Introduction
Colorectal carcinoma (CRC) is the fourth leading
cause of cancer-related mortality worldwide, accounting for
~900,000 deaths annually in 2018 (1). The pathogenesis of CRC starts as
low-grade dysplasia and often progresses slowly to form a polyp in
the colorectal lumen, which eventually develops into a tumor with
considerable size and symptoms (including pain, cramping and
bleeding) (2). Therefore, most
patients with CRC are at an advanced or metastatic stage when
diagnosed, at which point tumor cells have already invaded into
regional lymph nodes and spread into distal organs (such as the
liver, lung and pancreas), which accounts for the major comorbidity
in CRC (3,4). To overcome the aforementioned
problems and develop therapeutic interventions, understanding the
mechanism of CRC carcinogenesis (such as cell viability, invasion
and stemness) is necessary.
Circular (circ)RNA is a type of non-coding RNA
molecule, with a circular structure, which participates in gene
regulation under both physiological and pathological conditions
(5,6). Previous studies have demonstrated
that circRNA molecules [such as circular homeodomain interacting
protein kinase (circ-HIPK) 3, circular SWI/SNF-related,
matrix-associated, actin-dependent regulator of chromatin,
subfamily A, member 5 (circ-SMARCA5), circ_0026416 and circ_103809]
were associated with CRC pathogenesis (7–10).
circ_0001666, a recently identified circRNA derived from the
circulation of FAM120B gene, regulates multiple microRNA
(miRNA/miR) targets and downstream genes, such as ETS variant
transcription factor 4, to promote papillary thyroid carcinoma cell
proliferation and migration (11).
Furthermore, a previous bioinformatics study demonstrated that, in
CRC, circ_0001666 targeted miR-1229 as part of a circRNA/miRNA
network (12). In addition,
miR-1229 has been reported to regulate the Wnt/β-catenin signaling
pathway to promote the malignancy of several cancer cell types. The
Wnt/β-catenin signaling pathway is a key signaling pathway
associated with oncogenesis, including in breast cancer, liver
cancer and CRC (10,13–15).
Based on these aforementioned studies, it may be hypothesized that
circ_0001666 regulates the Wnt/β-catenin signaling pathway via
miR-1229 in CRC. However, to the best of our knowledge, this
hypothesis has yet to be investigated.
The aim of the present study was to examine the
interaction between circ_0001666, miR-1229 and the Wnt/β-catenin
signaling pathway, as well as the potential effect of this network
on cell viability, invasion and stemness in CRC.
Materials and methods
Cell culture and reagents
The human CRC cell lines (HT-29, HCT-116, SW480 and
SW-620) and the human intestinal epithelial cell line (HIEC)
(https://web.expasy.org/cellosaurus/CVCL_6C21) were
purchased from American Type Culture Collection. All the cell lines
used in the current study were authenticated using short tandem
repeat profiling. The cells were cultured in McCoy's 5a medium
(HyClone; Cytiva), containing 10% FBS (HyClone; Cytiva). The
circ_0001666 and negative control (NC) overexpression plasmids were
constructed using a pCD5-ciR backbone, and were purchased from
Guangzhou Geneseed Biotech Co., Ltd. The circ_0001666, miR-1229 and
NC knockdown plasmids were constructed using a pGPH1 backbone
purchased from Shanghai GenePharma Co., Ltd. In addition, the NC
and miR-1229 overexpression plasmids were constructed using a pGPU6
backbone purchased from Shanghai GenePharma Co., Ltd.
TRIzol®, Lipofectamine® 3000 Transfection
reagent and PVDF Membranes were purchased from Thermo Fisher
Scientific, Inc. RNase R was purchased from Epicentre®
Biotechnologies (Illumina, Inc.). The reverse transcription (RT)
kit and Fast quantitative (q)PCR Mix were purchased from Takara
Biotechnology Co., Ltd. The Cell Counting Kit (CCK)-8, One Step
TUNEL Apoptosis Assay kit, RIPA lysis buffer, BCA Protein assay
kit, BeyoECL kit, 293T cells, pGL6 plasmid and Luciferase Reporter
Gene Assay kit were purchased from Beyotime Institute of
Biotechnology. The 293T cell line was cultured in DMEM containing
10% FBS (both from HyClone; Cytiva). The Matrigel®
Basement Membrane Matrix and Transwell chambers were purchased from
BD Biosciences. The antibodies were purchased from Abcam.
circ_0001666 plasmid transfection
The 0.8 µg pCD5-ciR-NC (empty vector) (16), 0.8 µg pCD5-ciR-circ (the forward
cloning primer, 5′-CGGAATTCACTGCTATACTCTCTTGTCCTTGTGA-3′ and
reverse, 5′-CGGGATCCCAGCAACATGCGCTGACCAC-3′; expected product
length, 13,733 bp), 0.8 µg pGPH1-NC
5′-CACCGAGTGAAACAGTGCAGCTGCGAACAGCTGCACTGTTTCAC-3′) and 0.8 µg
pGPH1-circ
(5′-CACCGATGACCATTCCAGATCCTTTCGAAAAAGGATCTGGAATGGTCATC-3′) plasmids
were mixed with Lipofectamine® 3000 and transfected into
the HT-29 and HCT-116 cell lines for 6 h at 37°C. Untransfected
HT-29 and HCT-116 cell lines were also used as a control. At 24 h
post-transfection, circ_0001666 and miR-1229 expression, cell
proliferation, apoptosis and invasion, as well as the number of
CD133+ cells were evaluated.
miR-1229 plasmid transfection
The 0.8 µg pGPU6-NC (5′-AACACCGAACGAGACAGGATT-3′),
0.8 µg pGPU6-miR (5′-CTCTCACCACTGCCCTCCCACAG-3′), 0.8 µg pGPH1-NC
(5′-AAGAACAACACAAAAGAACAG-3′) and 0.8 µg pGPH1-miR
(5′-CTGTGGGAGGGCAGTGGTGAGAG-3′) plasmids were transfected into the
HT-29 and HCT-116 cell lines using Lipofectamine® 3000
for 6 h at 37°C, according to the manufacturer's instructions.
Untransfected HT-29 and HCT-116 cell lines were also used as a
control. At 24 h post-transfection, circ_0001666 and miR-1229
expression was evaluated.
Rescue experiments
For the rescue experiments, the following
co-transfections were performed in the HT-29 cell line: i) 0.8 µg
pCD5-ciR-NC + 0.8 µg pGPU6-NC; ii) 0.8 µg pCD5-ciR-circ plasmid +
0.8 µg pGPU6-NC; iii) 0.8 µg pCD5-ciR-NC + 0.8 µg pGPU6-miR; and
iv) 0.8 µg pCD5-ciR-circ + 0.8 µg pGPU6-miR. To perform these
transfections, Lipofectamine® 3000 was used according to
the manufacturer's protocol for 6 h at 37°C. At 24 h
post-transfection, circ_0001666 and miR-1229 expression, cell
proliferation, apoptosis and invasion, as well as the number of
CD133+ cells were evaluated.
