Introduction
As the most frequently diagnosed type of cancer,
lung cancer has the highest cancer-related incidence and mortality
worldwide (1). It is reported that
there were 220,000 newly diagnosed lung cancer cases and
>140,000 mortalities due to lung cancer in USA in 2019 (2). Lung adenocarcinoma (LUAD), which
accounts for >40% of lung cancer cases, is the most common
histological subtype of lung cancer (3). Although significant progress has been
made in the diagnostic and treatment methods in recent years, the
average 5-year relative survival rate among patients with lung
cancer is only ~18% (4). Thus, it
is important to explore the mechanism of LUAD and to identify an
optimal therapy to improve the treatment of LUAD.
As a member of SEC61 translocon, SEC61 translocon
subunit γ (SEC61G) consists of three subunits in mammals, namely
Sec61α, Sec61β and Sec61γ (5). The
SEC61 complex, which serves as the core component of the protein
translocation apparatus of the endoplasmic reticulum (ER) membrane
(6), is involved in protein
folding, modification and translocation and in the unfolded protein
response, particularly under conditions of hypoxia and nutrient
deprivation in the tumor microenvironment (7,8).
SEC61 serves a critical role in numerous types of cancer. For
example, SEC61 is overexpressed in hepatocellular carcinoma and its
silencing can suppress cell proliferation and induce apoptosis
(9). Meng et al (10) report that SEC61 is highly expressed
in human kidney tumor tissues, whereas its knockdown exerts
inhibitory effects on cell proliferation, migration and invasion in
kidney cancer. According to the UALCAN database, SEC61 is
upregulated in LUAD and SEC61 upregulation is associated with poor
prognosis of LUAD patients, which suggests that SEC61G may be
involved in the progression of LUAD. However, the biological roles
and special mechanism of SEC61 in LUAD remains to be
elucidated.
Cyclic AMP-responsive element-binding protein 3
(CREB3), also named LZIP or LUMAN, is an ER stress-related protein
that is considered to be a transcriptional coregulator (10). According to the Biological General
Repository for Interaction Datasets (BioGRID) database (https://thebiogrid.org/), SEC61G, a membrane
transporter on the ER, may interact with multiple proteins.
Silencing the expression of CREB3, a central protein in ER stress,
is able to induce ER stress and cell apoptosis (11). Thus, it was hypothesized that
SEC61G may interact with CREB3 to induce ER stress. In addition,
since CREB3 had been found to be increased in LUAD, it was inferred
that SEC61G could participate in ER stress by interacting with
CREB3 to promote the malignant progression of LUAD.
Materials and methods
Bioinformatic analysis
The Gene Expression Profiling Interactive Analysis
(GEPIA) database (http://gepia.cancer-pku.cn) version 1.0 was used to
analyze the mRNA expression of SEC61G and CREB3 in LUAD samples
from The Cancer Genome Atlas (TCGA) database (https://www.tcga.org/). The BioGRID database
(https://thebiogrid.org/) version 4.4 was used to
predict potential proteins that interact with SEC61G. In addition,
the expression of CREB3 in LUAD was analyzed by the UALCAN database
(http://ualcan.path.uab.edu).
Cell culture, treatment and
transfection
Human LUAD cell line A549 was from the Cell Bank of
the Chinese Academy of Sciences. The cells were cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml
streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C
in a humidified incubator with 5% CO2. Subsequently,
4-phenylbutyric acid (4-PBA; 5 mmol/l), an inhibitor of ER stress,
was used to treat the A549 cells.
For transfection, small interfering RNA
(siRNA)-negative control (si-NC), overexpression plasmid (Oe)-NC,
si-SEC61G-1/2 and Oe-CREB3 at a concentration of 20 µM were
obtained from Shanghai GenePharma Co., Ltd.
Lipofectamine® 2000 transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) was used for transfection. Cells
were incubated with 5% CO2 at 37°C for 8 h and were used
in subsequent experiments 48 h post-transfection. The sequences of
siRNAs were: si-SEC61G-1: 5′-GCAAUAGGAUUUGCUAUAAUG-3′; si-SEC61G-2:
5′-CUUAGAGAUUGGUGAACAAGU-3′; si-NC: 5′-UUCUCCGAACGUGUCACGU-3′.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from A549 cells
(5×106 cells) using TRIzol® reagent (Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. Synthesis of complementary DNA was conducted with
PrimeScript RT Reagent kit (Takara Bio, Inc.) according to the
manufacturer's protocols. Next, SYBR Premix Ex Taq reagents (Takara
Bio, Inc.) were employed to perform qPCR on an Applied Biosystems
7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocols and all
reaction was repeated three times. The following thermocycling
conditions were used for qPCR: 95°C for 10 min; followed by 40
cycles of denaturation at 95°C for 10 sec and annealing/extension
at 60°C for 60 sec. The primer sequences for PCR were: SEC61G:
5′-AAAGGACTCCATTCGGCTGGTT-3′ (forward) and
5′-CAAAGAAGCCAATGAATCCC-3′ (reverse); CREB3:
5′-ACCCTTTCCGTAGTTGTCCC′ (forward) and 5′-GAATGTTCAACGACGCTGGG-3′
(reverse); GAPDH: 5′-GGGAAACTGTGGCGTGAT-3′ (forward) and
5′-GAGTGGGTGTCGCTGTTGA-3′ (reverse). GAPDH served as an internal
control for normalization and relative gene expression was
evaluated with the 2−ΔΔCq method (12).
