Introduction
Despite the recent progress in the development of
targeted therapies, lung cancer remains to be the most prevalent
malignancy and the leading cause of cancer-associated mortality
worldwide (1). Non-small cell lung
cancer (NSCLC) accounts for 80–85% of all types of lung cancer,
where lung adenocarcinoma (LAC) is the most prevalent pathological
subtype and accounts for ~40% of all lung cancer-associated
mortality (2). Although
comprehensive therapeutic strategies, including surgery and
targeted therapies, have improved clinical outcome over the past
number of decades, the 5-year survival rate of LAC remains at only
18% (3). The reasons for this poor
prognostic rate from advanced LAC include low diagnosis rates and
the absence of effective therapeutic targets. Therefore, it would
be of great value to identify novel specific and sensitive
biomarkers and therapeutic targets for patients with LAC.
Circular RNAs (circRNAs) are a novel class of
endogenously-spliced RNAs that have been previously found to
regulate cancer progression (4,5). Due
to their covalently closed loop without a 5′-3′poly-adenylation
end, circRNAs possess the ability to resist the activities of
exonucleases and remain highly stable, suggesting that they have
potential to serve as cancer biomarkers and therapeutic targets
(6–10). Accumulating evidence has
demonstrated that circRNAs are involved in the development of LAC
(11–13). One key molecular function of
circRNAs is that they can function as competing endogenous RNAs
(ceRNAs) to sponge microRNAs (miRNAs), leading to the enhancement
of the expression of genes normally suppressed by miRNAs. The
circRNA circ-cysteine-rich transmembrane bone morphogenetic protein
regulator 1 inhibits the invasion and metastasis of LAC cells by
serving as the ceRNA of miR-182/miR-93 (14). In addition, another circRNA,
has_circ_0001946, promotes LAC cell proliferation by sponging
miR-135a-5p (15). In another
study, circ-zinc finger protein 609 promotes the proliferation of
LAC by targeting miR-1224-3p/ETS translocation variant 1 signaling
(16). Taken together, these
previous findings suggest that circRNAs serve crucial roles in
regulating the physiology of LAC and have the potential to serve as
biomarkers and therapeutic targets of LAC.
In the present study, by analyzing the GEO database,
three circRNAs were identified to be highly expressed in LAC
tissues compared with the paired-matched adjacent non-cancerous
tissues. A retrospective clinical study revealed that higher
expression levels of these three circRNAs indicated poorer
prognoses in patients with LAC. To unravel the potential biological
mechanism of these circRNAs, their target miRNAs and possible mRNAs
affected downstream were screened, following which a
circRNA-miRNA-mRNA regulatory network was constructed. A
protein-protein interaction (PPI) network was subsequently
established and six hub genes were eventually identified. A
retrospective study using the Gene Expression Profiling Interactive
Analysis (GEPIA) database showed that higher expression levels of
each hub gene were associated with poorer overall survival in
patients with LAC. Taken together, through bioinformatic analysis
and retrospective clinical study, these findings revealed a number
of novel circRNAs that could be important for LAC pathophysiology.
In addition, they provided insights into the molecular mechanism
that regulate the progression of LAC with respect to the
circRNA-miRNA-mRNA network.
Materials and methods
Datasets
The circRNA expression profile of GSE101586 was
obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/), which included
five pairs of matched LAC tissues (GSM2706428, GSM2706429,
GSM2706430, GSM2706431 and GSM2706432) and adjacent non-cancerous
tissues (GSM2706423, GSM2706424, GSM2706425, GSM2706426 and
GSM2706427). The tissues were obtained from five female patients
with LAC with no smoking history, and the physiology of the tissues
were confirmed by pathologists. The mRNA data of the LAC tissues
was downloaded from The Cancer Genome Atlas (https://portal.gdc.cancer.gov).
Clinical samples
In total, 60 cases of LAC tissues in the tissue bank
were obtained from the patients who underwent surgery in the Fourth
hospital of Hebei Medical University (Shijiazhuang, China) between
January 2016 and June 2016. A total of 20 pairs of LAC and adjacent
normal tissue (5-cm from the tumor tissue) were collected from the
above 60 cases. The clinicopathological characteristics and
survival status of the patients with LAC were obtained from the
follow-up data. (The duration of follow-up was between January 2016
and June 2021). The inclusion criteria were: i) All patients were
diagnosed LAC through pathology and ii) All patients did not
receive any therapy before surgery. The clinicopathological data
for the patients are presented in Table SI. The human tissues were obtained
with written informed consent, and the present study was approved
by The Clinical Research Ethics Committee of The Fourth hospital of
Hebei Medical University (approval no. 2021KY157).
Differentially-expressed circRNAs and
mRNAs
The limma package in R (v3.4.1; http://bioconductor.org/packages/release/bioc/html/limma.html)
was used to identify the differentially expressed circRNAs between
the LAC tissues and the adjacent non-cancerous tissues from the
GEO101586 dataset using the criteria of P<0.05 and
|Log2 fold change (FC)|>1. The ‘edger’ package
(v3.52; http://bioconductor.org/packages/edgeR/) was used to
screen for the differentially-expressed mRNAs using thresholds of
|Log2FC|>3 and P<0.01. The miRNAs-targeted mRNAs
were predicted based on the miRWalk software (v2.0; http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/).
Cell culture
LAC cell lines H1299 and H1975 were obtained from
Chinese Academy of Sciences Cell Bank were cultured in RPMI-1640
media (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin
and 100 µg/ml streptomycin. Cells were cultured at 37°C under 5%
CO2.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA from 80% confluent cells and tissues were
isolated by TRIzol® Reagent (Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocols. The cDNA was
prepared from total RNA according to the GoScript Reverse
Transcription System (Promega Corporation) protocol. qPCR analysis
was performed using the Promega GoTaq qPCR Master Mix (Promega
Corporation). GAPDH was used as the mRNA internal reference. The
primer sequences for all qPCR reactions are shown in Table SII. The PCR cycling conditions was
as follows: Denaturation at 95°C for 15 sec, annealing at 58°C for
30 sec and extension at 72°C for 30 sec. Three repeat wells were
set. The relative expression levels were calculated using the
2−ΔΔCq method (17).
The sequencing of PCR products were obtained from Sangon Biotech
Co., Ltd. RNase R treatment was carried out for 15 min at 37°C
using RNase R 3 U/mg.
siRNA transfection
siRNA for negative control and siRNAs targeting the
junction sites of circ-0006220, circ-0072088, and circ-0001666 were
designed and synthesized by Guangzhou RiboBio Co., Ltd. The
sequences of these siRNAs are listed in Table SIII. H1299 cells were transfected
with above siRNAs when reached 70–80% confluence by using HiPerFect
Transfection Reagent (Qiagen GmbH) based on the manufacturer's
instruction. The final siRNA concentrations were 50 nmol/l.
Following transfection, cells were cultured at 37°C for 72 h.
Cell Counting Kit-8 (CCK-8)
(MCE®) assay
H1299 cells were first seeded at a cell density of
5×103 cells/well into 96-well plates. After attachment
overnight, CCK-8 assays were performed after 24, 48 and 72 h. In
brief, 10 µl CCK-8 solution (cat. no. HY-K030; MedChemExpress) was
added into each well. After 1 h incubation at 37°C, the absorbance
readings for each well were performed at 450 nm using the
microplate reader (Tecan Group, Ltd.).
Transwell migration and Matrigel
invasion assays
The migration and Matrigel invasion assays were
performed in 6.5-mm Transwell chambers for migration assays or
Matrigel pre-coated (at 37°C for 24 h) chambers for invasion assays
based on the manufacturer's protocol (BD Biosciences). The cell
suspensions of different groups were added to the upper chambers at
a density of 5×104 cells/well and incubated for 24 h.
The migratory and invasive cell numbers were then quantified in
five random fields per chamber under the inverted microscope at
×100 magnification.
Prediction of circRNA-miRNA binding
sites
Prediction of interactions between circRNAs and
miRNAs was performed using the CircInteractome database (https://circinteractome.irp.nia.nih.gov/). miRNAs with
a context score percentile ≥98 were eventually selected.
Construction of the circRNA-miRNA-mRNA
regulatory network
The ceRNA regulatory network was established
according the possible interactions among the four
differentially-expressed circRNAs, eight miRNAs predicted to be
targeted by these circRNAs and the 232 overlapped mRNAs from the
list of predicted target genes and the upregulated genes in LAC.
