Introduction
Breast cancer (BC) is the most frequently diagnosed
cancer globally, with >2 million incident cases in 2020
(1). In Turkey, 25,345 new cases
of BC were diagnosed in 2020, corresponding to 24.4% of all cancers
diagnosed in women (2). Although
hereditary factors contribute to 10–30% of BC pathogenesis, only a
small fraction of BCs (5–10%) can be explained by germline
mutations in cancer susceptibility genes (3,4),
which means other susceptibility loci are likely to exist (5). Identifying unaffected disease-causing
variant carriers and governing their risk has been shown to reduce
BC and all-cause mortality (6).
The selection of patients for BC risk assessment and counseling is
mainly performed using information about the personal and/or family
history (FH) of BC, usually following the guidelines and
recommendations of The National Comprehensive Cancer Network (NCCN)
(7). Although most patients with
hereditary BC are found to have mutations in BRCA1/2 genes, it is
estimated that as much as 70% of BC cases could be caused by
mutations in other genes (8).
Furthermore, recent studies have shown the importance of carefully
selected patients who will benefit from genetic testing (9).
The use of multigene panel testing for detecting
hereditary cancer risk has increased within the last few years,
aiming to provide information and tailored management for more
families in a convenient way regarding patients with a less typical
presentation for a given cancer syndrome and/or missing FH data and
to provide a cost-effective alternative to BRCA-only testing
(10,11).
Consanguineous marriage (CM) is still common in
various parts of the world (12)
and mainly occurs as a first cousin union (13). For example, in Turkey, the
prevalence of CMs is still relatively high at 20–25% (ranges from
11.5 to 46%) (14,15). However, the relationship between CM
and BC-predisposing genes is not clear.
The present study aimed to analyze 33 genes
implicated in hereditary cancer predisposition in Turkish patients
with BC and investigate the impact of CM and FH of BC on carrying
pathogenic mutations in genes associated with BC.
Materials and methods
Patients
In this prospective clinical study, patients who
were treated with BC between 2018 and 2020 in our clinic were
included. Patients who did not agree to participate in the study
and did not have any genetic test indication were excluded.
Patients were divided into two groups as CM [study group (SG)] and
those without CM [control group (CG)] and were directed to genetic
tests. All women received genetic counseling and psychological
assistance and signed an informed consent form before molecular
genetic testing and permission to use their data for research
purposes. The Biruni University Ethical Committee approved the
study (approval no. 2015-KAEK-43-18-11; 19/09/2018). Basic
demographics, personal and family histories were noted, and
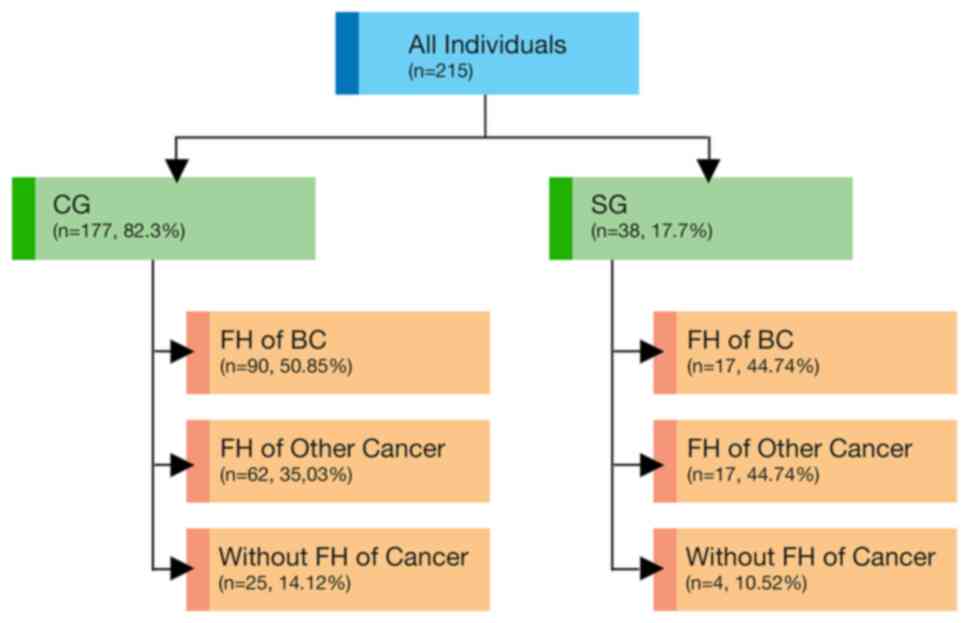
pedigrees were drawn by ordering clinicians (Fig. 1).
Two next-generation sequencing (NGS) multigene
panels, including 26 and 33 genes related to hereditary cancer
predisposition, were performed in both the SG and CG as previously
described (16). The panel genes
were grouped according to the associated risk for BC [high-risk
genes, BRCA1, BRCA2, CDH1, partner and localizer of BRCA2 (PALB2),
PTEN, serine/threonine-protein kinase STK11 and TP53; moderate-risk
genes, serine-protein kinase ATM (ATM), serine/threonine-protein
kinase Chk2 (CHEK2) and nibrin (NBN); low/unknown-risk genes, APC,
bone morphogenetic protein receptor type-1A, CDK4, CDKN2A,
epithelial cell adhesion molecule (EPCAM), menin, DNA mismatch
repair protein Mlh1 (MLH1), DNA mismatch repair protein Msh2
(MSH2), DNA mismatch repair protein Msh6 (MSH6), adenine DNA
glycosylase (MUTYH), mismatch repair endonuclease PMS2 (PMS2),
proto-oncogene tyrosine-protein kinase receptor Ret, SMAD4, von
Hippel-Lindau disease tumor suppressor, Fanconi anemia group J
protein (BRIP1), DNA repair protein RAD51 homolog 3 (RAD51C), DNA
repair protein RAD51 homolog 4 (RAD51D), BRCA1-associated RING
domain protein 1, serine/threonine-protein kinase Chk1,
double-strand break repair protein MRE11, neurofibromin, DNA repair
protein RAD50 (RAD50) and DNA repair protein RAD51 homolog 2]
(16).
