally invasive epigenetic biomarker for the
diagnosis and prognosis of breast cancer. Determining whether
PCDHB15 methylation is involved in the early carcinogenesis
of breast cancer warrants further investigation.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Ministry of
Science and Technology, Taiwan (grant no. MOST
108-2314-B-194-003-MY2), the Ditmanson Medical Foundation Chiayi
Christian Hospital, Taiwan (grant no. RCN009) and the Center for
Innovative Research on Aging Society (grant no. 107-B128-09) from
The Featured Areas Research Center Program within the framework of
the Higher Education Sprout Project by Ministry of Education in
Taiwan.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CCC, GLL, CWT, CFW and YCH collected patient samples
and analyzed clinical data. GLL, WLH, FCW, PJL, WHH, YMC and YTL
performed experiments. YMC, SYY and MWYC performed the
bioinformatic analysis. GLL, SYY and MWYC wrote the manuscript.
CCY, YCH and MWYC designed the experiments. GLL, WLH and MWYC
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved (approval no.
IRB2019006 on 2019/3/5) by the Institutional Review Board of the
Ditmanson Medical Foundation Chiayi Christian Hospital (Chiayi,
Taiwan). Written informed consent was obtained from all
participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu FC, Lin HT, Kuo CF, See LC, Chiou MJ
and Yu HP: Epidemiology and survival outcome of breast cancer in a
nationwide study. Oncotarget. 8:16939–16950. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kuo CN, Liao YM, Kuo LN, Tsai HJ, Chang WC
and Yen Y: Cancers in Taiwan: Practical insight from epidemiology,
treatments, biomarkers, and cost. J Formos Med Assoc.
119:1731–1741. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen YP, Lu YW and Yang CC: Breast cancer
trend in Taiwan. MOJ Womens Health. 6:1532017.
|
|
4
|
Rossi S, Cinini C, Di Pietro C, Lombardi
CP, Crucitti A, Bellantone R and Crucitti F: Diagnostic delay in
breast cancer: Correlation with disease stage and prognosis.
Tumori. 76:559–562. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang R, Li X, Zhang HM, Wang K and He JJ:
Cell-free circulating tumor DNA analysis for breast cancer and its
clinical utilization as a biomarker. Oncotarget. 8:75742–75755.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jafari SH, Saadatpour Z, Salmaninejad A,
Momeni F, Mokhtari M, Nahand JS, Rahmati M, Mirzaei H and Kianmehr
M: Breast cancer diagnosis: Imaging techniques and biochemical
markers. J Cell Physiol. 233:5200–5213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
De Mattos-Arruda L and Caldas C: Cell-free
circulating tumour DNA as a liquid biopsy in breast cancer. Mol
Oncol. 10:464–474. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Seale KN and Tkaczuk KHR: Circulating
biomarkers in breast cancer. Clin Breast Cancer. Sep 22–2021.doi:
10.1016/j.clbc.2021.09.006 (Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schwarzenbach H, Hoon DS and Pantel K:
Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev
Cancer. 11:426–437. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Potter NT, Hurban P, White MN, Whitlock
KD, Lofton-Day CE, Tetzner R, Koenig T, Quigley NB and Weiss G:
Validation of a real-time PCR-based qualitative assay for the
detection of methylated SEPT9 DNA in human plasma. Clin Chem.
60:1183–1191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lamb YN and Dhillon S: Epi proColon
® 2.0 CE: A blood-based screening test for colorectal
cancer. Mol Diagn Ther. 21:225–232. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Simon R and Roychowdhury S: Implementing
personalized cancer genomics in clinical trials. Nat Rev Drug
Discov. 12:358–369. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jones PA, Issa JP and Baylin S: Targeting
the cancer epigenome for therapy. Nat Rev Genet. 17:630–641. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Locke WJ, Guanzon D, Ma C, Liew YJ,
Duesing KR, Fung KYC and Ross JP: DNA methylation cancer
biomarkers: Translation to the clinic. Front Genet. 10:11502019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee TL, Leung WK, Chan MW, Ng EK, Tong JH,
Lo KW, Chung SC, Sung JJ and To KF: Detection of gene promoter
hypermethylation in the tumor and serum of patients with gastric
carcinoma. Clin Cancer Res. 8:1761–1766. 2002.PubMed/NCBI
|
|
16
|
Tang Q, Cheng J, Cao X, Surowy H and
Burwinkel B: Blood-based DNA methylation as biomarker for breast
cancer: A systematic review. Clin Epigenetics. 8:1152016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei KL, Chou JL, Chen YC, Low JT, Lin GL,
Liu JL, Chang TS, Chen WM, Hsieh YY, Yan PS, et al: Epigenetic
silencing of STAT3-targeted miR-193a, by constitutive activation of
JAK/STAT signaling, leads to tumor progression through
overexpression of YWHAZ in gastric cancer. Front Oncol.
11:5756672021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fackler MJ, Umbricht CB, Williams D,
Argani P, Cruz LA, Merino VF, Teo WW, Zhang Z, Huang P,
Visvananthan K, et al: Genome-wide methylation analysis identifies
genes specific to breast cancer hormone receptor status and risk of
recurrence. Cancer Res. 71:6195–6207. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fleischer T, Frigessi A, Johnson KC,
Edvardsen H, Touleimat N, Klajic J, Riis ML, Haakensen VD, Wärnberg
F, Naume B, et al: Genome-wide DNA methylation profiles in
progression to in situ and invasive carcinoma of the breast with
impact on gene transcription and prognosis. Genome Biol.