Cytoscape 3.7.1 software (v.3.7.1; http://cytoscape.org/) was used to visualize the
established ceRNA regulatory network.
Establishment of the PPI regulatory
network and identification of hub genes
A PPI network was constructed using the Search Tool
for the Retrieval of Interacting Genes/Proteins (STRING) software
(version 11.5; http://string-db.org/) and visualized
using the Cytoscape 3.7.1 software (https://cytoscape.org/). Additionally, suspected hub
genes with high predicted degrees of interactions from the PPI
network were screened using the CytoHubba plugin in Cytoscape.
Verification of hub gene expression
and survival using the GEPIA database
The mRNA expression levels of the hub genes and
their association with survival were assessed using the web-based
GEPIA database (http://gepia.cancer-pku.cn/) with the settings of
P≤0.05 and |Log2FC|≥1.
Immunohistochemistry
The 5-µm, paraffin-embedded slides were
deparaffinized in xylene, rehydrated using a decreasing alcohol
gradient and washed with 1X PBS (pH 7.2) three times at 5 min each.
The sections were then heated in a microwave oven for 5 min in 10
mmol/l Na-citrate buffer (pH 6.0) for antigen retrieval and washed
with 1X PBS. The sections were immersed in 0.3% hydrogen peroxide
in methanol for 20 min to suppress endogenous peroxidase activity.
After further washing with 1X PBS, the sections were incubated in
10% normal goat serum (cat. no. SP-9001 or SP-9002; OriGene
Technologies, Inc.) at room temperature in a humidified chamber for
30 min to prevent nonspecific immunoglobulin binding. The sections
were then treated with the 1:100-diluted KIF2C antibody (cat. no.
TA503320; OriGene Technologies, Inc.), KIF18B (cat. no. ab121798;
Abcam), CKAP2L (cat. no. ab122617; Abcam), PLK1 antibody (cat. no.
TA500383; OriGene Technologies, Inc.), MELK antibody (cat. no.
ab129373; Abcam), BIRC5 antibody (cat. no. TA301427; OriGene
Technologies, Inc.) at 4°C overnight. Normal IgG in place of the
primary antibody served as the negative control. A
streptavidin-biotinylated HRP-based detection system was used to
reveal specific binding. The sections were counterstained for 2 min
at room temperature with hematoxylin for light microscopic review
and evaluation. The expression was ranked on the sum of intensity
and area from 0 to 7: 0–2, negative expression; 3–7, positive
staining (of those, 3–4, weak positive expression; and 5–7, strong
positive expression). Staining intensity was graded as follows: 0
for no staining; 1 for mild staining; 2 for moderate staining; and
3 for intense staining. The staining area was scored as follows: 0
for no staining; 1 for 1–25% area; 2 for 26–50% area; 3 for 51–75%
area; and 4 for 76–100% area.
Statistical analysis
Statistical analysis was performed using SPSS 22.0
software (IBM Corp). The data were presented as the mean ± standard
deviation and measured using the Student's t-test (paired t-tests
were used in Fig. 1C, and unpaired
t-tests were used in Fig. 2C-F).
ANOVA analysis followed by Dunnett's post hoc test was used for
2G-I. Chi-square test was used to analyze the association of
circRNA expression with the hub protein expression (Fig. 7C). Kaplan-Meier analysis was used
to evaluate the overall survival and log-rank test was performed to
estimate the differences among the various groups. P<0.05 was
considered to indicate a statistically significant difference.
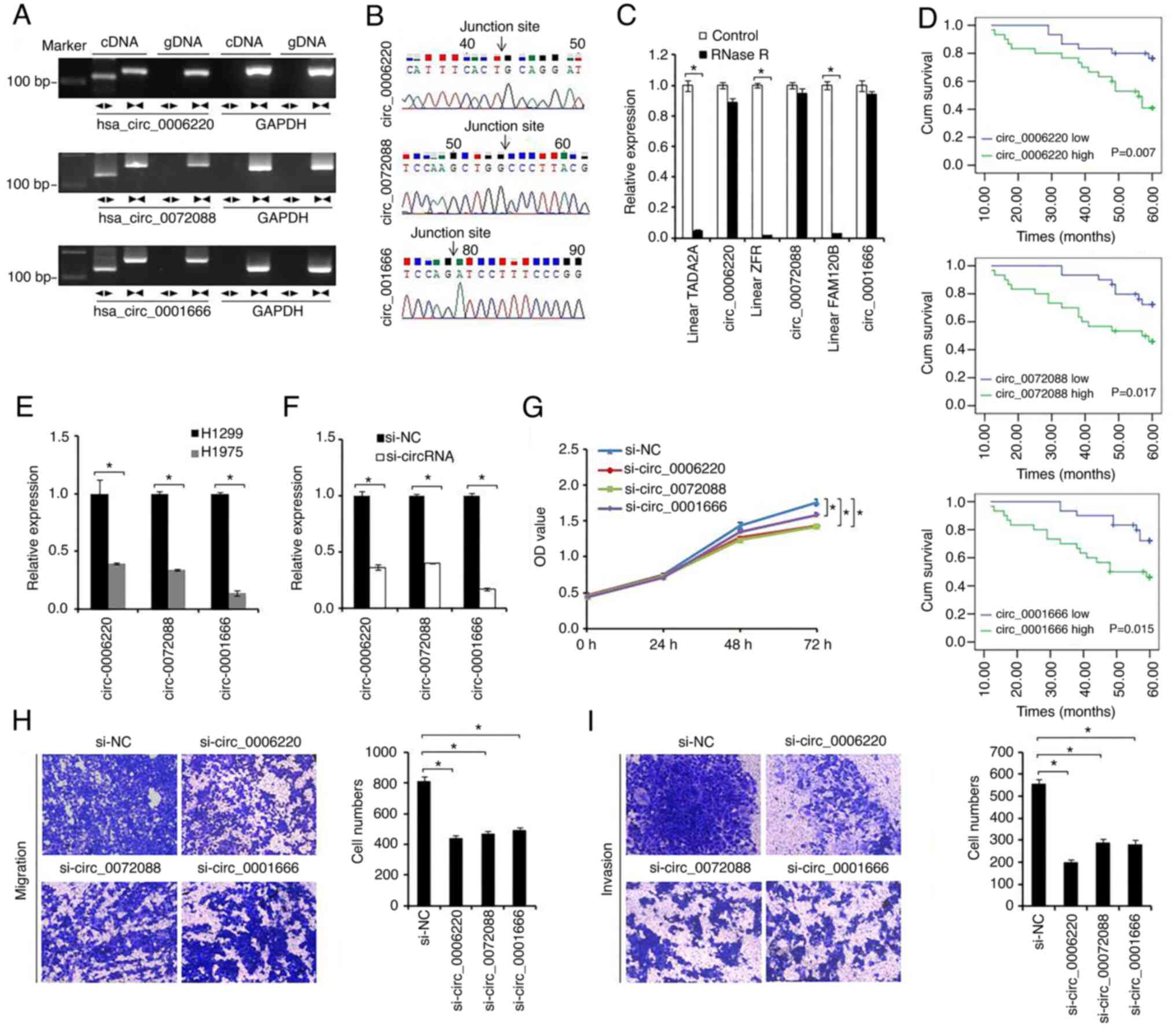 | Figure 2.Hsa_circ_0006220, hsa_circ_0072088
and hsa_circ_0001666 may serve oncogenic roles in LAC progression.
(A) The expression of hsa_circ_0006220, hsa_circ_0072088 and
hsa_circ_0001666 was validated in LAC tissues by RT-qPCR. Divergent
primers can amplify circRNAs in the cDNA but not gDNA. Convergent
primers can amplify linear RNAs in both cDNA and gDNA samples.
GAPDH was used as the negative control. (B) The sequence of the PCR
product revealed the junction sites of the circRNAs. (C) All three
of the circRNAs were resistant to RNase R treatment. (D) The
association between the expression these three circRNAs with the
survival of 60 patients with LAC. (E) The expression of the three
circRNAs in the LAC cell lines was measured using RT-qPCR. (F) The
knockdown efficiency of the circRNAs siRNAs in H1299 cells. (G)
Cell viability of H1299 cells was evaluated using MTT assay.
