Introduction
Lung cancer [World Health Organization
(WHO)/International Classification of Diseases 10th revision
(ICD-10) code of cancer, C34.901] is one of the leading causes of
cancer-associated morbidity and mortality worldwide. It is the
second most prevalent type of cancer (11.4%) after breast cancer
(WHO/ICD-10 C50.902; 11.7%). Globally, lung cancer contributes to
~1.8 million deaths per year (18%), which makes it the number one
cause of cancer-related deaths (1). Non-small cell lung cancer (NSCLC)
accounts for 80–85% of all lung cancers (2). The 5-year survival rate for patients
with stage IA lung cancer is 73%, while that for patients with
stage IV is only 13% (3). Based on
these statistics, it is imperative to develop efficient tools for
the early screening, diagnosis and treatment of lung cancer, to
lower the magnitude of lung cancer-related deaths and prolong
patient survival.
Circular RNAs (circRNAs) are non-coding RNAs that
differ from linear RNAs due to the presence of a special structure
that contains neither a 5′-cap structure nor a 3′-adenosine tail
(4). This closed loop structure is
formed by the reverse splicing of precursor mRNAs, and not only
confers resistance to hydrolysis by exonucleases, but also makes
the molecule more stable (5).
CircRNAs were first identified in the 1970s in an RNA virus. Since
then, they have been regarded as erroneous transcriptional products
of RNA, and have thus far been ignored by researchers (6). The development and advancement in
high-throughput sequencing and bioinformatics technologies have led
to the identification of multiple circRNAs in human tissues, blood,
urine and saliva (7), where they
regulate a multitude of biological processes. For example, they
have been shown to participate in the occurrence and development of
lung cancer, by regulating proliferation, differentiation,
metastasis and apoptosis of tumor cells (8). Consequently, researchers have
employed circRNAs in the screening, diagnosis, treatment and
prognosis of lung cancer (9).
Several adjuvant treatments for advanced
unresectable lung cancer have been developed, including
radiotherapy, chemotherapy, targeted therapy and immunotherapy
(10). Antitumor drugs targeting
epidermal growth factor receptor (EGFR), anaplastic lymphoma
kinase, proto-oncogene tyrosine-protein kinase ROS, programmed cell
death protein 1/programmed death-ligand 1 (PD-L1) and proton
therapy have been shown to prolong the survival of patients, but
the efficacy of these interventions is reduced by the emergence of
resistance in advanced lung cancer (11). In the present study, the biological
characteristics, biological functions, diagnostic and prognostic
value of circRNAs in lung cancer are reviewed, and the relationship
between circRNAs and resistance against adjuvant drugs (such as
chemotherapy resistance, targeted drug resistance, immunotherapy
resistance and radiation resistance) is discussed.