RT-qPCR
The cells were collected 48 h following transfection
for RT-qPCR. Total RNA was isolated from the cells using TRIzol.
Subsequently, for the determination of circ_0001666 expression
levels, total RNA was digested using RNase R. The RT reagent kit
was then used to reverse transcribe RNA to cDNA with thermal
cycling conditions of 37°C for 15 min and 85°C for 5 sec. qPCR was
performed using the TB Green Fast qPCR Mix (Takara Biotechnology
Co., Ltd.) with an Applied Biosystems 7500 Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions for qPCR were as follows: 95°C for 30 sec
for 1 cycle; followed by 95°C for 5 sec and 61°C for 15 sec for 40
cycles. The 2−ΔΔCq method was used to calculate the
relative expression levels of the targets (17). The following primer sequences were
used: i) circ_0001666 forward, 5′-TCTACTCACTCTTACTGGAGGACTG-3′ and
reverse, 5′-CTTGCTTCGGTGGTGCTCTG-3′; expected product length, 252
bp; ii) GAPDH forward, 5′-GAGTCCACTGGCGTCTTCAC-3′ and reverse,
5′-ATCTTGAGGCTGTTGTCATACTTCT-3′; expected product length, 149 bp;
iii miR-1229 forward, 5′-ACACTCCAGCTGGGCTCTCACCACTGCCCT-3′ and
reverse, 5′-TGTCGTGGAGTCGGCAATTC-3′; expected product length, 64
bp; and iv) U6 forward, 5′-GCTCGCTTCGGCAGCACATA-3′ and reverse,
5′-AATATGGAACGCTTCACGAATTTGC-3′; expected product length, 102 bp.
The detailed procedure of designing circ_0001666 primers were in
accordance with a previous study (18). In addition, the forward and reverse
primers for miR-1229 were design according to previous studies
(19,20).
Western blot analysis
The cells were collected 48 h following transfection
for western blot analysis and was performed using RIPA lysis
buffer, BCA protein assay kit and BeyoECL kit, as described in a
previous study (21). The
following antibodies were used: i) Anti-GSK3β (rabbit; monoclonal;
1:10,000; cat. no. ab32391); ii) anti-phosphorylated (p)-GSK3β
(rabbit; monoclonal; 1:15,000; cat. no. ab131097); iii)
anti-β-catenin (rabbit; monoclonal; 1:5,000; cat. no. ab32572); iv)
anti-GAPDH (rabbit; polyclonal; 1:3,000; cat. no. ab9485); v)
anti-C-X-C motif chemokine receptor 5 (CXCR5; rabbit; monoclonal;
1:1,000; cat. no. ab254415); and vi) HRP-conjugated goat
anti-rabbit IgG H&L (1:10,000; cat. no. ab181662). ImageJ
(version 1.8.0; National Institutes of Health) was used for
semi-quantification and analysis of the western blot bands.
Cell proliferation assay
CCK-8 assay solution was added and incubated with
the cells 0, 24, 48 and 72 h post-transfection. After a 2-h
incubation, the optical density was measured using a microplate
reader.
Apoptosis detection
Apoptosis was detected using a One Step TUNEL
Apoptosis assay kit 48 h following transfection. Briefly, the cells
were fixed with 4% paraformaldehyde for 30 min at room temperature,
permeabilized with 0.1% Triton X-100 for 5 min at room temperature,
incubated with TUNEL solution at 37°C for 60 min, then
counterstained with 0.5 µg/ml DAPI for 5 min at room temperature.
The cells were then observed under an inverted fluorescence
microscope and the TUNEL-positive cells were counted. The stained
cells were counted in five fields of view.
Matrigel assay
A total of 5×104 cells were harvested 48
h following transfection and counted. The cells in FBS-free McCoy's
5a medium were then seeded in a Transwell chamber (cat. no. 353097,
Corning, Inc.) pre-coated with Matrigel Basement Membrane Matrix
(37°C for 60 min) and incubated for 24 h at 37°C. The lower chamber
was filled with 10% FBS-containing McCoy's 5a medium. After being
fixed with 4% paraformaldehyde for 30 min at room temperature, the
cells on the bottom surface of the chamber were stained with
crystal violet for 30 min at room temperature and counted under a
light microscope. The stained cells were counted in five fields of
view.
Flow cytometry
The cells were harvested and stained in the dark for
60 min on ice with Alexa Fluor® 488-conjugated rabbit
monoclonal antibody against CD133 (cat. no. ab271092; 1:2,500) or
Alexa Fluor® 488-conjugated rabbit IgG isotype control
(cat. no. ab172730; 1:500) 48 h following transfection. The numbers
of CD133+ cells were analyzed using a FACSCalibur flow
cytometer (BD Biosciences) and Flowjo 7.6 (BD Biosciences). A
general viable cell gate was used to exclude apoptotic and dying
cells, and to distinguish cells based on size and granularity
(22).
Luciferase reporter gene assay
The binding site of circ_001666 and miR-1229 was
predicted by RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid).
The wild-type (WT) or mutant type (MT) circ_0001666 DNA fragments
were cloned into the pGL6 plasmid. The plasmid constructs were
co-transfected with pGPU6-NC/pGPU6-miR into the 293T cell line
(American Type Culture Collection) using Lipofectamine®
3000, according to the manufacturer's instructions. The cells were
then lysed at 48 h after transfection and luciferase activity was
detected using a Luciferase Reporter Gene Assay kit. The luciferase
activity of the circ_0001666 WT/pGPU6 group was set as 1. The
luciferase activity was normalized to Renilla luciferase
activity. The 293T cells were cultured in 10% FBS-containing DMEM
(HyClone; Cytiva) under 5% CO2 at 37°C.
Fluorescence in situ hybridization
(FISH)
FISH was used to detect the location of circ_0001666
expression in the CRC cell lines and to visualize circ_0001666 and
miR-1229 binding. Briefly, the CRC cell lines were fixed at room
temperature using 4% paraformaldehyde solution for 10 min at room
temperature. Subsequently, the cells were permeabilized by 0.5%
Tween X-100 at 4°C for 5 min and hybridized with 50 pM Cy3-labeled
circ_0001666 probes (5′-AGATGACCATTCCAGATCCTT-3′) (Sangon Biotech
Co., Ltd.) overnight at 37°C. After incubation, the cells were
washed by 0.1% Tween 20 containing 4X saline-sodium citrate (SSC)
for 5 min at 42°C, 2X SSC for 5 min at 42°C and 1X SSC for 5 min at
42°C, successively. DAPI was added for nuclear staining at room
temperature for 10 min and the images were visualized under a
fluorescence microscope. All procedures were conducted according to
the Ribo™ Fluorescent In Situ Hybridization kit
instructions (Guangzhou RiboBio Co., Ltd.).
Statistical analysis
All the data are presented as the mean ± SD and were
analyzed using GraphPad Prism v7.01 (GraphPad Software, Inc.). To
compare the differences between multiple groups one way ANOVA
followed by Tukey's post hoc test was used. All experiments were
conducted in triplicate. P<0.05 was considered to indicate a
statistically significant difference.