*P<0.05. H1299 cell (H) migration (magnification, ×100) and (I)
invasion (magnification, ×100) was evaluated by Transwell migration
and Matrigel invasion assays. *P<0.01. circRNA, circular RNA;
LAC, lung adenocarcinoma; RT-qPCR, reverse
transcription-quantitative PCR; cDNA, complementary DNA; gDNA,
genomic DNA; si, small-interfering. |
Results
Identification of differentially
expressed circRNAs
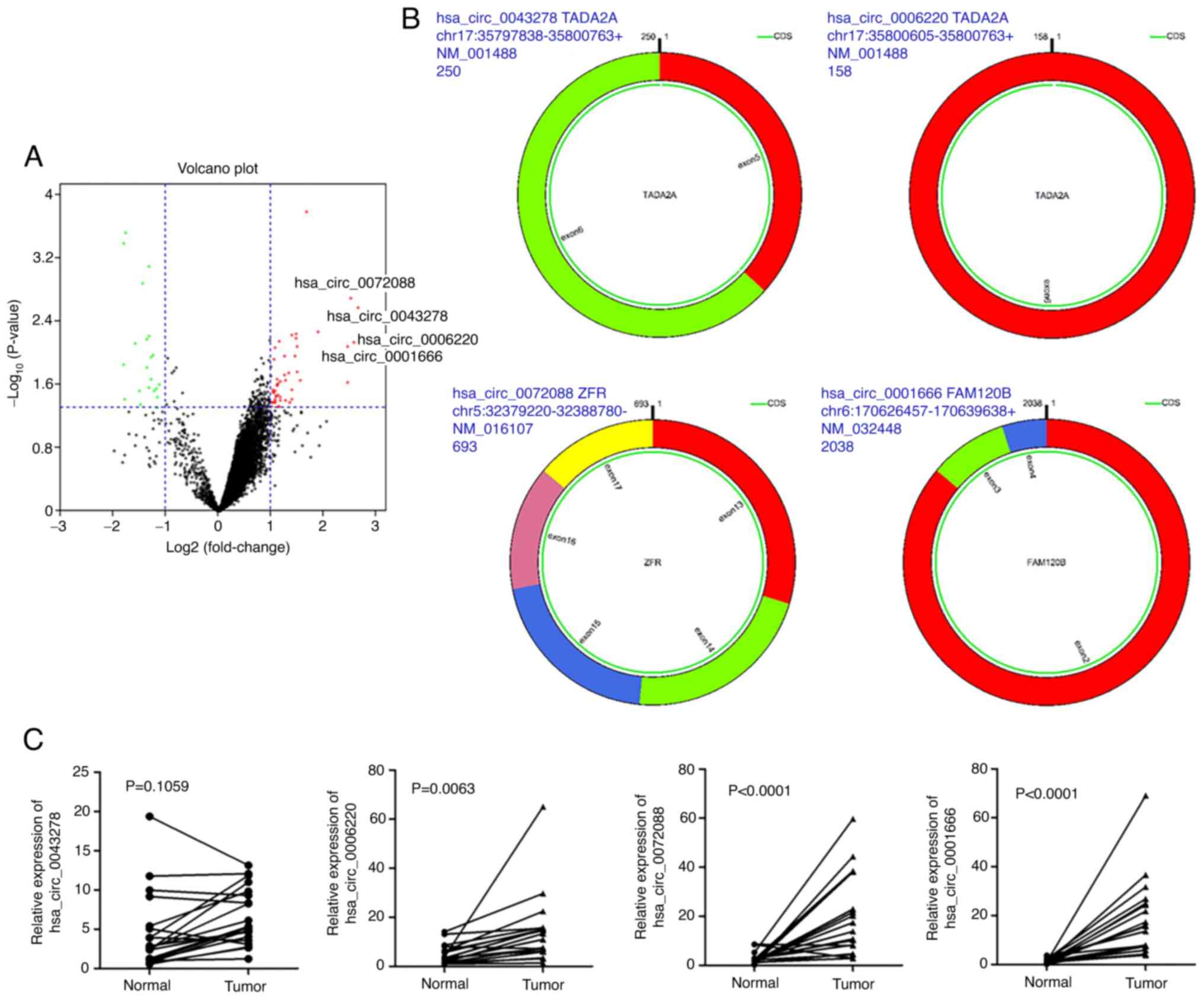
The GSE101586 dataset from GEO was analyzed using
the limma package in R (P<0.05 and |Log2FC|>1). In
total, 47 upregulated and 21 downregulated circRNAs were identified
in LAC tissues compared with adjacent non-cancerous tissues
(Table I). After tightening the
parameters to P<0.01 and |Log2FC|>2, four
circRNAs, hsa_circ_0043278, hsa_circ_0006220, hsa_circ_0072088 and
hsa_circ_0001666, were identified to be significantly upregulated
in LAC tissues compared with adjacent non-cancerous tissues
(Fig. 1A). The basic structural
profiles of these four circRNAs are shown in Fig. 1B. All four of these circRNAs are
derived from exons of their parent genes. Among them, three
circRNAs (hsa_circ_0006220, hsa_circ_0072088, hsa_circ_0001666)
were confirmed to be upregulated in LAC tissues compared with those
in the corresponding normal tissues by RT-qPCR (Fig. 1C).
 | Table I.The differential expressed circRNAs
between LAC tissues and the corresponding normal lung tissues. |
Table I.
The differential expressed circRNAs
between LAC tissues and the corresponding normal lung tissues.
| CircRNA ID | P-value | Log2
(fold-change) |
|---|
|
hsa_circ_0043278 | 0.002602 | 2.654576 |
|
hsa_circ_0006220 | 0.007278 | 2.567267 |
|
hsa_circ_0072088 | 0.001979 | 2.513804 |
|
hsa_circ_0000977 | 0.023568 | 2.450922 |
|
hsa_circ_0001666 | 0.008187 | 2.450695 |
|
hsa_circ_0022383 | 0.005301 | 1.898136 |
|
hsa_circ_0046263 | 0.00016 | 1.669339 |
|
hsa_circ_0000514 | 0.021638 | 1.553487 |
|
hsa_circ_0022392 | 0.016906 | 1.491607 |
|
hsa_circ_0005397 | 0.008214 | 1.480409 |
|
hsa_circ_0069086 | 0.006373 | 1.466104 |
|
hsa_circ_0036287 | 0.005599 | 1.460223 |
|
hsa_circ_0082564 | 0.0107 | 1.442822 |
|
hsa_circ_0027089 | 0.028481 | 1.394235 |
|
hsa_circ_0065214 | 0.005769 | 1.393836 |
|
hsa_circ_0018909 | 0.03837 | 1.389946 |
|
hsa_circ_0001998 | 0.041625 | 1.340807 |
|
hsa_circ_0004104 | 0.017903 | 1.324693 |
|
hsa_circ_0000519 | 0.02214 | 1.267825 |
|
hsa_circ_0003838 | 0.039573 | 1.267251 |
|
hsa_circ_0011385 | 0.01096 | 1.251054 |
|
hsa_circ_0002360 | 0.018803 | 1.249496 |
|
hsa_circ_0008583 | 0.035512 | 1.17612 |
|
hsa_circ_0055033 | 0.039073 | 1.169861 |
|
hsa_circ_0091710 | 0.022575 | 1.161549 |
|
hsa_circ_0067934 | 0.007101 | 1.143916 |
|
hsa_circ_0008539 | 0.047705 | 1.143884 |
|
hsa_circ_0084443 | 0.047543 | 1.136258 |
|
hsa_circ_0003028 | 0.03367 | 1.133201 |
|
hsa_circ_0017639 | 0.023814 | 1.123251 |
|
hsa_circ_0084429 | 0.021075 | 1.114951 |
|
hsa_circ_0072430 | 0.025704 | 1.103268 |
|
hsa_circ_0003958 | 0.024807 | 1.100155 |
|
hsa_circ_0001238 | 0.034705 | 1.099251 |
|
hsa_circ_0003528 | 0.034148 | 1.09902 |
|
hsa_circ_0040809 | 0.040398 | 1.077003 |
|
hsa_circ_0067971 | 0.03012 | 1.075765 |
|
hsa_circ_0007345 | 0.041766 | 1.075431 |
|
hsa_circ_0008274 | 0.038372 | 1.063426 |
|
hsa_circ_0025201 | 0.008555 | 1.062317 |
|
hsa_circ_0017109 | 0.011975 | 1.058244 |
|
hsa_circ_0069152 | 0.031401 | 1.051094 |
|
hsa_circ_0006948 | 0.028476 | 1.037828 |
|
hsa_circ_0000690 | 0.030795 | 1.033918 |
|
hsa_circ_0083054 | 0.041198 | 1.023474 |