NGS and mutation confirmation
The genomic DNA was isolated from peripheral blood
leukocytes according to manufacturers' protocol (Qiagen, Inc., and
RBC Bioscience) and was quantified by using NanoDrop 2000c
Spectrophotometer (Thermo Fisher Scientific, Inc.).
In the first protocol; a probe library (Roche
NimbleGen SeqCap EZ Choice) targeting all coding exons and 50 bp of
flanking intronic regions of the 33 genes was custom-designed, and
sample preparation was performed following the SeqCap EZ Choice
Library User's Guide (Roche NimbleGen, Inc.) as previously
described (16). Briefly, after
enzymatic fragmentation (Kappa Hyperplus kit), the library was
prepared according to the manufacturer's protocol (Roche NimbleGen,
Inc.). NGS was performed with MiSeq Reagent Kit v3 (600-cycle) on
Illumina MiSeq machine (Illumina, Inc.). Alignment to the reference
sequence (hg19) and variant calling was performed by the SeqNext
module of the SeqPilot suite (JSI Medical Systems GmbH).
In the second protocol; a commercially designed
panel, from SOPHiA Genetics™ was used: The SOPHiA Hereditary Cancer
Solutions (HCS). The library preparation was performed according to
manufacturer's protocol. Sequencing was performed on Illumina Next
Seq machine (Illumina, Inc.) with manufacturer's appropriate NGS
kits. Alignment to the reference sequence (hg19) and variant
calling was performed by the SOPHiA DDM platform of the same
manufacturer.
The annotation and interpretation of all identified
variants were performed using an in-house local knowledge base and
a proprietary bioinformatics pipeline. The clinical significance of
variants was examined using the standards and guidelines for the
interpretation of sequence variants recommended by the American
College of Medical Genetics and Genomics (ACMG Laboratory Quality
Assurance Committee) and the Association for Molecular Pathology
(AMP) (17).
All pathogenic, likely pathogenic variants (PVs)
obtained from the custom designed primers and VUS were confirmed by
Sanger Sequencing for further segregation analysis using Applied
Biosystems 3130 Genetic Analyzer (Thermo Fisher Scientific, Inc.).
Sanger sequences are presented in Figs. S1-S6. Family segregations were performed by
Sanger Sequencing for further segregation analysis also using
Applied Biosystems 3130 Genetic Analyzer (Thermo Fisher Scientific,
Inc.).
Analysis of large genomic
rearrangements (LGRs)
The copy number variation (CNV) module of the
software suite SeqPilot (JSI Medical Systems GmbH) and panels. MOPS
(18) was used for the
computational analysis of LGRs from NGS data for the following
genes/gene regions, including BRCA1, BRCA2, CHEK2, EPCAM (Exons 8,
9), MLH1, MSH2, MSH6, MUTYH, PALB2, RAD50 (Exons 1, 2, 4, 10, 14,
21, 23 and 25), RAD51C, RAD51D, and TP53. All predicted LGRs
detected with these algorithms were then experimentally studied
using the Multiplex Ligation-dependent Probe Amplification
technique as described previously (16,18).
Statistical analysis
Pearson correlation analysis with ‘N-1’ correction
was performed to correlate two parameters. In addition, chi-square
tests were conducted to compare the distribution of categorical
variables. Fischer's exact test was used when chi-square tests
assumptions did not hold due to low expected cell counts, All
analyses were performed with R software version 3.4.4. All
statistical tests were two-sided, P<0.05 was considered to
indicate a statistically significant difference.
Results
Patient characteristics
A total of 38 patients with BC with CM (SG) and 177
patients with BC without CM (CG) were referred for testing. The
detailed demographic and clinical features were summarized and
compared in Table I. The median
age of patients was 47 years (27–76 years). The median time from
diagnosis to testing was 1 year, and 67% (144/215) of patients
tested within 12 months following the diagnosis. A family history
(FH) of cancer constituted a primary reason for referral,
accounting for 86.4% (186/215). Almost half of women (49.7%,
107/215) reported FH of BC (44.7% in SG and 50.8% in CG). CM was
reported in 17.7% (38/215) of the patients. There were no
statistically significant differences in the patient
characteristics between the two groups (Tables I–III).
 | Table I.Overall characteristics and test
results in two groups. |
Table I.
Overall characteristics and test
results in two groups.
|
|
| Groups |
|
|---|
|
|
|
|
|
|---|
| Characteristic | Patients with
BC | SG | CG | P-value |
|---|
| Total patients | 215a | 38
(17.7)b | 177
(82.3)b |
|
| Age at testing,
years |
|
|
| 0.3173 |
| Mean ±
SD | 47.5±10.6 | 47.4±9.9 | 47.5±11.0 |
|
| Median,
range | 47 (27–76) | 47 (32–68) | 47 (27–76) |
|
| FH of cancer, n
(%) |
|
|
|
|
|
None | 29 (13.4) | 4 (10.5) | 25 (14.1) | 0.7900 |
| FH of
BC | 107 (49.7) | 17 (44.7) | 90 (50.8) | 0.5900 |
| FH of
other cancer(s) | 79 (36.7) | 17 (44.7) | 62 (35.0) | 0.2700 |
| Multigene test
result, n (%) |
|
|
|
|
|
Negative | 79 (36.7) | 13 (34.2) | 66 (37.3) | 0.8500 |
|
Positive | 34 (15.8) | 5 (13.2) | 29 (16.4) | 0.8000 |
| VUS
only | 102 (47.4) | 20 (52.6) | 82 (46.3) | 0.5900 |
| Positive in gene
categories, n (%) |
|
|
|
|
| NCCN
absolute risk category | 13 (37.1) | 1 (20.0) | 12 (41.4) | 0.6200 |
|
>60% |
|
|
|
|
| NCCN
absolute risk category | 2 (0.5) | 0 | 2 (6.9) | 1.0000 |
|
41-60% |
|
|
|
|
| NCCN
absolute risk category | 19 (55.8) | 4 (80.0) | 15 (51.7) | 0.6200 |
| 15-40%
and low/unknown risk |
|
|
|
|
 | Table III.Association between FH of BC and
genetic test results. |
Table III.