Results
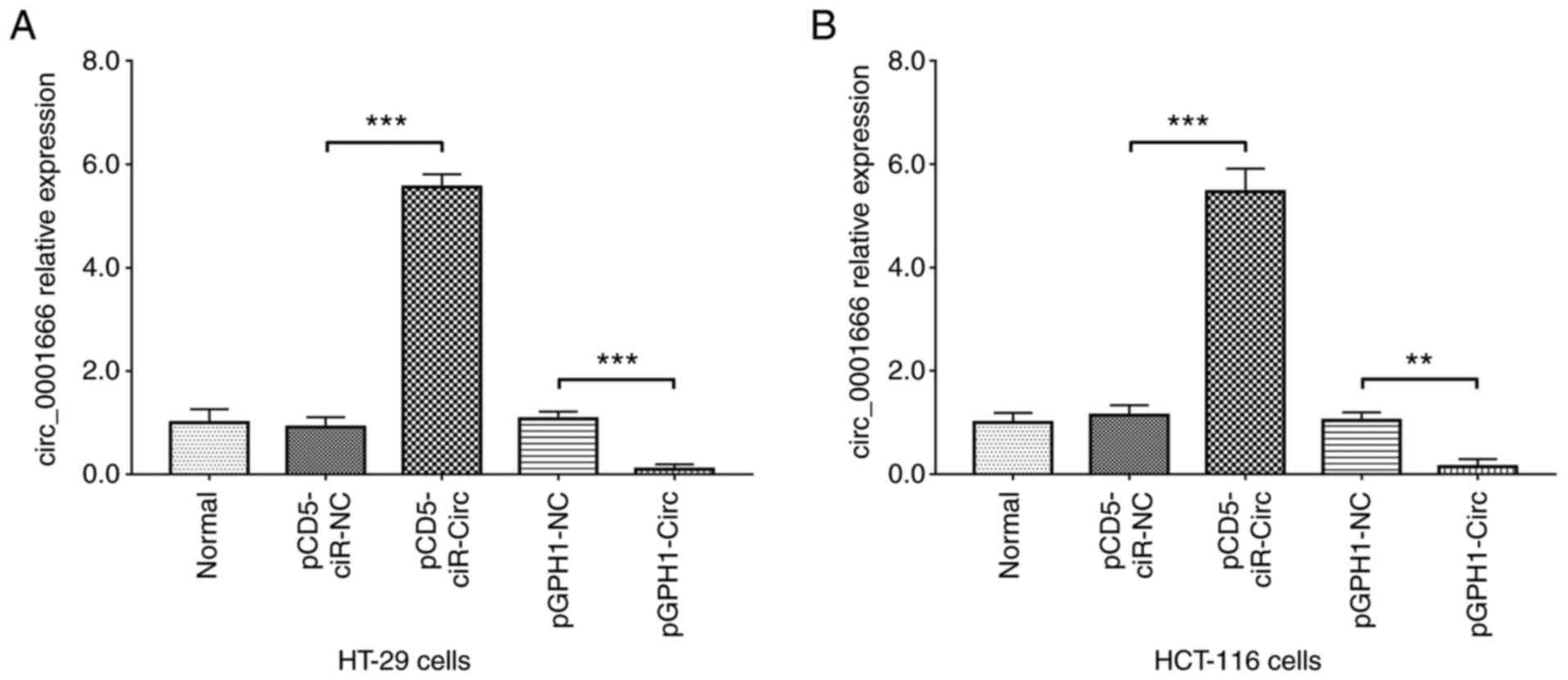
circ_0001666 expression following
transfection
circ_0001666 mRNA expression level was reduced,
while miR-1229 mRNA expression level was increased in the CRC cell
lines compared with that in the HIEC cell line (Fig. S1). The HT-29 and HCT-116 CRC cell
lines were transfected with pCD5-ciR-circ, pCD5-ciR-NC, pGPH1-circ
or pGPH1-NC plasmids. In both the cell lines, the relative mRNA
expression levels of circ_0001666 were upregulated in the
pCD5-ciR-circ group compared with that in the pCD5-ciR-NC group
(both P<0.001). In addition, relative mRNA expression levels of
circ_0001666 were downregulated in the pGPH1-circ group compared
with that in the pGPH1-NC group (both P<0.01) (Fig. 1). In addition, FISH was performed
to determine the cellular localization of circ_0001666. The results
demonstrated that circ_0001666 was expressed intracellularly in the
CRC cell lines (Fig. S2).
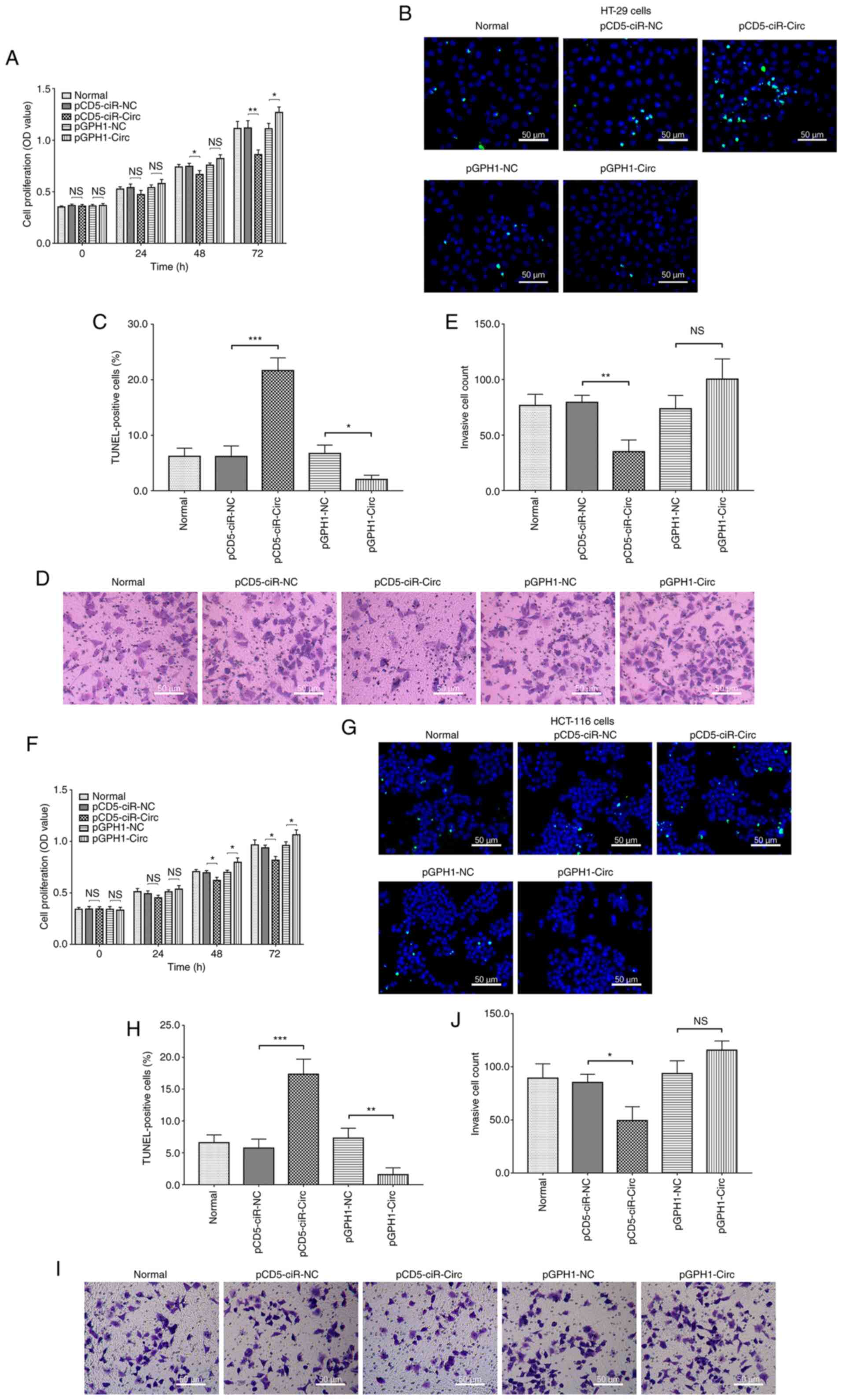
circ_0001666 suppresses CRC cell
proliferation, apoptosis and invasion
In the HT-29 cell line, circ_0001666 overexpression
reduced cell proliferation at 48 (P<0.05) and 72 h (P<0.01),
as well as the number of invading cells (P<0.01). In addition,
it also promoted apoptosis, as evidenced by the number of
TUNEL-positive cells (P<0.001; Fig.
