|
1
|
Fares J, Fares MY, Khachfe HH, Salhab HA
and Fares Y: Molecular principles of metastasis: A hallmark of
cancer revisited. Signal Transduct Target Ther. 5:282020.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weber J, Braun CJ, Saur D and Rad R: In
vivo functional screening for systems-level integrative cancer
genomics. Nat Rev Cancer. 20:573–593. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lin A and Sheltzer JM: Discovering and
validating cancer genetic dependencies: Approaches and pitfalls.
Nat Rev Genet. 21:671–682. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Allam M, Cai S and Coskun AF: Multiplex
bioimaging of single-cell spatial profiles for precision cancer
diagnostics and therapeutics. NPJ Precis Oncol. 4:112020.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Noble ME, Endicott JA and Johnson LN:
Protein kinase inhibitors: Insights into drug design from
structure. Science. 303:1800–1805. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Reubold TF and Eschenburg S: A molecular
view on signal transduction by the apoptosome. Cell Signal.
24:1420–1425. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Janse van Rensburg HJ and Yang X: The
roles of the Hippo pathway in cancer metastasis. Cell Signal.
28:1761–1772. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lytle JR, Yario TA and Steitz JA: Target
mRNAs are repressed as efficiently by microRNA-binding sites in the
5′UTR as in the 3′UTR. Proc Natl Acad Sci USA. 104:9667–9672. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Khraiwesh B, Arif MA, Seumel GI, Ossowski
S, Weigel D, Reski R and Frank W: Transcriptional control of gene
expression by microRNAs. Cell. 140:111–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cabili MN, Dunagin MC, McClanahan PD,
Biaesch A, Padovan-Merhar O, Regev A, Rinn JL and Raj A:
Localization and abundance analysis of human lncRNAs at single-cell
and single-molecule resolution. Genome Biol. 16:202015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chiu HS, Somvanshi S, Patel E, Chen TW,
Singh VP, Zorman B, Patil SL, Pan Y, Chatterjee SS; Cancer Genome
Atlas Research Network, ; et al: Pan-cancer analysis of lncRNA
regulation supports their targeting of cancer genes in each tumor
context. Cell Rep. 23:297–312.e12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shibata H, Oishi K, Yamagiwa A, Matsumoto
M, Mukai H and Ono Y: PKNbeta interacts with the SH3 domains of
Graf and a novel Graf related protein, Graf2, which are GTPase
activating proteins for Rho family. J Biochem. 130:23–31. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ishizaki T, Maekawa M, Fujisawa K, Okawa
K, Iwamatsu A, Fujita A, Watanabe N, Saito Y, Kakizuka A, Morii N
and Narumiya S: The small GTP-binding protein Rho binds to and
activates a 160 kDa Ser/Thr protein kinase homologous to myotonic
dystrophy kinase. EMBO J. 15:1885–1893. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Leung T, Manser E, Tan L and Lim L: A
novel serine/threonine kinase binding the Ras-related RhoA GTPase
which translocates the kinase to peripheral membranes. J Biol Chem.
