Introduction
Although lung cancer is a malignant tumor with the
highest rates of morbidity and mortality in the world (1), its pathogenesis remains unclear.
Recurrence and metastasis are the main hurdles in lung cancer
treatment. Angiogenesis plays a key role in tumor growth,
development, and metastasis (2).
Thus, blocking angiogenesis can inhibit tumor growth and metastasis
and induce tumor cell dormancy and apoptosis. However, the
mechanism underlying the regulation of lung cancer angiogenesis
remains unclear.
Gap junctional intercellular communication (GJIC)
plays an important role in the control of cell growth,
differentiation, homeostasis, and morphogenesis (3). Gap junction proteins, also called
connexins (Cxs), coordinate the functions of various cells by
forming gap junctions and affect cell growth and differentiation by
regulating gene expression (4).
Cxs are four-joint integral membrane proteins assembled by hexamers
to form a hemichannel, which connects cells in a head-to-head
manner to form gap junctions (5).
The functions of gap junctions, including exchange of metabolites
and electrical signals between cells, are highly diverse and also
include functions not related to cell-cell communication (6). Gap junctions allow inorganic ions
(Na+, K+, and Ca2+) and molecules
<1,000 Da to directly diffuse between adjacent cells (5,7–10),
which is important for the development of cells, tissues, and
organs as well as for normal metabolism and homeostasis (11–13).
Recently, the association between gap junctions and angiogenesis
has become a research hotspot. The activity of gap junction
channels is regulated through changes in voltage, calcium ion
concentration, pH, phosphorylation level, and protein interactions
(6,8). Among them, phosphorylation is deeply
involved in the regulation process during gap junction protein
synthesis, transport, assembly, membrane insertion,
internalization, and degradation as well as protection of gap
junction hemichannels and complete channels (7,8,12).
In a previous study by the authors, it was
determined that the expression of Cx43 was improved in vascular
endothelium of lung cancer compared with the normal control (Zhou
et al, unpublished data). Previous research has revealed
that Cx43 may participate in angiogenesis mainly by regulating
branching of capillaries (14,15),
and formed gap junctions play an important role in determining the
fate of endothelial cells, necessary for vascularization during the
process of damage repair (16–18).
Moreover, decreased expression of Cx43 can cause vascular
dysfunction and impair the process of neovascularization (18–20);
it can also directly inhibit endothelial cell migration,
proliferation, and angiogenesis and reduce vascular endothelial
growth factor (VEGF) transcription and translation (20). Recently, it was revealed that Cx43
plays an important role in angiogenesis (21). Based on these studies, it was
hypothesized that Cx43 was the key regulator factor involved in
angiogenesis. However, its specific mechanism in angiogenesis
especially in lung cancer angiogenesis remains to be
elucidated.
The C-terminal region is the primary region of Cx43
that is phosphorylated. It contains 21 serine and 2 tyrosine
residues, which are the targets of phosphorylation by several
protein kinases, such as mitogen-activated protein kinase (MAPK),
protein kinase A (PKA), protein kinase C (PKC), p34 (Cdc2)/cyclin B
kinase, casein kinase 1, and pp60 (src) kinase (12,22,23).
Tyr265 is phosphorylated by Src, Ser279 by MAPK, Ser368 by PKC, and
Ser373 by AKT/PKB (24).
Based on the available evidence, it was hypothesized
that several downstream proteins could be involved in angiogenesis,
and activation of the phosphorylation sites on the C-terminus of
Cx43 may trigger this regulatory pathway. In the present study, a
specific small-interfering RNA (siRNA) to inhibit Cx43
intracellularly and Cx43-overexpressing recombinant plasmid were
used to explore the role of Cx43 in regulating angiogenesis in lung
cancer. Furthermore, it was explored whether Cx43 modulates
angiogenesis depending on the activation of its C-terminal
phosphorylation sites and its possible association with downstream
zonula occludens-1 (ZO-1), E-cadherin, β-catenin, von Willebrand
factor (vWF), and plasminogen activator inhibitor-1 (PAI-1)
signaling proteins. Through in vitro experiments, the effect
and underlying action mechanism of Cx43 in angiogenesis was
explored. Finally, it was determined whether Cx43 is a
proangiogenic effector and the corresponding signaling pathway was
explored, which will provide new insight for the treatment of lung
adenocarcinoma by regulating Cx43-related angiogenesis.
Materials and methods
Reagents and instruments
The main reagents and instruments used the present
study included the following: Human pulmonary microvascular
endothelial cells (HPMECs; cat. no. HUM-iCell-a001; iCell
Bioscience, Inc.), Endothelial Cell Medium (ECM; ScienCell Research
Laboratories, Inc.); Lucifer Yellow CH (Invitrogen; Thermo Fisher
Scientific, Inc.), Matrigel (Corning, Inc.), Cell Lysis Buffer
(product no. 9803; Cell Signaling Technology, Inc.), vWF antibody
(product no. 65707; Cell Signaling Technology, Inc.), PAI-1
antibody (product code ab66705; Abcam), phosphorylated (p)-Cx43
(Tyr265) antibody (cat. no. AF2306; Affinity Biosciences), p-Cx43
(Ser279) antibody (cat. no. AF8228; Affinity Biosciences), p-Cx43
(Ser368) antibody (cat. no. AF3199; Affinity Biosciences), p-Cx43
(Ser373) antibody (cat. no. PA5-64670; Invitrogen; Thermo Fisher
Scientific, Inc.), Cx43 antibody (cat. no. AF0137; Affinity
Biosciences), ZO-1 antibody (product code ab96587; Abcam),
E-cadherin antibody (product no. 3195; Cell Signaling Technology,
Inc.), β-catenin antibody (product no. 8480; Cell Signaling
Technology, Inc.), Na+/K+-ATPase (product
code ab76020; Abcam), GAPDH (product code ab37168; Abcam), TRIpure
Total RNA Extraction Reagent (cat. no. EP013), EntiLink™ 1st Strand
cDNA Synthesis Kit (cat. no. EQ003), EntiLink™ PCR Master Mix (cat.
no. EQ004), Gel DNA Purification Kit (cat. no. EP006) and EndoFree
Plasmid Miniprep Kit [cat. no. EP004; all from Elk (Wuhan)
Biotechnology Co., Ltd.], an inverted microscope (OLYMPUS IX51;
Olympus Corporation), a fluorescence microscope (OLYMPUS BX51;
Olympus Corporation), a CO2 incubator (model no. SCO6WE;
SHEL LAB; Sheldon Manufacturing, Inc.), a biological safety cabinet
(model no. SW-CJ-1FD; Suzhou Antai Airtech Co., Ltd.), an imaging
system (MicroPublisher; QImaging), a microplate reader (model no.
DR-200Bs; Diatek), scanner (model no. LiDE110; Canon, Inc.), and a
real-time PCR system (StepOne™ Real-Time PCR System; Life
Technologies; Thermo Fisher Scientific, Inc.).
Plasmid constructs and transfection of
siRNA and plasmid-DNA
Design, synthesis, and silencing
efficiency of siRNA targeting Cx43 gene
siRNAs targeting the Cx43 gene (GenBank ID:
NM_000165.5) were designed. Multiple siRNA candidates were
obtained, of which three candidates with strong specificity were
selected for further analysis using the Basic Local Alignment
Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi). The three
candidate siRNAs [siRNA-1 base sequence (5′-3′): sense,
GCGCCUUAGGCAAACUCCUUGACAATT and antisense,
UUGUCAAGGAGUUUGCCUAAGGCGCTT; siRNA-2 base sequence (5′-3′): sense,
CCACACUCUUGUACCUGGCUCAUGUTT and antisense,
ACAUGAGCCAGGUACAAGAGUGUGGTT; siRNA-3 base sequence (5′-3′): sense,
AGUACGGUAUUGAAGAGCAUGGUAATT and antisense,
UUACCAUGCUCUUCAAUACCGUACUTT] and siRNAs with no homologous sequence
to the Cx43 fragment (i.e., siRNA control group; base sequence
(5′-3′): sense, GCGGAUUAACGCUCAGUUCACCCAATT and antisense,
UUGGGUGAACUGAGCGUUAAUCCGCTT) were chemically synthesized by HYcell
Biotechnology Company. The siRNAs were then individually
transfected into HPMECs cells, and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) was
used to detect the expression of Cx43 in the cells after 24 h. The
silencing efficiency of each candidate siRNA was determined, and
the siRNA sequence [base sequence (5′-3′): sense,
CCACACUCUUGUACCUGGCUCAUGUTT and antisense,
ACAUGAGCCAGGUACAAGAGUGUGGTT) with the highest interference
efficiency was selected for subsequent experiments.
Construction and identification of
recombinant plasmid
To biosynthesize the Cx43 gene, pcDNA3.1-CX43-HA
plasmid (GenScript) was constructed using the PCR-based Accurate
Synthesis method (25). The
plasmid was transferred to a TOP10 cloned strain, which was
cultured on Luria Bertani medium containing ampicillin overnight at
37°C. A single colony was selected and inoculated in Luria Bertani
broth containing ampicillin and cultured overnight at 37°C and
13,000 × g. Following incubation, Escherichia coli cells
containing the pcDNA3.1-CX43-HA plasmid were obtained [EndoFree
Plasmid Miniprep Kit; Elk (Wuhan) Biotechnology Co., Ltd.], and the
Cx43 fragment was amplified with bacterial plasmid as a template.
After the plasmid was extracted [Gel DNA Purification Kit, Elk
(Wuhan) Biotechnology Co., Ltd.] from the E. coli cells, it
was identified through XhoI and KpnI enzyme
digestion, and the whole recombinant plasmid was sequenced
[EntiLink™ PCR Master Mix, Elk (Wuhan) Biotechnology Co.,
Ltd.].