Association between FH of BC and
genetic test results.
|
|
| Groups |
|
|---|
|
|
|
|
|
|---|
| Genetic test
results | FH status | CG | SG | P-value |
|---|
| Negative | FH of BC |
|
| 0.022 |
|
| No | 33 (50.0) | 11 (84.6) |
|
|
| Yes | 33 (50.0) | 2 (15.4) |
|
|
| FH of any
cancer |
|
| 0.370 |
|
| No | 8 (12.0) | 3 (23.0) |
|
|
| Yes | 58 (88.0) | 10 (77.0) |
|
| VUS | FH of BC |
|
| 0.620 |
|
| No | 46 (56.1) | 10 (50.0) |
|
|
| Yes | 36 (43.9) | 10 (50.0) |
|
|
| FH of any
cancer |
|
| 0.170 |
|
| No | 14 (17.0) | 1 (5.0) |
|
|
| Yes | 68 (83.0) | 19 (95.0) |
|
| Negative,
PV+VUS | Only patients with
FH of BC |
|
| 0.045 |
| Negative |
| 33 (36.7) | 2 (11.8) |
|
| PV+VUS |
| 57 (63.3) | 15 (88.2) |
|
Multigene panel testing results
Identified pathogenic variants (PVs) were in 10/33
genes in 34 patients; 38.2% in BRCA1/2 genes; 6, 24 and, 14.3% in
other high, moderate, and low-risk genes, respectively (Table II). Specifically, the positive
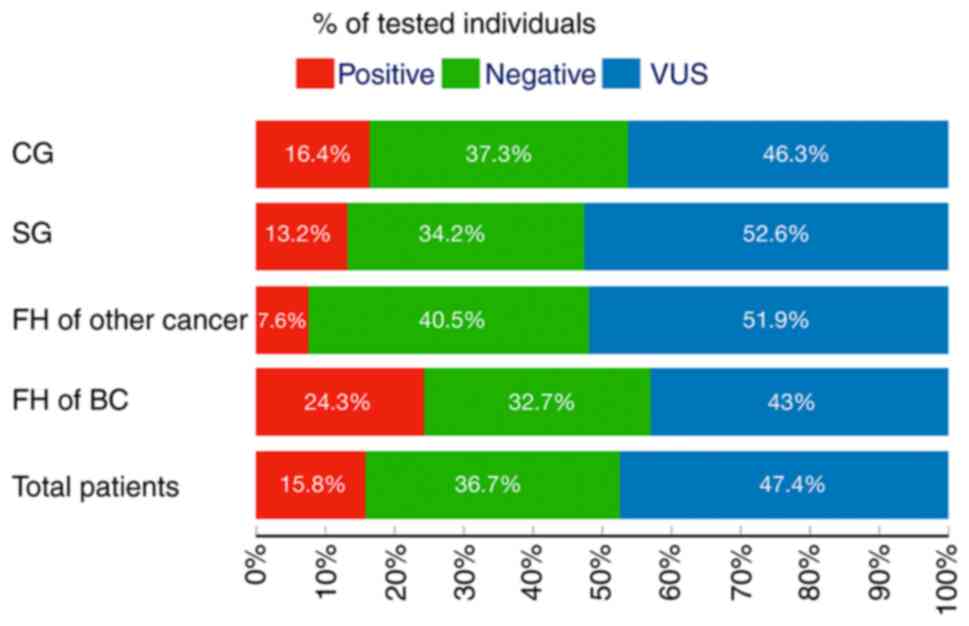
rate was 13.2 and 16.4% in the SG and the CG, respectively (P=0.80;
Table I and Fig. 2).
 | Table II.Multigene panel testing results among
the categories of tested individuals. |
Table II.
Multigene panel testing results among
the categories of tested individuals.