Classification and characteristics of
circRNAs
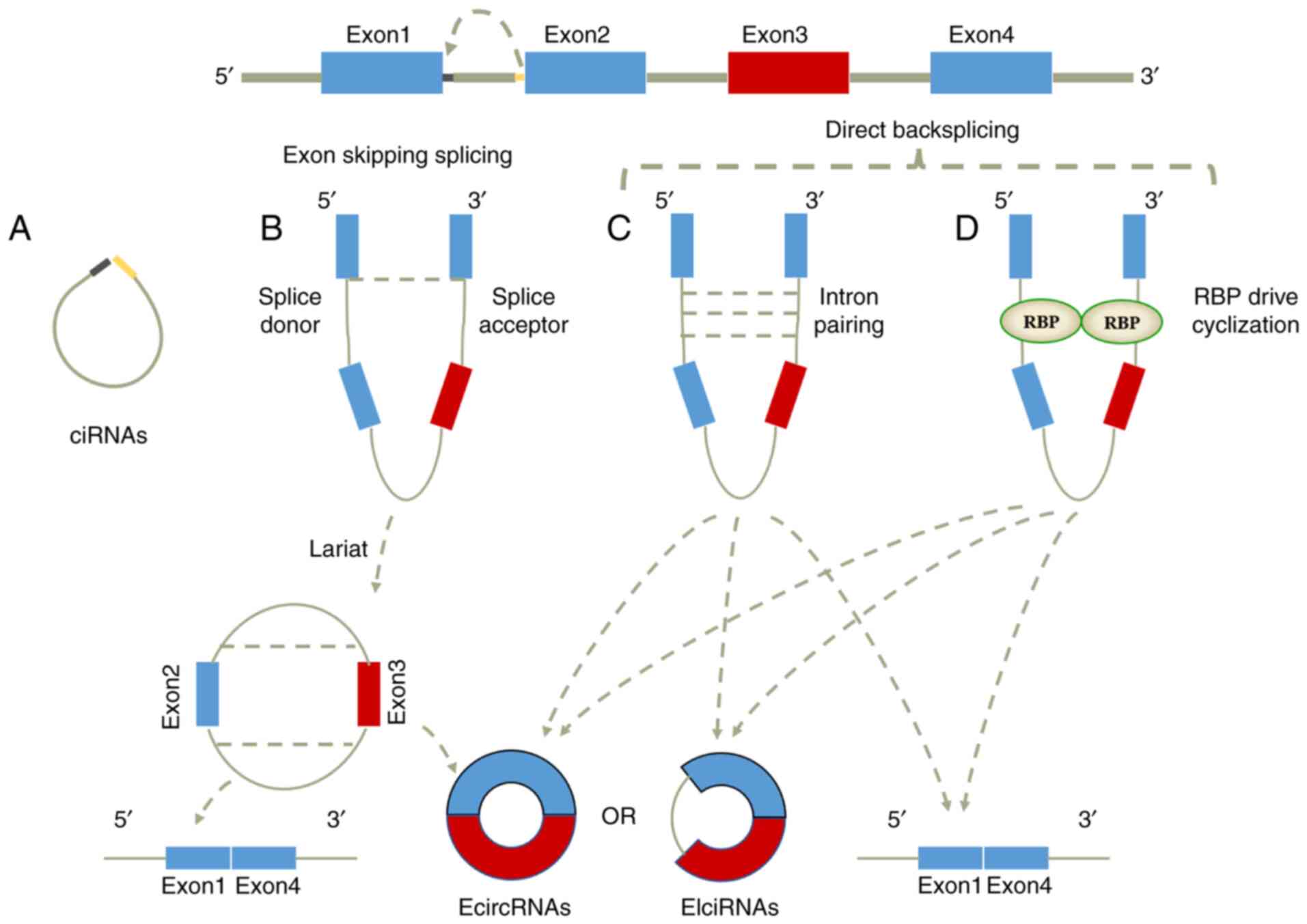
Based on their origin, circRNAs can be divided into
four types, namely intron circRNAs (ciRNAs), exon circRNAs
(EcircRNAs), exon-intron circRNAs (EIciRNAs) and intergenic
circRNAs (12), among which
EcircRNAs is the most extensively studied group. Mechanistically,
circRNAs actions include lasso-driven cyclization, intron
pair-driven cyclization and RNA binding protein (RBP)-driven
cyclization. Based on these, the actions of circRNAs can be divided
into two main modes, direct reverse splicing and exon jump
(13). Lasso-driven cyclization is
a mechanism of exon jump in which the 5′ end of one exon connects
to the 3′ end of another exon to form an exon jump. At this point,
it represents an RNA lasso composed of exons and introns. However,
intron removal leads to EcircRNA formation, while exon-intron
circRNAs (EIciRNAs) are formed when introns are cycled between
exons and exons are also retained. Direct reverse splicing
mechanisms include intron pairing-driven cyclization and RBP driven
cyclization (14). Notably, ciRNAs
depend on introns with conserved motifs at both ends, namely, the
GU-rich element near the 5′ splice site and the C-rich element near
the 3′ branch site. Additionally, intron lasso is cyclized by
connecting free hydroxyl terminal groups to 5′ splicing sites after
releasing the 3′ exon (15)
(Fig. 1).
Biological characteristics of circRNAs
Stability
CircRNAs are resistant to degradation by RNase R
enzymes, which is attributed to their circular configurations that
are a result of special 5′-3′ phosphodiester bonds (16). One study comparing the stability of
circRNAs and their related linear mRNAs revealed that the half-life
of circRNAs is >48 h, while the half-life of related mRNAs is
only 20 h, indicating that circRNAs are more stable than mRNAs
(5).
Universality
CircRNAs are abundant in human tissues, serum,
saliva and urine. Therefore, they are ideal diagnostic markers for
malignant tumors (17). For
instance, Jeck et al (5)
extracted total RNA from immortalized human fibroblast cell lines
and prepared cDNA using RNase R-treated samples and untreated
samples. The results showed that cANRIL (low-copy RNA) in RNase R
treated samples was higher than that in untreated samples Circle
>10-fold enrichment. CircRNAs are more abundant than linear
mRNAs (18).
Conservativeness
At the sequence level, circRNAs are highly conserved
between species, such as between species of the Drosophila
genus. Furthermore, certain circRNAs in Drosophila brains
are also expressed in mammalian brains (19). An estimated 5–10% of circRNAs are
homologous in human and porcine brains (20).
Specificity
Generally, circRNAs exhibit cell, tissue and
developmental stage-specific expression patterns (21). For example, circRNAs are expressed
at increased levels in developing organs, such as the heart, lungs
and brain (22).