Transfection
Log-phase HPMECs were seeded in a 6-well plate at
2×105 cells/well and cultured at 37°C in a 5%
CO2 incubator. After the cells were observed to adhere
to the walls for 24 h and reached 80-90% confluence (inverted
microscope; Olympus Corporation), they were transfected for 6 h at
37°C following the Lipofectamine™ 2000 transfection procedure of
Invitrogen; Thermo Fisher Scientific, Inc.
Cell culture and experimental
design
Frozen vials of primary cells were removed from a
liquid nitrogen storage tank and thawed quickly by placing the
vials in a 37°C water bath and simultaneously shaking them
vigorously. The cell suspensions (1 ml) from the vials were
transferred to 175-cm2 culture flasks containing ECM
(ScienCell Research Laboratories, Inc.). The flasks were incubated
at 37°C and 5% CO2. Subsequently, the medium was changed
to fresh standard cell culture medium to remove dimethyl sulfoxide
and incubated overnight. The medium was replaced every other day
until the cells reached 80-90% confluence (inverted microscope;
Olympus Corporation). Then, the cells were digested with
trypsin/EDTA solution and neutralized using complete medium
containing 5% fetal bovine serum (ScienCell Research Laboratories,
Inc.). Next, the cells were transferred to a 50-ml centrifuge tube
and centrifuged for 3 min at 220 × g at room temperature, and the
supernatant was discarded. The cells were resuspended in standard
cell culture medium and counted using disposable hemocytometers.
The cells were seeded at 30,000 cells per cm2 at 37°C in
a 5% CO2 humidified incubator for rapid growth. Passage
numbers 3-5 were used for subsequent experiments. RT-qPCR, western
blotting, Scrape loading/dye transfer assay, Cell Counting Kit-8
(CCK-8) assay, Transwell migration assay, and immunofluorescence
assay were performed in a biological safety cabinet (Suzhou Antai
Airtech Co., Ltd.). An angiogenesis 96-well microplate was used for
the tube formation assay.
Tube formation assay
Matrigel (Corning, Inc.) was placed on ice and
refrigerated (−4°C) overnight the day before cells were seeded.
Matrigel (10 µl) was then applied to each inner well, and the slide
was covered with a lid. Next, the slide was placed in an incubator
for polymerization (60 min). In addition, the cell suspension was
prepared. For a final density of 15,000 cells/well, a cell
suspension of 3×105 cells/ml was used from each group
(normal control group, connexin 43 group and small-interfering RNA
group; resuspended in antibiotic-free standard medium) and pipetted
up and down to mix thoroughly. The slide was removed from the
incubator and placed on a rack, and 50 µl of cell suspension was
added to each top well of the slide (repeated six times for each
group). The slide was covered with the lid and observed for
well-proportioned cells. The cells were then incubated at 37°C for
20 h in a humidified incubator. Finally, images were acquired using
a microscope (inverted microscope; Olympus Corporation;
magnification, ×50) and preserved. Tube network formation was
assessed by measuring all parameters with ImageJ Angiogenesis
Analyzer (ImageJ bundled with 64-bit Java 1.8.0_172; National
Institutes of Health), including the number of branches and nodes
and the length and spacing of the branches, which were selected for
evaluating the angiogenic potential.
Scrape loading/dye transfer and
characterization of gap junction channel function
Cells were cultured in a 6-well plate to 100%
confluency (inverted microscope; Olympus Corporation) as
aforementioned. The medium was discarded via suction with a
pipette, and Dulbecco's phosphate-buffered saline (DPBS) (Ca/Mg)
(product no. 14040133; Gibco; Thermo Fisher Scientific, Inc.) was
used to wash the cells three times (2 ml/well). Cells in each well
were then covered with LY dye solution (Lucifer Yellow CH;
Invitrogen; Thermo Fisher Scientific, Inc.; 1 ml); the dye was
loaded onto the cells by scratching one line with a blade tip of a
no. 15 surgical scalpel. Subsequently, the plates were kept on a
clean bench at room temperature for up to 4 min, allowing the LY
dye to pass across several adjacent cell layers through functional
gap junctions. To minimize fluorescence photobleaching of the dye,
the plates were concurrently covered with aluminum foil. LY dye
solution was then discarded from the plate wells. The cells were
rinsed twice with DPBS (Ca/Mg) to discard the extracellular dye,
and 2 ml of normal medium was then added into every well. The cells
were immediately observed using a fluorescence microscope (Olympus
Corporation) at a 100-fold magnification. The fluorescence distance
of the dye spread was measured using ImageJ (ImageJ bundled with
64-bit Java 1.8.0_172; National Institutes of Health) This
experiment was performed eight times. Four images and associated
results were randomly selected from the normal control group,
connexin 43 group and small-interfering RNA group.
CCK-8 assay
Once the cells reached 90% confluence (inverted
microscope; Olympus Corporation), conventional hemocytometer-based
cell counting was performed. Each of the 60 wells in the center of
a 96-well plate was pipetted with 100 µl of cells (concentration
2×105 cells/ml), and the surrounding wells were pipetted
with an equal volume of PBS (100 µl). Cells were labeled and
incubated at 37°C for 24 h. After 24 h of serum deprivation in an
equal volume of serum-free medium, the cells were washed once with
PBS. Then cells were grouped (normal control group, connexin 43
group and small-interfering RNA group). Six replicates were
performed for each of the groups, and the cells were cultured for
24 h after treatment. A 10% CCK-8 solution was prepared by adding
1.2 ml of CCK-8 (Dojindo Molecular Technologies, Inc.) into 12 ml
of Endothelial Cell Medium (ScienCell Research Laboratories, Inc.).
The treated cells were washed with PBS, and 100 µl of 10% CCK-8 was
added to each well. Following incubation for 3 h, the absorbance
(OD value) at 450 nm was measured using a microplate reader.
Transwell migration assays
Cells were counted using a hemocytometer, and the
concentration was adjusted to 1×105 cells/ml. In a
Transwell plate (pore size, 3 µm; thickness, 10 µm; diameter, 12
mm), 200 µl of solution of each group (normal control group,
connexin 43 group and small-interfering RNA group) was respectively
added to corresponding chamber; 800 µl complete medium was added to
each of the three wells. Cells were labeled and cultured for 24 h;
after a single PBS wash, the non-migrated cells were then swabbed
with cotton; whereas, migrated cells were fixed using
paraformaldehyde (concentration, 4%; duration, 30 min; room
temperature). After 20 min of 0.1% crystal violet staining at room
temperature, the migrated cells were washed thrice with PBS and the
entire chamber was cleaned to remove any residual dye. The filter
membrane was air-dried, cut and examined under a microscope
(inverted microscope; Olympus Corporation) on a glass slide. Three
fields were selected at random for cell counting.
Western blotting
Total protein was extracted from the three groups of
cells using Cell Lysis Buffer (Cell Signaling Technology, Inc.),
and the protein concentration was measured using a BCA protein
assay kit (cat. no. AS1086; Wuhan Aspen Biotechnology Co., Ltd.).
Subsequently, 50 µg of total protein from each sample was added to
10% polyacrylamide gel electrophoresis gels. According to the
pre-stained marker display, after judging that the target protein
was sufficiently separated, electrophoresis was terminated. The
proteins were then electroblotted onto a PVDF membrane and the
membrane was blocked with 1% western blocking reagent (product no.
11921681001; Roche Diagnostics GmbH) at room temperature for 1 h.
Subsequently, the membrane was successively incubated in the
primary antibody solutions for the targeted proteins [p-Cx43
(Tyr265) antibody (Affinity Biosciences), p-Cx43 (Ser279) antibody
(Affinity Biosciences), p-Cx43 (Ser368) antibody (Affinity
Biosciences), p-Cx43 (Ser373) antibody (Invitrogen; Thermo Fisher
Scientific, Inc.), Cx43 antibody (Affinity Biosciences), ZO-1
antibody (Abcam), E-cadherin antibody (Cell Signaling Technology,
Inc.), β-catenin antibody (Cell Signaling Technology, Inc.), vWF
antibody (Cell Signaling Technology, Inc.), PAI-1 antibody (Abcam),
Na+/K+-ATPase (Abcam) and GAPDH (Abcam); the
antibodies were diluted [p-Cx43 (Tyr265), 5% skim milk, 1:500;
p-Cx43 (Ser279), 5% skim milk, 1:500; p-Cx43 (Ser368), 5% skim
milk, 1:500; p-Cx43 (Ser373), 5% skim milk, 1:500; Cx43, 5% skim
milk, 1:3,000; ZO-1, 5% skim milk, 1:500; E-cadherin, 5% skim milk,
1:2,000; β-catenin, 5% skim milk, 1:1,000; vWF, 5% skim milk,
1:1,000; PAI-1, 5% skim milk, 1:1,000;
Na+/K+-ATPase, 5% skim milk, 1:1,000; GAPDH,
5% skim milk, 1:10,000] for one night at 4°C and then incubated in
the HRP-conjugated secondary antibody solution [HRP-Goat anti
Rabbit (cat. no. AS1107; Wuhan Aspen Biotechnology Co., Ltd.; the
antibody was diluted (5% skim milk, 1:10,000)] for 30 min at 37°C.
An appropriate amount of ECL substrate solution (cat. no. AS1059;
Aspen Biotechnology Co., Ltd.) was then added to each membrane, and
the membrane was incubated for several minutes. After the bands
appeared, filter paper was used to absorb the excess substrate
solution. The membrane was then exposed to an X-ray film. The film
was placed in the developer solution for development and fixing
solution for fixing (scanner; Canon, Inc.). Densitometry was
performed using AlphaEaseFC™ 4.0 software (ProteinSimple).