15:4352014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jovanovic J, Ronneberg JA, Tost J and
Kristensen V: The epigenetics of breast cancer. Mol Oncol.
4:242–254. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu J, Wilfred P, Korbie D and Trau M:
Regulation of canonical oncogenic signaling pathways in cancer via
DNA methylation. Cancers (Basel). 12:31992020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tao S, Li H, Ma X, Lian B, He J, Gao Y and
Li J: Methylation-mediated silencing of MicroRNA-497 promotes
breast cancer progression through up-regulation of Mucin1. Front
Oncol. 10:5520992020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang C, Zhao H, Li J, Liu H, Wang F, Wei
Y, Su J, Zhang D, Liu T and Zhang Y: The identification of specific
methylation patterns across different cancers. PLoS One.
10:e01203612015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu Q and Maniatis T: A striking
organization of a large family of human neural cadherin-like cell
adhesion genes. Cell. 97:779–790. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hulpiau P and van Roy F: Molecular
evolution of the cadherin superfamily. Int J Biochem Cell Biol.
41:349–369. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Serrano-Gomez SJ, Maziveyi M and Alahari
SK: Regulation of epithelial-mesenchymal transition through
epigenetic and post-translational modifications. Mol Cancer.
15:182016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zou D, Yoon HS, Perez D, Weeks RJ,
Guilford P and Humar B: Epigenetic silencing in non-neoplastic
epithelia identifies E-cadherin (CDH1) as a target for
chemoprevention of lobular neoplasia. J Pathol. 218:265–272. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Asiaf A, Ahmad ST, Aziz SA, Malik AA,
Rasool Z, Masood A and Zargar MA: Loss of expression and aberrant
methylation of the CDH1 (E-cadherin) gene in breast cancer patients
from Kashmir. Asian Pac J Cancer Prev. 15:6397–6403. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
de Ruijter TC, van der Heide F, Smits KM,
Aarts MJ, van Engeland M and Heijnen VCG: Prognostic DNA
methylation markers for hormone receptor breast cancer: A
systematic review. Breast Cancer Res. 22:132020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chou JL, Huang RL, Shay J, Chen LY, Lin
SJ, Yan PS, Chao WT, Lai YH, Lai YL, Chao TK, et al:
Hypermethylation of the TGF-β target, ABCA1 is associated with poor
prognosis in ovarian cancer patients. Clin Epigenetics. 7:12015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen PC, Tsai MH, Yip SK, Jou YC, Ng CF,
Chen Y, Wang X, Huang W, Tung CL, Chen GC, et al: Distinct DNA
methylation epigenotypes in bladder cancer from different Chinese
sub-populations and its implication in cancer detection using
voided urine. BMC Med Genomics. 4:452011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tseng KC, Chou JL, Huang HB, Tseng CW, Wu
SF and Chan MW: SOCS-1 promoter methylation and treatment response
in chronic hepatitis C patients receiving
pegylated-interferon/ribavirin. J Clin Immunol. 33:1110–1116. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wong CC, Xu J, Bian X, Wu JL, Kang W, Qian
Y, Li W, Chen H, Gou H, Liu D, et al: In colorectal cancer cells
with mutant KRAS, SLC25A22-mediated glutaminolysis reduces DNA
demethylation to increase WNT signaling, stemness, and drug
resistance. Gastroenterology. 159:2163–2180.e6. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Teschendorff AE, Gao Y, Jones A, Ruebner
M, Beckmann MW, Wachter DL, Fasching PA and Widschwendter M: DNA
methylation outliers in normal breast tissue identify field defects
that are enriched in cancer. Nat Commun. 7:104782016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Du T, Liu B, Wang Z, Wan X and Wu Y: CpG
methylation signature predicts prognosis in breast cancer. Breast
Cancer Res Treat. 178:565–572. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Peng Y, Shui L, Xie J and Liu S:
Development and validation of a novel 15-CpG-based signature for
predicting prognosis in triple-negative breast cancer. J Cell Mol
Med. 24:9378–9387. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mao XH, Ye Q, Zhang GB, Jiang JY, Zhao HY,
Shao YF, Ye ZQ, Xuan ZX and Huang P: Identification of
differentially methylated genes as diagnostic and prognostic
biomarkers of breast cancer. World J Surg Oncol. 19:292021.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chou JL, Chen LY, Lai HC and Chan MW:
TGF-β: Friend or foe? The role of TGF-β/SMAD signaling in
epigenetic silencing of ovarian cancer and its implication in
epigenetic therapy. Expert Opin Ther Targets. 14:1213–1223. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bloushtain-Qimron N, Yao J, Snyder EL,
Shipitsin M, Campbell LL, Mani SA, Hu M and Chen H: Cell
type-specific DNA methylation patterns in the human breast. Proc
Natl Acad Sci USA. 105:14076–14081. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Novak P, Stampfer MR, Munoz-Rodriguez JL,
Garbe JC, Ehrich M, Futscher BW and Jensen TJ: Cell-type specific
DNA methylation patterns define human breast cellular identity.
PLoS One. 7:e522992012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Beltran-Garcia J, Osca-Verdegal R,
Mena-Molla S and Garcia-Gimenez JL: Epigenetic IVD tests for
personalized precision medicine in cancer. Front Genet. 10:6212019.
View Article : Google Scholar : PubMed/NCBI
|