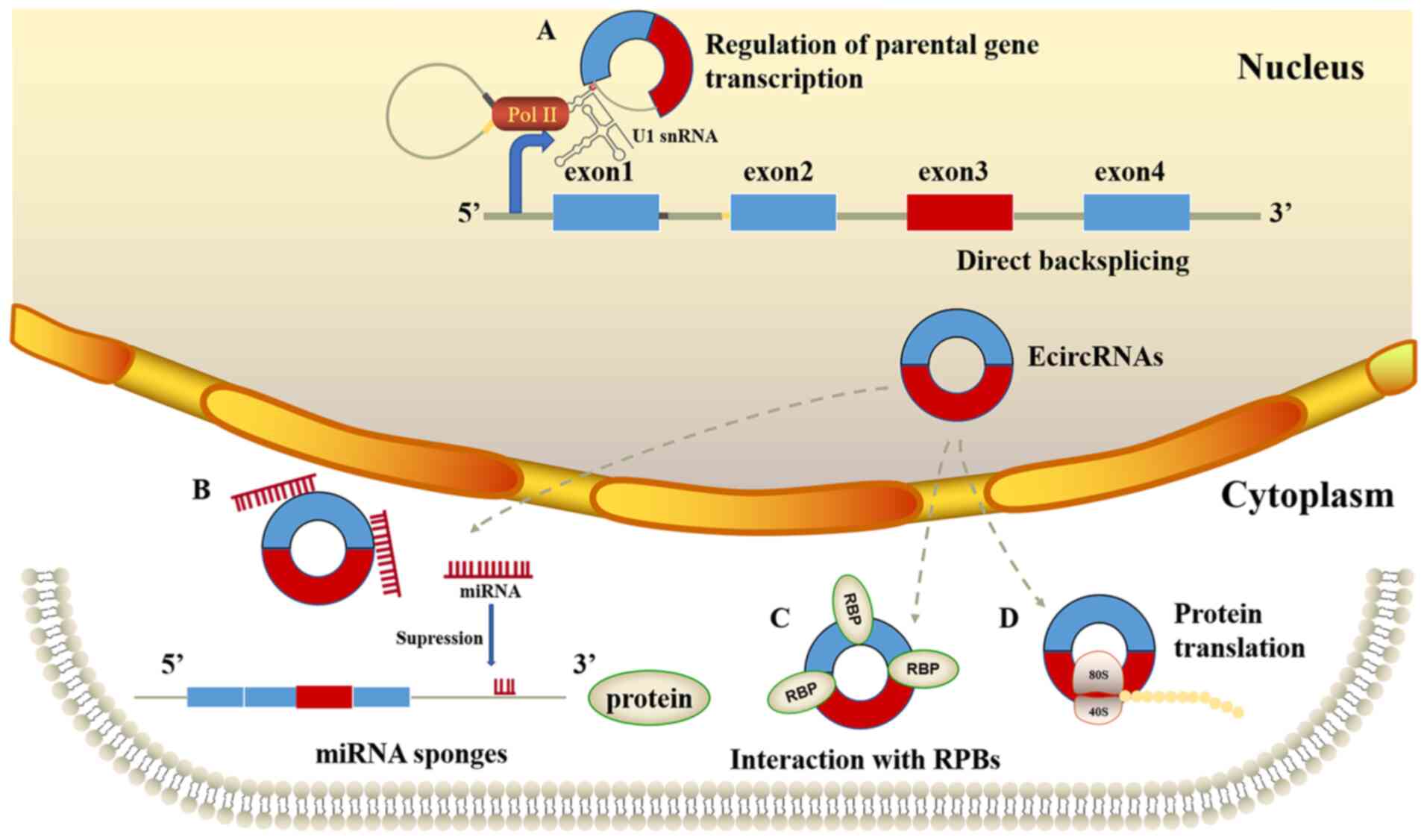
Biological functions of circRNAs
Various studies have been performed on the
biological functions of circRNAs. Firstly, the indirect regulation
of downstream target gene expression has been studied using
microRNA (miRNA/miR) ‘sponges’. miRNAs are non-coding RNAs that
complement and pair with mRNA bases to indirectly regulate gene
expression by inhibiting mRNA translation or promoting their
degradation at the transcriptional level. For instance, miR-330-5p
exerts anti-tumor effects in lung and prostate cancer
(International Classification of Diseases code C61), while
circ_0000517 targets miR-330-5p, thereby attenuating the inhibitory
effect on the expression of the downstream target protein YY1. This
results in promoting the proliferation, glycolysis and glutamine
decomposition of NSCLC cells (23).
Secondly, the role of circRNAs has been studied in
parental gene transcription regulation. EIciRNAs and ciRNAs are
largely localized in the nucleus, where they regulate
transcription. For example, EIciRNAs interact with U1 small nuclear
ribonucleoprotein to promote the transcription of their parent
genes (24).