|
|
|
| Positive in gene
categories |
|
|---|
|
|
|
|
|
|
|---|
| Individuals | N | Positive, % | BRCA1 and BRCA2,
% | Other
high-riska, % | Moderate
riskb, % | Low/unknown V
riskc, % | US, % |
|---|
| Total
individuals | 215 | 15.8 | 6.0 | 0.9 | 3.7 | 2.2 | 47.4 |
| SG | 38 | 13.2 | 2.6 | 0.0 | 5.2 | 5.2 | 52.6 |
| CG | 177 | 16.4 | 6.7 | 1.1 | 3.3 | 5.0 | 46.3 |
| FH of BC | 107 | 24.3 | 10.2 | 0.9 | 4.6 | 8.4 | 21.4 |
| SG | 17 | 29.4 | 5.8 | 0.0 | 11.7 | 11.7 | 58.8 |
| CG | 90 | 23.3 | 12.2 | 1.1 | 3.3 | 7.7 | 40.0 |
| FH of any
cancer | 187 | 17.1 | 6.9 | 1.0 | 3.7 | 5.3 | 46.5 |
| SG | 34 | 14.7 | 2.9 | 0.0 | 5.8 | 5.8 | 55.9 |
| CG | 153 | 17.6 | 7.8 | 1.3 | 3.2 | 5.2 | 44.6 |
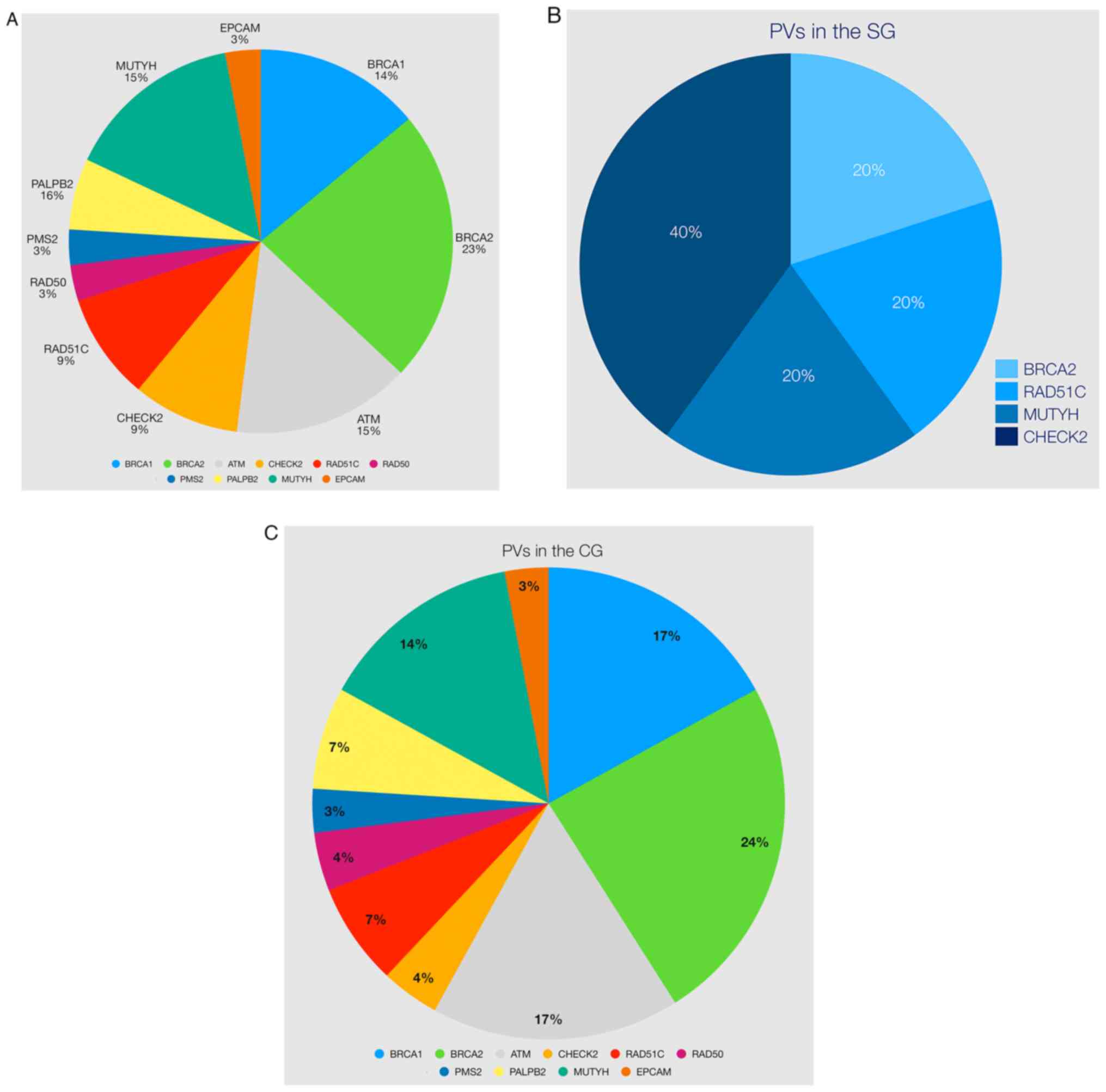
The distribution of PV in all patients was as
follows: BRCA1 (14%), ATM (15%), MUTYH (15%), BRCA2 (23%), CHECK2
(9%), RAD51C (9%), PALBB2 (6%), RAD50 (3%), PMS2 (3%) and EPCAM
(3%) (Table IV and Fig. 3A). There were five PVs in the SG
(BRCA2, CHECK2, RAD51C, and MUTYH) and 29 PVs in the CG,
respectively (Table IV and
Fig. 3B and C). Variants
classified as pathogenic or likely pathogenic were small deletions
and insertions, missense, and splice site variants with decreasing
frequency (Table IV).
 | Table IV.List of PVs/LPVs identified in this
study. |
Table IV.
List of PVs/LPVs identified in this
study.
| Variant | Clinical
significance | Personal history of
cancer | FH of cancer | CM status |
|---|
|
NM_007194(CHEK2): | LPV | Breast | Breast, colon | Yes |
| c.1427C>T, p.
(Thr476Met) |
|
| |
|
|
NM_007194(CHEK2): | LPV | Breast | Breast,
colorectal, |
|
| c.1427C>T, p.
(Thr476Met) |
| | kidney |
|
|
NM_001128425(MUTYH): | PV
(monoallelic) | Breast | Breast | Yes |
| c.1171C>T, p.
(Gln391*) |
|
|
|
|
|
NM_007294(BRCA1): | PV | Breast | Breast, lung | Yes |
| c.4035delA, p.
(Glu1346Lysfs*20) |
|
| |
|
|
NM_000059(BRCA2): | PV | Breast | Breast,
ovarian | No |
| c.9682delA, p.
(Ser3228Valfs*21) |
|
| |
|
|
NM_000059(BRCA2): | PV | Breast | Breast, prostate,
lung, gastric | No |
| c.8087T>A, p.
(Leu2696*) |
|
|
|
|
|
NM_024675(PALB2): | PV | Breast | Breast | No |
| c.3271C>T, p.
(Gln1091*) |
|
|
|
|
|
NM_000059(BRCA2): | PV | Breast | Breast | No |
| c.5557dupT, p.
(Cys1853Leufs*5) |
|
|
|
|
|
NM_007294(BRCA1): | PV | Breast | Breast, gastric,
skin, lung | No |
| c.5266dupC, p.