Western blot assay
Total proteins were extracted from A549 cells with
RIPA lysis buffer (Beijing Solarbio Science & Technology Co.,
Ltd.) and were quantified with a BCA protein assay kit (Thermo
Fisher Scientific, Inc.). After being subjected to 10% SDS-PAGE,
the proteins (30 µg per lane) were transferred onto PVDF membranes,
which were then blocked with 5% non-fat milk for 2 h at room
temperature and then incubated at 4°C overnight with primary
antibodies: anti-SEC61G (1:500; cat. no. 11147-2-AP; ProteinTech
Group, Inc.), anti-phosphorylated (p)-PERK (1:1,000; cat. no. 3179;
Cell Signaling Technology, Inc.), anti-PERK (1:1,000; cat. no.
ab229912; Abcam), anti-p-eukaryotic initiation factor 2 α (EIF2α;
1:1,000; cat. no. 3398; Cell Signaling Technology, Inc.),
anti-EIF2α (1:1,000; cat. no. ab169528; Abcam), anti-Activating
transcription factor 4 (ATF4; 1:1,000; cat. no. ab184909; Abcam),
anti-B cell lymphoma-2 (Bcl-2; 1:1,000; cat. no. ab32124; Abcam),
anti-BCL-2-associated X (Bax; 1:1,000; cat. no. ab32503; Abcam),
anti-C-Caspase 3 (1:500; cat. no. ab32042; Abcam), anti-Caspase 3
(1:5,000; cat. no. ab32351; Abcam), anti-matrix metallopeptidase 2
(MMP2; 1:1,000; cat. no. ab92536; Abcam), anti-matrix
metallopeptidase 2 (MMP9; 1:1,000; cat. no. ab76003; Abcam),
anti-CREB3 (1:2,000; cat. no. ab180119; Abcam) and anti-GAPDH
(1:2,500; cat. no. ab9485; Abcam). Subsequently, the membranes were
incubated with horseradish peroxidase-conjugated goat anti-rabbit
IgG secondary antibody for 2 h at room temperature (1:2,000; cat.
no. ab6721; Abcam). Finally, the protein bands were visualized by
using enhanced chemiluminescence (MilliporeSigma) and quantified
using Image-Pro Plus software (version 6.0; Media Cybernetics,
Inc.).
Cell counting kit-8 (CCK-8) assay
A549 cells were inoculated into 96-well plates and
cultured overnight at 37°C before being treated with 5 mmol/l
4-PBA. After 24, 48 and 72 h, 10 µl CCK-8 reagent (Beyotime
Institute of Biotechnology) was added into each well and the cells
were further incubated for additional 2 h. Finally, the absorbance
at 450 nm was determined with a microplate reader (Thermo Fisher
Scientific, Inc.).
Colony formation assay
A549 cells were inoculated in 6-well plates and
incubated for 10 days. Subsequently, the cell colonies were fixed
with 4% paraformaldehyde at room temperature for 15 min and stained
with 0.5% crystal violet solution, for 30 min at room temperature.
Finally, the number of colonies (defined as >50 cells) was
counted under an inverted microscope (magnification, ×10).
TUNEL assay
Following the corresponding treatment and
transfection, A549 cells were fixed with 4% paraformaldehyde for 15
min at room temperature and permeabilized with 0.25% Triton X-100
for 20 min at room temperature. Cells were incubated with 5% bovine
serum albumin (Yeasen Biotech Co., Ltd.) and then stained with
TUNEL reagent. Subsequently, DAPI staining solution (Beyotime
Institute of Biotechnology) was used to counterstain the sections
in dark. Images of apoptotic cells were captured under a
fluorescence microscope (Nikon Corporation; magnification,
×200).
Wound healing assay
A549 cells were seeded into 6-well plates and
incubated until reaching a confluence of 90–100%. A linear scratch
in the cell monolayer was created with a 10 µl pipette tip.
Subsequently, PBS was applied to wash the cells three times to
remove cell debris. The cells were then incubated at 37°C in the
presence of 5% CO2 and recorded at 0 and 24 h. ImageJ
software (version 1.46; National Institutes of Health) was used to
evaluate the area occupied by the migrated cells.
Transwell assay
The invasiveness of A549 cells was detected with
Transwell assay. The upper chamber was used to inoculate and
incubate A549 cells, while 10% FBS was added to the lower chamber.
After 24 h, fixation and staining of A549 cells were conducted with
4% paraformaldehyde for 10 min and 0.1% crystal violet for 10 min
at room temperature, respectively. The invaded cells across the
filter were observed in three random fields under a light
microscope (magnification, ×100).
Co-immunoprecipitation (Co-IP)
assay
According to the BioGRID and UALCAN databases, CREB3
was found to interact with SEC61G. Thus, Co-IP assay was used to
verify this hypothesis. Total proteins that had been isolated with
RIPA lysis buffer (Beijing Solarbio Science & Technology Co.,
Ltd.), centrifuged at 1,000 × g at 4°C for 10 min and quantified
with a BCA kit (Beyotime Institute of Biotechnology) were incubated
overnight at 4°C with 2 µg appropriate antibodies: Anti-SEC61G
(cat. no. PA5-106309; Thermo Fisher Scientific, Inc.) and
anti-CREB3 (cat. no. LS-C664784; LifeSpan BioSciences, Inc.).