Thirdly, protein translation has been studied in
relation to circRNAs. Endogenous circRNAs can be translated into
proteins or peptides. For example, circ_FAM188B is a stable circRNA
that is differentially expressed in skeletal muscles during the
development of broiler and layer embryos. Circ_FAM188B encodes the
novel protein, circ_FAM188B-103AA, as well as several other
peptides. Circ_FAM188B-103AA promotes the proliferation of
satellite cells in the skeletal muscles of chickens, as well as
inhibiting their differentiation (25).
Finally, circRNAs have been studied in relation to
RNA-binding proteins. CircRNAs can bind Argonaute proteins,
mannose-binding lectin, eukaryotic initiation factor 4A-III, RNA
polymerases and other RNA-binding proteins to form RNA-protein
complexes that subsequently affect gene expression and participate
in cell proliferation, differentiation and apoptosis (26) (Fig.
2).
Lab/experimental protocols for the detection
of circRNAs
The detection conditions of circRNAs are similar to
those of conventional RNA. At present, the detection process
adopted by several studies is as follows: Total RNA extraction, RNA
quality inspection and finally reverse transcription (RT) into
cDNA. Primers are designed according to the circRNAs requiring to
be detected and the expression level of circRNAs are detected by
RT-quantitative PCR (27).
Diagnostic significance of circRNAs in lung
cancer
Patients with stage IA and IV lung cancer have
5-year survival rates of 85 and 6%, respectively. Therefore, early
lung cancer diagnosis is crucial for effective patient prognosis
and treatment. If the presence of pulmonary nodules is confirmed by
imaging techniques, such as CT or chest radiography, pathological
results will lead to the next step of diagnosis and treatment.
However, CT-guided percutaneous lung puncture and electronic
bronchoscopy biopsy are invasive examinations. Thus, they are not
suitable as a common means of screening for pulmonary nodules
(28). Therefore, it is important
to identify efficient detection methods with low invasiveness,
fewer risks, improved cost effectiveness, improved support, strong
operability and easy acceptance by patients. The conservativeness
and stability of circRNAs make them promising biomarkers for tumor
diagnosis. In addition, most of the studies on the diagnostic value
of circRNAs in lung cancer have focused on tissues, with only a
small number of studies performed using blood (29).
Diagnostic value of circRNAs in lung
cancer tissues
Lin et al (30) demonstrated that hsa_circ_0007385 is
significantly upregulated in cancerous tissues compared with
paracancerous tissues (P<0.001), while receiver operating
characteristic (ROC) curve analysis revealed an area under the
curve (AUC) of 0.922 (95% CI, 0.890-0.953), suggesting that
hsa_circ_0007385 expression in tissues has a high value in cancer
diagnosis. Fu et al (31)
sequenced three cancerous tissues alongside paracancerous tissues
and validated the expression using smaller and larger sample sizes
comprising 20 and 60 cases of cancerous and paracancerous tissues,
respectively. They found that hsa_circ_012515 was significantly
upregulated in NSCLC and was positively correlated with late-stage
tumor development. The AUC value of the ROC curve was 0.89
(P<0.0001), suggesting that hsa_circ_012515 also has a high
diagnostic value. Therefore, it can be used as a specific,
sensitive diagnostic biomarker for patients with NSCLC (31). Geng et al (32) performed RNA sequencing of NSCLC
cases and found that circ-MTHFD2 was significantly elevated in
cancerous tissues compared with paracancerous tissues (P<0.001).
ROC curves revealed a critical value of 3.534 (for the relative
expression of the circ-MTHFD2 gene), with a sensitivity and
specificity of 90 and 71%, respectively. Furthermore, Li et
al (33) found that
hsa_circ_0000729 was upregulated in cancerous, relative to
paracancerous tissues, with further analysis revealing an AUC value
of 0.815 (95% CI, 0.805-0.862), which indicated a high accuracy and
confirmed its value as a diagnostic marker for lung adenocarcinoma.
The diagnostic accuracy of circRNAs in lung cancer could be
associated with the disease stage. For example, Zhu et al
(34) sequenced 49 cancerous
samples alongside paracancerous samples, including 12 stage I
(24.49%), 16 stage II (32.65%), 20 stage III patients (40.82%) and
1 stage IV (2.04%) patients. They found that hsa_circ_0013958 was
significantly upregulated in cancerous relative to paracancerous
tissues (P<0.001), with AUC values of 0.750, 0.766 and 0.874 for
patients at stage I, II and III + IV, respectively. This
demonstrated that hsa_circ_0013958 has an excellent diagnostic
value in advanced lung adenocarcinoma (34).