270:29051–29054. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang X, Sun D, Tai J, Chen S, Yu M, Ren D
and Wang L: TFAP2C promotes stemness and chemotherapeutic
resistance in colorectal cancer via inactivating hippo signaling
pathway. J Exp Clin Cancer Res. 37:272018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
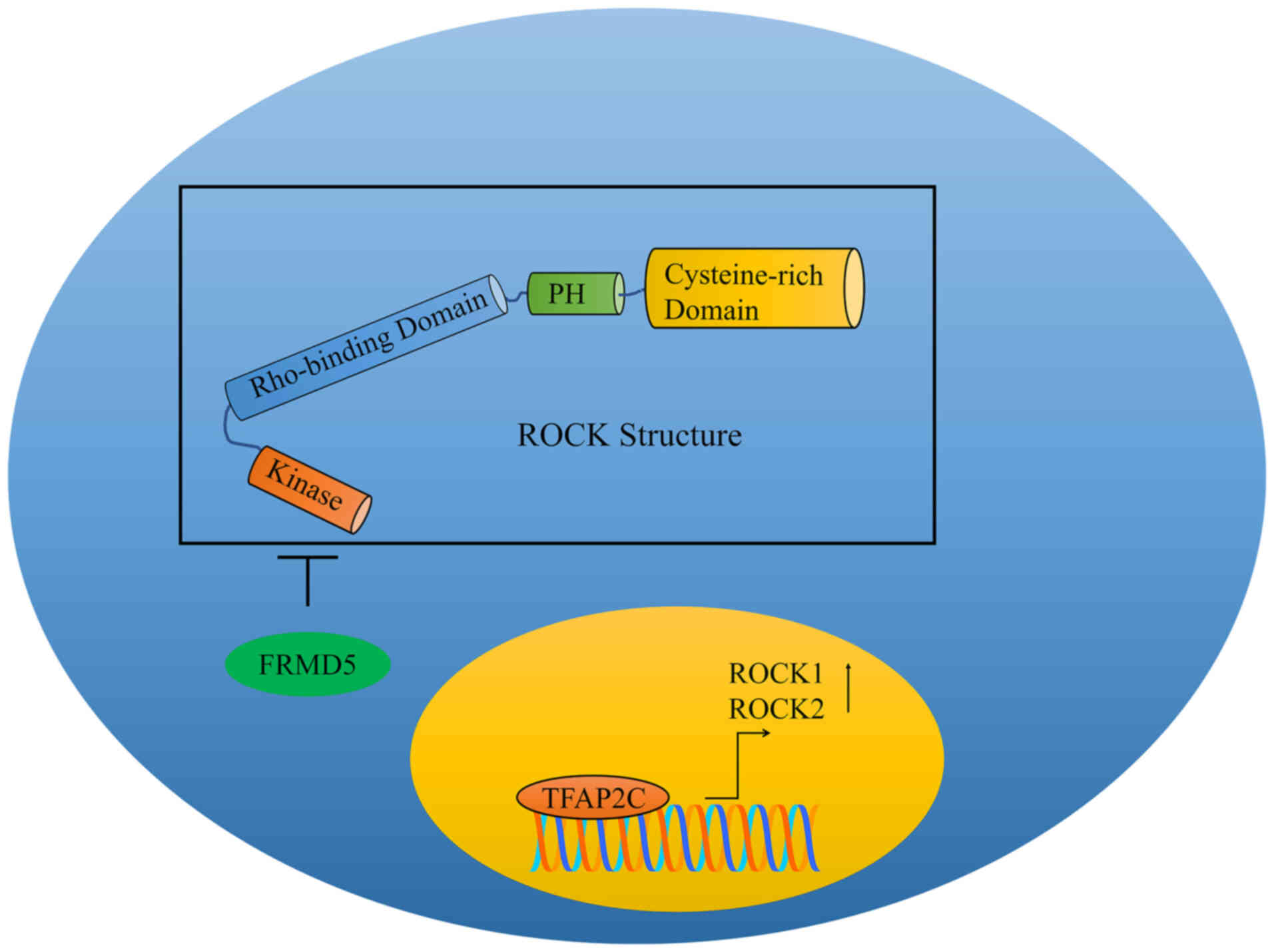
Hu J, Niu M, Li X, Lu D, Cui J, Xu W, Li
G, Zhan J and Zhang H: FERM domain-containing protein FRMD5
regulates cell motility via binding to integrin β5 subunit and
ROCK1. FEBS Lett. 588:4348–4356. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Deng X, Yi X, Deng J, Zou Y, Wang S, Shan
W, Liu P, Zhang Z, Chen L and Hao L: ROCK2 promotes osteosarcoma
growth and metastasis by modifying PFKFB3 ubiquitination and
degradation. Exp Cell Res. 385:1116892019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang D, Du X, Yuan R, Chen L, Liu T, Wen
C, Huang M, Li M, Hao L and Shao J: Rock2 promotes the invasion and
metastasis of hepatocellular carcinoma by modifying MMP2
ubiquitination and degradation. Biochem Biophys Res Commun.