RT-qPCR
TRIpure Total RNA Extraction Reagent [cat. no.
EP013; Elk (Wuhan) Biotechnology Co., Ltd.] was used to extract the
total RNA of the cells from each group (normal control group,
connexin 43 group, and small-interfering RNA group), and 3.75 µg of
total RNA was used for reverse transcription of RNA to cDNA
[EntiLink™ 1st Strand cDNA Synthesis Kit; cat. no. EQ003; Elk
(Wuhan) Biotechnology Co., Ltd.], and the cDNA was subjected to PCR
to detect the target gene. GAPDH was used as an reference gene. The
conditions of the reverse transcription reaction were: 42°C for 60
min and 95°C for 5 min. The semi-quantitative reaction conditions
were: 94°C for 4 min, 94°C for 30 sec, 56°C for 30 sec, and 72°C
for 25 sec for 30 cycles. Real-time PCR was performed again, cDNA
was diluted 10 times, and the reaction conditions were: 50°C for 2
min, 95°C for 10 min, 95°C for 30 sec, and 60°C for 30 sec for 40
cycles. The PCR products were analyzed by electrophoresis in
agarose gels (1% gel) stained with ethidium bromide, and the gel
was imaged using a gel imaging system and then scanned in
grayscale. Integral optical density (IOD) was used to represent the
expression level (real-time PCR system; Thermo Fisher Scientific,
Inc.). 6-carboxyfluorescein (product code c1360; Invitrogen; Thermo
Fisher Scientific, Inc.) was used as a fluorophore in the process.
The ratio of the expression of target gene to that of GAPDH was
analyzed, and the relative expression level of the target gene was
calculated using 2−ΔΔCq method (26). The sequences of the primers are
listed in Table I. The experiment
was performed in triplicate.
 | Table I.Sequences of the primers and lengths
of the corresponding PCR products. |
Table I.
Sequences of the primers and lengths
of the corresponding PCR products.
| Gene | Primer | Base sequence
(5′-3′) |
| Tm | GC (%) | Product size
(bp) |
|---|
| GAPDH | NM_001256799.2 | Sense |
CATCATCCCTGCCTCTACTGG | 59.4 | 57.1 | 259 |
|
|
| Antisense |
GTGGGTGTCGCTGTTGAAGTC | 60.1 | 57.1 |
|
| CX43 | NM_000165.5 | Sense |
AAGGTTCAAGCCTACTCAACTGC | 59.9 | 47.8 | 188 |
|
|
| Antisense |
ACATGAGAGATTGGGAAAGACTTG | 59.4 | 41.7 |
|
| ZO-1 | NM_001301025.1 | Sense |
CAAGCCTGCAGAGTCCAAGC | 60.7 | 60 | 139 |
|
|
| Antisense |
TGAAGGTATCAGCGGAGGGA | 60.3 | 55 |
|
| E-CAD | NM_001317184.1 | Sense |
TTCTTCGGAGGAGAGCGG | 58.3 | 61.1 | 234 |
|
|
| Antisense |
CAATTTCATCGGGATTGGC | 58.5 | 47.4 |
|
| β-catenin | NM_001098209.1 | Sense |
GCCAAGTGGGTGGTATAGAGG | 58.7 | 57.1 | 192 |
|
|
| Antisense |
GGGATGGTGGGTGTAAGAGC | 59 | 60 |
|
| vWF | NM_000552.5 | Sense |
TGGAGAAACAGTGAAGATTGGC | 59.4 | 45.5 | 168 |
|
|
| Antisense |
ACCAGAACGTACTGGCACTCC | 59 | 57.1 |
|
| PAI-1 | NM_000602.4 | Sense |
ATTACTACGACATCCTGGAACTGC | 59.6 | 45.8 | 106 |
|
|
| Antisense |
AGAATGTTGGTGAGGGCAGAG | 59.1 | 52.4 |
|
Statistical analysis
SPSS 22.0 (IBM Corp.) software was utilized for
statistical analysis. The data are presented as the mean ± standard
deviation. Experimental groups were compared using one-way analysis
of variance followed by Student-Newman-Keuls post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of Cx43 and other
angiogenesis-related mRNAs in HPMECs
Following transfection of pcDNA3.1-CX43-HA plasmid,
the mRNA level of Cx43 in the Cx43 overexpression group (i.e, Cx43
group) increased by approximately two-fold, whereas following the
transfection of Cx43-specific siRNA in the Cx43-knockdown group
(i.e, Cx43 siRNA group), it decreased to 48.62±0.50% compared with
that in the normal control group (i.e, NC group) (P<0.01)
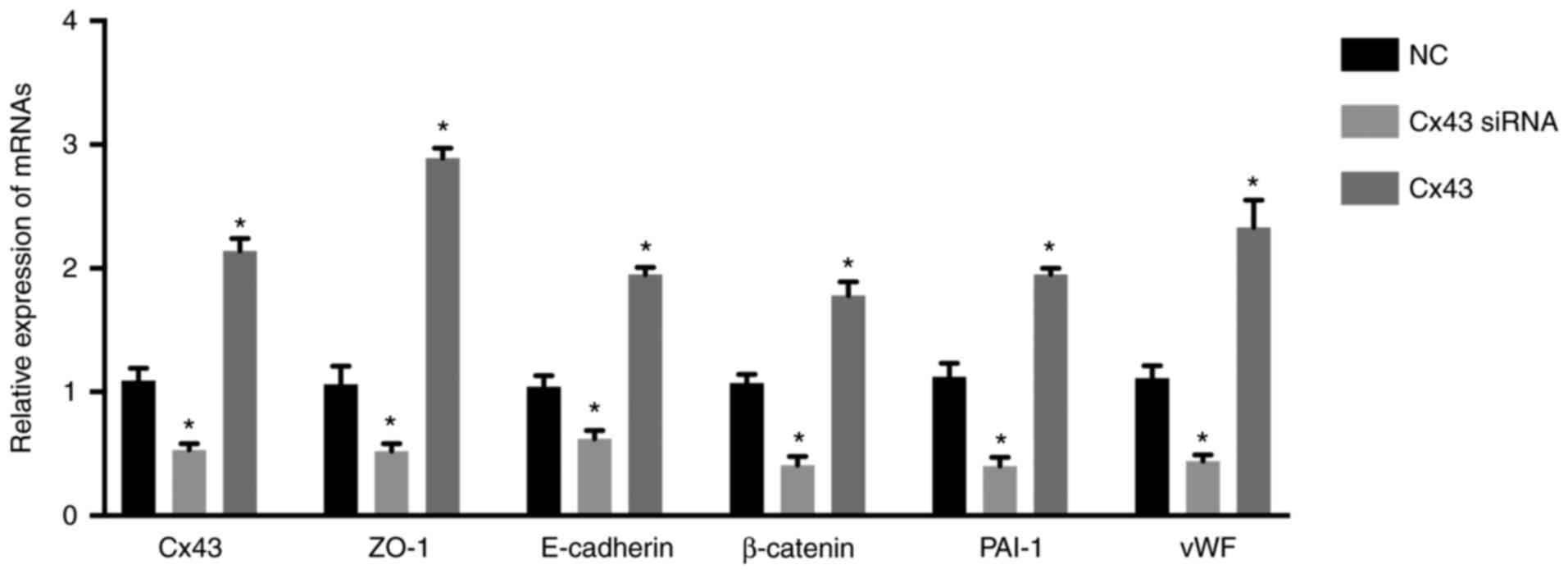
(Fig. 1). The mRNA levels of CX43,
ZO-1, E-cadherin, β-catenin, PAI-1, and vWF were significantly
increased in the Cx43 group and significantly decreased in the
Cx43-siRNA group compared with those in the normal control group
(P<0.01) (Fig. 1). These
results suggested that overexpression of Cx43 stimulated the mRNA
expression levels of ZO-1, E-cadherin, β-catenin, PAI-1, and vWF,
while silencing of Cx43 suppressed their expression levels.