(Gln1756Profs*74) |
|
|
|
|
|
NM_058216(RAD51C): | LPV | Breast,
ovarian | Breast, ovarian
prostate | No |
| c.904+5G>T |
|
|
|
|
|
NM_000051(ATM):c.6527delT, p. | PV | Breast | Breast, lung,
uterine | No |
|
(Leu2176Cysfs*59) |
|
|
|
|
|
NM_007294(BRCA1): | PV | Breast | Breast, skin,
melanoma, uterine | No |
|
c.3700_3704delGTAAA, p. |
|
|
|
|
|
(Val1234Glnfs*8) |
|
|
|
|
|
NM_005732(RAD50): | PV | Breast | Breast, skin,
melanoma, uterine | No |
| c.326_329delCAGA,
p. (Thr109Asnfs*20) |
|
|
|
|
|
NM_002485(NBN): c.657_661delACAAA,
p. (Lys219Asnfs*16) | PV | Breast | lymphoma | No |
|
NM_007194(CHEK2): c.1427C>T, p.
(Thr476Met) | LPV | Breast | None | No |
|
NM_001128425(MUTYH): c.884C>T,
p. (Pro295Leu) | PV
(monoallelic) | None | Pancreatic | NA |
|
NM_000059(BRCA2):
c.4936_4939delGAAA, p.Glu1646Glnfs*23 | PV | None | Breast,
ovarian | NA |
|
NM_000179(MSH6):c.2764C>T,
p.(Arg922*) | PV | None | Breast,
intestine | NA |
|
NM_007194(CHEK2):c.793-1G>A | PV | None | Breast, ovarian,
lung | NA |
|
NM_001128425.1(MUTYH):c.1187G>A | LPV | Breast | No | No |
|
NM_002878.3(RAD51D):c.480+1G>A | PV | Breast | Breast | Yes |
|
NM_000059.3(BRCA2):c.8940delA | PV | Breast | Prostate, lung | No |
|
NM_007294.4(BRCA1):c.1621C>T | PV | Breast | Breast,
ovarian | No |
|
NM_000051.3(ATM):c.2922-2A>G | PV | Breast | Pancreas, lung | No |
| NM_000059.3(BRCA2):
c.2307dupT | PV | Breast | Colon | No |
|
NM_000059.3(BRCA2):c.5073dupA | PV | Breast | Breast,
prostate | Yes |
|
NM_001128425.1(MUTYH): | PV | Breast | Breast, colon | No |
|
c.1437_1439delGGA |
|
|
|
|
|
NM_000051.3(ATM):c.2284_2285delCT | PV | Breast | Breast | No |
|
NM_058216.3(RAD51C):c.181_182delCT | PV | Breast | No | No |
|
NM_000051.3(ATM):c.6527delT | PV | Breast | Breast | No |
|
NM_024675.4(PALB2):c.932_933insC | PV | Breast | Lung | No |
|
NM_000535.7(PMS2):c.988+1del | PV | Breast | Breast | No |
|
NM_000314.8(PTEN):c.93delC | LPV | Breast | Breast, gastric,
prostate, lung | No |
|
NM_005732.4(RAD50):c.1873delT | LPV | Breast | Breast, gastric,
prostate, lung | No |
|
NM_007294.4(BRCA1):c.5266dupC | PV | Breast | Breast, ovarian,
lung | No |
|
NM_000051.3(ATM):c.3576G>A | PV | Breast | NA | NA |
|
NM_000059.3(BRCA2):c.4376dupA | LPV | Breast | Breast, ovarian,
endometrium | No |
|
NM_002354.2(EPCAM):c.556-14A>G | PV | Breast | Breast,
gastric | No |
|
NM_000059.3(BRCA2):rsa13q13.1 | LPV | Breast | Breast | No |
| (BRCA2exon3)x1 |
|
|
|
|
|
NM_007294.4(BRCA1):c.5176A>T | PV | Breast | Breast, ovarian,
CNS | No |
|
NM_001128425.1(MUTYH):c.1187G>A | PV | Breast | Colon, lung | No |
|
NM_001128425.1(MUTYH):c.734G>A | PV | Breast | Breast | No |
The incidence of PV in FH of BC was slightly higher
in the SG (29.4 vs. 23.3%, P=0.51; Tables II and III). Variants with uncertain
significance (VUS) were identified in 47.4% of women. The VUS rates
ranged from 21.4 to 55.9% (Table
II). Among the women with a PV, 38.2% of them had BRCA1/2 gene
mutations. The high mutation frequency of the BRCA1/2 genes was
19.6% in the SG and 40.8% in the CG, respectively (P=0.48). In
addition, distributions of PVs in patients were identified in other
high- (6%,), moderate (24%) or low risk (14.3%) genes apart from
the BRCA1/2 genes, PVs were identified in CHEK2, ATM, NBN, PALB2,
RAD50, RAD51C and MUTYH (monoallelic) (Fig. 3A and Table IV).
The negative genetic test result was significantly
higher in those in the SG who did not have FH of BC (84.6 vs. 50%,
P=0.022; Table III). However,
when only patients with FH of BC were considered, the total PV and
VUS rates were significantly higher in the SG compared with the CG
(88.2 vs. 63.3%, P=0.045; Table
III).
Discussion
Consanguineous marriage (CM) between biological
relatives is a social custom with a long history in various parts
of the world (12). Today,
hundreds of millions of individuals live in consanguineous families
(12–14). The offspring of consanguineous
parents are more likely to have the same two alleles (homozygosity)
by descent. The prevalence of CM is still relatively high at 20–25%
(ranges from 11.5 to 46%) (14,15)
in Turkey. However, the relationship between CM and BC-predisposing
genes is not clear. In this prospective clinical study, the effect
of CM on cancer-related genes in patients with BC was
investigated.
The rate of pathogenic variants (PVs) identified
with multigene NGS panel testing in Turkish patients with BC was
15.8% in our study. Most of these PVs were BRCA1 and BRCA2 genes
(~39%), which is consistent with previous literature (16,19).