Diagnostic value of blood circRNAs for
lung cancer
In clinical diagnosis, plasma is the most easily
accessible material. Plasma samples can be obtained via less
invasive approaches than tissue samples and are, therefore, most
widely used for prospecting for potential diagnostic markers in
lung cancer. Due to their high stability and wide distribution, the
significance of circRNAs as diagnostic markers in lung cancer has
been evaluated. Exosomes, which contain proteins, RNAs and other
components, are widely distributed in blood, saliva, milk and urine
(35). Exosome-derived circRNAs
have attracted widespread attention as potential diagnostic markers
(36). Using high-throughput
sequencing, Wang et al (37) analyzed the expression patterns of
circRNAs in plasma exosomes from six pairs of patients with lung
squamous cell carcinoma and obtained 252 differentially expressed
circRNAs. Validation of expression via RT-qPCR confirmed that
hsa_circ_0014235 and hsa_circ_0025580 were significantly
upregulated in lung squamous cell carcinoma compared with the
normal control group. ROC curves revealed that hsa_circ_0014235
(AUC=0.8254; 95% CI, 0.762-0.889) and hsa_circ_0025580 (AUC=0.8003;
95% CI, 0.741-0.862) have good diagnostic value for lung squamous
cell carcinoma (37).
CircRNA_002178 has also been reported to be significantly
upregulated in plasma exosomes from patients with lung
adenocarcinoma, compared with healthy controls (P<0.001), with
an AUC of 0.9956 affirming its excellent diagnostic value (38). Ding et al (39) isolated exosomes from the serum of
patients with NSCLC (n=30) and healthy subjects (n=25). They
established that circ_MEMO1 was significantly upregulated in
diseased relative to healthy controls (P<0.05), with an AUC
value of 56.67 and specificity of 96%, suggesting the potential for
serum circ_MEMO1 to be a new biomarker for NSCLC diagnosis.
In plasma, circRNA levels are a potential diagnostic
index for lung cancer and also tumor metastasis. Zhang et al
(40) reported that circ_SATB2 is
significantly upregulated in serum exosomes of patients with lung
cancer, relative to non-cancerous controls (P<0.01), and these
expression levels were significantly higher in serum exosomes of
patients with metastatic lung cancer than in non-metastatic lung
cancer counterparts (P<0.01). The respective AUC values were
0.660 and 0.797, suggesting that this circRNA has a potential
diagnostic value. Other circRNAs have also shown potential as
diagnostic markers for various cancers. For instance,
hsa_circ_0001821 exhibited a diagnostic value in colorectal, liver
and lung cancers, as evidenced by AUC values of 0.815 (95% CI,
0.751-0.869), 0.692 (95% CI, 0.617-0.760) and 0.792 (95% CI,
0.724-0.850), respectively (41).
CircRNAs exhibit different diagnostic abilities across various
stages of NSCLC development and expression. Hsa_circ_0102533
exhibited a higher diagnostic value (AUC=0.774; 95%, CI,
0.624-0.923) in stage I–II NSCLC than at stage III–IV (AUC=0.728;
95% CI, 0.588-0.869). This suggests that it has a higher diagnostic
accuracy during early lung cancer stages and has potential as a
blood-derived tumor marker for the early detection of NSCLC
(42).
The significance of circRNAs in lung cancer
diagnosis has not been fully elucidated, with associated studies
exhibiting certain limitations. Firstly, sample sizes in most
studies were insufficient, necessitating further expansion. In
addition, most studies have only proven differential expression of
certain circRNAs in cancer, relative to paracancerous tissues,
without further clarifying the corresponding changes in expression
across developmental stages. Thirdly, most studies on the
diagnostic value of circular RNAs in both blood and lung cancer
tissues have only revealed differential expression of certain
circRNAs between patients with lung cancer and non-cancerous
subjects, without a control group (namely, patients with benign
tumors). Specifically, the levels of circRNAs in patients with
benign lung tumors are unknown. If there are benign and malignant
masses in the lung, the expression of corresponding circular RNAs
have been found to be significantly high, relative to non-cancerous
subjects. Further studies are required to elucidate the diagnostic
value of circRNAs in lung cancer.