453:49–56. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang X, Zhang L, Du Y, Zheng H, Zhang P,
Sun Y, Wang Y, Chen J, Ding P, Wang N, et al: A novel FOXM1
isoform, FOXM1D, promotes epithelial-mesenchymal transition and
metastasis through ROCKs activation in colorectal cancer. Oncogene.
36:807–819. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhong Y, Yang S, Wang W, Wei P, He S, Ma
H, Yang J, Wang Q, Cao L, Xiong W, et al: The interaction of
Lin28A/Rho associated coiled-coil containing protein kinase2
accelerates the malignancy of ovarian cancer. Oncogene.
38:1381–1397. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hsu PW, Huang HD, Hsu SD, Lin LZ, Tsou AP,
Tseng CP, Stadler PF, Washietl S and Hofacker IL: miRNAMap: Genomic
maps of microRNA genes and their target genes in mammalian genomes.
Nucleic Acids Res. 34:(Database Issue). D135–D139. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Du Z, Sun T, Hacisuleyman E, Fei T, Wang
X, Brown M, Rinn JL, Lee MG, Chen Y, Kantoff PW and Liu XS:
Integrative analyses reveal a long noncoding RNA-mediated sponge
regulatory network in prostate cancer. Nat Commun. 7:109822016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kumar MS, Lu J, Mercer KL, Golub TR and
Jacks T: Impaired microRNA processing enhances cellular
transformation and tumorigenesis. Nat Genet. 39:673–677. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Treiber T, Treiber N and Meister G:
Regulation of microRNA biogenesis and its crosstalk with other
cellular pathways. Nat Rev Mol Cell Biol. 20:5–20. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nair L, Chung H and Basu U: Regulation of
long non-coding RNAs and genome dynamics by the RNA surveillance
machinery. Nat Rev Mol Cell Biol. 21:123–136. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Geisler S and Coller J: RNA in unexpected
places: Long non-coding RNA functions in diverse cellular contexts.
Nat Rev Mol Cell Biol. 14:699–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Santer L, Bär C and Thum T: Circular RNAs:
A novel class of functional RNA molecules with a therapeutic
perspective. Mol Ther. 27:1350–1363. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
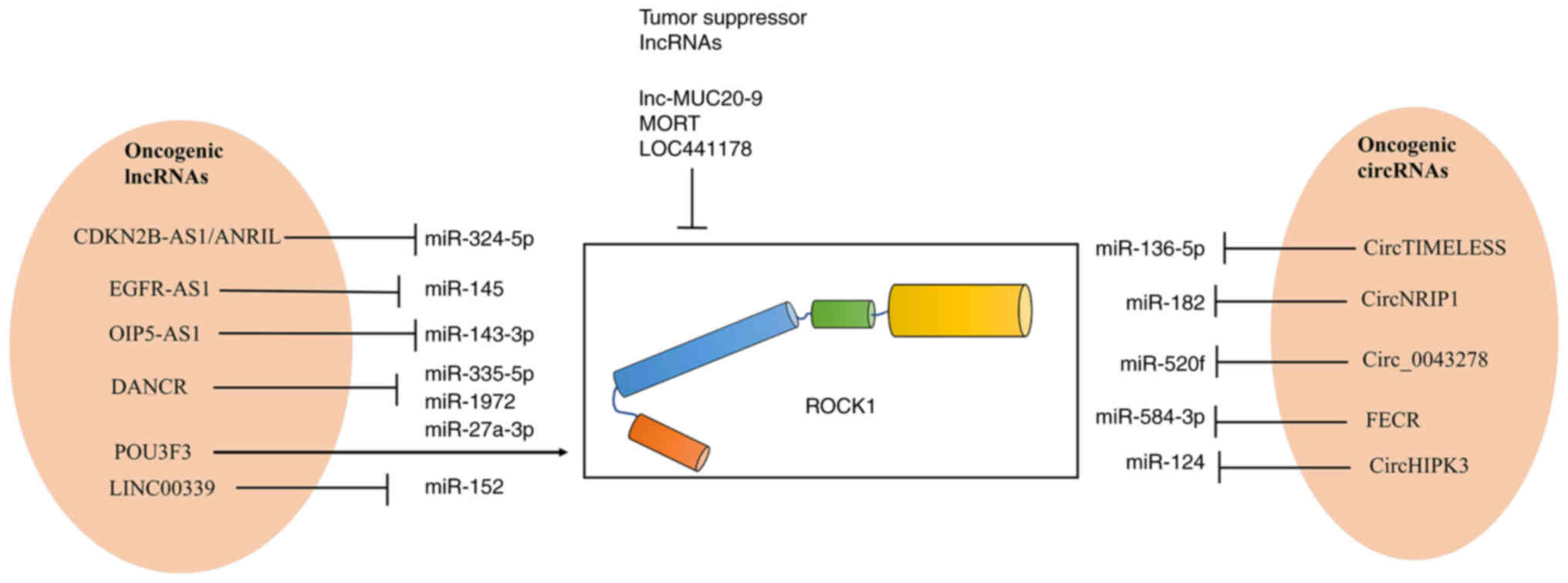
Liu F, Xiao Y, Ma L and Wang J: Regulating
of cell cycle progression by the lncRNA CDKN2B-AS1/miR-324-5p/ROCK1
axis in laryngeal squamous cell cancer. Int J Biol Markers.
35:47–56. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Feng Z, Li X, Qiu M, Luo R, Lin J and Liu
B: LncRNA EGFR-AS1 Upregulates ROCK1 by sponging miR-145 to promote
esophageal squamous cell carcinoma cell invasion and migration.
Cancer Biother Radiopharm. 35:66–71. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Song L, Wang L, Pan X and Yang C: lncRNA
OIP5-AS1 targets ROCK1 to promote cell proliferation and inhibit
cell apoptosis through a mechanism involving miR-143-3p in cervical
cancer. Braz J Med Biol Res. 53:e88832020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liang H, Zhang C, Guan H, Liu J and Cui Y:
LncRNA DANCR promotes cervical cancer progression by upregulating
ROCK1 via sponging miR-335-5p. J Cell Physiol. 234:7266–7278. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang Y, Zeng X, Wang N, Zhao W, Zhang X,
Teng S, Zhang Y and Lu Z: Long noncoding RNA DANCR, working as a
competitive endogenous RNA, promotes ROCK1-mediated proliferation
and metastasis via decoying of miR-335-5p and miR-1972 in
osteosarcoma. Mol Cancer. 17:892018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Guo D, Li Y, Chen Y, Zhang D, Wang X, Lu
G, Ren M, Lu X and He S: DANCR promotes HCC progression and
regulates EMT by sponging miR-27a-3p via ROCK1/LIMK1/COFILIN1
pathway. Cell Prolif. 52:e126282019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wan X, Xiang J, Zhang Q and Bian C: Long
noncoding RNA POU3F3 promotes cancer cell proliferation in prostate
carcinoma by upregulating rho-associated protein kinase 1. J Cell
Biochem. Nov 26–2018.(Epub ahead of print). doi:
10.1002/jcb.28101.