Furthermore, the prevalence of BRCA1/2 mutations in the CG was 6.7%
and in those with FH of BC was 14.1%, which was similar to the
literature in which the prevalence ranges from 9 to 21% (20–22).
Analysis of positive gene mutations showed that more
than half of the patients with BC (61.8%) having a PV would have
been missed if genetic testing was restricted to BRCA1/2 or
high-risk genes included in the NCCN guidelines for
Genetic/Familiar high-risk assessment for breast and ovarian cancer
and could enable personalized management decisions for these
patients (23).
It is noteworthy that most of the variants
classified as pathogenic or likely pathogenic in the current study
were deleterious (variants with small deletions, insertions, and
splice sites). This may be related to the fact that most missense
variants remain in the VUS class upon compliance with the ACMG
classification. A notable feature in the cohort of the present
study was that no large deletions and duplications (CNVs) were
observed. In cases where it is impossible to calculate CNVs by the
NGS method, additional testing may be considered to detect
pathological CNVs, especially in BRCA1 and BRCA2 genes. Thus, it is
clear that VUS monitoring will continue to maintain its
importance.
Our previous study analyzed genes involved in
hereditary cancer predisposition using an NGS approach in 1,197
individuals from Greece, Romania, and Turkey (16). A PV was identified in 264 of the
individuals (22.1%) analyzed, while a VUS was identified in 34.8%
of cases. Nevertheless, the PV rate was lower (15.8%) and the VUS
rate was higher (43.7%) in Turkish individuals. Moreover, as a PV,
the BRCA1/2 ratio was 10.1%, while other high-risk, moderate-risk,
and low/unknown risk ratios were 4.4, 0.6, and 1.9%, respectively,
for these individuals. Similar to these findings, the PV and VUS
rates were 12.8 and 47.4%, respectively, in the current study. This
study also found similarities regarding BRCA1/2 and other
high-risk, moderate-risk, and low/unknown risk ratios (Table I). There were no significant
differences between the two groups regarding PV rates (Fig. 3A-C).
The Turkish public health system covered hereditary
cancer predisposition gene tests on a routine basis, which has been
covered by for several years. However, only two recent studies with
multigene panels were performed in Turkey. The first was a
multinational study, and the pathogenic/likely pathogenic mutation
rate was found to be similar (15.8%) to our study (16). In the second study conducted by
Akcay et al (19), cancer
susceptibility genes were analyzed in breast and colorectal cancer
patients. High BC risk genes were found in 17.2% of patients and, a
low BC risk gene in 3.9%.
Due to the sample size of the present study, no LGRs
were found by any of the bioinformatics approaches, which is
essential to evaluate the full mutation spectrum of tested genes,
especially for BRCA1/2. In Turkey, Large Genomic Rearrangements
(LGRs). rates are between 1–4% (19,24,25)
comparable with different populations around the world (0.1-12.7%)
(16,26–30).
The likelihood of identifying variants of uncertain
significance (VUS) increases with multigene panel testing as high
as 40% (31). In the current
study, the VUS rate was found to be 47.4%. Most VUS
reclassifications involve a downgrade to a benign variant; a small
proportion may be reclassified as pathogenic (32). Therefore, the VUS should not be
used to guide medical management until the clinical significance of
these findings is determined (33).
Multigene panel testing is a powerful tool that
detects BRCA and non-BRCA germline mutations in individuals with a
family history (FH) of BC. When FH of BC was used as a
stratification factor among patients with BC, almost half of women
(49.7%, 107/215) reported FH of BC (44.7% in SG and 50.8% in CG).
The patients' suitability can explain this high FH of BC rate in
this study for genetic testing.
In populations where CMs are prevalent, patients are
at risk for homozygous/compound heterozygous germline mutations in
BRCA2, BRIP1 and PALB2 result in Fanconi's anemia (34–36).
In a previous study, the consanguinity rate in Southern
Mediterranean populations was inversely correlated with families
with BRCA1 germline deleterious mutations (37).
In a study by Medimegh et al (38), it was demonstrated that the
parental consanguinity was protective against BC, especially over
the age of 50. However, the results of the present study showed no
significant relationships between patients with BC with or without
CM and pathogenic mutations. Thus, although thousands of multigene
panel tests have been performed worldwide (39), there is still an important fraction
of BC cases that remain undiagnosed, underlying the predisposition
to BC (40).
In a retrospective review of the multi-institutional
tumor registry, including 2,237 patients with BC, 509 (60.7%) had
negative results, 108 (12.8%) had deleterious mutations and 221
(26.3%) had VUS (41). In the
present study, 15.8% of the patients had PVs, 47.4% had VUS and
36.7% had negative results. The PV rate did not differ between the
two groups (13.2 and 16.4% in SG and CG, respectively), and the VUS
rate was higher in the CM group (52.6 vs. 46.3% in the SG and CG,
respectively, P=0.59). However, when the sum of PV and VUS results
in patients with FH of BC in two groups were compared, it was found
that the sum of PV and VUS was significantly higher in the SG (63.3
vs. 88.2% in the CG and SG, respectively, P=0.045). This
significant result may suggest that genes of unknown importance in
VUS may be important in the future.
In conclusion, the present study demonstrated the
importance of using multigene panels in a multi-ethnic population
and contributed to the knowledge of hereditary BC in Turkey. The
pathogenic mutation rate was 24.3% in patients with FH of BC. The
PV rate in patients with FH of BC reached 29.4% in the SG.
Therefore, multigene panel testing should be considered for these
patients. The CM did not significantly increase the pathogenic
mutation rate, but the VUS rate was higher in this group. This
observation should be confirmed with further prospective clinical
trials with more extensive series.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the ClinVar respiratory,
ncbi.nlm.nih.gov/clinvar/?term=SUB10439877 and
ncbi.nlm.nih.gov/clinvar/?term=SUB10447902
Authors' contributions
GNT, EP and AS drafted the manuscript. GNT performed
bioinformatics and statistical analysis, analyzed and interpreted
the variant data and prepared all supporting materials and figures.