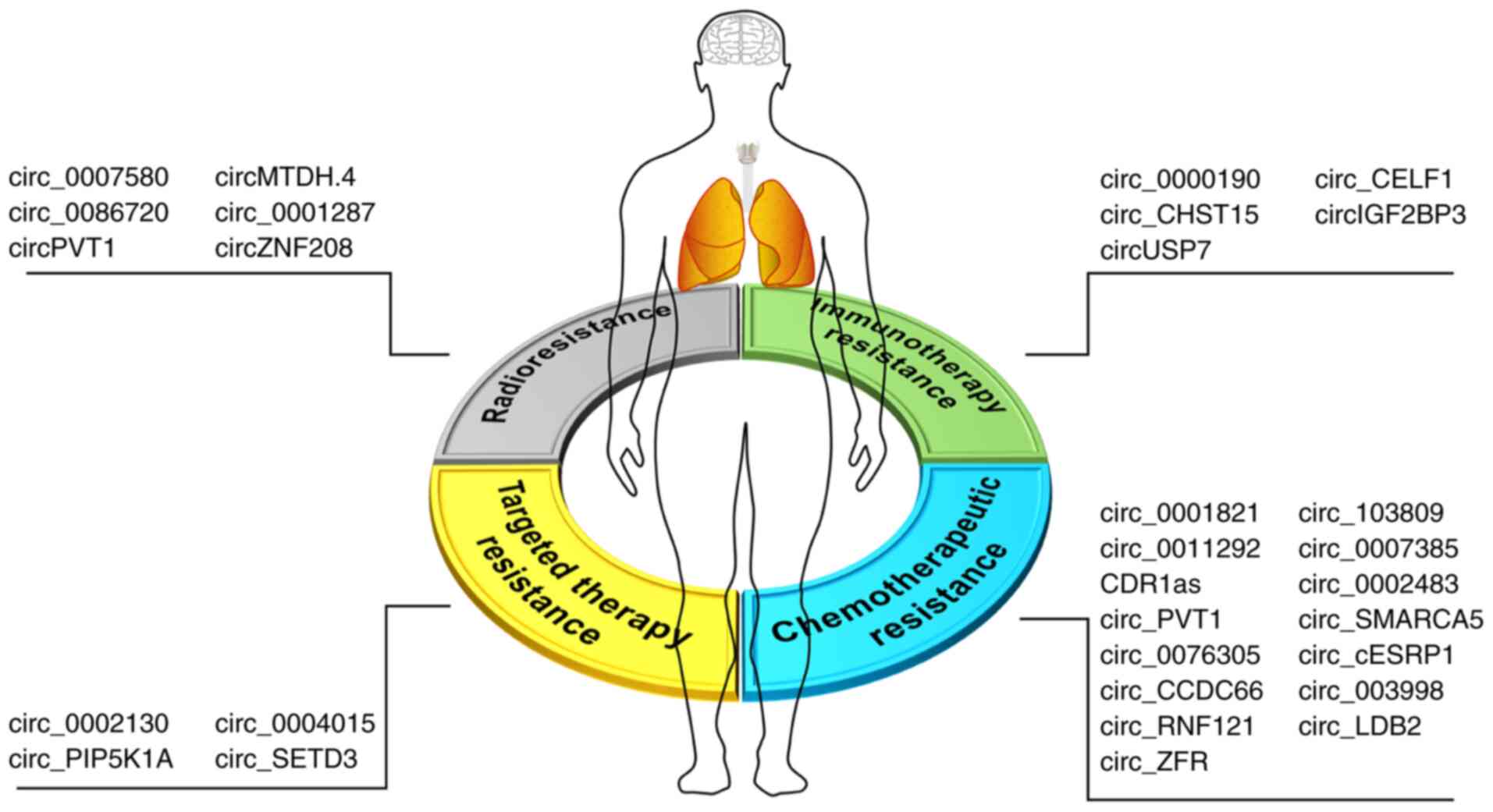
CircRNAs and resistance to adjuvant therapy
in lung cancer
Common adjuvant therapies for lung cancer include
chemotherapy, targeted therapy and immunotherapy. However, they are
associated with drug resistance risks, which subsequently
compromise their efficacies. CircRNAs regulate cancer cell invasion
as well as proliferation, and play an important role in
chemotherapy, immunity and targeted drug resistance (Table I) (43–65).
 | Table I.CircRNAs and lung cancer drug
resistance. |
Table I.
CircRNAs and lung cancer drug
resistance.
