Introduction
Colorectal cancer (CRC) is the fourth most common
human malignancy, with ~900,000 cases of mortality worldwide
annually (1). In addition, it is
the second most common cause of cancer-associated mortality in the
United States (2). Regarding the
molecular mechanism underlying CRC carcinogenesis, a model was
previously proposed, where the steps required for its development
involve the activating mutation of KRAS combined with the
inactivating mutations of tumor suppressor genes, such as
adenomatous polyposis coli (APC), SMAD4 and
TP53 (3). When devising
strategies for developing molecular target-based drugs,
intertumoral and intratumoral genetic heterogeneity are garnering
research interest due to their key role in cancer evolution
(4), in which CRC is of no
exception (5). CRC has been
reported to harbor genetic alterations in components of the Wnt
signaling pathway in ~90% of the cases (6,7).
Furthermore, the Wnt signaling pathway has been reported to serve a
key role in the activation of not only CRC cells but also
cancer-associated fibroblasts (CAFs) (8). Models of organoids and CAFs (9), in addition to those of
patient-derived xenografts (PDX), were previously established using
surgical specimens isolated from each corresponding patient. This
was conducted in parallel with the targeted sequencing of
cancer-associated gene mutations to evaluate efficacy of anticancer
drugs to individual patient in a partially-reproduced tumor
microenvironment.
E7386 is a selective inhibitor of the interaction
between β-catenin and the cAMP response element binding
protein-binding protein, which forms a part of the Wnt/β-catenin
signaling pathway (10).
Therefore, E7386 is expected to affect CRC cells, particularly
those with aberrant activation of the Wnt/β-catenin signaling
pathway. In the present study, the effects of E7386 treatment on
the in vitro models of CRC-derived organoids and a
co-culture model of these organoids with corresponding CAFs were
comprehensively analyzed on a molecular level. Subsequently, the
effects of E7386 on the in vivo PDX model were also
investigated, to screen for potential pharmacodynamic biomarkers
that are applicable for clinical trials. In addition, the
pharmacological effects of E7386 were examined.
Materials and methods
Chemicals
A stock solution of E7386 dissolved in DMSO to 10 mM
and E7386 powder (Eisai Co., Ltd.) diluted with 100 mM hydrochloric
acid was used for in vitro and in vivo studies,
respectively.
Surgical tissue specimens
A total of 45 surgical CRC specimens collected after
the pathological examination of patients with CRC at the National
Cancer Center Hospital (Tokyo, Japan) were received between
February 2016 and February 2018 and used to prepare
organoids/fibroblast culture and implantation into mice for the
establishment of PDX. Of the 45 cancer patients, 60% were males and
40% were females, and average ages were 62 years old, ranging from
36 years old to 88 years old. All experiments were performed
following the relevant domestic guidelines and regulations in
Japan, including Ethical Guidelines for Medical and Health Research
Involving Human Subjects. The use of surgical specimens in this
study was approved by the Ethics Committee of the National Cancer
Center (approval no. 2015-108) and written informed consent was
obtained from all patients. In the present study, three cases of
typical gene mutational statuses [case 21: KRAS, APC and
TP53; case 28: PI3K catalytic subunit (PIK3CA)
and TP53; and case 32: APC and TP53] were used
(9). Clinical and pathological
information has been previously reported (9).
Culture of organoids and fibroblasts
and co-culture of organoids with fibroblasts using a chamber
system
The organoids and fibroblasts used in the present
study were established from surgical specimens. The culturing of
organoids and fibroblasts was performed as previously described
(9). Briefly, for co-culture,
organoids and fibroblasts were cultured in inserts with porous
membranes and carrier plates (pore size, 1 µm; cat. nos. 353104 and
353504; Corning, Inc.). In total, 1 day before co-culture,
fibroblasts (1×104 cells/well) were seeded into 24-well
plates with RPMI-1640 (cat. no. 189-02025; Fujifilm Wako Pure
chemical Corporation) containing 10% FBS (cat. no. SH30075;
Hyclone; Cytiva). Organoids (1×104 cells/insert) were
seeded into polymerized Matrigel (cat. no. 354234; Corning, Inc.)
in cell culture inserts, followed by setting the inserts in
companion carrier plates, and a medium for organoid culture was
added into the basal compartments. The next day, the organoids were
covered with Matrigel, and the inserts containing the organoids
were transferred to the 24-well plates containing the fibroblasts.
During co-culturing, the organoid culture medium was applied to
both organoids and fibroblasts. After E7386 incubation for 6 or 24
h, organoids and fibroblasts in each insert and well were collected
separately for the analysis of gene expression.
Establishment of the PDX model
Surgical specimens were cut 2 or 3-mm cubes or
established CRC organoids were implanted into the subcutis of
female
NOD.Cg-PrkdcscidIl2rgtm1Sug/ShiJic
(NOG) mice at 7 weeks of age (In-Vivo Science, Inc.) under
isoflurane anesthesia (4% for initiation and 2% for maintenance),
and their body weights were 20.8±0.8 g at 14–16 weeks of age (at
the start of drug administration). The mice were housed in plastic
cages (n≤5 per cage) with recycled paper bedding in a
specific-pathogen-free environment maintained at 22±1°C and 55±10%
relative humidity, with 12-h light/dark cycle and were provided
free access to the CE2 standard chow diet (CLEA Japan, Inc.) and
tap water. Mouse experiments were performed at the Animal Facility
of the National Cancer Center Research Institute (Tokyo, Japan)
according to institutional guidelines. Ethics approval was obtained
from the National Cancer Center Animal Ethics Committee (approval
no. T17-006).
Treatment of E7386 in the in vitro and
in vivo systems
For the in vitro studies, E7386 at
concentrations of 0, 10, 30, 100 and 300 nM in culture media was
used, before cell viability was measured using the RealTime-Glo™ MT
Cell Viability Assay solution kit (cat. no. G9711; Promega
Corporation). Organoids/CAF samples planned for RNA analysis were
collected 6 and 24 h after treatment, whereas those planned for
metabolome analysis were collected 24 h after the addition of
E7386. Triplicate samples of organoids and corresponding CAFs from
each case were used for the measurement of cell viability.
For the in vivo PDX mouse model, E7386
solution was administered at 0 and 50 mg/kg body weight by gavage
twice a day for 14 days. The number of mice used in the experiments
were five of case 21, five of case 28 and three of case 32 per the
control and E7386-treated group each. The size of the implanted
tissue was measured using a caliper. In total, 6 h after the last
drug administration, all mice were sacrificed by cervical
dislocation under 4% isoflurane anesthesia before subcutaneous PDX
tissues were excised cut into pieces, frozen and stored at −80°C or
fixed with 10% neutral buffer formalin.
Gene expression analyses for in vitro
experiments using DNA microarrays and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated using the RNeasy Micro Kit
(cat. no. 74104; Qiagen GmbH). Triplicate samples were prepared for
the RNA cocktail used for DNA microarray analysis from E7386-0, 30,
and 100 nM groups. The quality of RNA samples and DNA microarray
analysis [SurePrint G3 Human GE Microarray GE 8×60 K Ver.3.0 (cat.
no. G4851C; Agilent Technologies, Inc.)] were evaluated as
previously described (9). For
RT-qPCR analysis, aliquots of total RNA were subjected to a reverse
transcription with random primers using the High-Capacity cDNA
Reverse Transcription Kit (cat. no. 4368814; Applied Biosystems;
Thermo Fisher Scientific, Inc.). qPCR in duplicate in three
independent experiments was performed using the SsoAdvanced™
Universal SYBR-Green Supermix (cat. no. 172-5271; Bio-Rad
Laboratories, Inc.) in the DNA Engine Opticon® 2 system
(MJ Research, Inc.; Bio-Rad Laboratories, Inc.). The primer
sequences are shown in Table SI.