|
hsa_circ_0005962 | 0.046675 | 1.002101 |
|
hsa_circ_0062389 | 0.04119 | 1.00164 |
|
hsa_circ_0005394 | 0.025027 | −1.12294 |
|
hsa_circ_0005139 | 0.035675 | −1.16601 |
|
hsa_circ_0092367 | 0.048694 | −1.17796 |
|
hsa_circ_0076092 | 0.028292 | −1.18494 |
|
hsa_circ_0000662 | 0.03011 | −1.23507 |
|
hsa_circ_0061749 | 0.01061 | −1.24893 |
|
hsa_circ_0001644 | 0.011159 | −1.29063 |
|
hsa_circ_0002404 | 0.021288 | −1.29969 |
|
hsa_circ_0001936 | 0.006073 | −1.32711 |
|
hsa_circ_0000979 | 0.027511 | −1.32785 |
|
hsa_circ_0029426 | 0.000788 | −1.32894 |
|
hsa_circ_0000253 | 0.015088 | −1.3675 |
|
hsa_circ_0030569 | 0.006596 | −1.37248 |
|
hsa_circ_0003162 | 0.001318 | −1.44889 |
|
hsa_circ_0031027 | 0.044118 | −1.49911 |
|
hsa_circ_0043256 | 0.029359 | −1.51818 |
|
hsa_circ_0019390 | 0.007575 | −1.58187 |
|
hsa_circ_0007518 | 0.000301 | −1.76043 |
|
hsa_circ_0049271 | 0.037986 | −1.78724 |
|
hsa_circ_0076798 | 0.000399 | −1.81001 |
|
hsa_circ_0015278 | 0.013596 | −1.81839 |
Survival analysis and biological
functions of the four differentially-expressed circRNAs
To further explore the characteristics of three
differentially-expressed circRNAs, two sets of primers for each
circRNA were first designed. Divergent primers were expected to
amplify the circular forms of the RNAs, whereas convergent primers
were expected to amplify the linear forms of RNAs. Using cDNA and
genomic DNA (gDNA) from LAC tissues as templates, all three of the
differentially-expressed circRNAs were amplified by the divergent
primers in cDNA, but no amplification products were observed in the
gDNA samples (Fig. 2A). All three
linear forms of the circRNAs were amplified in both cDNA and gDNA
samples. The sequences of the PCR products from all three circRNAs
were verified by sequencing (Fig.
2B). By using RT-qPCR, it was verified further that all three
of these circRNAs were resistant to RNase R, whilst the levels of
their corresponding linear forms were significantly reduced after
RNase R treatment (Fig. 2C).
Subsequently, the expression of these three circRNAs
were observed in the 60 LAC tissue samples. In addition, higher
expression levels of hsa_circ_0006220, hsa_circ_0072088 and
hsa_circ_0001666 indicated poor prognosis in patients with LAC
(Fig. 2D). To further explore the
biological functions of these differentially-expressed circRNAs,
their expression was then measured in the LAC cell lines (Fig. 2E). siRNA-mediated knockdown of
these circRNAs was found to suppress proliferation, migration and
invasion in LAC cells (Fig. 2F-I).
Taken together, these results suggest that hsa_circ_0006220,
hsa_circ_0072088 and hsa_circ_0001666 can serve oncogenic roles in
LAC progression and have potential as biomarkers and therapeutic
targets for LAC.
Identification of miRNAs that can
interact with the circRNAs
Accumulating evidence has demonstrated that several
circRNAs derived from exons can exert biological functions by
acting as ‘sponges’ or ‘decoys’ to sequester miRNAs. To assess if
these three aforementioned circRNAs performed similar functions in
LAC, miRNAs that can potentially interact with these three circRNAs
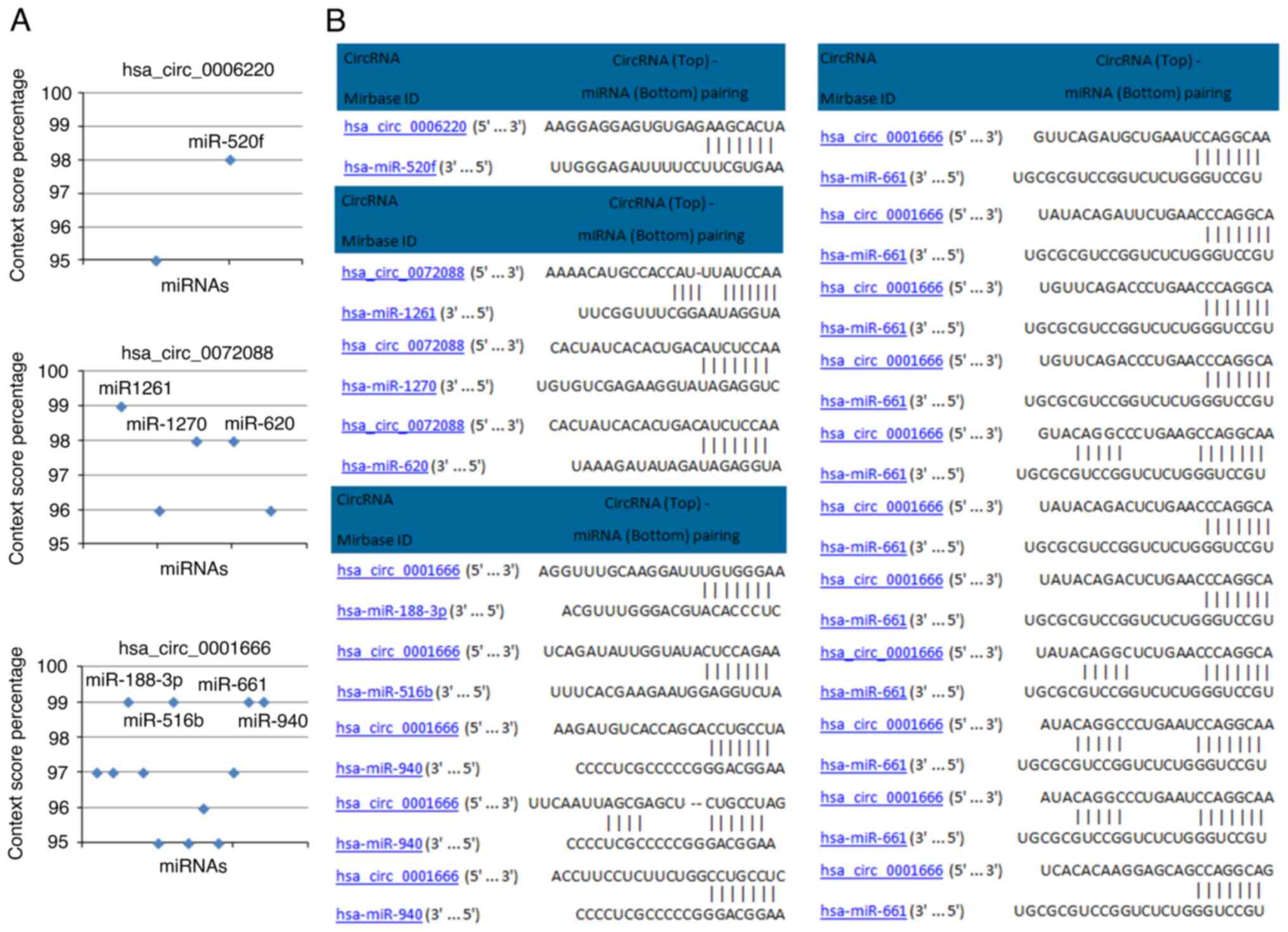
were screened using the CircInteractome database. In total, eight
miRNAs (miR-520f, miR-1261, miR-1270, miR-620, miR-188-3p,
miR-516b, miR-940 and miR-661) with context score percentiles ≥98
were selected (Fig. 3A). The
predicted binding sites between the circRNAs and miRNAs of interest
are shown in Fig. 3B.