2A-E). However, circ_0001666 knockdown had the opposite effect
on proliferation and apoptosis; however, it did not affect cell
invasion. In addition, the same results were observed in the
HCT-116 cell line following circ_0001666 overexpression and
knockdown (Fig. 2F-J). These data
suggested that circ_0001666 inhibited cell proliferation and
invasion, whilst promoting apoptosis, in the CRC cell lines.
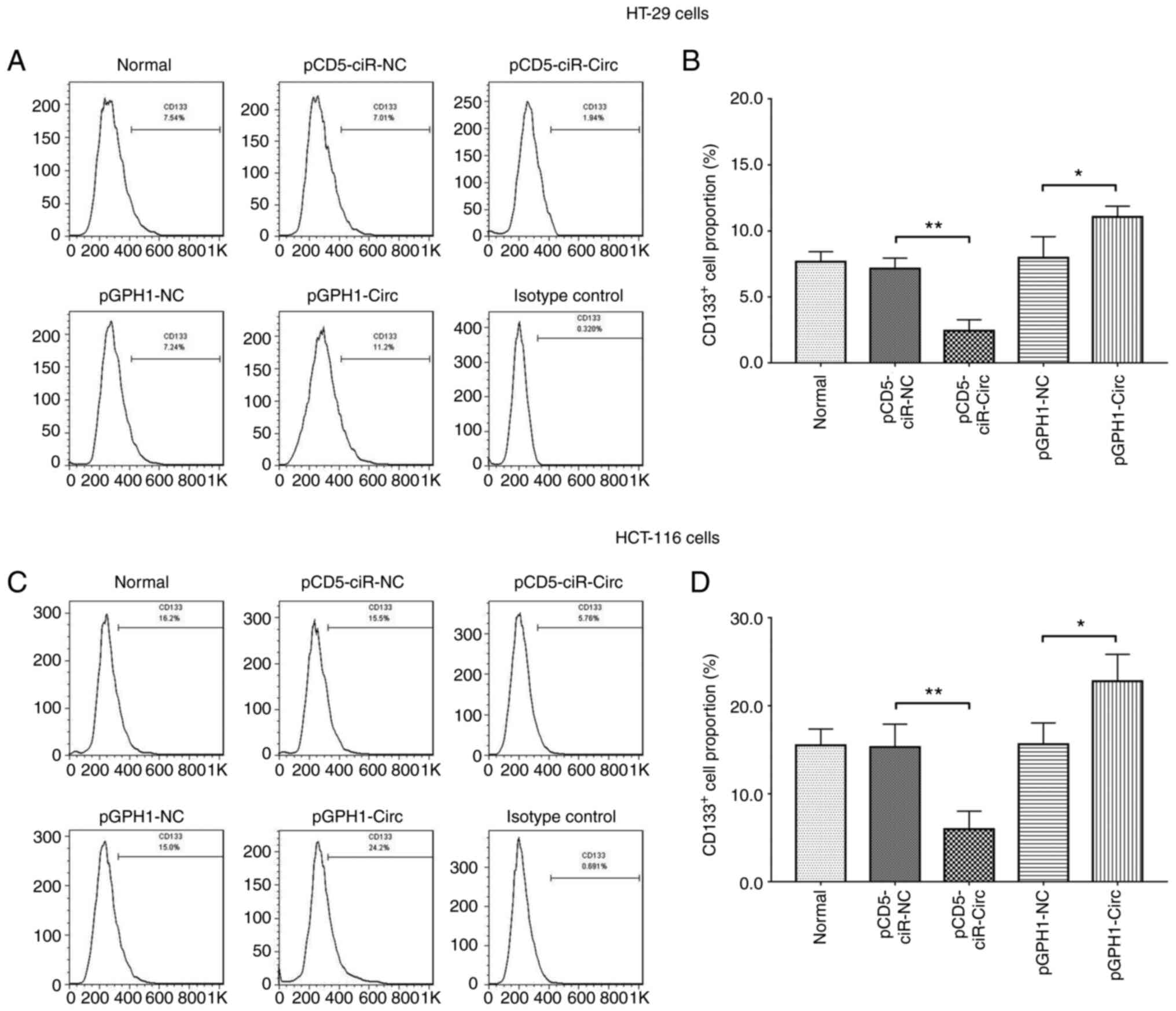
circ_0001666 inhibits cancer
stemness
In the HT-29 cell line, the numbers of
CD133+ cells were reduced following circ_0001666
overexpression (P<0.01), but was increased following knockdown
of expression (P<0.05) (Fig. 3A and
B). Furthermore, the same results were observed in the HCT-116
cell line (Fig. 3C and D). The
forward-side scatter cryptogram of CD133+ cell is
displayed in Fig. S3. As CD133 is
a biomarker for cancer stem cells, these findings indicated that
circ_0001666 inhibited cancer stemness in the CRC cell lines
(23).
circ_001666 directly binds to
miR-1229
In both the HT-29 and HCT-16 cell lines,
circ_0001666 overexpression downregulated miR-1229 mRNA expression
level, while knockdown of circ_0001666 expression upregulated
miR-1229 mRNA expression level (both P<0.01) (Fig. 4A and C). In addition, miR-1229 mRNA
expression level was increased in the HT-29 and HCT-16 cell lines
following transfection with miR-1229 overexpression plasmid, while
miR-1229 expression level was decreased in the HT-29 and HCT-16
cell lines following transfection with miR-1229 knockdown plasmid,
indicating a successful transfection (all P<0.01) (Fig. S4A and B). However, neither
miR-1229 overexpression nor knockdown affected circ_0001666
expression (both P>0.05) (Fig. 4B
and D). Furthermore, the results from a luciferase reporter
assay and FISH suggested that miR-1229 could bind to circ_0001666
(Figs. 4E and F, and S5). These data suggested that
circ_0001666 negatively regulated miR-1229 expression level;
however, miR-1229 expression did not affect the expression level of
circ_0001666 in the CRC cell lines.