Silencing of Cx43 suppresses and
overexpression of Cx43 induces ZO-1, E-cadherin, β-catenin, vWF,
and PAI-1 expression in HPMECs
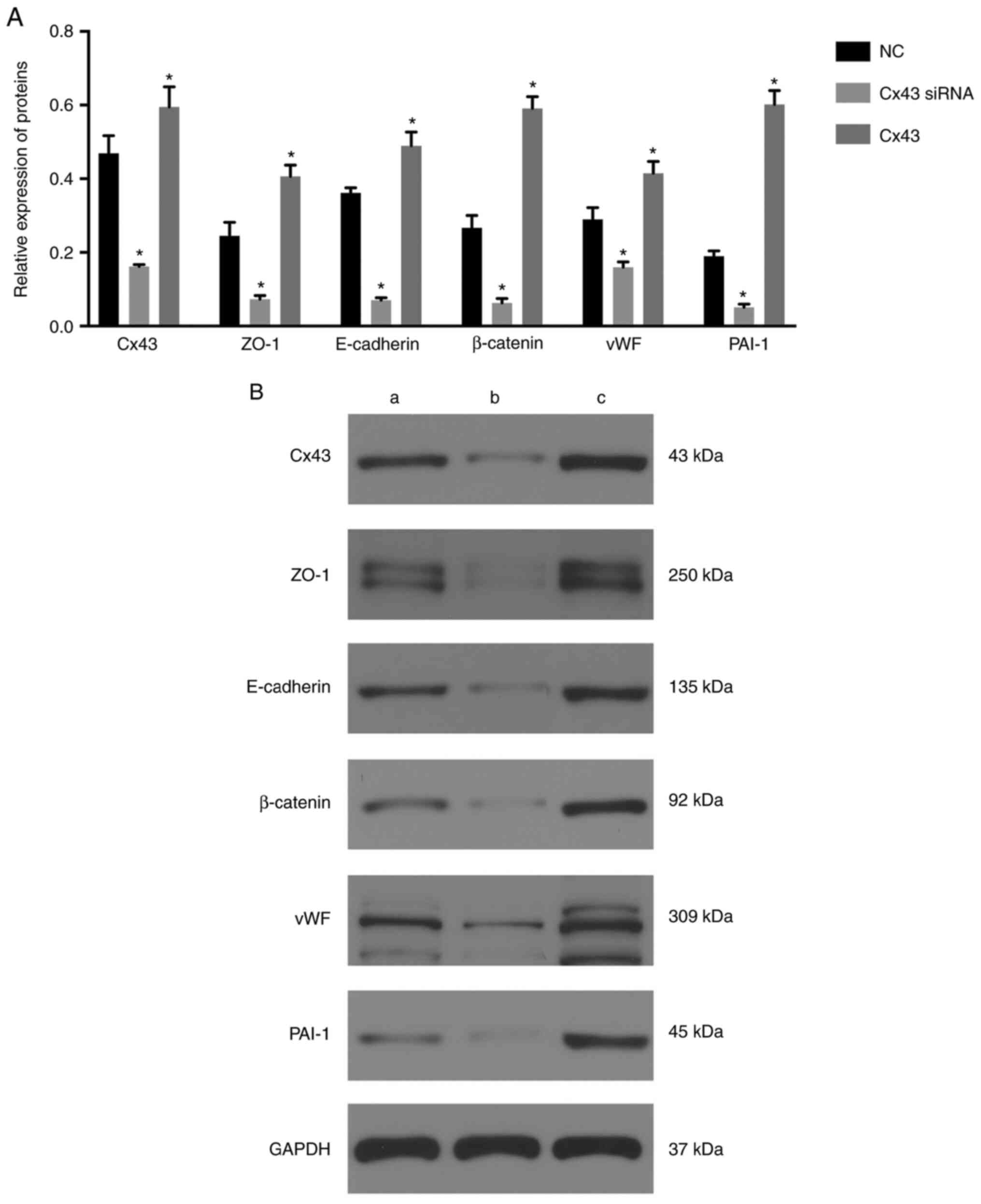
The expression levels of Cx43, ZO-1, E-cadherin,
β-catenin, vWF, and PAI-1 were significantly decreased in the Cx43
siRNA group and significantly increased in the Cx43 group compared
with those in the normal control group (P≤0.01) (Fig. 2). These results suggested that
overexpression of Cx43 stimulated the expression of ZO-1,
E-cadherin, β-catenin, vWF, and PAI-1, whereas silencing of Cx43
suppressed the expression of these molecules compared with that in
the normal control group.
Silencing of Cx43 suppresses and
overexpression of Cx43 induces VEGF-associated Ser279 expression in
HPMECs
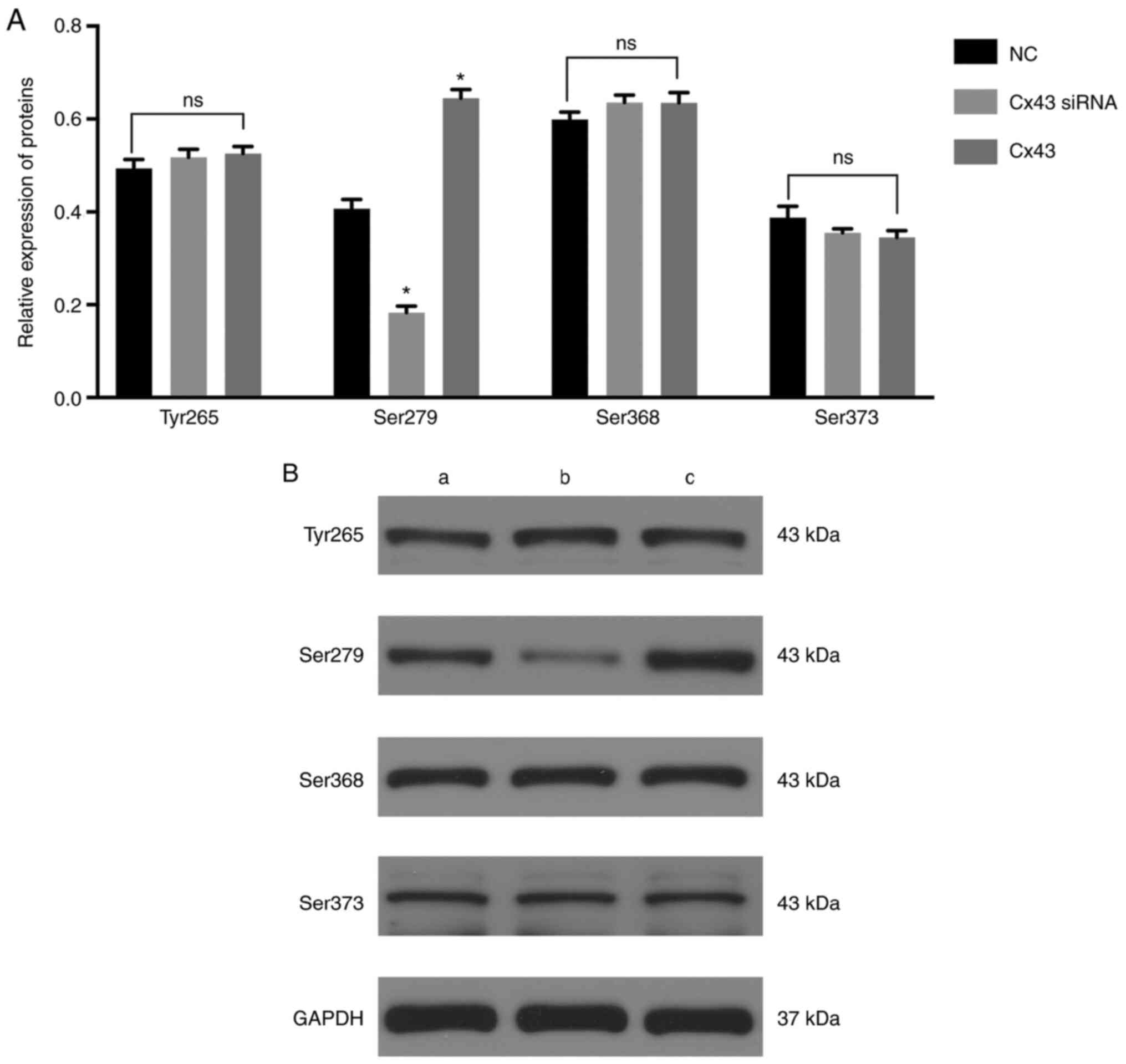
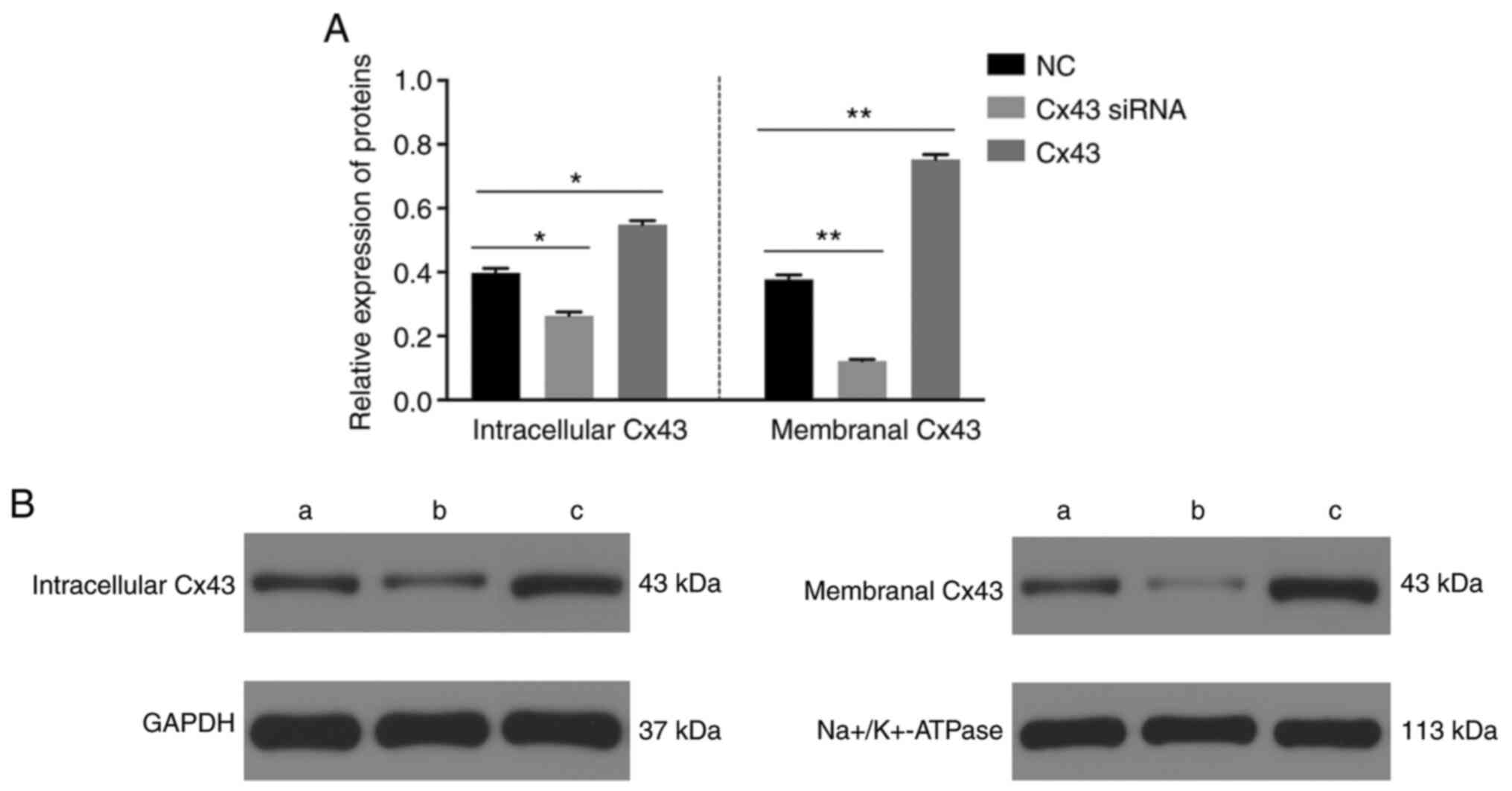
The levels of Ser279 (Fig. 3) and intracellular and membranal
levels of Cx43 were significantly decreased in the Cx43-knockdown
group, particularly the membranal levels of Cx43 (Fig. 4), whereas the levels were
significantly increased in the Cx43 group compared with those in
the normal control group (P<0.01) (Figs. 3 and 4). Conversely, the levels of Tyr265,
Ser368, and Ser373 in the Cx43-knockdown and overexpression groups
were not significantly different from those in the normal control
group (Fig. 3). These results
suggested that the overexpression of Cx43 stimulated the levels of
Ser279 and intracellular and membranal levels of Cx43, particularly
the membranal levels of Cx43.