|
|
43
|
Dong P, Xiong Y, Yue J, Xu D, Ihira K,
Konno Y, Kobayashi N, Todo Y and Watari H: Long noncoding RNA NEAT1
drives aggressive endometrial cancer progression via
miR-361-regulated networks involving STAT3 and tumor
microenvironment-related genes. J Exp Clin Cancer Res. 38:2952019.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen K and Zhang L: LINC00339 regulates
ROCK1 by miR-152 to promote cell proliferation and migration in
hepatocellular carcinoma. J Cell Biochem. 120:14431–14443. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Deng R, Zhang J and Chen J: lncRNA SNHG1
negatively regulates miRNA-101-3p to enhance the expression of
ROCK1 and promote cell proliferation, migration and invasion in
osteosarcoma. Int J Mol Med. 43:1157–1166. 2019.PubMed/NCBI
|
|
46
|
Zhuang S, Liu F and Wu P: Upregulation of
long noncoding RNA TUG1 contributes to the development of
laryngocarcinoma by targeting miR-145-5p/ROCK1 axis. J Cell
Biochem. 120:13392–13402. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen S, Liu Z, Lu S and Hu B: EPEL
promotes the migration and invasion of osteosarcoma cells by
upregulating ROCK1. Oncol Lett. 17:3133–3140. 2019.PubMed/NCBI
|
|
48
|
Hu M, Han Y, Zhang Y, Zhou Y and Ye L:
lncRNA TINCR sponges miR-214-5p to upregulate ROCK1 in
hepatocellular carcinoma. BMC Med Genet. 21:22020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yang J, Wang WG and Zhang KQ: LINC00452
promotes ovarian carcinogenesis through increasing ROCK1 by
sponging miR-501-3p and suppressing ubiquitin-mediated degradation.
Aging (Albany NY). 12:21129–21146. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
She JK, Fu DN, Zhen D, Gong GH and Zhang
B: LINC01087 is highly expressed in breast cancer and regulates the
malignant behavior of cancer cells through miR-335-5p/Rock1. Onco
Targets Ther. 13:9771–9783. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yang H and Wang Z and Wang Z: Long
noncoding RNA KCNMB2-AS1 increases ROCK1 expression by sponging
microRNA-374a-3p to facilitate the progression of non-small-cell
lung cancer. Cancer Manag Res. 12:12679–12695. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen X, Li D, Chen L, Hao B, Gao Y, Li L,
Zhou C, He X and Cao Y: Long noncoding RNA LINC00346 promotes
glioma cell migration, invasion and proliferation by up-regulating
ROCK1. J Cell Mol Med. 24:13010–13019. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang J, He Z, Xu J, Chen P and Jiang J:
Long noncoding RNA LINC00941 promotes pancreatic cancer progression
by competitively binding miR-335-5p to regulate ROCK1-mediated
LIMK1/Cofilin-1 signaling. Cell Death Dis. 12:362021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Dai R, Zhou Y, Chen Z, Zou Z, Pan Z, Liu P
and Gao X: Lnc-MUC20-9 binds to ROCK1 and functions as a tumor
suppressor in bladder cancer. J Cell Biochem. 121:4214–4225. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Jin Z, Jiang S, Jian S and Shang Z: Long
noncoding RNA MORT overexpression inhibits cancer cell
proliferation in oral squamous cell carcinoma by downregulating
ROCK1. J Cell Biochem. Feb 25–2019.(Epub ahead of print). doi:
10.1002/jcb.28449. View Article : Google Scholar
|
|
56
|
Xu K, Tian H, Zhao S, Yuan D, Jiang L, Liu
X, Zou B and Zhang J: Long Noncoding RNA LOC441178 reduces the
invasion and migration of squamous carcinoma cells by targeting
ROCK1. Biomed Res Int. 2018:43576472018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hu J, Wang L, Zhao W, Huang Y, Wang Z and
Shen H: mi-R4435-2HG promotes proliferation and inhibits apoptosis
of cancer cells in ovarian carcinoma by upregulating ROCK2. Oncol
Lett. 19:1305–1309. 2020.PubMed/NCBI
|
|
58
|
Liu X, Li Y, Wen J, Qi T and Wang Y: Long
non-coding RNA TTN-AS1 promotes tumorigenesis of ovarian cancer
through modulating the miR-139-5p/ROCK2 axis. Biomed Pharmacother.