Normal rabbit IgG (cat. no. sc-2027; Santa Cruz Biotechnology,
Inc.) were used as control. Next, the cell lysates were incubated
with 40 µl Protein G/A Agarose Beads (Invitrogen; Thermo Fisher
Scientific, Inc.). Following centrifugation at 600 × g at 4°C for
10 min, PBS was then used to wash the beads three times and the
precipitated proteins were then re-suspended in 2X SDS-PAGE loading
buffer, boiled in Laemmli buffer for 5 min and eluted from the
beads. Finally, the immunoprecipitated products were determined
using western blotting.
Statistical analysis
Data were expressed as the mean ± standard
deviation. SPSS 22.0 (IBM Corp.) was employed for data analysis.
One-way ANOVA followed by Tukey's post hoc test was used for the
comparisons of multiple groups, while Student's t-test was employed
for comparisons between two groups. Mantel-Cox test was to
determine the overall survival rate of LUAD patients. P<0.05 was
considered to indicate a statistically significant difference.
Results
SEC61G is upregulated in LUAD and its
downregulation can trigger ER stress
According to the GEPIA database, SEC61G expression
was greatly increased in LUAD (Fig. 1A
and B). The RT-qPCR and western blot results revealed that the
mRNA and protein expression levels of SEC61G were increased in LUAD
A549 cells (Fig. 1C and D).
Compared with those of si-NC, the mRNA and protein expression
levels of SEC61G were greatly decreased after transfection with
SEC61G interfering plasmids (Fig. 1E
and F). In addition, the expression levels of ER stress-related
proteins, including phosphorylated (p)-protein kinase RNA-like ER
kinase (PERK), p-eukaryotic initiation factor-2α (eIF2α) and
activating transcription factor 4 (ATF4), were significantly
increased upon SEC61G knockdown (Fig.
1G).
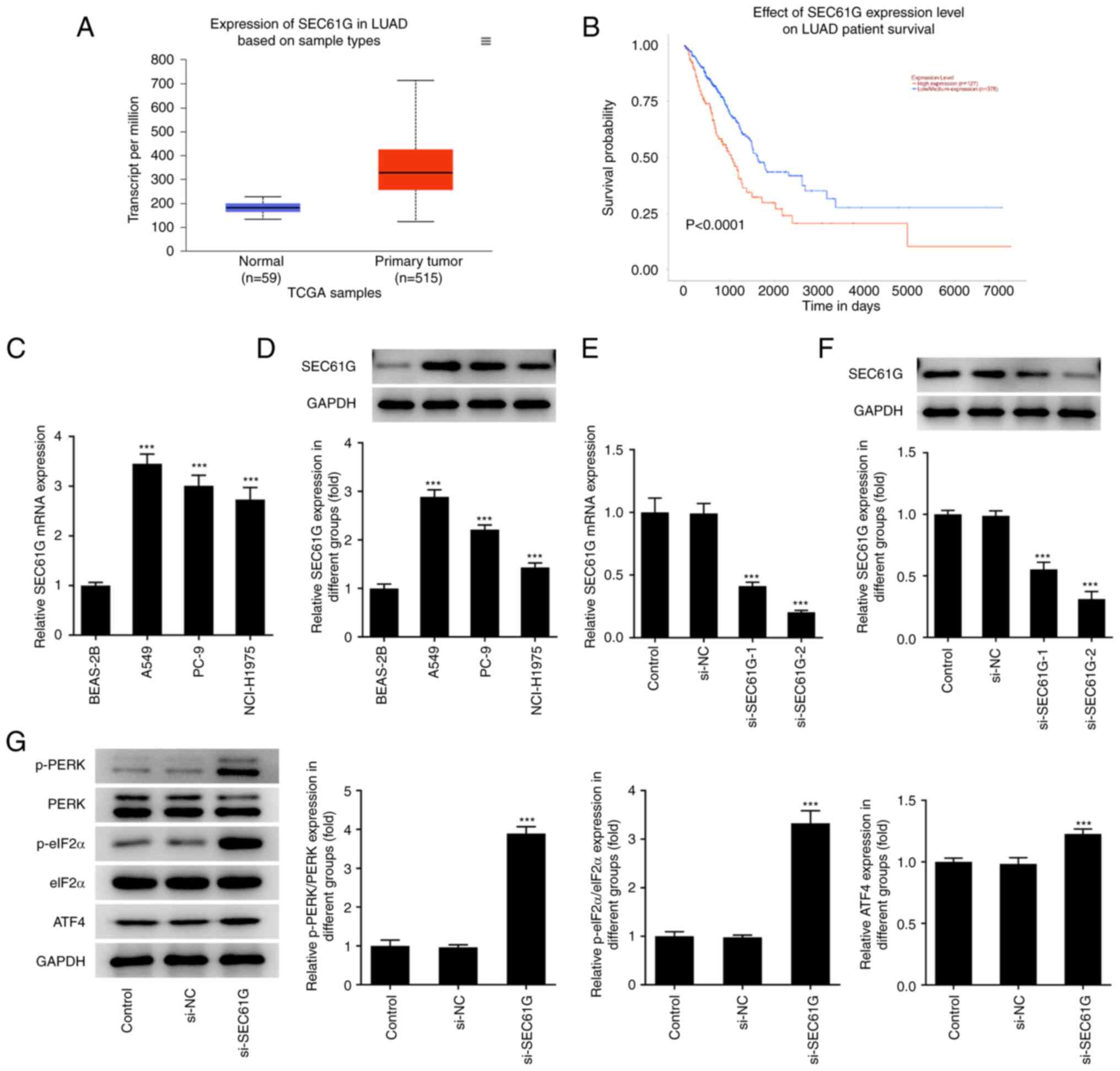 | Figure 1.SEC61G is upregulated in lung
adenocarcinoma and its downregulation can activate endoplasmic
reticulum stress. (A and B) According to GEPIA database, SEC61G was
found to be increased in LUAD. mRNA and protein expressions of
SEC61G in lung adenocarcinoma cells were measured by (C) RT-qPCR
and (D) western blotting, respectively. (E) mRNA and (F) protein
expressions of SEC61G were measured by RT-qPCR and western
blotting. (G) The expressions of ER stress-related proteins were
measured by western blotting. Data are expressed as mean ± standard
deviation. ***P<0.001 vs. BEAS-2B or si-NC. SEC61G, SEC61
translocon subunit γ; GEPIA, Gene Expression Profiling Interactive
Analysis; LUAD, lung adenocarcinoma; RT-qPCR, reverse
transcription-quantitative PCR; ER, endoplasmic reticulum; si,
small interfering; NC, negative control; p-, phosphorylated; PERK,
protein kinase RNA-like ER kinase; eIF2α, eukaryotic initiation
factor-2α; TCGA, The Cancer Genome Atlas. |
SEC61G silencing inhibits the
malignant progression of LUAD via ER stress
As depicted in Fig.