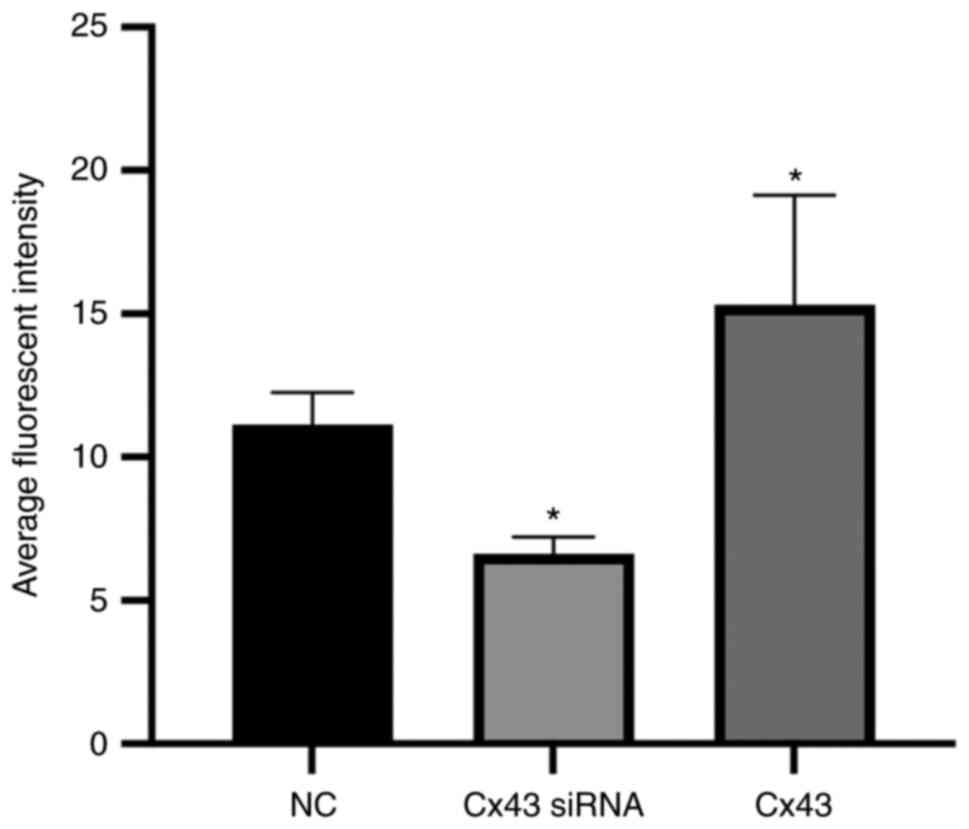
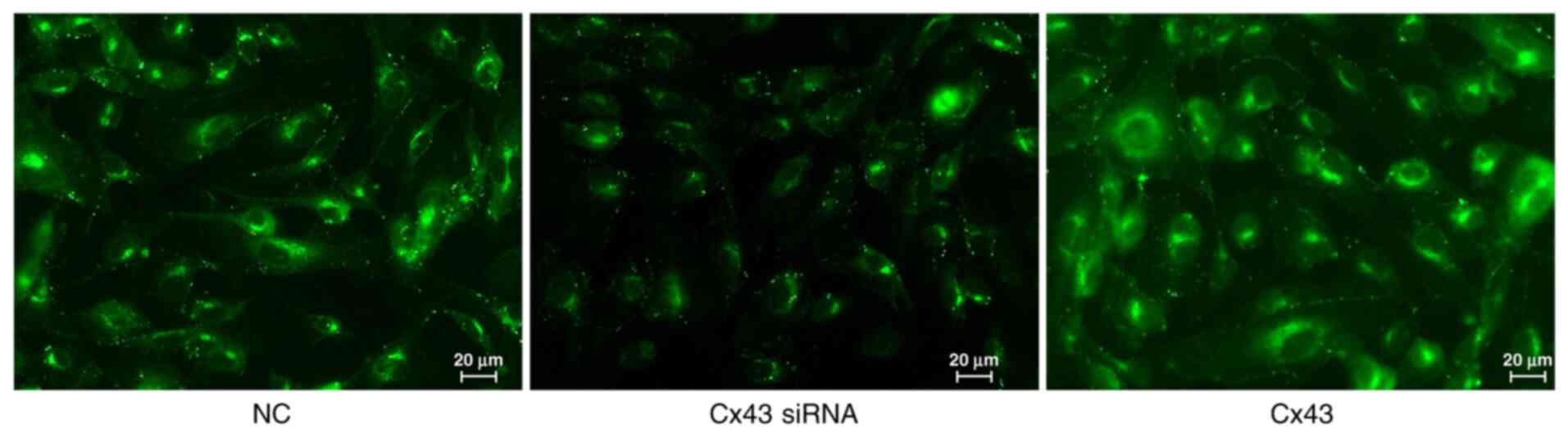
Average fluorescence intensity of
different groups
Immunofluorescence results showed that the average
fluorescence intensity for the Cx43 overexpression group was
higher, whereas for the Cx43-knockdown group it was lower compared
with that for the normal control group (×400; P<0.01). The
average fluorescence intensity in the Cx43 overexpression group was
increased by 1.4-fold, whereas in the Cx43-knockdown group it was
decreased by approximately 0.6-fold compared with that in the
normal control group (Figs. 5 and
6).