AOC, EP, KA, GP, SK and KY designed the study and experiments,
collected the demographic data, and coordinated the study and all
mutational analyses. GNT, EP, KA, GP, SK, GN, VO, TO, KY and AOC
participated in the writing of the manuscript, performed DNA
extraction, sequencing and Multiplex Ligation-dependent Probe
Amplification experiments, and contributed to the analysis and
interpretation of the variant data. VO, AOC, KY, CO, FA, TO, ASI,
GS, GA, GNT, EP, KA, GP, SK, GN, ES and AS provided the patient
material, diagnosis and management. VO, TO and AS conceived the
study and participated in its design and coordination. GNT and KY
confirm the authenticity of all raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
All tested individuals provided signed informed
consent form before molecular genetic testing and permission for
the anonymous use of their data for research purposes and/or
scientific publications. The study was approved by The Biruni
University Ethical Committee (approval no. 2015-KAEK-43-18-11;
date, 19/09/2018).
Patient consent for publication
Present within informed consent forms.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Colombet M, Soerjomataram I,
Parkin DM, Piñeros M, Znaor A and Bray F: Cancer statistics for the
year 2020: An overview. Int J Cancer. 149:778–789. 2021. View Article : Google Scholar
|
|
2
|
Ferlay J, Ervik M, Lam F, Colombet M, Mery
L, Piñeros M, Znaor A, Soerjomataram I and Bray F: (2018). Global
cancer observatory: Cancer today. Lyon, France: International
Agency for Research on Cancer. Available from. https://gco.iarc.fr/today28–May. 2019
|
|
3
|
Newman B, Austin MA, Lee M and King MC:
Inheritance of human breast cancer: evidence for autosomal dominant
transmission in high-risk families. Proc Natl Acad Sci USA.
85:3044–3048. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Claus EB, Risch N and Thompson WD: Genetic
analysis of breast cancer in the cancer and steroid hormone study.
Am J Hum Genet. 48:232–242. 1991.PubMed/NCBI
|
|
5
|
Walsh T and King MC: Ten genes for
inherited breast cancer. Cancer Cell. 11:103–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Domchek SM, Friebel TM, Singer CF, Evans
DG, Lynch HT, Isaacs C, Garber JE, Neuhausen SL, Matloff E, Eeles
R, et al: Association of risk-reducing surgery in BRCA1 or BRCA2
mutation carriers with cancer risk and mortality. JAMA.
304:967–975. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
The National Comprehensive Cancer Network,
. https://www.nccn.org30–October. 2020
|
|
8
|
Valencia OM, Samuel SE, Viscusi RK, Riall
TS, Neumayer LA and Aziz H: The role of genetic testing in patients
with breast cancer: A review. JAMA Surg. 152:589–94. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Beitsch PD, Whitworth PW, Hughes K, Patel
R, Rosen B, Compagnoni G, Baron P, Simmons R, Smith LA, Grady I, et
al: Underdiagnosis of hereditary breast cancer: Are genetic testing
guidelines a tool or an obstacle? J Clin Oncol. 37:453–460. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Asphaug L and Melberg HO: The
cost-effectiveness of multigene panel testing for hereditary breast
and ovarian cancer in Norway. MDM Policy Pract. Feb 1–2019.(Epub
ahead of print). doi: 10.1177/2381468318821103. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stanislaw C, Xue Y and Wilcox WR: Genetic
evaluation and testing for hereditary forms of cancer in the era of
next-generation sequencing. Cancer Biol Med. 13:55–67. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Romeo G and Bittles AH: Consanguinity in
the contemporary world. Hum Hered. 77:6–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hamamy H: Consanguineous marriages:
Preconception consultation in primary health care settings. J
Community Genet. 3:185–192. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Alper OM, Erengin H, Manguoğlu AE, Bilgen
T, Cetin Z, Dedeoğlu N and Lüleci G: Consanguineous marriages in
the province of Antalya, Turkey. Annales de Genetique. 47:129–138.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Erdem Y and Tekşen F: Genetic screening
services provided in Turkey. J Genet Couns. 22:858–864. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tsaousis GN, Papadopoulou E, Apessos A,
Agiannitopoulos K, Pepe G, Kampouri S, Diamantopoulos N, Floros T,
Iosifidou R, Katopodi O, et al: Analysis of hereditary cancer
syndromes by using a panel of genes: Novel and multiple pathogenic
mutations. BMC Cancer. 19:5352019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Richards S, Aziz N, Bale S, Bick D, Das S,
Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al:
Standards and guidelines for the interpretation of sequence
variants: A joint consensus recommendation of the american college
of medical genetics and genomics and the association for molecular
pathology. Genet Med. 17:405–424. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Povysil G, Tzika A, Vogt J, Haunschmid V,
Messiaen L, Zschocke J, Klambauer G, Hochreiter S and Wimmer K:
panelcn.MOPS: Copy-number detection in targeted NGS panel data for
clinical diagnostics. Hum Mutat. 38:889–897. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Akcay IM, Celik E, Agaoglu NB, Alkurt G,
Kizilboga Akgun T, Yildiz J, Enc F, Kir G, Canbek S, Kilic A, et
al: Germline pathogenic variant spectrum in 25 cancer
susceptibility genes in Turkish breast and colorectal cancer
patients and elderly controls. Int J Cancer. 148:285–295. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lang GT, Shi JX, Hu X, Zhang CH, Shan L,
Song CG, Zhuang ZG, Cao AY, Ling H, Yu KD, et al: The spectrum of
BRCA mutations and characteristics of BRCA-associated breast
cancers in China: Screening of 2,991 patients and 1,043 controls by
next-generation sequencing. Int J Cancer. 141:129–142. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Walsh T, Casadei S, Coats KH, Swisher E,
Stray SM, Higgins J, Roach KC, Mandell J, Lee MK, Ciernikova S, et
al: Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in
families at high risk of breast cancer. JAMA. 295:1379–1388. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han SA, Kim SW, Kang E, Park SK, Ahn SH,
Lee MH, Nam SJ, Han W, Bae YT, Kim HA, et al: The prevalence of
BRCA mutations among familial breast cancer patients in Korea:
Results of the Korean hereditary breast cancer study. Fam Cancer.