Data were normalized to the housekeeping gene β-actin,
calculated using the 2−ΔΔCq method (11) and are presented as the mean ±
standard deviation (1.0-fold was as control).
Gene ontology (GO) enrichment
analysis
To clarify the biological meaning of the key
molecules, gene information obtained in the DNA microarray analysis
was loaded into Metascape 3.5 (https://metascape.org) for GO enrichment analysis
(12). Terms with a P<0.01, ≥3
and an enrichment factor >1.5 were collected before being
grouped into clusters based on their membership similarity (κ-score
>0.3).
Hydrophilic metabolome analysis of the
in vitro data
For hydrophilic metabolite analysis, capillary
electrophoresis time-of-flight mass spectrometry (CE-TOFMS) was
used for cationic and anionic metabolite analyses. To extract the
metabolites, cultured organoids from E7386-0, 30, and 100 nM groups
were collected after adding 1.0 ml methanol containing internal
standards (25 µM each of methionine sulfone and
D-camphor-10-sulfonic acid). The suspension (400 µl) was then mixed
with Milli-Q water and chloroform at a volumetric ratio of 5:2:5
and centrifuged at 9,100 × g for 10 min at 20°C. Subsequently, the
aqueous layer (400 µl) was centrifugally filtered through a 5 kDa
cut-off filter (cat. no. UFC3LCCNB-HMT; Human Metabolome
Technologies, Inc.) to remove proteins. The filtrate was
centrifugally concentrated and dissolved in 25 µl Milli-Q water
containing the reference compounds (200 µM each of
3-aminopyrrolidine and trimesic acid) immediately prior to CE-TOFMS
analysis [Agilent 1100 isocratic HPLC pump, Agilent G1603ACE-MS
adapter kit, and Agilent G1607A CE-electrospray ionization (ESI)-MS
sprayer; Agilent Technologies, Inc.] (13,14).
The quantified data were standardized with the internal standards
before being normalized to the DNA concentrations of each
sample.
Histopathological and
immunohistochemical analyses of in vivo experiments
PDX tissues fixed with formalin were routinely
processed into paraffin-embedded (FFPE) sections and stained with
hematoxylin and eosin for histopathological evaluation using a
transmission light microscope (BX51; Olympus Corporation).
Immunohistochemistry was performed using the FFPE
sections and primary antibodies against β-catenin (cat. no. 610154;
clone 14; BD Biosciences), phosphoenolpyruvate carboxykinase 2
(PCK2; (cat. no. 14892-1-AP; ProteinTech Group, Inc.), α-smooth
muscle actin (αSMA; cat. no. M0851; clone 1A4; Agilent
Technologies, Inc.) and very low density lipoprotein receptor
(VLDLR; cat. no. ab203271; Abcam). Antigen retrieval was performed
by autoclaving at 121°C for 10 min in citric acid buffer (pH 6.0)
or proteinase K (FUJIFILM Wako Pure Chemical Corporation)
treatment. For signal detection, Histofine MAX PO solution (cat.
no. 424131; Nichirei Corporation) was applied, visualized with
3,3′-diaminobenzidine followed by counterstaining with hematoxylin.
Negative controls without primary antibody addition was set for
each antigen. Quantitative imaging analysis of αSMA-positive areas
was performed using the HALO Imaging Analysis Platform
(001-WS-HALO; Indica Labs, Inc.).
For the immunoblotting of β-catenin and PCK2,
protein samples (10 µg) extracted with RIPA Lysis kit (cat. no.
WSE-7420; ATTO Corporation) from the PDX tissues were subjected to
SDS-PAGE. Immunoblotting and detection of chemiluminescence signals
(cat. no. WSE-7120; ATTO Corporation) were performed as previously
reported (15). Quantitative