Construction of the circRNA-miRNA-mRNA
regulatory network in LAC
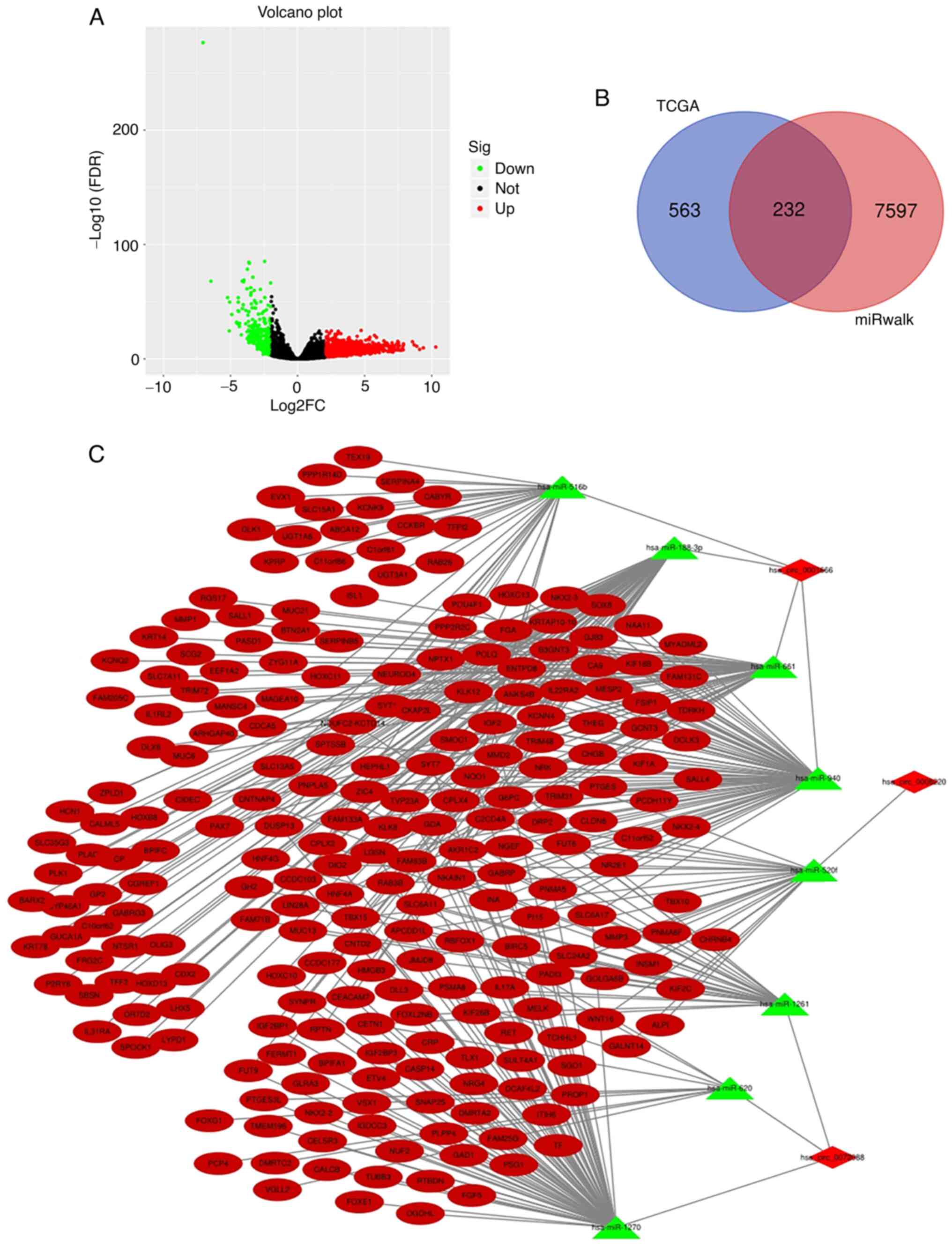
A total of 7,829 target genes of the eight
aforementioned miRNAs were obtained from the miRWalk software.
Additionally, 795 upregulated genes in LAC were obtained from TCGA,
according to the thresholds of false discovery rate value <0.01
and Log2|FC|>3 (Fig.
4A). By overlapping the predicted target genes and the
upregulated genes in LAC, 232 target genes that may serve key roles
in LAC were identified (Fig. 4B).
From this, a circRNA-miRNA-mRNA network was constructed by
integrating the circRNA-miRNA interactions and miRNA-mRNA
interactions (Fig. 4C), which
provided a preliminary insight into the links among the three
differentially-expressed circRNAs (hsa_circ_0006220,
hsa_circ_0072088 and hsa_circ_0001666), their eight suspected
target miRNAs (miR-520f, miR-1261, miR-1270, miR-620, miR-188-3p,
miR-516b, miR-940 and miR-661) and the 232 mRNAs.
PPI network construction and module
selection
After removing the unconnected nodes, a PPI network
consisting of 158 nodes and 287 edges was constructed to view the
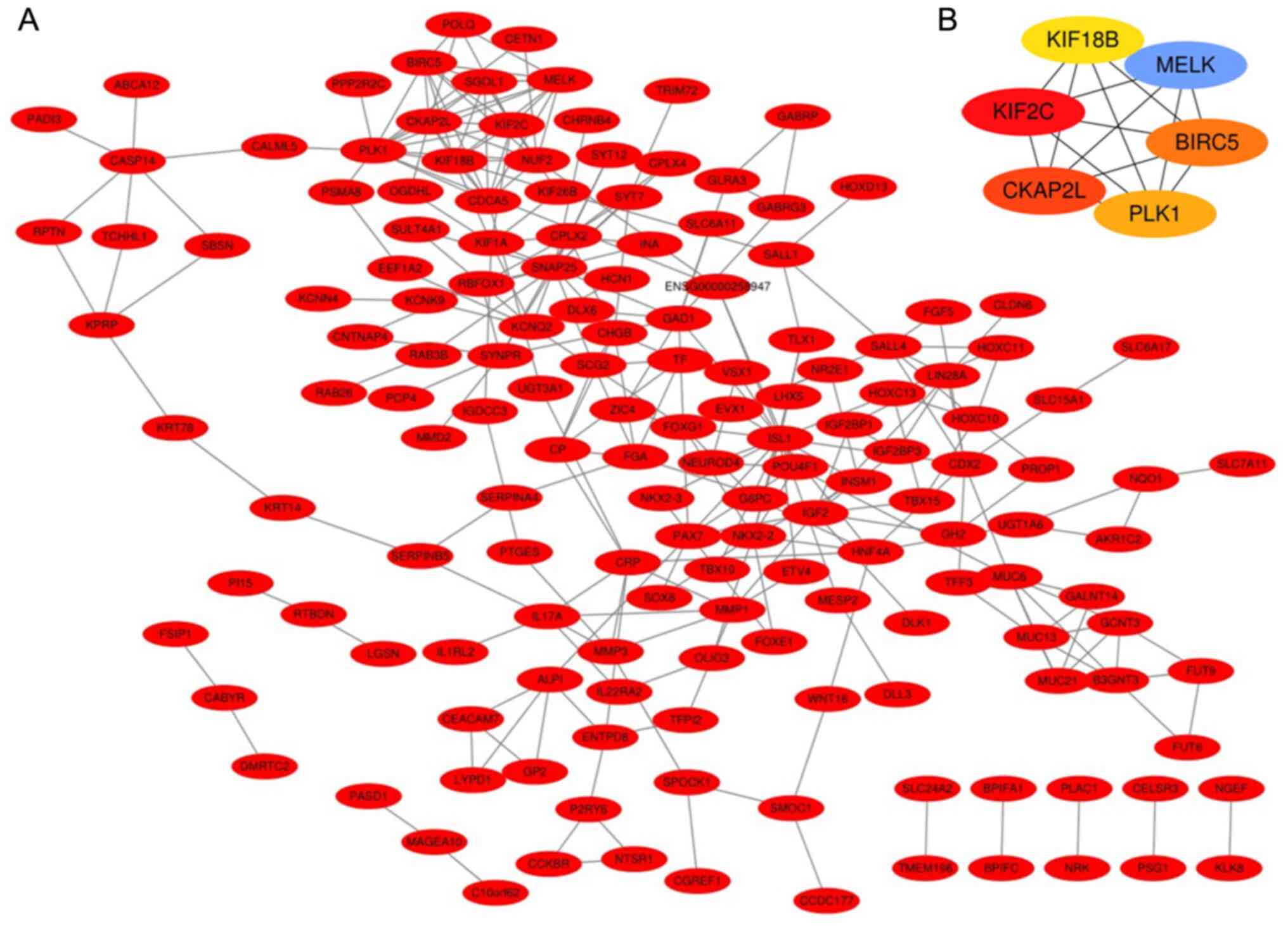
interaction among the 232 target genes (Fig. 5A). Subsequently, a pivotal module
of six hub genes, kinesin family member (KIF) 2C, KIF18B, maternal
embryonic leucine zipper kinase (MELK), baculoviral IAP
repeat-containing 5 (BIRC5), polo-like kinase 1 (PLK1) and
cytoskeleton-associated protein 2-like (CKAP2L), were identified
from the PPI network using the CytoHubba plugin in Cytoscape
(Fig. 5B).