2A, cell viability was greatly decreased by SEC61G silencing,
while 4-PBA treatment partially recovered the viability of A549
cells. The decreased colony number in SEC61G-silenced A549 cells
was significantly increased after treatment with 4-PBA (Fig. 2B). The results from Fig. 2C and D showed that the increased
apoptosis rate caused by SEC61G knockdown was decreased by 4-PBA
treatment, which indicated that 4-PBA abolished the promoting
effects of SEC61G knockdown on the apoptosis rate of A549 cells.
Additionally, SEC61G silencing increased Bcl-2 expression but
decreased Bax and cleaved caspase 3 expression in comparison with
the results of si-NC and these findings were subsequently reversed
by 4-PBA treatment (Fig. 2E).
Compared with those of si-NC, the migration and invasion rate of
A549 cells were reduced after transfection with SEC61G interfering
plasmids, while 4-PBA treatment reversed the inhibitory effects of
SEC61G silencing on cell migration and invasion (Fig. 2F and G). Furthermore, the decreased
expression levels of MMP9 and MMP2 in SEC61G-silenced A549 cells
were increased after 4-PBA treatment (Fig. 2H).
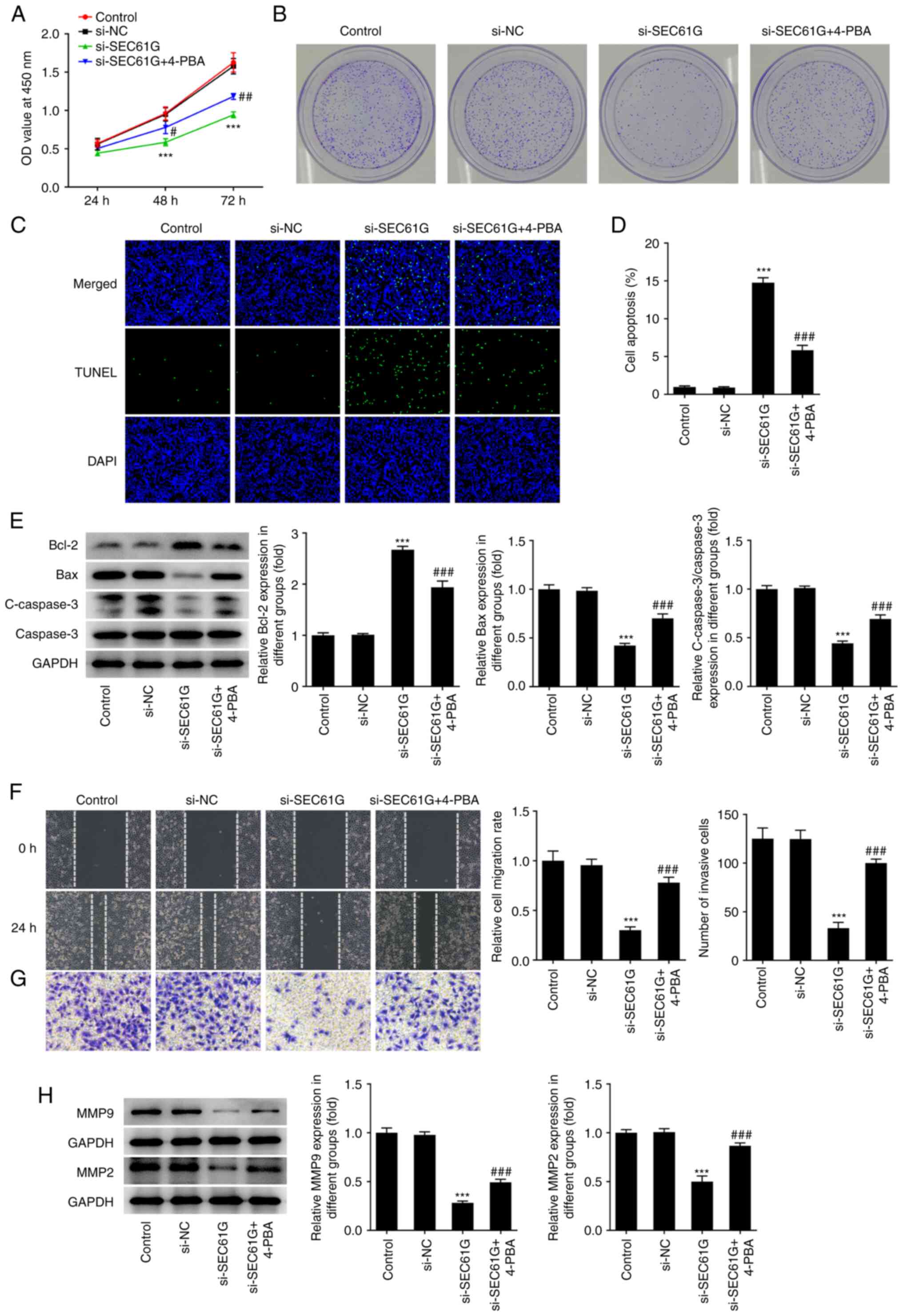 | Figure 2.SEC61G silence inhibits malignant
development of lung adenocarcinoma via endoplasmic reticulum
stress. (A) The cell viability was detected using CCK-8. (B) The
colony formation was detected using colony formation assay
(magnification, ×10). (C and D) The apoptosis was detected using
TUNEL (magnification, ×200). (E) The expressions of
apoptosis-related proteins were measured by western blotting. The
(F) migration and (G) invasiveness were evaluated by wound healing
and Transwell assays (magnification, ×100). (H) The expressions of
MMP9 and MMP2 were measured using western blotting. Data are
expressed as mean ± standard deviation. ***P<0.001 vs. si-NC.