analysis was performed using Science Lab. 2005, Multi Gauge Ver.
3.0 software (FUJIFILM Wako Pure Chemical Corporation). For the
internal control, a primary antibody against β-actin (cat. no.
A1978; clone AC-15; Sigma-Aldrich; Merck KGaA) was used.
Statistical analysis
Quantitative data from in vitro and in
vivo experiments, including cell viability assay, hydrophilic
metabolomics, RT-qPCR and immunoblotting analysis results were
presented as the mean ± SD. Homogeneity of variance in data was
checked by Bartlett's test. When the data were homogenous,
differences among the control and treatment groups were analyzed by
one-way analysis of variance (ANOVA) or two-way ANOVA followed by
Tukey's test using Easy R (EZR ver. 4.1.2) (16). In the heterogenous cases,
Kruskal-Wallis test followed by Bonferroni test was applied.
Immunohistochemical grade data were tested using Fisher's exact
test in EZR. P-values of <0.05 were considered significant.
Results
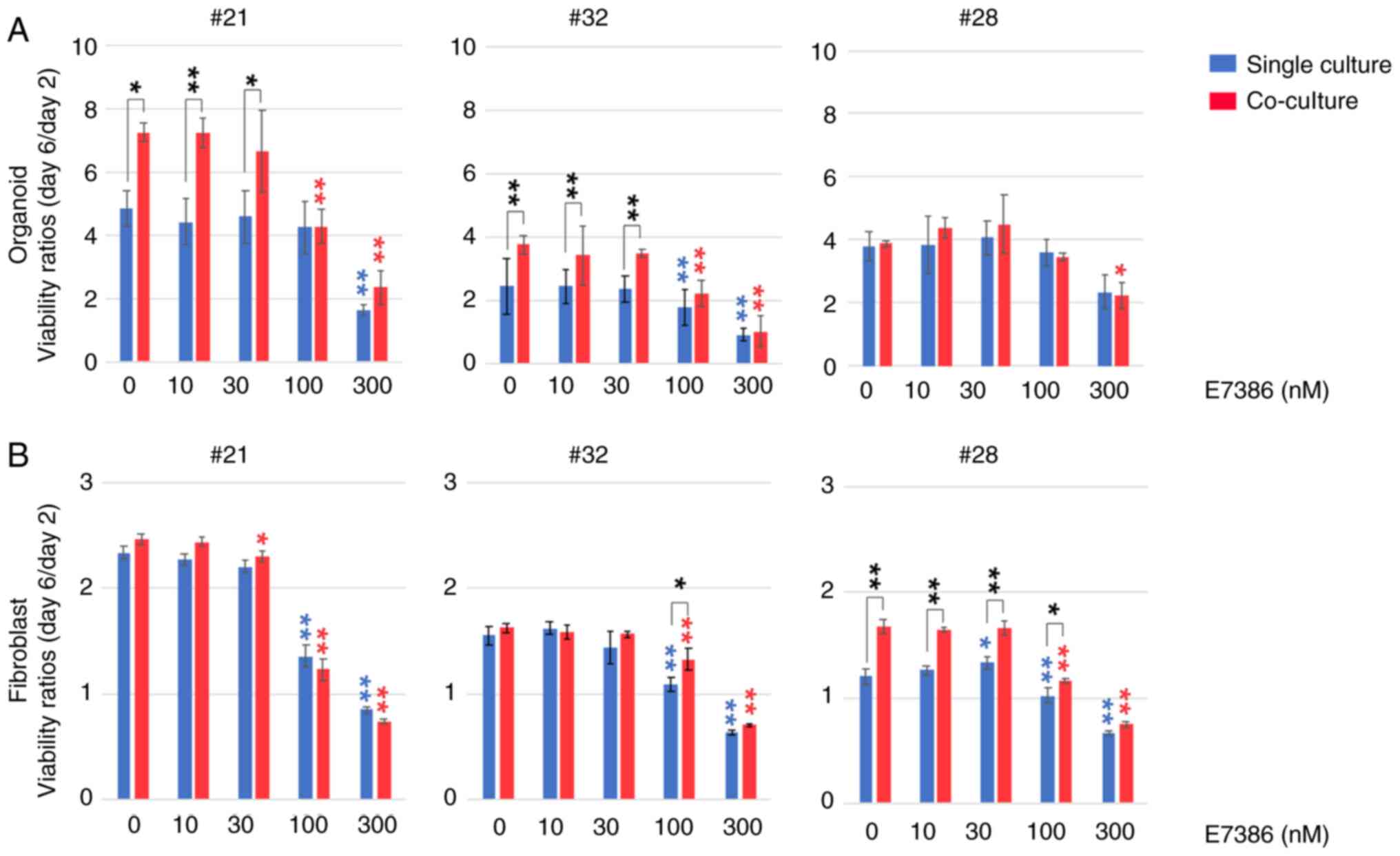
Effects of E7386 on the viability of
CRC organoids and CAFs varies among the cases
In total, three types of organoids, each harboring
the following gene mutational status, were prepared (Fig. 1): i) APC, KRAS and
TP53 mutations (case 21); ii) APC and TP53
mutations (case 32); or iii) PIK3CA and TP53
mutations (case 28). After comparison with the baseline cell
viability of organoids or CAF monoculture in the E7386 (0 nM)
group, cell viability was found to vary among the cases dependent
on their origin. Increased cell viability of organoids following
co-culture with CAFs was observed in cases 21 (P<0.05) and 32
(P<0.01) but not in case 28, which was reported previously
(9). By contrast, an inhibitory
effect of E7386 at 100 nM on the organoids from cases 21 and 32 was
observed, which was more potent in the co-cultured groups
(P<0.01). However, these inhibitory effects were diminished in
the single-cultured groups from case 21 at 100 nM E7386. In case
28, inhibitory effects of E7386 on the organoids were observed in
the co-culture group at 300 nM (P<0.05). In terms of CAFs,
potentiating effects of co-culturing with organoids on viability
were observed in case 28 (P<0.01) but not in cases 21 and 32.
However, the degree of this enhancement was smaller compared with
that in the organoids. Inhibitory effects of E7386 on the CAFs
could be found at 100 nM, which was more potent in the
single-culture groups from all three cases (P<0.01). There were
slight differences in the sensitivity of the CAFs to E7386 among
the co-cultured groups.
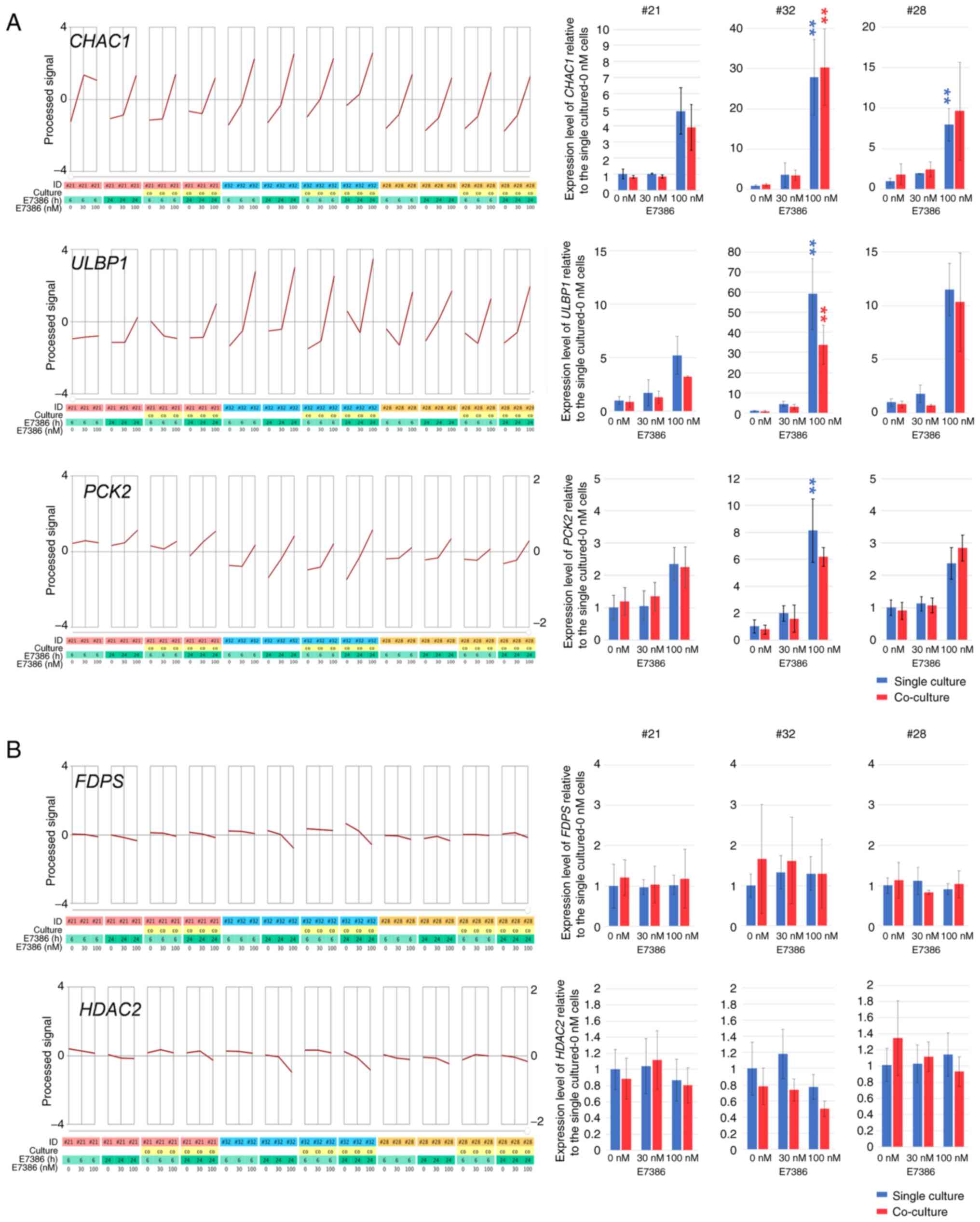
Changes in the expression of genes
regulating stress responses and cellular metabolism after E7386
treatment in CRC organoids and/or CAFs
The effects of E7386 on gene expression in the
organoids and CAFs were next evaluated using DNA microarrays. In
the organoids, 46 genes were found to be upregulated associated
with E7386 concentration (correlation coefficient >0.7) 24 h
after E7386 treatment (Table
SII). GO term enrichment analysis of these upregulated genes
revealed that genes associated with stimulation, such as
glutathione-specific γ-glutamylcyclotransferase 1
(CHAC1) and growth factor receptor bound protein 10
(GRB10), in addition to those associated with
trans-sulfuration and one-carbon metabolism, including
phosphoserine aminotransferase 1 (PSAT1),
cystathionine γ-lyase and phosphoglycerate
dehydrogenase (PHGDH), were amongst the most
significantly enriched genes (Fig.