| Effect on drug | Antitumor drug | CircRNA | Target
molecule | Target
gene/pathway | Expression level in
tumor tissue | (Refs.) |
|---|
| Enhance
resistance | PTX | circ_0001821 | miR-526b-5p | GRK5 | Upregulated | (43) |
| Enhance
resistance | PTX | circ_0011292 | miR-379-5p | TRIM65 | Upregulated | (44) |
| Enhance
resistance | Cisplatin | CDR1as | miR-641 | HOXA9 | Upregulated | (45) |
| Enhance
resistance | Cisplatin | circ_PVT1 | miR-145-5p | ABCC1 | Upregulated | (46) |
| Enhance
resistance | Cisplatin | circ_0076305 | miR-296-5p | STAT3 | Upregulated | (47) |
| Enhance
resistance | Osimertinib | circ_0002130 | miR-498 | GLUT1 | Upregulated | (48) |
|
|
|
|
| HK2 |
|
|
|
|
|
|
| LDHA | |
|
| Enhance
resistance | Gefitinib | circ_PIP5K1A | miR-101 | ABCC1 | Upregulated | (49) |
| Enhance
resistance | Gefitinib | circ_0004015 | miR-1183 | PDPK1 | Upregulated | (50) |
| Enhance
resistance | Gefitinib | circ_SETD3 | miR-520h | ABCG2 | Upregulated | (51) |
| Enhance
resistance | Cisplatin | circ_CCDC66 | - | EMT | Upregulated | (52) |
| Enhance
resistance | Cisplatin | circ_RNF121 | miR-646 | SOX4 | Upregulated | (53) |
| Enhance
resistance | PTX, Cisplatin | circ_ZFR | miR-195-5p | KPNA4 | Upregulated | (54) |
| Enhance
resistance | Cisplatin | circRNA_103809 | miR-377-3p | GOT1 | Upregulated | (55) |
| Enhance
resistance | Cisplatin | circ_0007385 | miR-519d-3p | HMGB1 | Upregulated | (56) |
| Enhance
resistance | Anti-PD-1 | circUSP7 | miR-934 | SHP2 | Upregulated | (57) |
| Enhance
resistance | Anti-PD-1 | circ_CELF1 | miR-491-5p | EGFR | Upregulated | (58) |
| Enhance
resistance | Anti-PD-1 | circIGF2BP3 | miR-328-3p,
miR-3173-5p | PKP3 | Upregulated | (59) |
| Enhance
resistance | Anti-PD-1 | circCHST15 | miR-155-5p,
miR-194-5p | PD-L1 | Upregulate | (60) |
| Increase
sensitivity | PTX | circ_0002483 | miR182-5p |
GrB2/FOXO_1/FOXO_3 | Downregulated | (61) |
| Increase
sensitivity | Cisplatin | circ_SMARCA5 | miR-670-5p | RBM24 | Downregulated | (62) |
| Increase
sensitivity | Cisplatin | circ_cESRP1 | miR-93-5p | CDKN1A | Downregulated | (63) |
| Increase
sensitivity | PTX | circ_003998 | miR-558 | MMP1 | Downregulated | (64) |
|
|
|
|
| MMP17 |
|
|
| Increase
sensitivity | Cisplatin | circ_LDB2 | miR-346 | LIMCH1 | Downregulated | (65) |
Upregulated circRNAs in tumor tissues
and cells are associated with drug resistance
CircRNAs can influence NSCLC drug resistance through
the ‘sponge’ miRNA model, binding to RBPs and/or regulating the
translation of their parental genes (66).
CircRNAs and lung cancer targeted
therapy resistance
Gefitinib, an inhibitor of EGFR tyrosine kinase
(EGFR-TK), is commonly used to treat NSCLC. However, most patients
develop acquired drug resistance after using gefitinib for 9-11
months (67). The ATP-binding
cassette (ABC) transporters (ABCB1 and ABCG2) are a class of drug
resistance-related proteins that hydrolyze ATP to generate energy
and pump out endogenous as well as exogenous substances, including
antineoplastic drugs, against a concentration gradient (68). Circ_SETD3, which is localized in
the cytoplasm and overexpressed in drug-resistant tumor tissues,
downregulates miR-520h expression, inhibits transporter ABCG2
degradation, pumps gefitinib out of cells, and promotes tumor drug
resistance through the circ_SETD3/miR-520h/ABCG2 axis (51). Wen et al (69) found that hsa_circ_0000567 and
hsa_circ_0006867 were upregulated and downregulated, respectively,
in two drug-resistant gefitinib cell lines, suggesting that changes
in circRNA expression play an important role in the development of
NSCLC-acquired resistance to EGFR-TK inhibitors (EGFR-TKI).
Exosomes, which are messengers of intercellular communication and
signal transduction, contain biological macromolecules, such as
proteins and nucleic acids (70).
Yang et al (71) found that
circ_102481 was significantly upregulated in EGFR-TKIs
drug-resistant exosomes, relative to sensitive ones. Notably, its
overexpression promoted the proliferation of EGFR-TKIs-sensitive
cells, thereby making them resistant to drugs and inhibiting cell
apoptosis (71).
CircRNAs and chemotherapeutic
resistance in lung cancer
CircRNAs are associated with both targeted and
chemotherapeutic drug resistance. For instance, circ_PIP5K1A was
indicated to be upregulated in the exosomes of NSCLC tissues, serum
and cells, and was also associated with inhibition of NSCLC cell
proliferation, migration and invasion, as well as the promotion of
cell sensitivity to cisplatin and elevated apoptosis (49). Epithelial-mesenchymal transition
(EMT), which plays an important role in embryogenesis, wound
healing, tumor progression and endogeneity of tumor cells with
invasion and metastasis potential, is closely associated with tumor
cell drug resistance (72).
Circ_CCDC66 knockout downregulated the expression and invasive
abilities of EMT markers, and significantly increased the
resistance of human H23 cells to cisplatin (52). Besides, circ_0003998 (64), circ_ZFR (54), circ_0001821 (43) and circ_0011292 (44) were significantly upregulated in
paclitaxel (PTX)-resistant NSCLC tissues and cells, and their
silencing promoted the sensitivity of NSCLC to PTX. These findings
indicate that circRNAs play a role in tumor drug resistance.