Survival analysis of the six hub
genes
The six hub genes in the PPI network were next
evaluated for their expression levels and prognostic value using
the GEPIA database. As shown in Fig.
6A-F, the expression of these six hub genes in LAC tissues was
significantly higher compared with that in the normal tissues. All
of these six hub genes exhibited their potential in the prediction
of overall survival based on their expression. High expression
levels of each hub gene were associated with poorer overall
survival in patients with LAC (Fig.
6A-F).
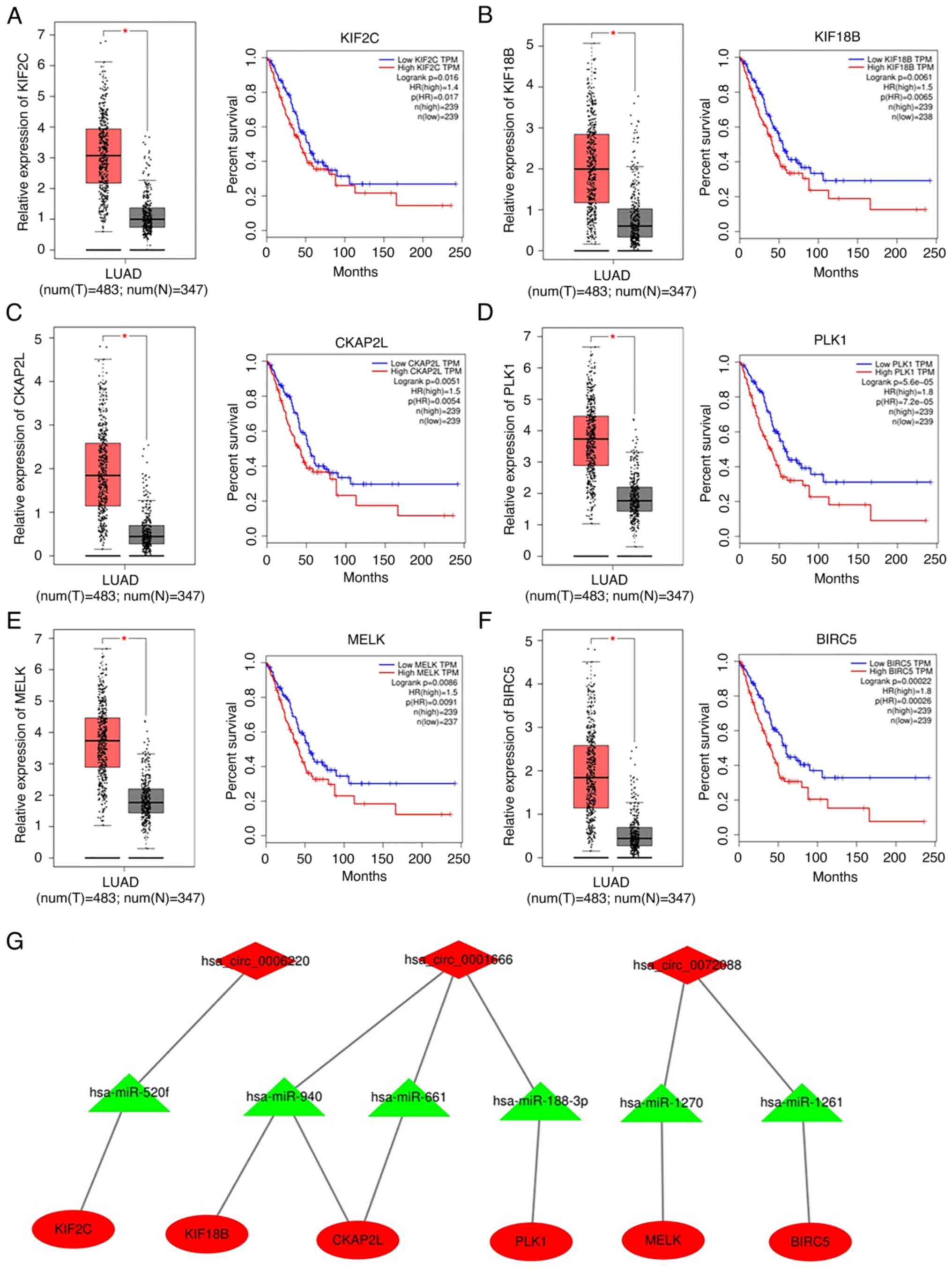 | Figure 6.Relative expression and survival
analysis of the six hub genes in LAC using the GEPIA database.
Expression of (A) KIF2C, (B) KIF18B, (C) cytoskeleton-associated
protein 2-like, (D) polo-like kinase 1, (E) maternal embryonic
leucine zipper kinase and (F) and baculoviral IAP repeat-containing
5 was measured. Red color represents tumor tissues, and gray color
represents normal tissues. (G) The circRNA-miRNA-hubgene
sub-network was constructed. Diamonds represent circRNAs, triangles
represent miRNAs and ovals represent mRNAs. *P<0.05. LAC, lung
adenocarcinoma; GEPIA, Gene Expression Profiling Interactive
Analysis; KIF, kinesin family member; circRNA, circular RNA; miRNA,
microRNA. |
A circRNA-miRNA-hub gene sub-network was then built
to delineate the links among the differentially-expressed circRNAs,
target miRNAs and hub genes (Fig.
6G). Of note, the hsa_circ_0006220/hsa-miR-520f/KIF2C,
has_circ_0001666/hsa-miR-940/KIF18B,
has_circ_0001666/hsamiR-940/CKAP2L,
has_circ_0001666/hsa-miR-661/CKAP2L,
has_circ_0001666/hsa-miR-188-3p/PLK1,
has_circ_0072088/hsamiR-1270/MELK and
has_circ_0072088-hsa-miR-1261-BIRC5 regulatory axes were found from
this network. Taken together, these results suggest that
hsa_circ_0006220, hsa_circ_0072088 and hsa_circ_0001666 may serve
oncogenic roles by regulating the expression of these six hub
genes.