S1A). In addition, UL16-binding protein
(ULBP)1, which enhances susceptibility to natural
killer (NK)-cell-mediated lysis of cancer cells (17) and PCK2, which regulates
metabolic adaptation and enables glucose-independent tumor growth
(18), were included amongst the
upregulated genes. A number of the upregulated genes associated
with E7386 concentrations were also categorized as ≥ two increased
genes with significance (P<0.05) at 100 nM E7386, but
associations between the gene expression changes and inhibition of
the viability of CRC organoids by E7386 were uncertain (Figs. S2A and S3A). Among the genes that were
significantly decreased by two-fold at 100 nM E7386, the
cancer-related genes were not enriched (data not shown). RT-qPCR
analysis was subsequently performed for CHAC1, ULBP1 and
PCK2 to verify the results from the DNA microarray analysis.
Elevated expression (P<0.01) of the three genes was observed in
the single-culture organoids and after they were co-cultured with
CAFs in the three cases (Fig. 2A).
As a transport of small molecule-related genes (Fig. S1A), VLDLR expression was
also found to be upregulated in association with E7386
concentrations according to results from DNA microarrays, which was
confirmed by RT-qPCR in both single-culture organoids and
co-cultures consisting of organoids and CAFs from case 32 but cases
21 and 28 (data not shown). The expression of 17 genes were
downregulated associated with E7386 concentrations according DNA
microarrays in the organoids following E7386 treatment (Table SIII). The results from the GO term
enrichment analysis are shown in Fig.
S1B. The downregulated genes included GRPEL1, histone
deacetylase 2 (HDAC2) and farnesyl diphosphate
synthase (FDPS). For the expression of FDPS and
HDAC2, RT-qPCR analysis was performed to verify the results
from the DNA microarray. No clear changes in FDPS expression
could be found, whereas a trend toward a reduction in HDAC2
expression was observed in the organoids co-cultured with CAFs from
cases 32 and 28 (Fig. 2B). For
HDAC and FDPS expression, suggestive relationships
with the Wnt/β-catenin signaling pathway were introduced (19,20).
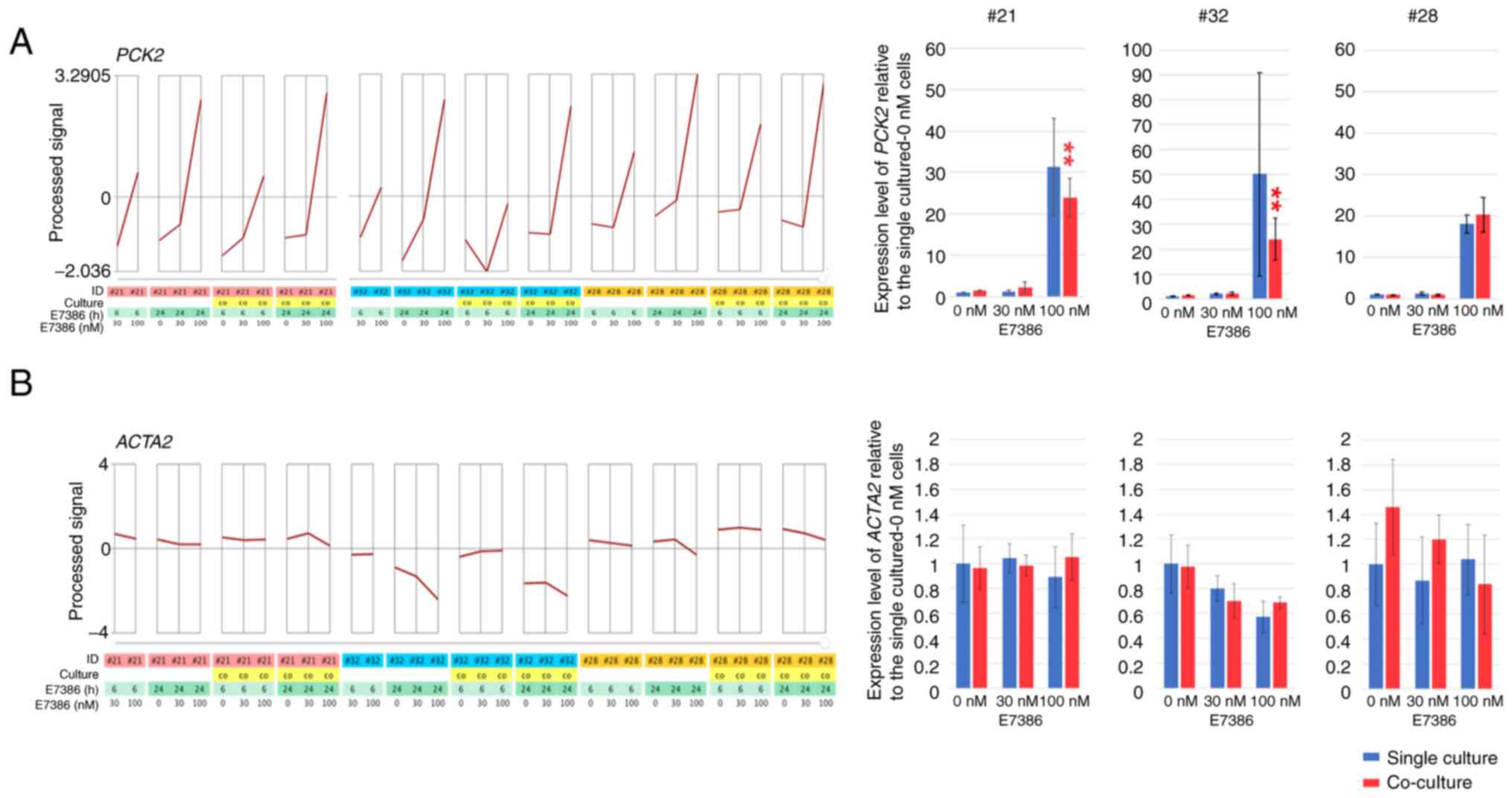
In the corresponding CAFs for each organoid, 52
upregulated and 21 downregulated genes related to E7386
concentrations (correlation coefficient >0.9 and <-0.9,
respectively) were found 24 h after the addition of E7386 (Tables SIV and SV). After they were evaluated at
correlation coefficients of >0.7 and <-0.7, 458 upregulated
and 431 downregulated genes were found (data not shown). GO term
enrichment analysis of the upregulated genes revealed that those
associated with cellular amino acid metabolism, including
asparagine synthetase (glutamine-hydrolyzing; ASNS),
PSAT1, PHGDH and glycyl-tRNA synthetase 1
(GARS), were amongst the most significantly enriched genes
(Fig. S1C). In addition, those
associated with gluconeogenesis, such as PCK2, activating
transcription factor 4 and sestrin 2, were upregulated
in the E7386-treated CAFs. Various upregulated genes related to
E7386 concentrations were also categorized as ≥ two increased genes
with a significance of P<0.05 at 100 nM E7386, but associations
between the gene expression changes and inhibition of the viability
of CAFs by E7386 were uncertain (Figs. S2B and S3B). Among these genes associated with
glucose and amino acid metabolism, ASNS, PSAT1, PHGDH and
PCK2 expression were also increased in the organoids. The
genes downregulated associated with E7386 concentrations included
those associated with the resolution of sister chromatid cohesion,
such as centromere protein O, spindle pole body component 25
and structural maintenance of chromosomes 1A (Fig. S1D). In total, ≥ two decreased
genes with significance at 100 nM E7386 included those involved in
mitotic cell cycle processes, such as cyclin E1 and
MYBL2 and in DNA replication, such as ATPase family AAA
domain-containing 5 and zinc finger GRF-type-containing
1 (Fig. S3B). Upregulation of
PCK2 (P<0.01) expression was then confirmed by RT-qPCR in the
single-culture CAFs and in the co-culture with organoids in each
case (Fig. 3A). As a gene
decreased by E7386, diaphanous related formin 1
(DIAPH1) was reported to be associated with the
differentiation of hepatic stellate cells into tumor-promoting
myofibroblasts (21). Therefore,
to evaluate the alterations in the general activity of CAFs, the
expression levels of ACTA2, which is a classical gene
associated with an activated status of fibroblasts (8), were examined. They were found to be
partially downregulated in cases 32 and 28 (Fig. 3B).