125:1098822020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liu J, Zhu Y and Ge C: LncRNA ZFAS1
promotes pancreatic adenocarcinoma metastasis via the RHOA/ROCK2
pathway by sponging miR-3924. Cancer Cell Int. 20:2492020.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yuan S, Luan X, Chen H, Shi X and Zhang X:
Long non-coding RNA EGFR-AS1 sponges micorRNA-381 to upregulate
ROCK2 in bladder cancer. Oncol Lett. 19:1899–1905. 2020.PubMed/NCBI
|
|
61
|
Yuan S, Luan X, Han G, Guo K, Wang S and
Zhang X: LINC01638 lncRNA mediates the postoperative distant
recurrence of bladder cancer by upregulating ROCK2. Oncol Lett.
18:5392–5398. 2019.PubMed/NCBI
|
|
62
|
Fang G, Wang J, Sun X, Xu R, Zhao X, Shao
L, Sun C and Wang Y: LncRNA MAGI2-AS3 is downregulated in the
distant recurrence of hepatocellular carcinoma after surgical
resection and affects migration and invasion via ROCK2. Ann
Hepatol. 19:535–540. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jiang L, He Y, Shen G, Ni J, Xia Z, Liu H,
Cao Y and Li X: lncRNA HAND2-AS1 mediates the downregulation of
ROCK2 in hepatocellular carcinoma and inhibits cancer cell
proliferation, migration and invasion. Mol Med Rep. 21:1304–1309.
2020.PubMed/NCBI
|
|
64
|
Zhang W, Shi J, Cheng C and Wang H:
CircTIMELESS regulates the proliferation and invasion of lung
squamous cell carcinoma cells via the miR-136-5p/ROCK1 axis. J Cell
Physiol. 235:5962–5971. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liang L and Li L: Down-regulation of
circNRIP1 promotes the apoptosis and inhibits the migration and
invasion of gastric cancer cells by miR-182/ROCK1 Axis. Onco
Targets Ther. 13:6279–6288. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Cui J, Li W, Liu G, Chen X, Gao X, Lu H
and Lin D: A novel circular RNA, hsa_circ_0043278, acts as a
potential biomarker and promotes non-small cell lung cancer cell
proliferation and migration by regulating miR-520f. Artif Cells
Nanomed Biotechnol. 47:810–821. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Li L, Li W, Chen N, Zhao H, Xu G, Zhao Y,
Pan X, Zhang X, Zhou L, Yu D, et al: FLI1 Exonic circular RNAs as a
novel oncogenic driver to promote tumor metastasis in small cell
lung cancer. Clin Cancer Res. 25:1302–1317. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kai D, Yannian L, Yitian C, Dinghao G, Xin
Z and Wu J: Circular RNA HIPK3 promotes gallbladder cancer cell
growth by sponging microRNA-124. Biochem Biophys Res Commun.
503:863–869. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yin D, Wei G, Yang F and Sun X: Circular
RNA has circ 0001591 promoted cell proliferation and metastasis of
human melanoma via ROCK1/PI3K/AKT by targeting miR-431-5p. Hum Exp
Toxicol. 40:310–324. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang T, Zhang L, Han D, Tursun K and Lu
X: Circular RNA hsa_Circ_101141 as a competing endogenous RNA
facilitates Tumorigenesis of hepatocellular carcinoma by regulating
miR-1297/ROCK1 pathway. Cell Transplant. 29:9636897209480162020.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Peng L, Sang H, Wei S, Li Y, Jin D, Zhu X,
Li X, Dang Y and Zhang G: circCUL2 regulates gastric cancer
malignant transformation and cisplatin resistance by modulating
autophagy activation via miR-142-3p/ROCK2. Mol Cancer. 19:1562020.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang D, Jiang X, Liu Y, Cao G, Zhang X and
Kuang Y: Circular RNA circ_HN1 facilitates gastric cancer
progression through modulation of the miR-302b-3p/ROCK2 axis. Mol
Cell Biochem. 476:199–212. 2021. View Article : Google Scholar : PubMed/NCBI
|