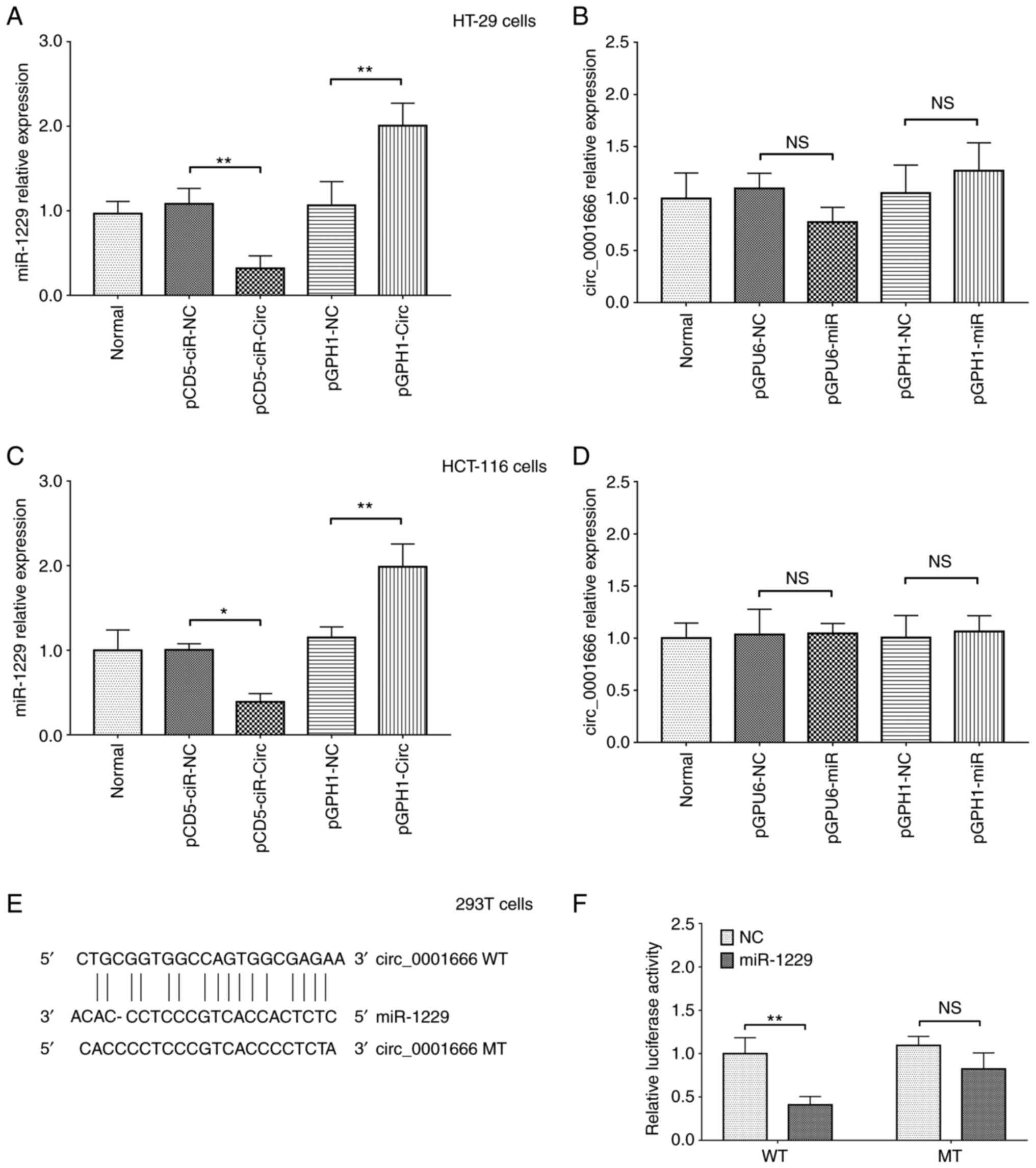 | Figure 4.Regulatory mechanism between
circ_0001666 and miR-1229. miR-1229 mRNA expression levels in the
(A) HT-29 and (C) HCT-116 cell lines following circ_0001666
overexpression and knockdown. circ_0001666 mRNA expression level in
the (B) HT-29 and (D) HCT-16 cell lines following miR-1229
overexpression and knockdown. (E) Binding sequences between
circ_0001666 and mir-1229. (F) Effect of miR-1229 on the luciferase
activity of circ_0001666 WT and circ_0001666 MT in the 293T cell
line. *P<0.05, **P<0.01. NS, not significant; WT, wild-type;
MT, mutant; circ, circular RNA; NC, negative control; miR,
microRNA. pCD5-ciR-Circ represented circ_0001666 overexpression
plasmids, pCD5-ciR-NC represented overexpression NC plasmids.
pGPH1-Circ represented circ_0001666 knockdown plasmids, pGPH1-miR
represented miR-1229 knockdown plasmids, pGPH1-NC represented
knockdown NC plasmids. pGPU6-miR represented miR-1229
overexpression plasmids, pGPU6-NC represented overexpression NC
plasmids. |
Analysis of circ_0001666, miR-1229,
CXCR5 and the Wnt/β-catenin pathway in rescue experiments
To determine whether circ_0001666 regulated CRC cell
function by targeting miR-1229, rescue experiments were performed.
The transfection efficiencies of each plasmid in these experiments
are shown in Fig. 5A and B.
Western blot analysis indicated that, in the HT-29 cell line,
miR-1229 overexpression reduced p-GSK-3β protein expression levels
and increased those of β-catenin. In addition, it also attenuated
the effect of circ_0001666 overexpression on the Wnt/β-catenin
pathway (Fig. 5C and D). These
data suggested that circ_0001666 regulated the Wnt/β-catenin
pathway by targeting miR-1229 in CRC cells.