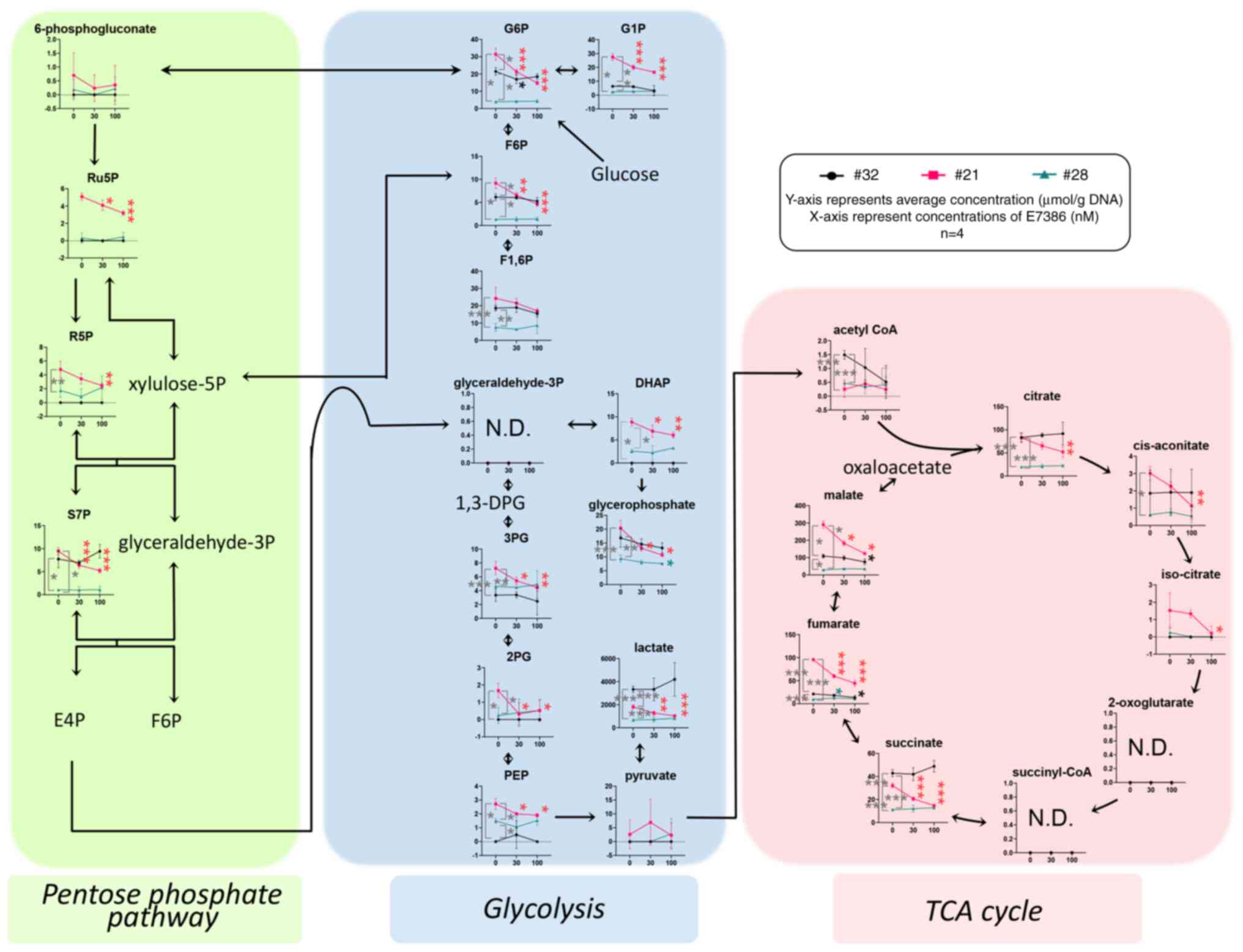
Depletion of glycolysis-associated
metabolites by E7386 treatment in the CRC organoids
The quantified levels of metabolites produced by the
pentose phosphate pathway, glycolysis, tricarboxylic acid (TCA)
cycle, amino acid metabolism and their derivatives were obtained
from the organoid samples in triplicate after treatment with E7386
for 24 h. The data are presented graphically on a large-scale
metabolomic map (Fig. S4) and
summarized in Fig. 4. The
concentrations of glucose-6-phosphate (G6P) and glucose-1-phosphate
(G1P) in the control (0 nM E7386) of case 21 were higher compared
with those of cases 32 and 28 (P≤<0.05). In addition, G6P and
G1P levels in case 21 were reduced by E7386 treatment
(P≤<0.001), but those of cases 32 and 28 were unchanged,
suggestive of E7386-induced glucose depletion in case 21. As a
response to this effect, gluconeogenesis followed by the reduced
levels of metabolites in the TCA cycle was observed in organoids
from case 21. In terms of amino acids, the majority of essential
amino acids, including histidine, isoleucine, leucine, methionine,
phenylalanine, threonine, tryptophan and valine, in addition to a
number of nonessential amino acids, including alanine and
glutamate, were found to be at higher levels in the control
conditions from case 21 compared with those from cases 32 and 28.
Furthermore, those of case 21 were reduced by E7386 treatment
(Fig. S4).
Changes in the expression levels of
PCK2 and VLDLR in carcinoma cells and α-SMA in CAFs are partly
verified in the PDX models
PDX using samples from case 21 was successfully
established, whereas organoids from case 28, which were engrafted
and passaged in the subcutis of NOG mice, were used as PDXs. For
case 32, organoids implanted into the subcutis of NOG mice,
followed by confirmation of their engraftment, were used. No
obvious changes after the administration of E7386 could be found in
the size of tumor tissues (Figs.
S5 and S6) or
histopathological characteristics of PDX from cases 21 and 28 (data
not shown). Evaluation could not be performed in case 32.
