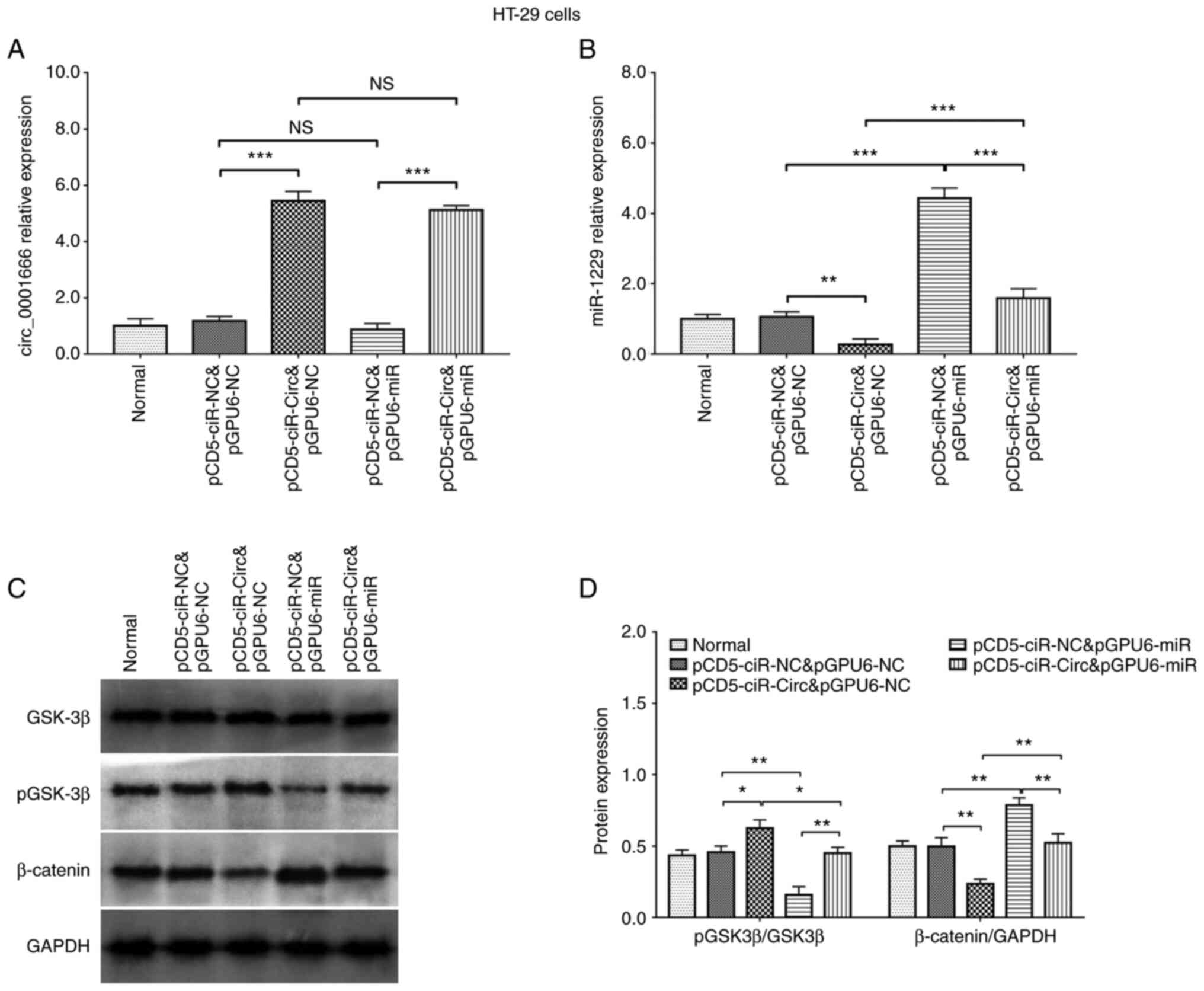
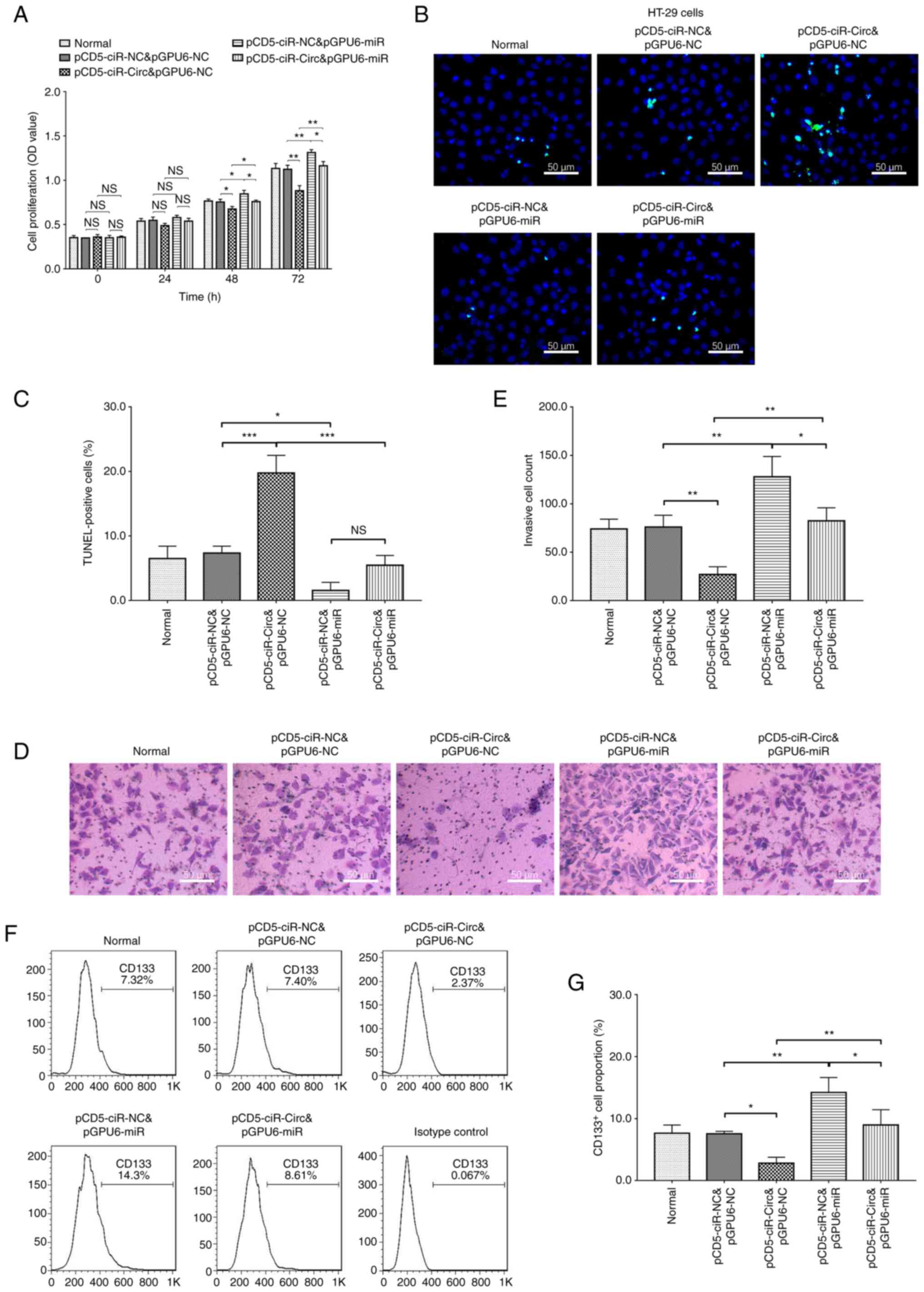
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Simon K: Colorectal cancer development and
advances in screening. Clin Interv Aging. 11:967–976. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nappi A, Berretta M, Romano C, Tafuto S,
Cassata A, Casaretti R, Silvestro L, Divitiis C, Alessandrini L,
Fiorica F, et al: Metastatic colorectal cancer: Role of target
therapies and future perspectives. Curr Cancer Drug Targets.
18:421–429. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Messersmith WA: NCCN guidelines updates:
Management of metastatic colorectal cancer. J Natl Compr Canc Netw.