#P<0.05, ##P<0.01,
###P<0.001 vs. si-SEC61G. SEC61G, SEC61 translocon
subunit γ; si, small interfering; NC, negative control; 4-PBA,
4-phenylbutyric acid; C-, cleaved. |
SEC61G participates in ER stress via
CREB3 in LUAD
According to the BioGRID database, SEC61G is
predicted to interact with CREB3 (Fig.
3A). The UALCAN database revealed that CREB3 expression was
significantly increased in LUAD (Fig.
3B). As shown in Fig. 3C and
D, the mRNA and protein expression levels of CREB3 were
markedly high in A549 cells. A Co-IP assay was adopted to further
verify the binding of SEC61G and CREB3. As shown in Fig. 3E, CREB3 existed in anti-SEC61G and
SEC61G existed in anti-CREB3, revealing that SEC61G could bind to
CREB3 in LUAD. Compared with the si-NC group, CREB3 expression was
greatly decreased by SEC61G silencing (Fig. 3F). In addition, Fig. 3G and H revealed that the expression
of CREB3 markedly increased after transfection with CREB3
overexpression plasmids. Furthermore, the increased expression
levels of p-PERK, p-eIF2α and ATF4 in SEC61G-silenced A549 cells
were decreased upon CREB3 overexpression (Fig. 3I).
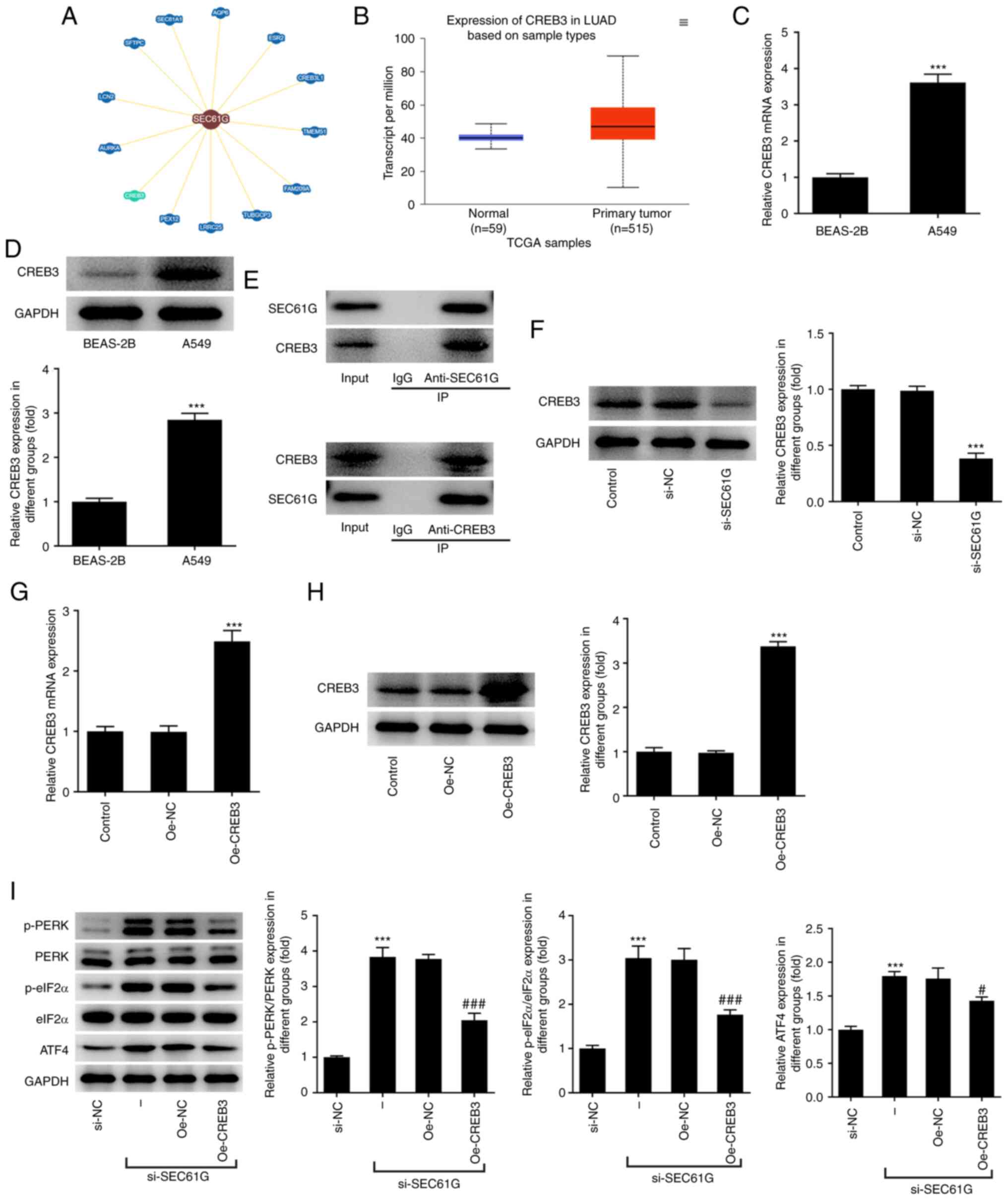 | Figure 3.SEC61G participates in endoplasmic
reticulum stress via CREB3 in lung adenocarcinoma. (A) According to
BioGRID database, SEC61G could interact with CREB3. (B) According
to UALCAN database, CREB3 was upregulated in LUAD. (C and D) The
mRNA and protein expressions of CREB3 were measured by RT-qPCR and
western blotting, respectively. ***P<0.001 vs. BEAS-2B. (E) The
binding of SEC61G and CREB3 was verified by Co-immunoprecipitation.
(F) The expression of CREB3 was measured by western blotting.
***P<0.001 vs. si-NC. (G and H) The mRNA and protein expressions
of CREB3 were measured by RT-qPCR and western blotting,