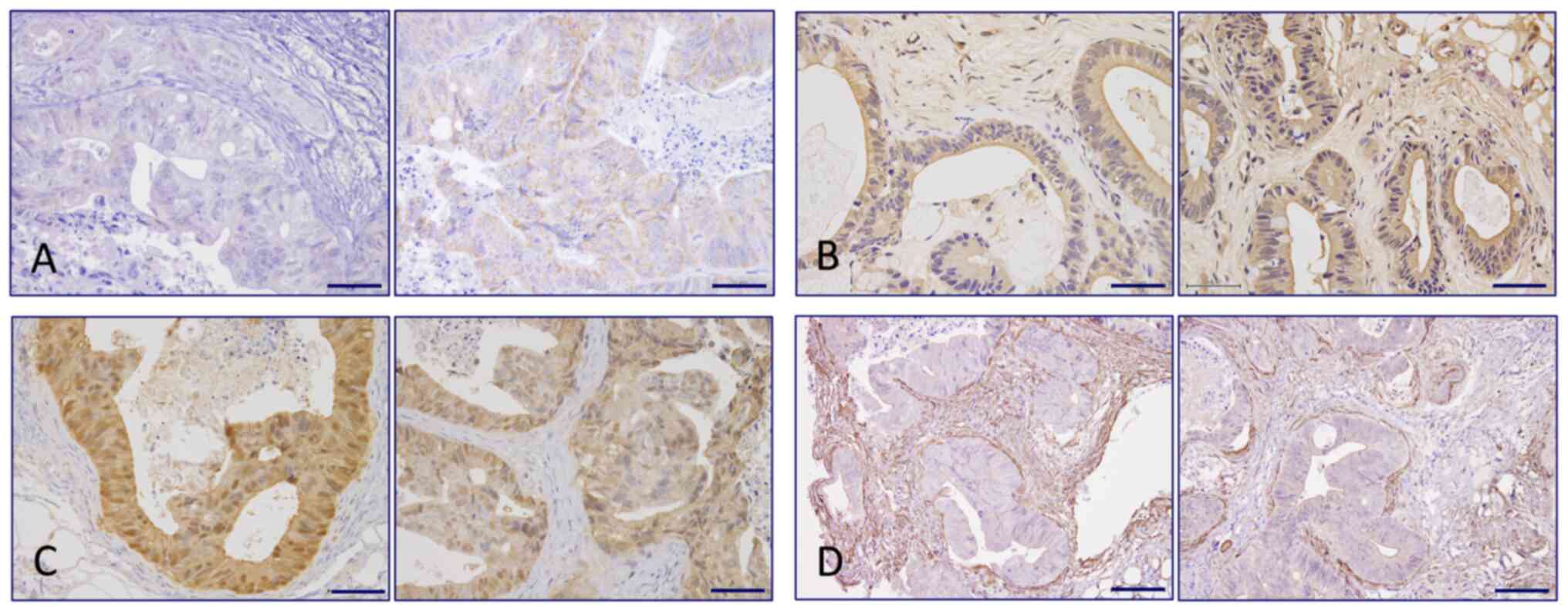
Immunohistochemistry staining for PCK2 revealed an increased
tendency for cytoplasmic granular positivity in the carcinoma cells
of case 21 in the E7386 group (Fig.
5A), which was confirmed by immunoblotting (Figs. S7A and S8A), but not in case 28 (Figs. S7D and S8A). No clear changes in PCK2 positivity
could be observed between the E7386 and control groups from cases
28 and 32 (Table SVI). VLDLR
staining could be detected on the apical surface of the carcinoma
cells in the control and E7386 groups of cases 21 and 28. However,
in case 32, they were strongly visible not only on the apical
surface but also on the intercellular membranes in the E7386 group
(P=0.014; Fig. 5B; Table SVI). In terms of β-catenin,
nuclear positivity in the carcinoma cells of case 28 tended to
decrease in the E7386 group compared with that in the control group
(Fig. 3C) but not in cases 21 and
32 (Table SVI). According to
β-catenin immunoblotting, no notable changes in expression could be
observed between the E7386 and control groups in cases 21 and 28
(Figs. S7B, S7E and S8B). As a marker for the activation of
CAFs, immunohistochemistry staining for αSMA was performed. αSMA
expression in the CAFs surrounding the CRC foci in the E7386 group
from cases 21 or 28 showed a tendency to decrease compared with
that in the control group (Figs.
5D and S9; Table SVI). The visually judged data was
not necessarily reflected to the quantitative data.
Discussion
In the present study, the following were found: i)
E7386 inhibited the viability of both CRC organoids and CAFs; ii)
upregulation of genes associated with gluconeogenesis and cellular
amino acid metabolism in the both organoids and CAFs following
E7386 treatment were found; iii) hydrophilic metabolomic analysis
in the organoid samples revealed the depletion of glycolytic
metabolites after E7386 treatment; and iv) in the PDX model,
membranous positivity for VLDLR and nuclear positivity for
β-catenin in carcinoma cells were increased, but in turn showed a
tendency to decrease after E7386 treatment, in of one of three
cases. By contrast, the expression of αSMA in CAFs showed a
tendency to decrease in two of the three cases after E7386
treatment. In addition, it was noteworthy that in organoids, the
expression of genes associated with stress responses and NK-cell
mediated cytotoxicity were also increased. By contrast, in CAFs
ACTA2/αSMA expression were partially decreased.
The Wnt/β-catenin signaling pathway has been widely
reported to regulate the expression of target genes downstream,
primarily those associated with cell proliferation and
differentiation (22).
Furthermore, the Wnt signaling pathway has been documented to serve
an important role in the activation of not only CRC cells but also
CAFs (8). Therefore, E7386, a
selective inhibitor of the interaction between β-catenin and the
CREB binding protein, was hypothesized to exert a physiological
effect on CRC cells. This was likely particularly in those with
constitutively active Wnt/β-catenin signaling but without mutations
in the driver oncogenes of key signaling pathways. Inhibitory
effects on the viability of organoids were observed after treatment
with 100 nM E7386, which was more potent in the co-cultured groups
of cases 21 (harboring APC, KRAS and TP53 mutations)
and 32 (harboring APC and TP53 mutations). However,
the effects of E3786 were diminished in the single-culture groups
of case 21. In case 28, which harbored PIK3CA and
TP53 mutations, inhibitory effects of E7386 on the viability
of organoids were observed at 300 nM. The inhibitory effects of
E7386 on CAF viability were also observed at 100 nM, which were
more potent in the single-culture group from the three cases. These
results suggest that E7386 affected not only the CRC organoids but
can also the CAFs differently depending on each individual case.
There was a report supporting the present results, where CAFs
derived from CRC tissues revealed a differential transcriptomic
profile compared with that in fibroblasts derived from the paired
adjacent normal mucosal tissue of each case (8). Wnt signaling, focal adhesion and cell
cycle progression were proposed to be involved (8). The effect of E7386 on the viability
of organoids and CAFs was evaluated according to the minimum
inhibitory concentrations (MIC), not IC50, since
organoids/CAFs with low response rates were found among the cases
in the present experiment.
To clarify the molecular events underlying the
inhibitory effects of E7386 on CRC organoids and CAFs, a DNA
microarray analysis was performed. No obvious changes in the direct
target genes of Wnt signaling pathway were observed in the present
experiments. The cause of these results could be that E7386 is a
selective inhibitor of the interaction between β-catenin and the
CREB binding protein, whilst not being a direct inhibitor of
β-catenin. One of the main findings was that the number of
upregulated and downregulated genes in CAFs associated E7386
concentrations was higher (458 and 431, respectively), compared
with those (46 and 17, respectively) in organoids, suggesting that
E7386 can exert effects on not only in CRC organoids but also in
CAFs. The upregulated genes in CAFs at 100 nM included those in
cellular amino acid metabolism and gluconeogenesis, suggesting that
the inhibition of CAF viability was partly associated with the
disruption of glucose and amino acid metabolism. Furthermore,
DIAPH1, which was previously reported to be associated with
the differentiation of hepatic stellate cells into tumor-promoting
myofibroblasts (21), was
decreased after E7386 treatment. The expression levels of
ACTA2/αSMA, which indicates the general phenotype of CAFs,
were also found to be decreased on both gene and protein expression
levels after treatment with E7386, suggesting that E7386 inhibited
the activation of CAFs.