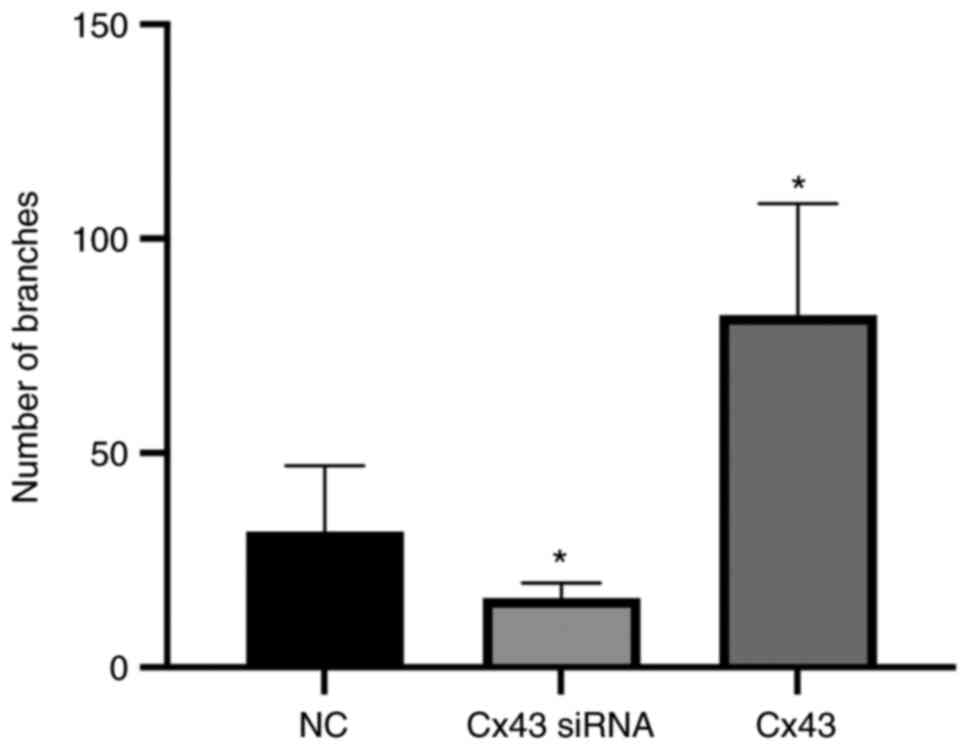
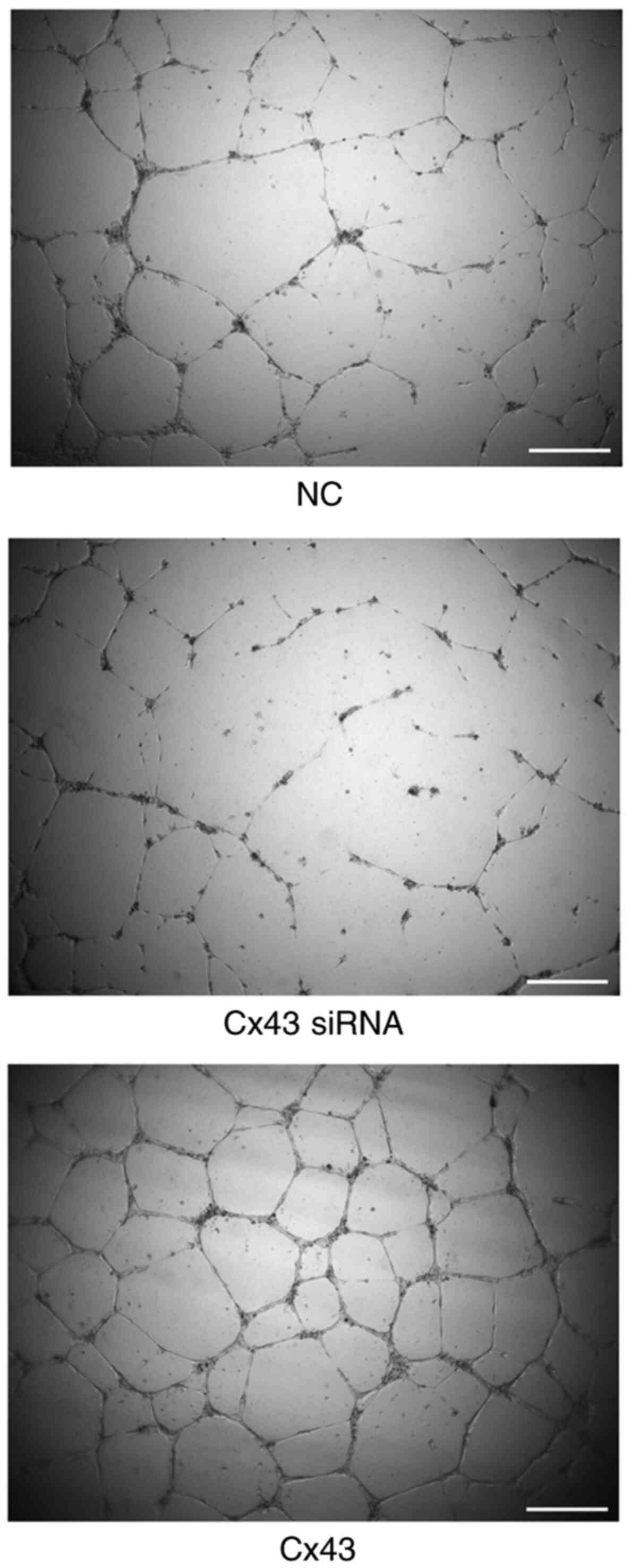
Cx43 overexpression improves
angiogenic potential of HPMECs
Using tube formation assay, the angiogenic potential
(number of segments) was analyzed. The angiogenic potential of
HPMECs was increased in the Cx43 overexpression group, whereas it
was decreased in the Cx43-knockdown group compared with that in the
normal control group (P<0.01). The angiogenic potential in the
Cx43 overexpression group was increased by two-fold, whereas that
in the Cx43-knockdown group was decreased by approximately 75%
compared with that in the normal control group (Figs. 7 and 8).
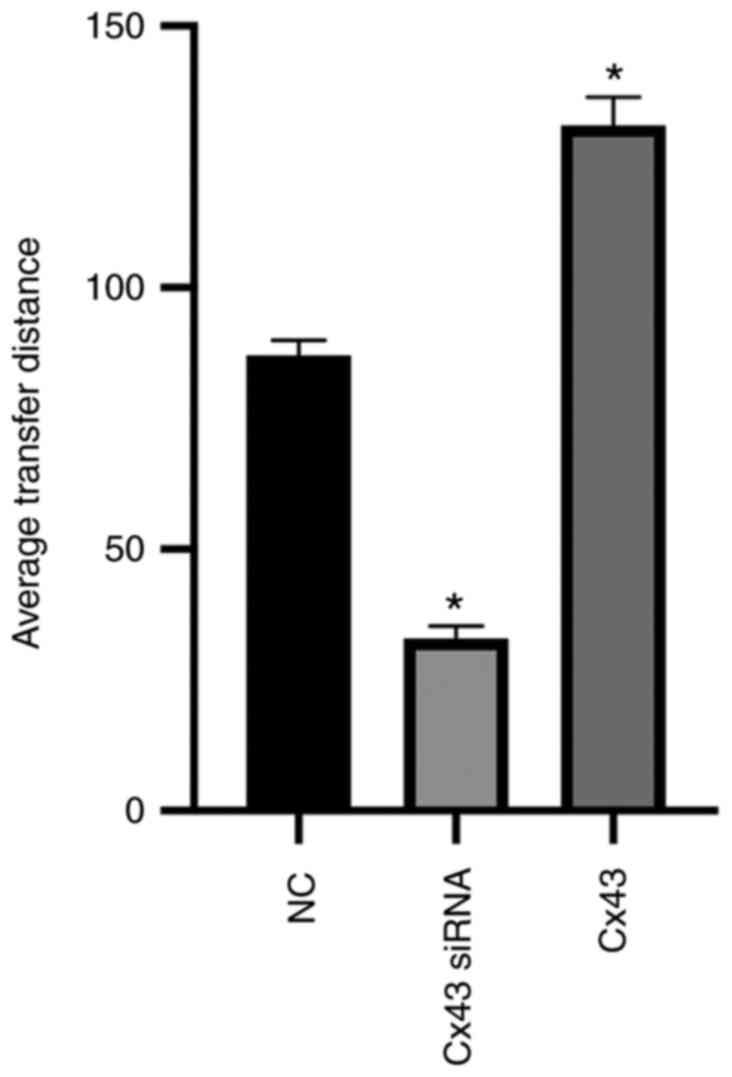
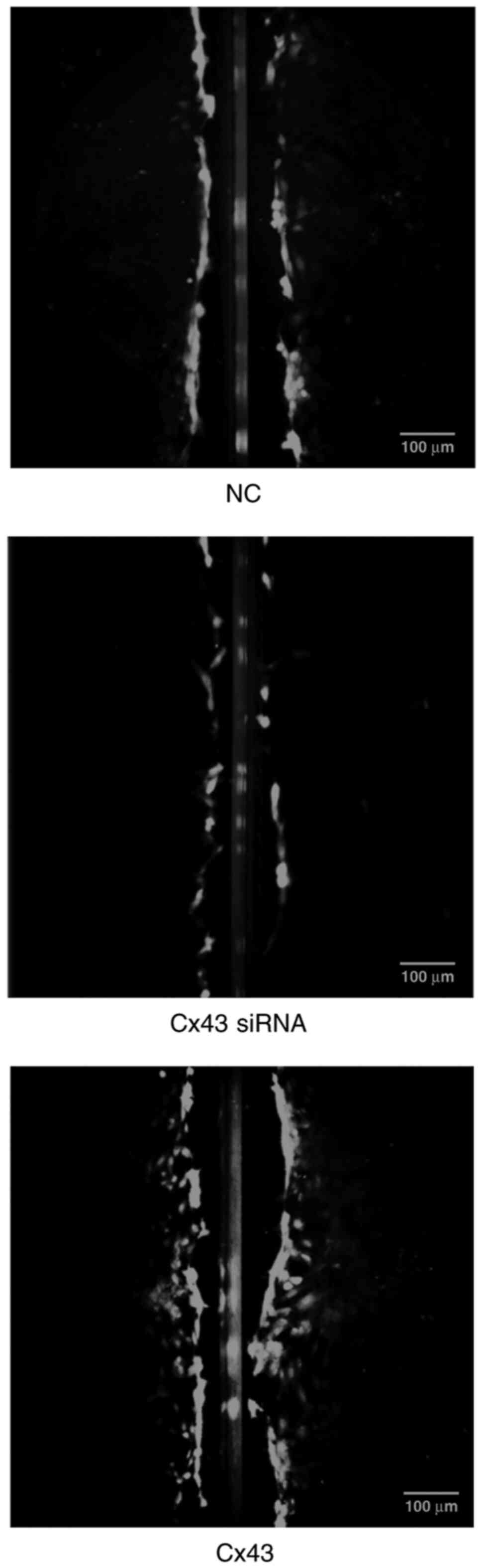
Cx43 promotes scrape loading dye
transfer in HPMECs
Scrape loading/dye transfer assay revealed that the
average dye transfer distance of the Cx43 overexpression group was
the highest among all groups. It was approximately 1.5-fold higher
than that of the normal control group. Conversely, the average dye
transfer distance of the Cx43-knockdown group was lower than the
other groups; it was approximately 60% lower than that of the
normal control group (P<0.01) (Figs. 9 and 10).