17:599–601. 2019.PubMed/NCBI
|
|
5
|
Li Z, Ruan Y, Zhang H, Shen Y, Li T and
Xiao B: Tumor-suppressive circular RNAs: Mechanisms underlying
their suppression of tumor occurrence and use as therapeutic
targets. Cancer Sci. 110:3630–3638. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lei B, Tian Z, Fan W and Ni B: Circular
RNA: A novel biomarker and therapeutic target for human cancers.
Int J Med Sci. 16:292–301. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Miao X, Xi Z, Zhang Y, Li Z, Huang L, Xin
T, Shen R and Wang T: Circ-SMARCA5 suppresses colorectal cancer
progression via downregulating miR-39-3p and upregulating ARID4B.
Dig Liver Dis. 52:1494–1502. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liang Y, Shi J, He Q, Sun G, Gao L, Ye J,
Tang X and Qu H: Hsa_circ_0026416 promotes proliferation and
migration in colorectal cancer via miR-346/NFIB axis. Cancer Cell
Int. 20:4942020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T,
Sun H, Pan Y, He B and Wang S: CircHIPK3 promotes colorectal cancer
growth and metastasis by sponging miR-7. Cell Death Dis. 9:4172018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nusse R and Clevers H: Wnt/β-catenin
signaling, disease, and emerging therapeutic modalities. Cell.
169:985–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qi Y, He J, Zhang Y, Wang L, Yu Y, Yao B
and Tian Z: Circular RNA hsa_circ_0001666 sponges miR-330-5p,
miR-193a-5p and miR-326, and promotes papillary thyroid carcinoma
progression via upregulation of ETV4. Oncol Rep. 45:502021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Song W and Fu T: Circular RNA-associated
competing endogenous RNA network and prognostic nomogram for
patients with colorectal cancer. Front Oncol. 9:11812019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tan Z, Zheng H, Liu X, Zhang W, Zhu J, Wu
G, Cao L, Song J, Wu S, Song L and Li J: MicroRNA-1229
overexpression promotes cell proliferation and tumorigenicity and
activates Wnt/β-catenin signaling in breast cancer. Oncotarget.
7:24076–24087. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Taciak B, Pruszynska I, Kiraga L, Bialasek
M and Krol M: Wnt signaling pathway in development and cancer. J
Physiol Pharmacol. 69:85–196. 2018.PubMed/NCBI
|
|
15
|
Cheng X, Xu X, Chen D, Zhao F and Wang W:
Therapeutic potential of targeting the Wnt/β-catenin signaling
pathway in colorectal cancer. Biomed Pharmacother. 110:473–481.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li H, Xu JD, Fang XH, Zhu JN, Yang J, Pan
R, Yuan SJ, Zeng N, Yang ZZ, Yang H, et al: Circular RNA
circRNA_000203 aggravates cardiac hypertrophy via suppressing
miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc Res.
116:1323–1334. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Panda AC and Gorospe M: Detection and
analysis of circular RNAs by RT-PCR. Bio Protoc. 8:e27752018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kramer MF: Stem-loop RT-qPCR for miRNAs.
Curr Protoc Mol Biol. Chapter 15: Unit 15 10. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu HY, Yu CH, Zhang HH, Zhang SZ, Yu WY,
Yang Y and Chen Q: Exosomal miR-1229 derived from colorectal cancer
cells promotes angiogenesis by targeting HIPK2. Int J Biol
Macromol. 132:470–477. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Flynn J and Gorry P: Flow cytometry
analysis to identify human CD4+ T cell subsets. Methods
Mol Biol. 2048:15–25. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu Y and Wu PY: CD133 as a marker for
cancer stem cells: Progresses and concerns. Stem Cells Dev.
18:1127–1134. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Song W, Ren J, Wang C, Ge Y and Fu T:
Analysis of circular RNA-related competing endogenous RNA
identifies the immune-related risk signature for colorectal cancer.
Front Genet. 11:5052020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kourtidis A, Lu R, Pence LJ and
Anastasiadis PZ: A central role for cadherin signaling in cancer.
Exp Cell Res. 358:78–85. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Panda AC: Circular RNAs Act as miRNA
Sponges. Adv Exp Med Biol. 1087:67–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saliminejad K, Khorram Khorshid HR,
Soleymani Fard S and Ghaffari SH: An overview of microRNAs:
Biology, functions, therapeutics, and analysis methods. J Cell
Physiol. 234:5451–5465. 2019. View Article : Google Scholar : PubMed/NCBI
|