Among the genes in the glucose and amino acid
metabolic pathways, the expression of ASNS, PSAT1, PHGDH and
PCK2 were increased in organoids after treatment with 100 nM
E7386, which also decreased cell viability. Hydrophilic metabolomic
analysis of E7386-treated organoids revealed the depletion of
glucose, essential and several non-essential amino acids, which
supported the transcriptomic data, particularly in case 21. In the
tumor environment, metabolic adaptations can occur, which may
reflect the survival responses of not only cancer cells but also
the surrounding fibroblasts (23,24).
These adaptations include metabolite sharing and nutrient
competition (23,24).
In addition to those directly related to glucose and
amino acid metabolism, upregulation of CHAC1 expression in
organoids by E7386 was suggested to act by the pharmacological
inhibition of antiporter system Xc−,
resulting in cystine-glutamate exchange and the induction of
endoplasmic reticulum stress and ferroptosis (25). Upregulation of ULBP1
expression suggested that E7386 may activate natural killer group
2, member D (NKG2D). It has been extensively reported that
induction of the NKG2D ligands, ULBP1-6 and major
histocompatibility complex class I chain-related genes A/B), can
enhance NK cell recognition of cancer cells to mediate the lysis of
the latter (26).
E7386 suppresses the β-catenin/cAMP-binding protein
axis by inhibiting their interaction without affecting the
β-catenin/p300 complex (10).
Therefore, E7386 was considered to function as a Wnt signaling
modulator (10). The precise
molecular mechanisms underlying the E7386-induced phenotypes remain
poorly understood. Wnt signaling activity is closely associated
with glucose level in tumor cells, such that glucose can promote
the formation of the lymphoid enhancer binding factor 1
(LEF1)/β-catenin complex, leading to β-catenin acetylation and
increased transcriptional activity (27). The close interaction among the
signals related to LEF1/β-catenin-driven gene expression changes
raises the possibility of a positive feedback loop, where Wnt
suppression may cause alterations in glucose metabolism. β-catenin
has been reported to serve a role in NK T-cell development and
function (28). A recent study
showed that suppression of β-catenin activity by ICG-001, an
inhibitor of the interaction between cAMP-binding protein and
β-catenin, enhanced the early IFN-γ responses of NKT cells
stimulated with a-galactosylceramide (29). These results suggest that
upregulation of ULBP1 may be involved in the activation.
To confirm the transcriptomic data found after the
in vitro experiments, immunohistochemical staining analysis
was performed using E7386-treated PDX models. PCK2-positivity in
the CRC cells was increased in the E7386-treated PDX group from
case 21, which was consistent with results from immunoblotting
analysis. However, PCK2 protein expression levels remained
unchanged in cases 32 and 28. In the hydrophilic metabolomic
analysis of the in vitro experiments, the concentrations of
G6P and G1P in the control of case 21 were higher compared with
those of cases 32 and 28, whereas those of case 21 were decreased
by treatment with E7386. This suggested that E7386 treatment
induced glucose depletion. The cause of this higher potency of
E7386 on glucose metabolism in case 21 warrants further
investigation. Enhancement of immunoreactivity to VLDLR in
E7386-administered PDXs from case 32 was consistent with the DNA
microarray data. VLDLR is a multifunctional receptor that regulates
cellular signaling by binding numerous ligands. Its expression has
been reported to be downregulated in CRC tissues compared with that
in their corresponding paired adjacent non-tumor tissues.
Additionally, VLDLR overexpression has been shown to inhibit the
proliferation and migration of CRC cells (30). VLDLR was proposed to mediate
effects on the Wnt/β-catenin signaling pathway (31). Although changes in the expression
of β-catenin on both gene and protein levels could not be detected
in the present study, a tendency for its reduction in the nucleus
according to immunohistochemical staining was observed in the
E7386-administered PDXs from case 28. There is a possibility that
the β-catenin-negative regions in the nucleus were predominantly
grown by E7386 administration in PDXs. To verify the transcriptomic
data obtained in the in vitro experiments using CAFs,
immunohistochemical staining for αSMA using PDX models was
conducted. αSMA expression in the fibroblasts surrounding the CRC
cell foci in the E7386-administered groups from cases 28 and 21
showed a tendency to decrease compared with that in the
corresponding control group. This was consistent with the
transcriptomic data.
In conclusion, the present comprehensive molecular
analysis of E7386-treated CRC organoids, CAFs and PDXs revealed
that several independent molecular mechanisms may be involved
upstream of the reduction of cell viability. Alterations in the
expression of genes associated with the glucose and amino acid
metabolic pathways, namely PCK2, ASNS, PSAT1 and PHGDH, were
observed. In addition, genes related to the stimulation of stress
responses and NK-cell mediated cytotoxicity, specifically CHAC1 and
ULBP1, were found to be altered. Modifications in the expression
and localization of components in the Wnt/β-catenin signaling
pathway, VLDLR and β-catenin, were also found. Further studies
using preclinical models are required to clarify the molecular
mechanisms underlying the E7386-induced reactions, with focus on
the Wnt/β-catenin signaling pathway. Furthermore, association
studies and analyses between preclinical data and clinical
sample-based biomarker need to be performed to evaluate the
accuracy of the preclinical models used in the present study.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Ms Yurika Shiotani
(The Omics Core and Animal Core Facilities of the National Cancer
Center Research Institute, Tokyo, Japan) for her technical support
in histopathological evaluation. In addition, the authors would
like to thank Mr Ryoichi Masui and Ms Ruri Nakanishi (Central
Animal Division, National Cancer Center Research Institute, Tokyo,
Japan) for their technical assistance.
Funding
The present study was supported by a grant for Research on
Development of New Drugs (Funding for Research to Expedite
Effective Drug Discovery by Government, Academia and Private
Partnership; grant no. JP19ak0101043h0105) from the Japan Agency
for Medical Research and Development. The core facilities were
supported by the National Cancer Center Research and Development
Fund (grant no. 2020-J-002).
Availability of data and materials
The datasets presented in this study can be found in
online repositories. The names of the repository/repositories and
accession number(s) can be found below: DDBJ database: https://ddbj.nig.ac.jp/public/ddbj_database/gea/experiment/E-GEAD-000/E-GEAD-492
(GEA accession number, E-GEAD-492).
Authors' contributions
MO, SI, MK and YK performed the experiments. MN, KM,
AH, TS, YH and AY contributed to the analysis and interpretation of
data. AO conceptualized the study. TI was responsible for the study
design and data analysis. All authors confirm the authenticity of
the raw data and have read and approved the final manuscript.
Ethics approval and consent to
participate
The use of surgical specimens in this study was
approved by the Ethics Committee of the National Cancer Center
(approval no. 2015-108) and written informed consent was obtained
from all patients. Ethics approval was obtained from the National
Cancer Center Animal Ethics Committee (approval no. T17-006) for
the animal studies.
Patient consent for publication
Not applicable.
Competing interests
YH and AY report personal fees from Eisai Co., Ltd.
for the duration of the present study and outside of the present
submitted work. The other authors declare that they have no
competing interests.