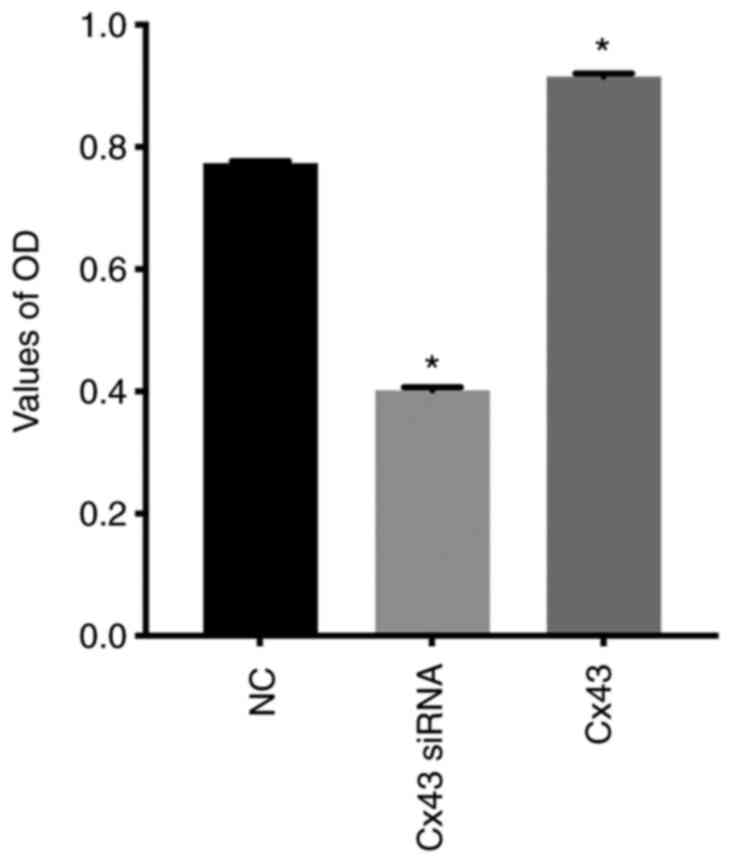
Cx43 stimulates proliferation of
HPMECs
CCK-8 assay showed that the relative absorbance
value (OD value) in the Cx43 overexpression group was increased by
approximately 30% compared with that in the normal control group,
whereas in the Cx43-knockdown group it was decreased by
approximately 25% compared with that in the normal control group
(P<0.01) (Fig. 11). These
results indicated that overexpression of Cx43 significantly
promoted the proliferation of HPMECs, whereas silencing of Cx43
significantly inhibited the proliferation of the cells.
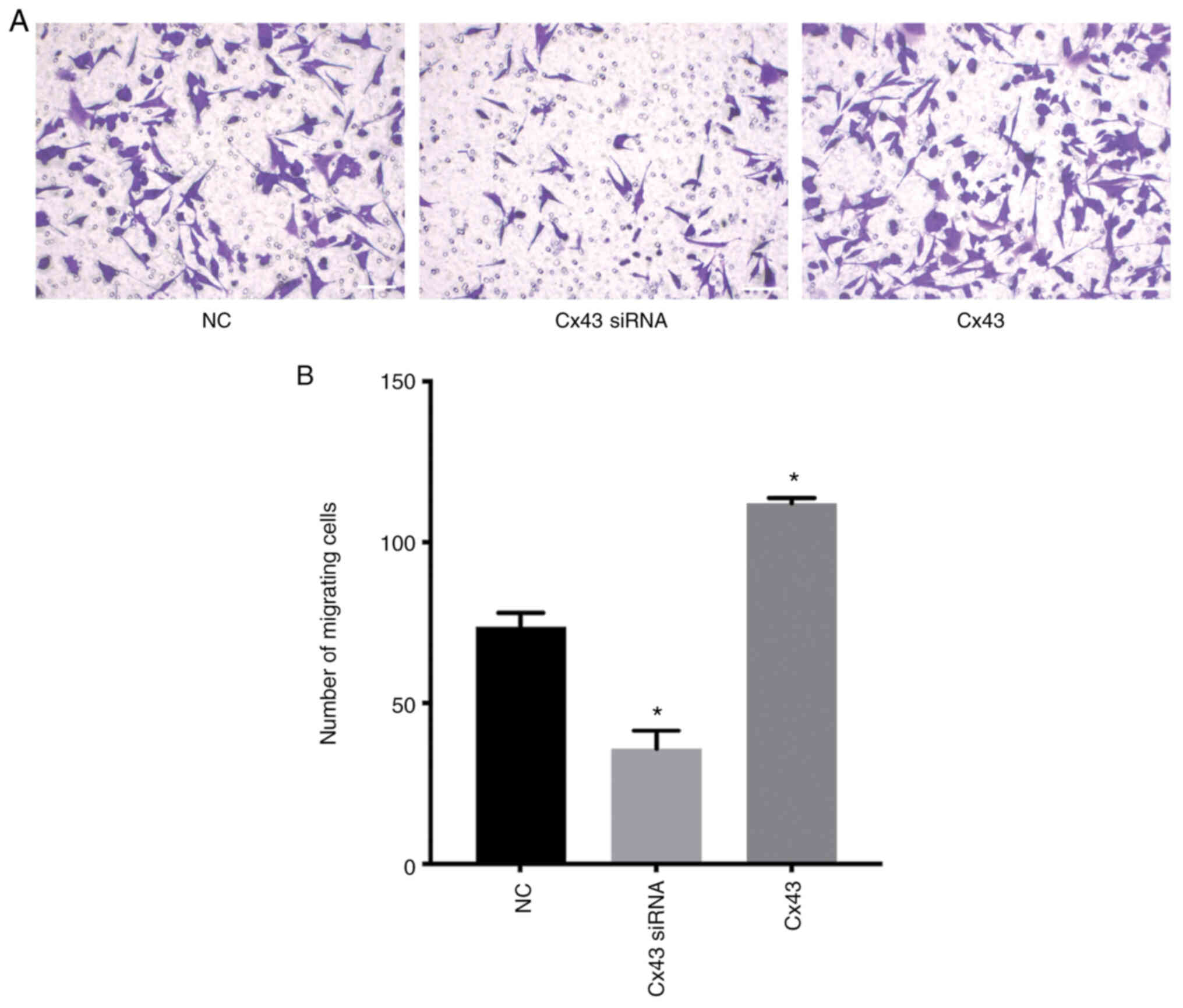
Cx43 stimulates migration of
HPMECs
Transwell migration assay revealed that the number
of migrating cells in the Cx43 overexpression group was increased,
whereas in the Cx43-knockdown group it was decreased compared with
that in the normal control group (P<0.01) (Fig. 12). These results indicated that
Cx43 overexpression significantly promoted the migration of HPMECs,
while silencing of Cx43 significantly inhibited cell migration.
Discussion
Angiogenesis is the process through which blood
vessels sprout from pre-existing blood vessels into avascular
tissues, following which they undergo reconstruction such that the
original irregular blood vessel network is developed into
functional branch blood vessels with complex three-dimensional
structures (27). It is jointly
regulated by growth factors, pro-angiogenic cytokines, and
neovascularization inhibitors (28,29).
As an indispensable link in the vascularization process,
angiogenesis involves proliferation, survival, migration, and
differentiation of endothelial cells (30), which play a major role in tumor
growth and metastasis (27,31).
Moreover, angiogenesis is a major hurdle in the treatment of lung
cancer. Therefore, preventing angiogenesis plays an important role
in successfully controlling the progression of lung cancer.
Consequently, exploring new methods of anti-angiogenesis therapy is
essential for lung cancer treatment.
The phosphorylation site of Cx43 is located at the
C-terminus of the cytoplasm, which contains multiple serine,
threonine, and tyrosine residues, hence can be phosphorylated by
various protein kinases, such as MAPK, PKA, PKC, p34 (Cdc2)/cyclin
B kinase, CK1, and pp60 (src) kinase (7,12,22,23).
Phosphorylation at specific sites may play a regulatory role in the
internalization and degradation of Cx43 channels, thereby affecting
gap junctions (10,32). In addition, during normal
development and disease processes, protein-protein interactions and
modification of the C-terminal of Cx43 are key determinants of gap
junction function, size, distribution, and organization (33). Phosphorylation is the primary way
to modify the carboxyl end of Cx43. Therefore, phosphorylation of
Cx43 may become a new focus of future research.
In the present study it was determined that after
mature HPMECs were transfected with the Cx43-overexpressed gene,
the expression of adhesion marker proteins ZO-1, E-cadherin, and
β-catenin, pro-thrombotic factor vWF, and coagulation factor PAI-1
increased, and the changes in the protein levels of these molecules
were consistent with the changes in their mRNA levels. Following
intracellular inhibition of Cx43 using a specific siRNA, the mRNA
and protein levels of the aforementioned molecules were
significantly reduced. Therefore, overexpression of Cx43 was
capable of stimulating the downstream ZO-1, E-cadherin, β-catenin,
vWF, and PAI-1 signaling proteins at the molecular and gene levels,
which was associated with angiogenesis. Moreover, the proliferation
and migration of cells in the overexpression group were
significantly increased, whereas those in the silenced group were
significantly reduced, suggesting that overexpression of Cx43 can
promote the proliferation and migration of HPMECs, which is
inconsistent with the results from a study by Koepple et al,
in which the proliferation level was unchanged (21). It is therefore theorized that the
reason for this inconsistency, is the diverse impact of Cx43 on
different cell types. However, this hypothesis warrants further
investigation. The tube formation experiments indicated that Cx43
can promote angiogenesis while silencing of the Cx43 gene can
inhibit the process. This previous study by Koepple et al
(21) found that using Cx43
mimetic peptide Gap27, after transfection of Cx43 cDNA to block
Cx43-dependent gap junctional communication, did not affect
angiogenic potential of endothelial cells compared to cells only
transfected with Cx43 cDNA, and dye transfer was significantly
attenuated in cells treated with Gap27 compared with the solvent
control, which demonstrated that the expression of Cx43 stimulated
endothelial angiogenesis independently of gap junctional
communication in vitro. In the present study, it was also
determined that the overexpression of Cx43 could improve the dye
transfer, while the transfection of Cx43-specific siRNA reduced dye
transfer distance distinctly, and these were consistent with their
results (21). However, further
experiments to verify the specific function of gap junction
communication during angiogenesis should be performed. In addition,
the expression of Ser279 was increased in the Cx43 overexpression
group, and decreased in the silenced group, which demonstrated that
Cx43 modulates angiogenesis depending on the activation of its
C-terminal phosphate site-Ser279 and the downstream ZO-1,
E-cadherin, β-catenin, vWF, and PAI-1 signaling proteins. Following
overexpression of Cx43, the intracellular and membranal content of
Cx43 was increased; in particular, Cx43 expression in the cell
membrane was significantly increased. Conversely, after silencing
of Cx43, the Cx43 protein level was significantly decreased,
particularly in the cell membrane. These results suggest that the
angiogenesis effect of Cx43 may rely on the membranal Cx43
expression level.