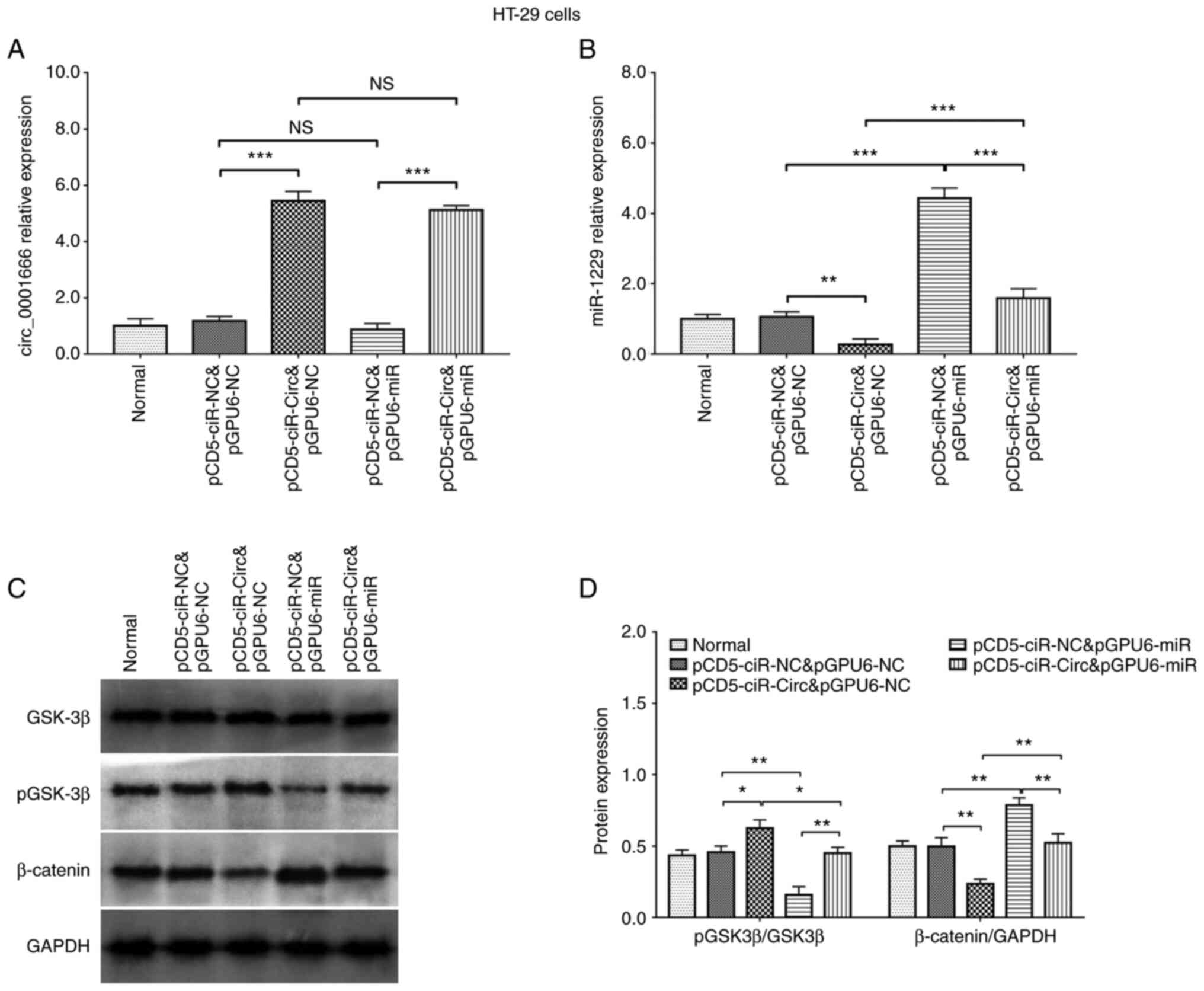 | Figure 5.Analysis of circ_0001666 and miR-1229
expression, and the Wnt/β-catenin pathway in rescue experiments.
(A) circ_0001666 and (B) miR-1229 mRNA expression level and (C and
D) protein expression level of proteins in the Wnt/β-catenin
signaling in the HT-29 cells line in rescue experiments.
*P<0.05, **P<0.01, ***P<0.001. NS, not significant; p,
phosphorylated; miR, microRNA; circ, circular RNA; NC, negative
control. pCD5-ciR-Circ represented circ_0001666 overexpression
plasmids, pCD5-ciR-NC represented overexpression NC plasmids.
pGPU6-miR represented miR-1229 overexpression plasmids, pGPU6-NC
represented overexpression NC plasmids. |
To investigate the effect of circ_0001666 and
miR-1229 on CXCR5, the mRNA and protein expression level of CXCR5
in rescue experiments was determined. In the HT-29 cell line,
circ_0001666 suppressed CXCR5 mRNA and protein expression level,
while miR-1229 increased CXCR5 mRNA and protein expression level.
In addition, miR-1229 attenuated the effect of circ_0001666
overexpression on regulating CXCR5 mRNA and protein expression
level (Fig. S6). This data
suggested that circ_0001666 regulated the expression level of
miR-1229, which was mediated by CXCR5 in the CRC cell lines.
Cell proliferation, apoptosis,
invasion and stemness in rescue experiments
In the HT-29 cell line, miR-1229 overexpression
promoted cell proliferation at 48 (P<0.05) and 72 h (P<0.01),
the numbers of invading cells (P<0.01), and the number of
CD133+ cells (P<0.01). By contrast, miR-1229
overexpression inhibited apoptosis, as evidenced by the number of
TUNEL-positive cells (P<0.01; Fig.
6). In addition, miR-1229 overexpression abolished the effect
of circ_0001666 overexpression on CRC cell proliferation,
apoptosis, invasion and stemness (Fig.
6). These data suggested that circ_0001666 regulated CRC cell
malignancy by targeting miR-1229.