References
|
1
|
Dekker E, Tanis PJ, Vleugels JL, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD, Goding Sauer A,
Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA and Jemal
A: Colorectal cancer statistics, 2020. CA Cancer J Clin.
70:145–164. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fearon ER and Vogelstein B: A genetic
model for colorectal tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar
|
|
4
|
Burrell RA, McGranahan N, Bartek J and
Swanton C: The causes and consequences of genetic heterogeneity in
cancer evolution. Nature. 501:338–345. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Normanno N, Rachiglio AM, Lambiase M,
Martinelli E, Fenizia F, Esposito C, Roma C, Troiani T, Rizzi D,
Tatangelo F, et al: Heterogeneity of KRAS, NRAS, BRAF and PIK3CA
mutations in metastatic colorectal cancer and potential effects on
therapy in the CAPRI GOIM trial. Ann Oncol. 26:1710–1714. 2015.
View Article : Google Scholar
|
|
6
|
Kim TM, Lee SH and Chung YJ: Clinical
applications of next-generation sequencing in colorectal cancers.
World J Gastroenterol. 19:6784–6793. 2013. View Article : Google Scholar
|
|
7
|
Cancer Genome Atlas Network, .
Comprehensive molecular characterization of human colon and rectal
cancer. Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Berdiel-Acer M, Sanz-Pamplona R, Calon A,
Cuadras D, Berenguer A, Sanjuan X, Paules MJ, Salazar R, Moreno V,
Batlle E, et al: Differences between CAFs and their paired NCF from
adjacent colonic mucosa reveal functional heterogeneity of CAFs,
providing prognostic information. Mol Oncol. 8:1290–1305. 2014.
View Article : Google Scholar
|
|
9
|
Naruse M, Ochiai M, Sekine S, Taniguchi H,
Yoshida T, Ichikawa H, Sakamoto H, Kubo T, Matsumoto K, Ochiai A
and Imai T: Re-expression of REG family and DUOXs genes in CRC
organoids by co-culturing with CAFs. Sci Rep. 11:20772021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamada K, Hori Y, Inoue S, Yamamoto Y, Iso
K, Kamiyama H, Yamaguchi A, Kimura T, Uesugi M, Ito J, et al:
E7386, a selective inhibitor of the interaction between
beta-catenin and CBP, exerts antitumor activity in tumor models
with activated canonical Wnt signaling. Cancer Res. 81:1052–1062.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou Y, Zhou B, Pache L, Chang M,
Khodabakhshi AH, Tanaseichuk O, Benner C and Chanda SK: Metascape
provides a biologist-oriented resource for the analysis of
systems-level datasets. Nat Commun. 10:15232019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Soga T, Baran R, Suematsu M, Ueno Y, Ikeda
S, Sakurakawa T, Kakazu Y, Ishikawa T, Robert M, Nishioka T and
Tomita M: Differential metabolomics reveals ophthalmic acid as an
oxidative stress biomarker indicating hepatic glutathione
consumption. J Biol Chem. 281:16768–16776. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hirayama A, Kami K, Sugimoto M, Sugawara
M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, et
al: Quantitative metabolome profiling of colon and stomach cancer
microenvironment by capillary electrophoresis time-of-flight mass
spectrometry. Cancer Res. 69:4918–4925. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Naruse M, Ishigamori R and Imai T: The
unique genetic and histological characteristics of DMBA-induced
mammary tumors in an organoid-based carcinogenesis model. Front
Genet. 12:7687312021. View Article : Google Scholar
|
|
16
|
Kanda Y: Investigation of the freely
available easy-to-use software ‘EZR’ for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar
|
|
17
|
Bae JH, Kim SJ, Kim MJ, Oh SO, Chung JS,
Kim SH and Kang CD: Susceptibility to natural killer cell-mediated
lysis of colon cancer cells is enhanced by treatment with epidermal
growth factor receptor inhibitors through UL16-binding protein-1
induction. Cancer Sci. 103:7–16. 2012. View Article : Google Scholar
|
|
18
|
Vincent EE, Sergushichev A, Griss T,
Gingras MC, Samborska B, Ntimbane T, Coelho PP, Blagih J, Raissi
TC, Choiniere L, et al: Mitochondrial phosphoenolpyruvate
carboxykinase regulates metabolic adaptation and enables
glucose-independent tumor growth. Mol Cell. 60:195–207. 2015.
View Article : Google Scholar
|
|
19
|
Arce L, Pate KT and Waterman ML: Groucho
binds two conserved regions of LEF-1 for HDAC-dependent repression.
BMC Cancer. 9:1592009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen Z, Chen G and Zhao H: FDPS promotes
glioma growth and macrophage recruitment by regulating CCL20 via
Wnt/β-catenin signalling pathway. J Cell Mol Med. 24:9055–9066.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu D, Fu X, Wang Y, Wang X, Wang H, Wen J
and Kang N: Protein diaphanous homolog 1 (Diaph1) promotes
myofibroblastic activation of hepatic stellate cells by regulating
Rab5a activity and TGFβ receptor endocytosis. FASEB J.
34:7345–7359. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Clevers H and Nusse R: Wnt/β-catenin
signaling and disease. Cell. 149:1192–1205. 2012. View Article : Google Scholar
|
|
23
|
Kalluri R: The biology and function of
fibroblasts in cancer. Nat Rev Cancer. 16:582–598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lyssiotis CA and Kimmelman AC: Metabolic
interactions in the tumor microenvironment. Trends Cell Biol.
27:863–875. 2017. View Article : Google Scholar
|
|
25
|
Dixon SJ, Patel DN, Welsch M, Skouta R,
Lee ED, Hayano M, Thomas AG, Gleason CE, Tatonetti NP, Slusher BS
and Stockbell BR: Pharmacological inhibition of cystine-glutamate
exchange induces endoplasmic reticulum stress and ferroptosis.
Elife. 3:e025232014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Raulet DH, Gasser S, Gowen BG, Deng W and
Jung H: Regulation of ligands for the NKG2D activating receptor.
Annu Rev Immunol. 31:413–441. 2013. View Article : Google Scholar
|
|
27
|
Chocarro-Calvo A, Garcia-Martinez JM,
Ardila-Gonzalez S, De la Vieja A and Garcia-Jimenez C:
Glucose-induced β-catenin acetylation enhances Wnt signaling in
cancer. Mol Cell. 49:474–486. 2013. View Article : Google Scholar
|
|
28
|
Kling JC and Blumenthal A: Roles of WNT,
NOTCH, and Hedgehog signaling in the differentiation and function
of innate and innate-like lymphocytes. J Leukoc Biol. 101:827–840.
2017. View Article : Google Scholar
|
|
29
|
Kling JC, Jordan MA, Pitt LA, Meiners J,
Thanh-Tran T, Tran LS, Nguyen TT, Mittal D, Villani R, Steptoe RJ,
et al: Temporal regulation of natural killer T Cell interferon
gamma responses by beta-catenin-dependent and -independent Wnt
signaling. Front Immunol. 9:4832018. View Article : Google Scholar
|
|
30
|
Kim BK, Yoo HI, Lee AR, Choi K and Yoon
SK: Decreased expression of VLDLR is inversely correlated with
miR-200c in human colorectal cancer. Mol Carcinog. 56:1620–1629.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lee K, Shin Y, Cheng R, Park K, Hu Y,
McBride J, He X, Takahashi Y and Ma JX: Receptor heterodimerization
as a novel mechanism for the regulation of Wnt/β-catenin signaling.
J Cell Sci. 127:4857–4869. 2014.
|