A siRNA targeting Cx43 and a recombinant vector that
overexpressed the Cx43 gene were successfully designed and
constructed. Following transfection of HPMECs, it was revealed that
overexpression of the Cx43 gene could induce pulmonary angiogenesis
in vitro. Cx43 overexpression induced angiogenesis by
promoting cell proliferation and migration and may be associated
with the activation of adhesion marker proteins ZO-1, E-cadherin,
and β-catenin, thrombosis-related factor vWF, and coagulation
factor PAI-1. A limitation of the present study is that it only
identified proteins ZO-1, E-cadherin, β-catenin, vWF, and PAI-1 as
participants in the process of Cx43-associated angiogenesis.
However, if these proteins play direct roles or are affected by
some other key intermediate proteins of the signaling pathway
requires further research. This pro-angiogenesis process may be
activated through activation of intracellular signaling site Ser279
on the C-terminal. In addition, the results demonstrated that
silencing of the Cx43 gene using Cx43-siRNA mainly affected the
integration of Cx43 and reduced the distribution of Cx43 in the
cell membrane, while overexpression of Cx43 had the opposite
effect. However, further research should be performed to confirm
this finding. In subsequent research, the construction of a Cx43
mutant body (Ser279), protein-protein interactions and other
molecular biotechnologies could be used to explore their specific
effects.
In summary, the present study demonstrated that
activation of the phosphorylation site Ser279 at the C-terminal of
Cx43 was an important pathway for Cx43 to regulate angiogenesis
in vitro. This also promoted angiogenesis by stimulating
cell proliferation, migration, and distribution of Cx43 to the cell
membrane, possibly via the activation of the downstream ZO-1,
E-cadherin, β-catenin, vWF, and PAI-1 signaling proteins. The
results indicated that Cx43 and its intracellular signaling site
Ser279 on the C-terminal are potential new targets for regulating
angiogenesis and exploring new treatment approaches for
angiogenesis-associated diseases.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science
Foundation of Shenzhen University General Hospital (grant no.
SUGH2019QD007 to ZZ, and grant no. SUGH2019QD014 to XZ) and the
Science and Technology Foundation of Nanshan District, Shenzhen
(grant no. NS2021167 to ZZ).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZZ, WC, YiL, YaL, HP, QW and XZ contributed to the
study concept, design, experiments, data collection, analysis, and
interpretation. ZZ wrote the manuscript. XZ revised the manuscript
and gave final approval of the version to be published. ZZ and XZ
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jiang X, Wang J, Deng X, Xiong F, Zhang S,
Gong Z, Li X, Cao K, Deng H, He Y, et al: The role of
microenvironment in tumor angiogenesis. J Exp Clin Cancer Res.
39:2042020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Saito T, Tanaka R, Wataba K, Kudo R and
Yamasaki H: Overexpression of estrogen receptor-alpha gene
suppresses gap junctional intercellular communication in
endometrial carcinoma cells. Oncogene. 23:1109–1116. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jiang JX and Gu S: Gap junction- and
hemichannel-independent actions of connexins. Biochim Biophys Acta.
1711:208–214. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goodenough DA and Paul DL: Gap junctions.
Cold Spring Harb Perspect Biol. 1:a0025762009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nielsen MS, Axelsen LN, Sorgen PL, Verma
V, Delmar M and Holstein-Rathlou NH: Gap junctions. Compr Physiol.
2:1981–2035. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Saez JC, Berthoud VM, Branes MC, Martinez
AD and Beyer EC: Plasma membrane channels formed by connexins:
Their regulation and functions. Physiol Rev. 83:1359–1400. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lampe PD and Lau AF: The effects of
connexin phosphorylation on gap junctional communication. Int J
Biochem Cell Biol. 36:1171–1186. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maeda S and Tsukihara T: Structure of the
gap junction channel and its implications for its biological
functions. Cell Mol Life Sci. 68:1115–1129. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Roy S, Jiang JX, Li AF and Kim D: Connexin
channel and its role in diabetic retinopathy. Prog Retin Eye Res.
61:35–59. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Söhl G and Willecke K: Gap junctions and
the connexin protein family. Cardiovasc Res. 62:228–232. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Solan JL and Lampe PD: Connexin
phosphorylation as a regulatory event linked to gap junction
channel assembly. Biochim Biophys Acta. 1711:154–163. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meşe G, Richard G and White TW: Gap
junctions: Basic structure and function. J Invest Dermatol.
127:2516–2524. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Laws MJ, Taylor RN, Sidell N, DeMayo FJ,
Lydon JP, Gutstein DE, Bagchi MK and Bagchi IC: Gap junction
communication between uterine stromal cells plays a critical role
in pregnancy-associated neovascularization and embryo survival.
Development. 135:2659–2668. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Buschmann I, Pries A, Styp-Rekowska B,
Hillmeister P, Loufrani L, Henrion D, Shi Y, Duelsner A, Hoefer I,
Gatzke N, et al: Pulsatile shear and Gja5 modulate arterial
identity and remodeling events during flow-driven arteriogenesis.
Development. 137:2187–2196. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Walker DL, Vacha SJ, Kirby ML and Lo CW:
Connexin43 deficiency causes dysregulation of coronary
vasculogenesis. Dev Biol. 284:479–498. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gärtner C, Ziegelhöffer B, Kostelka M,
Stepan H, Mohr FW and Dhein S: Knock-down of endothelial connexins
impairs angiogenesis. Pharmacol Res. 65:347–357. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang HH, Su CH, Wu YJ, Li JY, Tseng YM,
Lin YC, Hsieh CL, Tsai CH and Yeh HI: Reduction of connexin43 in
human endothelial progenitor cells impairs the angiogenic
potential. Angiogenesis. 16:553–560. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chou Y, Tsai CH, Ueng KC, Tian TY, Chen SC
and Yeh HI: Endothelial gap junctions are down-regulated by arsenic
trioxide. Eur J Pharmacol. 569:29–36. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang HH, Kung CI, Tseng YY, Lin YC, Chen
CH, Tsai CH and Yeh HI: Activation of endothelial cells to
pathological status by down-regulation of connexin43. Cardiovasc
Res. 79:509–518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Koepple C, Zhou Z, Huber L, Schulte M,
Schmidt K, Gloe T, Kneser U, Schmidt VJ and de Wit C: Expression of
Connexin43 stimulates endothelial angiogenesis independently of Gap
junctional communication in vitro. Int J Mol Sci. 22:74002021.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jansen JA, van Veen TA, de Bakker JM and
van Rijen HV: Cardiac connexins and impulse propagation. J Mol Cell
Cardiol. 48:76–82. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schulz R, Görge PM, Görbe A, Ferdinandy P,
Lampe PD and Leybaert L: Connexin 43 is an emerging therapeutic
target in ischemia/reperfusion injury, cardioprotection and
neuroprotection. Pharmacol Ther. 153:90–106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Omasits U, Ahrens CH, Müller S and
Wollscheid B: Protter: Interactive protein feature visualization
and integration with experimental proteomic data. Bioinformatics.
30:884–886. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Marsic D, Hughes RC, Byrne-Steele ML and
Ng JD: PCR-based gene synthesis to produce recombinant proteins for
crystallization. BMC Biotechnol. 8:442008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Distler O, Neidhart M, Gay RE and Gay S:
The molecular control of angiogenesis. Int Rev Immunol. 21:33–49.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Polverini PJ: The pathophysiology of
angiogenesis. Crit Rev Oral Biol Med. 6:230–247. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Goel S, Duda DG, Xu L, Munn LL, Boucher Y,
Fukumura D and Jain RK: Normalization of the vasculature for
treatment of cancer and other diseases. Physiol Rev. 91:1071–1121.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bir SC, Xiong Y, Kevil CG and Luo J:
Emerging role of PKA/eNOS pathway in therapeutic angiogenesis for
ischaemic tissue diseases. Cardiovasc Res. 95:7–18. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Carmeliet P and Jain RK: Angiogenesis in
cancer and other diseases. Nature. 407:249–257. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fernandes R, Girão H and Pereira P: High
glucose down-regulates intercellular communication in retinal
endothelial cells by enhancing degradation of connexin 43 by a
proteasome-dependent mechanism. J Biol Chem. 279:27219–27224. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Palatinus JA, Rhett JM and Gourdie RG: The
connexin43 carboxyl terminus and cardiac gap junction organization.
Biochim Biophys Acta. 1818:1831–1843. 2012. View Article : Google Scholar : PubMed/NCBI
|