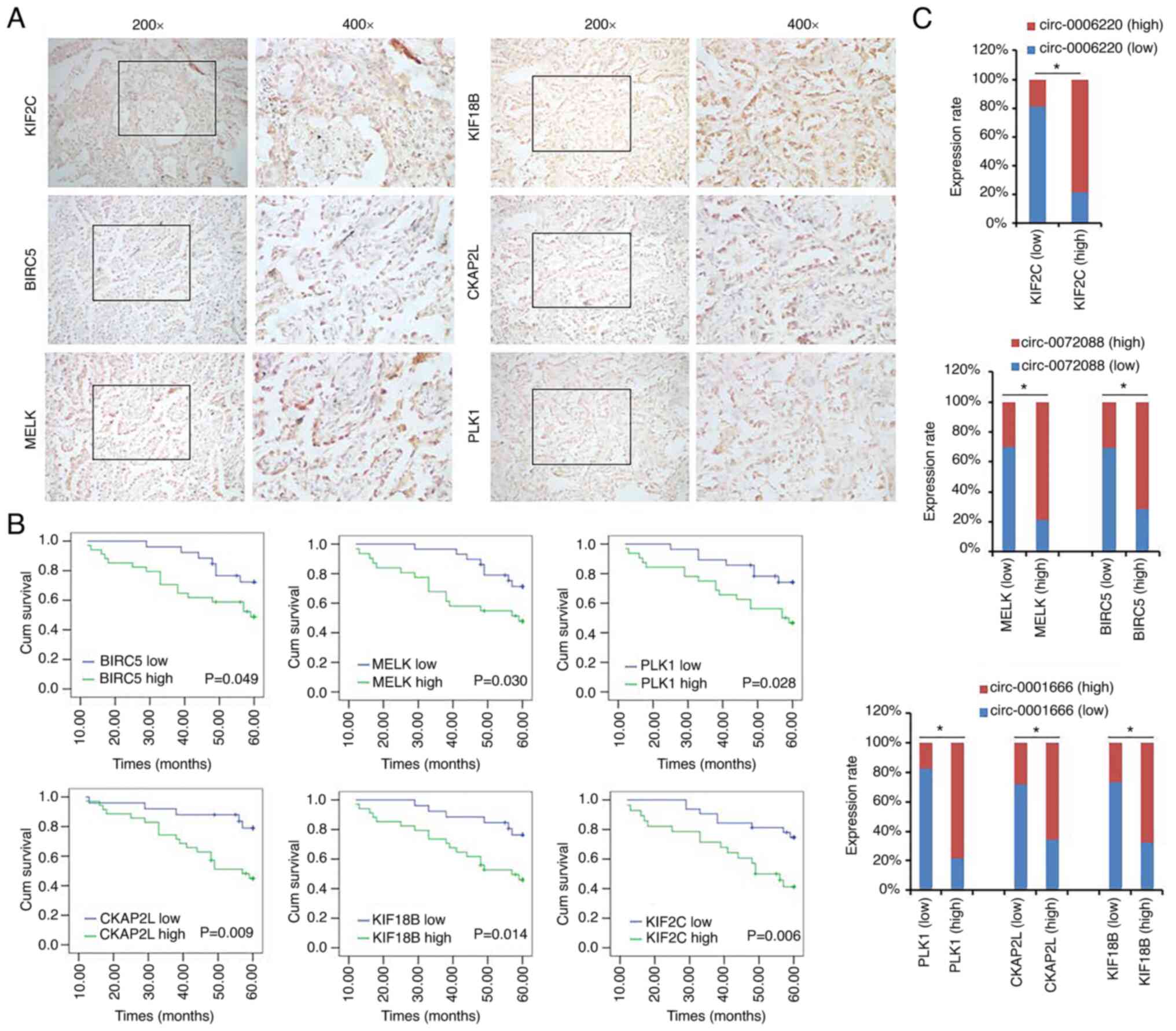
Validation in clinical samples
To further validate the findings,
immunohistochemistry was performed in the 60 LAC samples. The
statistical results showed that high expression levels of KIF2C,
KIF18B, MELK, BIRC5, PLK1 and CKAP2L were associated with the
poorer survival in patients with LAC (Fig. 7A and B), which was consistent with
the results of GEPIA database analysis. In addition, the expression
of hsa_circ_0006220 was positively correlated with the expression
of KIF2C, whereas the expression of has_circ_0001666 was positively
correlated with the expression of KIF18B, MELK and BIRC5. The
expression of has_circ_0072088 was positively correlated with the
expression of PLK1 and CKAP2L (Fig.
7C).
Discussion
Accumulating evidence indicates that circRNAs are
stable, abundant and highly conserved in eukaryotic cells, with
high disease specificity (18–20).
In lung cancer, an increasing number of circRNAs, including
circ-calcium/calmodulin dependent protein kinase IIα (21), circ-solute carrier family 25 member
16 (22), circ-SATB homeobox 2
(23), circ-ATP-binding cassette
subfamily B member 10 (24),
circHMCU (25) and
circ-phosphatidylinositol-4-phosphate 5-kinase type 1α (26), have been reported to serve
regulatory roles in disease progression. These previous findings
suggest that circRNAs are suitable markers for guiding clinical
diagnosis and therapy. However, a large number of potentially
beneficial circRNAs remain undiscovered.
In the present study, a GEO dataset was analyzed to
investigate the differentially-expressed circRNAs in LAC, which
found four circRNAs (hsa_circ_0043278, hsa_circ_0006220,
hsa_circ_0072088 and hsa_circ_0001666) to be highly expressed in
LAC tissues. These candidate circRNAs may be involved in LAC
pathogenesis and can serve as the biomarkers for targeted therapy.
By analyzing their expression in 20 paired LAC tissues and adjacent
normal lung tissues, it was found that the expression levels of
hsa_circ_0006220, hsa_circ_0072088 and hsa_circ_0001666 were
markedly higher in LAC tissues, where high expression levels of
these three circRNAs indicated poorer prognosis in patients with
LAC. In addition, siRNA-mediated knockdown of these three circRNAs
suppressed the proliferation, migration, and invasion of LAC cell
lines, suggesting their potential oncogenic functions in LAC. These
results suggest that hsa_circ_0006220, hsa_circ_0072088 and
hsa_circ_0001666 can serve oncogenic roles in LAC cells.
As conserved endogenous RNAs, circRNAs can sponge
miRNAs and serve key roles in regulating the expression of the
target genes of miRNAs. To explore the sponged miRNAs of these
three aforementioned circRNAs, the CircInteractome online database
(27,28) was used to predict the miRNAs that
have the ability to bind these circRNAs. In total, eight miRNAs
(miR-520f, miR-1261, miR-1270, miR-620, miR-188-3p, miR-516b,
miR-940 and miR-661) were determined to have high potential to bind
these circRNAs. Subsequently, potential mRNAs targeted by these
miRNAs were predicted using miRWalk before a ceRNA network was
constructed.
To verify the action of this ceRNA network, a PPI
network was next constructed, which yielded six hub genes, namely
KIF2C, KIF18B, MELK, BIRC5, PLK1 and CKAP2L. The oncogenic roles of
these six genes in cancer have also been demonstrated by previous
studies (29–39). In the present study, the prognostic
value of these hub genes was also analyzed in LAC based on the
GEPIA database. High expression levels of each hub gene were
associated with a poorer overall survival in patients with LAC. The
in vivo experiments showed that high expression levels of
KIF2C, KIF18B, MELK, BIRC5, PLK1 and CKAP2L were associated with
poorer survival in patients with LAC, which was in accordance with
the results from the GEPIA database. In addition, the expression of
hsa_circ_0006220 was positively correlated with the expression of
KIF2C, whereas the expression of has_circ_0001666 was positively
correlated with the expression of KIF18B, MELK and BIRC5. The
expression of has_circ_0072088 was positively correlated with the
expression of PLK1 and CKAP2L. These results suggested that
hsa_circ_0006220, hsa_circ_0072088 and hsa_circ_0001666 may serve
oncogenic roles in LAC by regulating the expression of these hub
genes. However, further studies are required to confirm that the
circRNAs affect the expression of the proteins encoded via the
above six miRNAs.
As there is no public circRNA data in TCGA database,
the GEO database was chosen to screen for the target circRNAs in
LAC, which provided guidance for the follow-up validation and
experiments. The retrospective clinical study and biological
experiments confirmed the oncogenic function of these selected
circRNAs. The ceRNA and PPI networks were then constructed using
bioinformatic analysis, using which the hub genes were revealed and
partially explained the molecular mechanism underlying the
aforementioned circRNAs. However, further experiments are required
to validate these findings. Conclusively, through the bioinformatic
analysis and retrospective clinical study, results from the present
study provided novel circRNAs biomarkers and therapeutic targets
for LAC. Further experiments in animals would be useful to validate
these findings.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Qianglin Duan, a
skilled English proofreader from Tongji University, for assistance
with the revision of this paper.
Funding
This study was supported by the Financial Supporting Program of
Hebei Province [grant nos. (2014)1257 and (2016)361006].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BS and MS contributed to the conception and design
of the work. YZ, FC, SL, LM, LG performed the experiments. FL and
HZ analyzed the data and performed the bioinformatical analysis. YZ
and BS confirm the authenticity of all the raw data. All authors
read and approved the final manuscript.
Ethical approval and consent to
participate
The human tissues were obtained with informed
consent and the present study was approved by The Clinical Research
Ethics Committee of The Fourth hospital of Hebei Medical University
(approval no. 2021KY157).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lin JJ, Cardarella S, Lydon CA, Dahlberg
SE, Jackman DM, Jänne PA and Johnson BE: Five-year survival in
EGFR-mutant metastatic lung adenocarcinoma treated with EGFR-TKIs.