respectively. ***P<0.001 vs. Oe-NC. (I) The expressions of ER
stress-related proteins were measured by western blotting.
***P<0.001 vs. si-NC. #P<0.05,
###P<0.001 vs. Oe-NC. Data are expressed as mean ±
standard deviation. SEC61G, SEC61 translocon subunit γ; CREB3,
cyclic AMP-responsive element-binding protein 3; LUAD, lung
adenocarcinoma; RT-qPCR, reverse transcription-quantitative PCR;
si, small interfering; NC, negative control; Oe, overexpression
plasmid; TCGA, The Cancer Genome Atlas; p-, phosphorylated; PERK,
protein kinase RNA-like ER kinase; eIF2α, eukaryotic initiation
factor-2α; ATF4, activating transcription factor 4. |
SEC61G knockdown inhibits the
malignant progression of LUAD via CREB3
As demonstrated in Fig.
4A, the decreased cell viability of SEC61G-silenced A549 cells
was increased by CREB3 overexpression. SEC61G silencing decreased
the number of colonies, while CREB3 overexpression partially
abolished the inhibitory effects of SEC61G silencing (Fig. 4B). SEC61G knockdown greatly
increased the apoptosis rate of A549 cells compared with that of
si-NC. However, CREB3 overexpression reversed the promoting effects
of SEC61G silencing on cell apoptosis, as evidenced by the
decreased apoptosis rate of si-SEC61G + Oe-CREB3 (Fig. 4C and D). Fig. 4E revealed that the increased
expression of Bcl-2 and the decreased expression of Bax and cleaved
caspase 3 were reversed by CREB3 overexpression. In addition, the
decreased cell migration and invasion rate caused by SEC61G
silencing were increased after transfection with Oe-CREB3 plasmids
(Fig. 4F and G). Furthermore, the
expression levels of MMP9 and MMP2 were markedly increased by CREB3
overexpression in comparison with those of si-SEC61G + Oe-NC
(Fig. 4H).