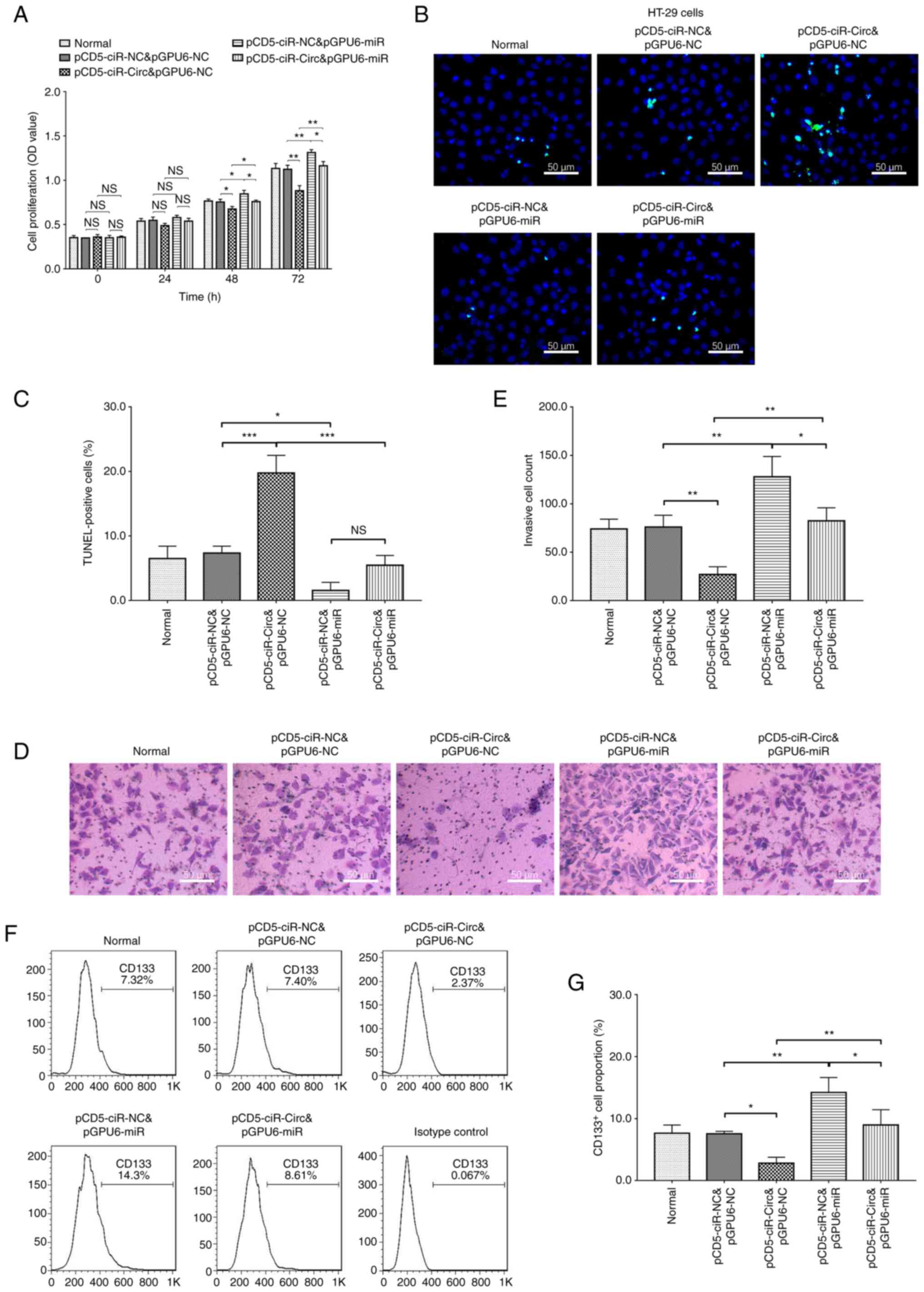 | Figure 6.Cell proliferation, apoptosis,
invasion and stemness in rescue experiments. (A) Cell
proliferation, (B and C) apoptosis and (D and E) invasion in rescue
experiments in the HT-29 cell line. (F and G) Number of
CD133+ cells in rescue experiments in the HT-29 cell
line. *P<0.05, **P<0.01, ***P<0.001. NS, not significant;
miR, microRNA; circ, circular RNA; NC, negative control.
pCD5-ciR-Circ represented circ_0001666 overexpression plasmids,
pCD5-ciR-NC represented overexpression NC plasmids. pGPU6-miR
represented miR-1229 overexpression plasmids, pGPU6-NC represented
overexpression NC plasmids. |
Discussion
CRC places a huge medical burden across the world.
The majority of patients with CRC are diagnosed at an advanced
stage and miss the therapeutic window (2,3).
Even after aggressive management, multiple recurrence and
metastasis are often reported in patients with CRC resulting in an
unfavourable survival profile (4).
To further improve the CRC prognosis and individualize the
management options are still a critical issue for clinicians.
Understanding the role of circRNA tumorgenicity for
therapeutic intervention has attracted increased attention from
researchers. Previous studies have demonstrated that several
circRNA molecules were associated with CRC tumorigenesis by
modulating cell proliferation and migration. For example,
circ-HIPK3 suppressed miR-7 mRNA expression level to promote
proliferation and migration in CRC cells (9). In addition, circ_0026416 regulated
the mRNA expression level of miR-346 and nuclear factor IB to
promote proliferation and migration in CRC cells (8). By contrast, other circRNA molecules
may serve as tumor suppressors in CRC. For example, circ-SMARCA5
was found to suppresses the mRNA expression level of miR-39-3p,
which resulted in reduced CRC cell proliferation and migration
(7). Another circRNA, circ_103809,
served as a sponge for miR-532-3p, leading to inhibition of cell
proliferation and migration in CRC cells (10). Altogether, this indicates that
several circRNA molecules have been associated with CRC
tumorigenesis and may serve as potential therapeutic targets.
As a newly identified circRNA, few studies have
described the role of circ_0001666 in carcinogenesis, to the best
of our knowledge. An in vitro study suggested that
circ_0001666 regulated the expression levels of several miRNA
targets (including miR-330-5p, miR-193a-5p and miR-326) to promote
papillary thyroid carcinoma cell proliferation and migration
(11). In addition, another study
identified an immune-related risk signature of CRC, in which
circ_0001666 was a critical component of a circRNA/miRNA/hub gene
network (24). Furthermore,
according to a bioinformatics study, circ_0001666 and miR-1229 may
be involved in CRC pathogenesis (12). Despite these bioinformatics studies
and the finding that circ_0001666 is part of an important competing
endogenous RNA (ceRNA) network in CRC, the molecular mechanism
underlying the role of circ_0001666 in CRC carcinogenesis has not
been determined.
The results from the present in vitro study
suggested that circ_0001666 inhibited cell proliferation, invasion
and stemness in CRC. A possible reason to explain this result is
that circ_0001666 overexpression reduces β-catenin protein
expression, which, in turn, would increase the protein expression
levels of E-cadherin, a key protein in intercellular junctions
(25). Therefore, increased
E-cadherin protein expression would lead to tightening of
intercellular junctions, and eventually prevent local invasion and
metastasis of CRC cells (25). It
was also hypothesized that overexpression of circ_0001666 decreases
cancer stemness, as evidenced by the reduced numbers of
CD133+ cells in the present study; further experiments
are required to validate this. Therefore, circ_0001666
overexpression would lead to reduced tumor cell formation and
reduced tumor cell numbers.
A major role of circRNA molecules is to serve as
ceRNA, which inhibit target miRNA expression and downstream gene
regulation (26). miRNA is a type
of small non-coding RNA involved in multiple cellular processes,
including cell proliferation, differentiation and apoptosis
(27). Accumulating evidence
suggests that miRNA molecules are also involved in tumorigenesis,
including CRC pathogenesis. A previous study demonstrated that
miR-1229 was a target of circ_0001666, based on a circRNA/miRNA
network analysis of CRC (12). In
addition, miR-1229 promoted breast cancer cell proliferation and
tumor growth by activating the Wnt/β-catenin signaling pathway
(13). Furthermore, an in
vivo study into CRC suggested that miR-1229 regulated the
protein expression levels of HIPK2 and promoted angiogenesis,
highlighting its oncogenic role in CRC (21). In the present study, miR-1229
promoted CRC cell proliferation, invasion and stemness.
Wnt/β-catenin is a signaling pathway with a critical
role in cancer biology (10). Wnt
binds to the surface receptor Frizzled and its coreceptor
low-density lipoprotein receptor-related protein group 5/6, causing
β-catenin to dissociate from its degradation complex, leading to
transactivation of target genes (14). In addition to its function in
normal physiological conditions, Wnt/β-catenin also plays a
critical role in CRC pathogenesis by promoting cell proliferation,
modulating epithelial-to-mesenchymal transition, disrupting
intercellular junctions and regulating angiogenesis (14,15).