12:75–81. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
The National Comprehensive Cancer Network.
Genetic/Familiar High-Risk Assessment, Breast and Ovarian (Version
3.2019), . https://www.nccn.org/professionals/physician_gls/pdf/genetics_screening.pdf30–October2020.
|
|
24
|
Yazıcı H, Kılıç S, Akdeniz D, Şükrüoğlu Ö,
Tuncer ŞB, Avşar M, Kuru G, Çelik B, Küçücük S and Saip P:
Frequency of rearrangements versus small indels mutations in BRCA1
and BRCA2 Genes in Turkish patients with high risk breast and
ovarian cancer. Eur J Breast Health. 14:93–99. 2018.PubMed/NCBI
|
|
25
|
Aktas D, Gultekin M, Kabacam S,
Alikasifoglu M, Turan AT, Tulunay G, Kose MF, Ortac F, Yüce K,
Tunçbilek E and Ayhan A: Identification of point mutations and
large rearrangements in the BRCA1 gene in 667 Turkish unselected
ovarian cancer patients. Gynecol Oncol. 119:131–135. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Judkins T, Rosenthal E, Arnell C, Burbidge
LA, Geary W, Barrus T, Schoenberger J, Trost J, Wenstrup RJ and Roa
BB: Clinical significance of large rearrangements in BRCA1 and
BRCA2. Cancer. 118:5210–5216. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Arnold AG, Otegbeye E, Fleischut MH,
Glogowski EA, Siegel B, Boyar SR, Salo-Mullen E, Amoroso K, Sheehan
M, Berliner JL, et al: Assessment of individuals with BRCA1 and
BRCA2 large rearrangements in high-risk breast and ovarian cancer
families. Breast Cancer Res Treat. 145:625–634. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Casilli F, Tournier I, Sinilnikova OM,
Coulet F, Soubrier F, Houdayer C, Hardouin A, Berthet P, Sobol H,
Bourdon V, et al: The contribution of germline rearrangements to
the spectrum of BRCA2 mutations. J Med Genet. 43:e492006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gonzalez-Hormazabal P, Gutierrez-Enriquez
S, Gaete D, Reyes JM, Peralta O, Waugh E, Gomez F, Margarit S,
Bravo T, Blanco R, et al: Spectrum of BRCA1/2 point mutations and
genomic rearrangements in high-risk breast/ovarian cancer Chilean
families. Breast Cancer Res Treat. 126:705–716. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pylkäs K, Erkko H, Nikkilä J, Sólyom S and
Winqvist R: Analysis of large deletions in BRCA1, BRCA2 and PALB2
genes in Finnish breast and ovarian cancer families. BMC Cancer.
8:1462008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yurgelun MB, Allen B, Kaldate RR, Bowles
KR, Judkins T, Kaushik P, Roa BB, Wenstrup RJ, Hartman AR and
Syngal S: Identification of a variety of mutations in cancer
predisposition genes in patients with suspected lynch syndrome.
Gastroenterology. 149:604–613.e20. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Macklin S, Durand N, Atwal P and Hines S:
Observed frequency and challenges of variant reclassification in a
hereditary cancer clinic. Genet Med. 20:346–350. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Moghadasi S, Eccles DM, Devilee P,
Vreeswijk MP and van Asperen CJ: Classification and clinical
management of variants of uncertain significance in high penetrance
cancer predisposition genes. Hum Mutat. 37:331–336. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Howlett NG, Taniguchi T, Olson S, Cox B,
Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals
G, et al: Biallelic inactivation of BRCA2 in Fanconi anemia.
Science. 297:606–609. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Levitus M, Waisfisz Q, Godthelp BC, de
Vries Y, Hussain S, Wiegant WW, Elghalbzouri-Maghrani E,
Steltenpool J, Rooimans MA, Pals G, et al: The DNA helicase BRIP1
is defective in Fanconi anemia complementation group J. Nat Genet.
37:934–935. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Reid S, Schindler D, Hanenberg H, Barker
K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, et al:
Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and
predispose to childhood cancer. Nat Genet. 39:162–164. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Belaiba F, Medimegh I, Bidet Y, Boussetta
S, Baroudi O, Mezlini A, Bignon YJ and Benammar El gaaied A:
BRCA1/BRCA2 Mutations Shaped by ancient consanguinity practice in
southern mediterranean populations. Asian Pac J Cancer Prev.
19:2963–2972. 2018.PubMed/NCBI
|
|
38
|
Medimegh I, Troudi W, Omrane I, Ayari H,
Uhrhummer N, Majoul H, Benayed F, Mezlini A, Bignon YJ, Sibille C
and Elgaaied AB: Consanguinity protecting effect against breast
cancer among tunisian women: Analysis of BRCA1 haplotypes. Asian
Pac J Cancer Prev. 16:4051–4055. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
LaDuca H, Polley EC, Yussuf A, Hoang L,
Gutierrez S, Hart SN, Yadav S, Hu C, Na J, Goldgar DE, et al: A
clinical guide to hereditary cancer panel testing: Evaluation of
gene-specific cancer associations and sensitivity of genetic
testing criteria in a cohort of 165,000 high-risk patients. Genet
Med. 22:407–415. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Couch FJ, Nathanson KL and Offit K: Two
decades after BRCA: Setting paradigms in personalized cancer care
and prevention. Science. 343:1466–1470. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bagwell AK, Sutton TL, Gardiner S and
Johnson N: Outcomes of large panel genetic evaluation of breast
cancer patients in a community-based cancer institute. Am J Surg.
221:1159–1163. 2021. View Article : Google Scholar : PubMed/NCBI
|