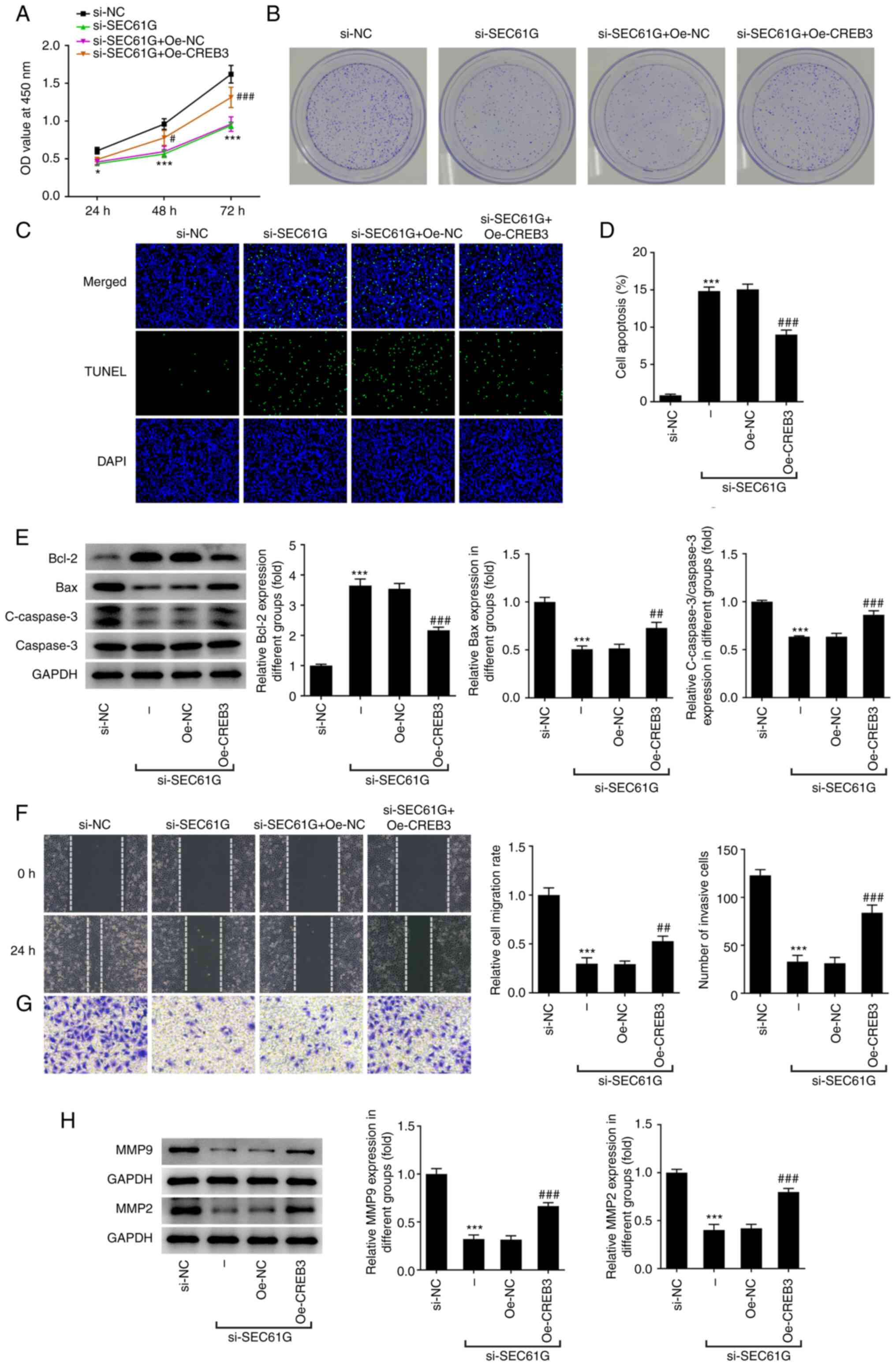 | Figure 4.SEC61G knockdown inhibits malignant
development of lung adenocarcinoma via CREB3. (A) The cell
viability was detected using CCK-8. *P<0.05, ***P<0.001 vs.
si-NC. #P<0.05, ###P<0.001 vs.
si-SEC61G + Oe-NC. (B) The colony formation was detected using
colony formation assay (magnification, ×10). (C and D) The
apoptosis was detected using TUNEL (magnification, ×200).
***P<0.001 vs. si-NC. ###P<0.001 vs. Oe-NC. (E)
The expressions of apoptosis-related proteins were measured by
western blotting. The (F) migration and (G) invasiveness were
evaluated by wound healing and Transwell assays (magnification,
×100). (H) The expressions of MMP9 and MMP2 were measured using
western blotting. ***P<0.001 vs. si-NC. ##P<0.01,
###P<0.001 vs. Oe-NC. Data are expressed as mean ±
standard deviation. SEC61G, SEC61 translocon subunit γ; CREB3,
cyclic AMP-responsive element-binding protein 3; si, small
interfering; Oe, overexpression plasmid; NC, negative control; C-,
cleaved. |
Discussion
As the type of cancer with the highest incidence
worldwide, LUAD remains the main cause of cancer-associated
morality globally (13). Effective
methods to improve the efficacy of LUAD treatment remain to be
explored (14). The present study
evaluated the expression of SEC61G and CREB3 in LUAD, as well as
their association. It was found that SEC61G was upregulated in A549
cells and its downregulation could trigger ER stress. SEC61G
silencing could suppress LUAD cell viability. Knockdown of SEC61G
inhibited colony formation, migration, invasiveness and the
expression of migration-related proteins, while it promoted the
apoptosis of A549 cells, which was reversed after treatment with
4-PBA. According to the BioGRID and UALCAN databases, CREB3 was
upregulated in LUAD and could interact with SEC61G. CREB3
overexpression partially reversed the effects of SEC61G silencing
on the viability, colony formation, apoptosis, migration and
invasiveness of A549 cells. The present study revealed that SEC61G
acted as an oncogene in the development of LUAD and that silencing
SEC61G could suppress LUAD progression via the regulation of
CREB3.
Various studies have demonstrated that SEC61G has an
abnormal expression in numerous cancer types. Shi et al
(15) reported that SEC61G
expression was increased in head and neck squamous cell carcinoma.
It has also been found that SEC61G serves an important role in the
progression and prognosis of head and neck squamous cell carcinoma,
thus serving as an effective biomarker to predict patient survival
(15). In addition, Sheu et
al (16) report that SEC61G is
important in oral squamous cell carcinoma. In the present study,
SEC61G expression was detected in A549 cells and it was found that
SEC61G levels were increased in LUAD cells. To explore the specific
role of SEC61G in LUAD, the expression of SEC61G in A549 cells was
silenced. The results indicated that SEC61G knockdown inhibited
cell viability, colony formation, migration and invasiveness and
promoted apoptosis in A549 cells via ER stress.