A previous study suggested that miR-1229 regulated the
Wnt/β-catenin signaling pathway to induce breast cancer cell
malignancy (13). In the present
study, circ_0001666 regulated the Wnt/β-catenin signaling pathway
by targeting miR-1229 in the CRC cell lines. This finding could be
explained as follows: miR-1229 overexpression upregulated p-GSK-3β
protein expression levels and p-GSK-3β targeted β-catenin to
regulate its activity (10). In
addition, circ_0001666 overexpression resulted in the regulation of
β-catenin protein expression. miR-1229 overexpression promoted cell
proliferation, invasion and stemness to a greater extent than
circ_0001666 overexpression in the CRC cell lines. These data
suggested that the circ_0001666/miR-1229/β-catenin signaling
pathway may represent a potential area for metastatic CRC
therapeutic intervention.
There are several limitations in the current study.
Firstly, circ_0001666 and miR-1229 mRNA expression levels were not
evaluated in human CRC samples. Secondly, RNA immunoprecipitation
experiment was not performed to determine the binding between
circ_0001666 and miR-1229 in the CRC cell lines.
In conclusion, circ_0001666 suppressed CRC cell
proliferation, invasion and stemness by targeting miR-1229 and the
Wnt/β-catenin signaling pathway, which may represent potential
targets for CRC treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by The Hunan Province Health Committee
of China (grant no. 20201427).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZY conceived and designed the study. FB and CZ
performed the experiments and data analysis. YO and KX contributed
to acquisition of data and interpretation of data. ZY and FB
confirm the authenticity of the raw data. ZH was involved in data
interpretation, and wrote the manuscript. All authors reviewed and
edited the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal carcinoma
|
|
circRNAs
|
circular RNAs
|
|
miRNAs
|
microRNAs
|
|
NC
|
negative control
|
|
CD133+
|
CD133 positive
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
WT
|
wild-type
|
|
MT
|
mutant type
|
|
ceRNA
|
competitive endogenous RNA
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Simon K: Colorectal cancer development and
advances in screening. Clin Interv Aging. 11:967–976. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nappi A, Berretta M, Romano C, Tafuto S,
Cassata A, Casaretti R, Silvestro L, Divitiis C, Alessandrini L,
Fiorica F, et al: Metastatic colorectal cancer: Role of target
therapies and future perspectives. Curr Cancer Drug Targets.
18:421–429. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Messersmith WA: NCCN guidelines updates:
Management of metastatic colorectal cancer. J Natl Compr Canc Netw.
17:599–601. 2019.PubMed/NCBI
|
|
5
|
Li Z, Ruan Y, Zhang H, Shen Y, Li T and
Xiao B: Tumor-suppressive circular RNAs: Mechanisms underlying
their suppression of tumor occurrence and use as therapeutic
targets. Cancer Sci. 110:3630–3638. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lei B, Tian Z, Fan W and Ni B: Circular
RNA: A novel biomarker and therapeutic target for human cancers.
Int J Med Sci. 16:292–301. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Miao X, Xi Z, Zhang Y, Li Z, Huang L, Xin
T, Shen R and Wang T: Circ-SMARCA5 suppresses colorectal cancer
progression via downregulating miR-39-3p and upregulating ARID4B.
Dig Liver Dis. 52:1494–1502. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liang Y, Shi J, He Q, Sun G, Gao L, Ye J,
Tang X and Qu H: Hsa_circ_0026416 promotes proliferation and
migration in colorectal cancer via miR-346/NFIB axis. Cancer Cell
Int. 20:4942020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T,
Sun H, Pan Y, He B and Wang S: CircHIPK3 promotes colorectal cancer
growth and metastasis by sponging miR-7. Cell Death Dis. 9:4172018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nusse R and Clevers H: Wnt/β-catenin
signaling, disease, and emerging therapeutic modalities. Cell.
169:985–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qi Y, He J, Zhang Y, Wang L, Yu Y, Yao B
and Tian Z: Circular RNA hsa_circ_0001666 sponges miR-330-5p,
miR-193a-5p and miR-326, and promotes papillary thyroid carcinoma
progression via upregulation of ETV4. Oncol Rep. 45:502021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Song W and Fu T: Circular RNA-associated
competing endogenous RNA network and prognostic nomogram for
patients with colorectal cancer. Front Oncol. 9:11812019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tan Z, Zheng H, Liu X, Zhang W, Zhu J, Wu
G, Cao L, Song J, Wu S, Song L and Li J: MicroRNA-1229
overexpression promotes cell proliferation and tumorigenicity and
activates Wnt/β-catenin signaling in breast cancer. Oncotarget.
7:24076–24087. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Taciak B, Pruszynska I, Kiraga L, Bialasek
M and Krol M: Wnt signaling pathway in development and cancer. J
Physiol Pharmacol. 69:85–196. 2018.PubMed/NCBI
|
|
15
|
Cheng X, Xu X, Chen D, Zhao F and Wang W:
Therapeutic potential of targeting the Wnt/β-catenin signaling
pathway in colorectal cancer. Biomed Pharmacother. 110:473–481.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li H, Xu JD, Fang XH, Zhu JN, Yang J, Pan
R, Yuan SJ, Zeng N, Yang ZZ, Yang H, et al: Circular RNA
circRNA_000203 aggravates cardiac hypertrophy via suppressing
miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc Res.
116:1323–1334. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Panda AC and Gorospe M: Detection and
analysis of circular RNAs by RT-PCR. Bio Protoc. 8:e27752018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kramer MF: Stem-loop RT-qPCR for miRNAs.
Curr Protoc Mol Biol. Chapter 15: Unit 15 10. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu HY, Yu CH, Zhang HH, Zhang SZ, Yu WY,
Yang Y and Chen Q: Exosomal miR-1229 derived from colorectal cancer
cells promotes angiogenesis by targeting HIPK2. Int J Biol
Macromol. 132:470–477. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Flynn J and Gorry P: Flow cytometry
analysis to identify human CD4+ T cell subsets. Methods
Mol Biol. 2048:15–25. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu Y and Wu PY: CD133 as a marker for
cancer stem cells: Progresses and concerns. Stem Cells Dev.
18:1127–1134. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Song W, Ren J, Wang C, Ge Y and Fu T:
Analysis of circular RNA-related competing endogenous RNA
identifies the immune-related risk signature for colorectal cancer.
Front Genet. 11:5052020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kourtidis A, Lu R, Pence LJ and
Anastasiadis PZ: A central role for cadherin signaling in cancer.
Exp Cell Res. 358:78–85. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Panda AC: Circular RNAs Act as miRNA
Sponges. Adv Exp Med Biol. 1087:67–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saliminejad K, Khorram Khorshid HR,
Soleymani Fard S and Ghaffari SH: An overview of microRNAs:
Biology, functions, therapeutics, and analysis methods. J Cell
Physiol. 234:5451–5465. 2019. View Article : Google Scholar : PubMed/NCBI
|