J Thorac Oncol. 11:556–565. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He J, Xie Q, Xu H, Li J and Li Y: Circular
RNAs and cancer. Cancer Lett. 396:138–144. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu G, Zhou W, Pan X, Sun Z, Sun Y, Xu H,
Shi P, Li J, Gao L and Tian X: Circular RNA profiling reveals
exosomal circ_0006156 as a novel biomarker in papillary thyroid
cancer. Mol Ther Nucleic Acids. 19:1134–1144. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fan L, Wang J, Cao Q, Ding X and Li B:
Aberrant miR-1246 expression promotes radioresistance in non-small
cell lung cancer: A potential prognostic biomarker and radiotherapy
sensitization target. Am J Cancer Res. 10:314–335. 2020.PubMed/NCBI
|
|
8
|
Lei B, Tian Z, Fan W and Ni B: Circular
RNA: A novel biomarker and therapeutic target for human cancers.
Int J Med Sci. 16:292–301. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cheng X, Qiu J, Wang S, Yang Y, Guo M,
Wang D, Luo Q and Xu L: Comprehensive circular RNA profiling
identifies CircFAM120A as a new biomarker of hypoxic lung
adenocarcinoma. Ann Transl Med. 7:4422019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen F, Huang C, Wu Q, Jiang L, Chen S and
Chen L: Circular RNAs expression profiles in plasma exosomes from
early-stage lung adenocarcinoma and the potential biomarkers. J
Cell Biochem. 121:2525–2533. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang C, Tan S, Liu WR, Lei Q, Qiao W, Wu
Y, Liu X, Cheng W, Wei YQ, Peng Y and Li W: RNA-Seq profiling of
circular RNA in human lung adenocarcinoma and squamous cell
carcinoma. Mol Cancer. 18:1342019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu XX, Yang YE, Liu X, Wang X, Liu XX and
Mai J: A two-circular RNA signature as a noninvasive diagnostic
biomarker for lung adenocarcinoma. J Transl Med. 17:502019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z,
Xu W, Zhang E, Wang J, Fang T, et al: The Circular RNA circPRKCI
promotes tumor growth in lung adenocarcinoma. Cancer Res.
78:2839–2851. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang L, Liang Y, Mao Q, Xia W, Chen B,
Shen H, Xu L, Jiang F and Dong G: Circular RNA circCRIM1 inhibits
invasion and metastasis in lung adenocarcinoma through the microRNA
(miR)-182/miR-93-leukemia inhibitory factor receptor pathway.
Cancer Sci. 110:2960–2972. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yao Y, Hua Q, Zhou Y and Shen H: CircRNA
has_circ_0001946 promotes cell growth in lung adenocarcinoma by
regulating miR-135a-5p/SIRT1 axis and activating Wnt/β-catenin
signaling pathway. Biomed Pharmacother. 111:1367–1375. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zuo Y, Shen W, Wang C, Niu N and Pu J:
Circular RNA Circ-ZNF609 promotes lung adenocarcinoma proliferation
by modulating miR-1224-3p/ETV1 signaling. Cancer Manag Res.
12:2471–2479. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Patop IL, Wüst S and Kadener S: Past,
present, and future of circRNAs. EMBO J. 38:e1008362019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bach DH, Lee SK and Sood AK: Circular RNAs
in cancer. Mol Ther Nucleic Acids. 16:118–129. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Du J, Zhang G, Qiu H, Yu H and Yuan W: The
Novel Circular RNA circ-CAMK2A enhances lung adenocarcinoma
metastasis by regulating the miR-615-5p/fibronectin 1 pathway. Cell
Mol Biol Lett. 24:722019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shangguan H, Feng H, Lv D, Wang J, Tian T
and Wang X: Circular RNA circSLC25A16 contributes to the glycolysis
of non-small-cell lung cancer through epigenetic modification. Cell
Death Dis. 11:4372020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu
M, Dai X, Zhou H, Zhu J, Zhang H and Jiang Y: Circular RNA
circSATB2 promotes progression of non-small cell lung cancer cells.
Mol Cancer. 19:1012020. View Article : Google Scholar : View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma D, Qin Y, Huang C, Chen Y, Han Z, Zhou
X and Liu H: Circular RNA ABCB10 promotes non-small cell lung
cancer progression by increasing E2F5 expression through sponging
miR-584-5p. Cell Cycle. 19:1611–1620. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song X, Liang Y, Sang Y, Li Y, Zhang H,
Chen B, Du L, Liu Y, Wang L, Zhao W, et al: circHMCU promotes
proliferation and metastasis of breast cancer by sponging the let-7
family. Mol Ther Nucleic Acids. 20:518–533. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma Y, Cong X, Zhang Y, Yin X, Zhu Z and
Xue Y: CircPIP5K1A facilitates gastric cancer progression via
miR-376c-3p/ZNF146 axis. Cancer Cell Int. 20:812020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bai S, Wu Y, Yan Y, Shao S, Zhang J, Liu
J, Hui B, Liu R, Ma H, Zhang X and Ren J: Construct a
circRNA/miRNA/mRNA regulatory network to explore potential
pathogenesis and therapy options of clear cell renal cell
carcinoma. Sci Rep. 10:136592020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu Y, Xu Y, Xiao F, Zhang J, Wang Y, Yao
Y and Yang J: Comprehensive analysis of a circRNA-miRNA-mRNA
network to reveal potential inflammation-related targets for
gastric adenocarcinoma. Mediators Inflamm. 2020:94356082020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wei S, Dai M, Zhang C, Teng K, Wang F, Li
H, Sun W, Feng Z, Kang T, Guan X, et al: KIF2C: A novel link
between Wnt/β-catenin and mTORC1 signaling in the pathogenesis of
hepatocellular carcinoma. Protein Cell. 12:788–809. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li X, Huang W, Huang W, Wei T, Zhu W, Chen
G and Zhang J: Kinesin family members KIF2C/4A/10/11/14/18B/20A/23
predict poor prognosis and promote cell proliferation in
hepatocellular carcinoma. Am J Transl Res. 12:1614–1639.
2020.PubMed/NCBI
|
|
31
|
Gan H, Lin L, Hu N, Yang Y, Gao Y, Pei Y,
Chen K and Sun B: KIF2C exerts an oncogenic role in nonsmall cell
lung cancer and is negatively regulated by miR-325-3p. Cell Biochem
Funct. 37:424–431. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bai Y, Xiong L, Zhu M, Yang Z, Zhao J and
Tang H: Co-expression network analysis identified KIF2C in
association with progression and prognosis in lung adenocarcinoma.
Cancer Biomark. 24:371–382. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gao T, Yu L, Fang Z, Liu J, Bai C, Li S,
Xue R, Zhang L, Tan Z and Fan Z: KIF18B promotes tumor progression
in osteosarcoma by activating β-catenin. Cancer Biol Med.
17:371–386. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chung S, Suzuki H, Miyamoto T, Takamatsu
N, Tatsuguchi A, Ueda K, Kijima K, Nakamura Y and Matsuo Y:
Development of an orally-administrative MELK-targeting inhibitor
that suppresses the growth of various types of human cancer.
Oncotarget. 3:1629–1640. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cao Y, Zhu W, Chen W, Wu J, Hou G and Li
Y: Prognostic value of BIRC5 in lung adenocarcinoma lacking EGFR,
KRAS, and ALK mutations by integrated bioinformatics analysis. Dis
Markers. 2019:54512902019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shin SB, Jang HR, Xu R, Won JY and Yim H:
Active PLK1-driven metastasis is amplified by TGF-beta signaling
that forms a positive feedback loop in non-small cell lung cancer.
Oncogene. 39:767–785. 2020. View Article : Google Scholar : View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nieto-Jimenez C, Galan-Moya EM,
Corrales-Sanchez V, Noblejas-Lopez MDM, Burgos M, Domingo B,
Montero JC, Gomez-Juarez M, Picazo-Martinez MG, Esparis-Ogando A,
et al: Inhibition of the mitotic kinase PLK1 overcomes therapeutic
resistance to BET inhibitors in triple negative breast cancer.
Cancer Lett. 491:50–59. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang P and He X: Oncogenic and prognostic
role of CKAP2L in hepatocellular carcinoma. Int J Clin Exp Pathol.
13:923–933. 2020.PubMed/NCBI
|
|
39
|
Xiong G, Li L, Chen X, Song S, Zhao Y, Cai
W and Peng J: Up-regulation of CKAP2L expression promotes lung
adenocarcinoma invasion and is associated with poor prognosis. Onco
Targets Ther. 12:1171–1180. 2019. View Article : Google Scholar : PubMed/NCBI
|