As an ER membrane-bound transcription factor
(17), CREB3 induces apoptosis by
activating ER stress (18). Penney
et al (19,20) suggest that CREB3 can alter stress
sensitivity, thus serving as a potential factor in the development
of stress-related pathologies. The present study found that CREB3
level was increased in LUAD cells. According to the BioGRID
database, SEC61G could bind to CREB3; therefore, further
experiments were conducted to verify this hypothesis. The results
revealed that the decreased cell viability, colony formation,
migration and invasiveness caused by SEC61G silencing were reversed
by CREB3 overexpression, which indicated that SEC61G knockdown
exerted inhibitory effects on the malignant progression of LUAD via
targeting CREB3. However, the present study only produced the
results for SEC61G knockdown and CREB3 overexpression. These
experiments of SEC61G overexpression and CREB3 silencing will
provide more evidence to support the hypothesis of the present
study and will supplement related experiments in further
studies.
In conclusion, SEC61G and CREB3 were upregulated in
LUAD cells and SEC61G could interact with CREB3. Therefore, SEC61G
silencing could inhibit LUAD progression via regulating CREB3.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural Science
Foundation of China, China (grant no. 82070076).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
QZ and ZG designed the research, performed the
experiments, drafted and revised the manuscript. QZ searched the
literature and analyzed the data. ZG guided the experiments. QZ and
ZG confirm the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Abe Y and Tanaka N: The hedgehog signaling
networks in lung cancer: The mechanisms and roles in tumor
progression and implications for cancer therapy. Biomed Res Int.
2016:79692862016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao Y, Varn FS, Cai G, Xiao F, Amos CI
and Cheng C: A P53-deficiency gene signature predicts recurrence
risk of patients with early-stage lung adenocarcinoma. Cancer
Epidemiol Biomarkers Prev. 27:86–95. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Greenfield JJ and High S: The Sec61
complex is located in both the ER and the ER-Golgi intermediate
compartment. J Cell Sci. 112:1477–1486. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liang L, Huang Q, Gan M, Jiang L, Yan H,
Lin Z, Zhu H, Wang R and Hu K: High SEC61G expression predicts poor
prognosis in patients with head and neck squamous cell carcinomas.
J Cancer. 12:3887–3899. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Oakes SA and Papa FR: The role of
endoplasmic reticulum stress in human pathology. Annu Rev Pathol.
10:173–194. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Y, Ji W, Shergalis A, Xu J, Delaney
AM, Calcaterra A, Pal A, Ljungman M, Neamati N and Rehemtulla A:
Activation of the unfolded protein response via inhibition of
protein disulfide isomerase decreases the capacity for DNA repair
to sensitize glioblastoma to radiotherapy. Cancer Res.
79:2923–2932. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gao H, Niu W, He Z, Gao C, Peng C and Niu
J: SEC61G plays an oncogenic role in hepatocellular carcinoma
cells. Cell Cycle. 19:3348–3361. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meng H, Jiang X, Wang J, Sang Z, Guo L,
Yin G and Wang Y: SEC61G is upregulated and required for tumor
progression in human kidney cancer. Mol Med Rep. 23:4272021.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sampieri L, Di Giusto P and Alvarez C:
CREB3 transcription factors: ER-Golgi stress transducers as hubs
for cellular homeostasis. Front Cell Dev Biol. 7:1232019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li W, Gao LN, Song PP and You CG:
Development and validation of a RNA binding protein-associated
prognostic model for lung adenocarcinoma. Aging (Albany NY).
12:3558–3573. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang C, Zhang Z, Zhang G, Zhang Z, Luo Y,
Wang F, Wang S, Che Y, Zeng Q, Sun N and He J: Clinical
significance and inflammatory landscapes of a novel
recurrence-associated immune signature in early-stage lung
adenocarcinoma. Cancer Lett. 479:31–41. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shi Y, Liang Y, Wang W and Zhang G: SEC61G
identified as a prognostic biomarker of head and neck squamous cell
carcinoma. Eur Arch Otorhinolaryngol. 279:2039–2048. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sheu JJ, Hua CH, Wan L, Lin YJ, Lai MT,
Tseng HC, Jinawath N, Tsai MH, Chang NW, Lin CF, et al: Functional
genomic analysis identified epidermal growth factor receptor
activation as the most common genetic event in oral squamous cell
carcinoma. Cancer Res. 69:2568–2576. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Oh-Hashi K, Soga A, Naruse Y, Takahashi K,
Kiuchi K and Hirata Y: Elucidating post-translational regulation of
mouse CREB3 in Neuro2a cells. Mol Cell Biochem. 448:287–297. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu Y, Chu L, Liu J, Yu L, Song SB, Yang H
and Han F: Knockdown of CREB3 activates endoplasmic reticulum
stress and induces apoptosis in glioblastoma. Aging (Albany NY).
11:8156–8168. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Penney J, Taylor T, MacLusky N and Lu R:
LUMAN/CREB3 plays a dual role in stress responses as a cofactor of
the glucocorticoid receptor and a regulator of secretion. Front Mol
Neurosci. 11:3522018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Penney J, Mendell A, Zeng M, Tran K, Lymer
J, Turner PV, Choleris E, MacLusky N and Lu R: LUMAN/CREB3 is a key
regulator of glucocorticoid-mediated stress responses. Mol Cell
Endocrinol. 439:95–104. 2017. View Article : Google Scholar : PubMed/NCBI
|


