CircRNAs and immunotherapeutic
resistance in lung cancer
Lung cancer immunotherapy is currently a popular
research topic. Although circRNAs are associated with immune
escape, only a handful of studies have described this phenomenon.
Luo et al (73) found
upregulated plasma hsa_circ_0000190 levels in lung cancer patients
compared with healthy subjects (P<0.0001), which were
significantly associated with higher expression of PD-L1 in tumor
tissues (P=0.0283). Long-term follow-up of immunotherapy cases
showed that upregulation of hsa_circ_0000190 was significantly
associated with systemic and immunotherapeutic adverse reactions
(P=0.0002 and P=0.0058, respectively) (73). miR-155-5p and miR-194-5p in
circ_CHST15 (hsa_circ_0109320) ‘sponges’ have been indicated to
upregulate PD-L1 expression, thereby promoting PD-L1-mediated
immune escape from lung cancer and increasing drug resistance to
PD-L1 inhibitors. The significance of circ_CHST15 in promoting
immune escape was indicated by inhibiting CD8+ T cell
activities and inducing CD8+ T cell apoptosis (60). Patterns of PD-L1 expression are not
necessarily associated with lung cancer drug resistance. Findings
from a previous clinical trial, comprising 305 patients with
advanced NSCLC showed that upregulated PD-L1 levels enhanced the
sensitivity of NSCLC cells to immunotherapy (74). These findings suggest that the
relationship between PD-L1 levels and tumor drug resistance is not
simply ‘promotion’ or ‘inhibition’, while the mechanism underlying
the actions of circRNAs in tumor immune escape should be further
evaluated.
A previous study demonstrated that other circRNAs
are downregulated in drug-resistant tumor tissues and cells and
have a role in enhancing the sensitivity of tumor drug-assisted
therapy. For instance, circ_0002483 was downregulated in
drug-resistant NSCLC tissues, and its overexpression was found to
not only inhibit proliferation and invasion of NSCLC cells via the
miR182-5p/growth factor receptor-bound protein 2/FOXO1/FOXO3
signaling pathway, but also promote their sensitivity to PTX
chemotherapeutic drugs (61).
CircRNAs and radioresistance in lung
cancer
Radiotherapy is clinically used as an adjunct
therapy in the treatment of lung cancer. This therapy destroys the
DNA of tumor cells leading to the death of cancer cells (75). The development of radioresistance,
on the other hand, negates the benefits of radiotherapy (76). The mechanism leading to the
development of tumor radioresistance is complex and heterogeneous.
It involves cancer-associated fibroblasts, tumor stem cells,
non-coding RNA, DNA damage response, proinflammatory cytokines and
regulatory enzymes among other molecules (77). CircRNAs, as a type of non-coding
RNA, have been implicated in the development of radiation
resistance in lung cancer (78).
In a previous study, certain circRNAs were
positively associated with the occurrence of radioresistance in
NSCLC. Based on the response of patients to radiotherapy, NSCLC
tissues were divided into radioresistance and radiation-sensitive
groups (44). circ_0007580 and
circ_0086720 were highly expressed in radioresistant NSCLC tissues.
Additionally, knockdown of the target circRNAs enhanced the
radiotherapy sensitivity of NSCLC cells (44). Huang et al (79) induced radioresistance in NSCLC cell
lines and found a positive correlation between circPVT1 expression
level and the occurrence of radioresistance in NSCLC cell lines. In
addition, circPVT1 knockdown enhanced the radiosensitivity of NSCLC
cell lines.
Other circRNAs have been reported to be negatively
associated with the occurrence of radioresistance in NSCLC. For
example, Zhang et al (80)
discovered that the expression level of circ_0001287 was
significantly lower in cancerous tissues from 87 patients with
NSCLC than in paracancerous tissues. Circ_0001287 overexpression
inhibited the proliferation, migration and invasion of
radioresistant H1299 cells. Clinicopathological analysis revealed
that patients with low circ_0001287 expression were more likely to
have positive lymph node metastasis and poorly differentiated
cancer, implying that circRNAs may enhance the sensitivity to
radiotherapy in lung cancer cells.
CircRNAs can also be used as reference targets for
differential radiotherapy in lung cancer. In a series of
experiments conducted by Jin et al (81), it was found that radioresistant
A549-R11 cells were significantly more resistant to conventional
X-ray radiotherapy than their paternal A549 cells, although both
cell types demonstrated comparable sensitivity to carbon-ion
radiotherapy (CIRT). CircZNF208 is highly expressed in
radioresistant cells, and inhibiting its expression can improve the
sensitivity of A549 and A549-R11 cells to X-ray irradiation. Since
CIRT is costly, few hospitals have X-ray irradiation therapy
equipment, and therefore the treatment is mainly covered by
commercial insurance companies (78). CircZNF208 is expected to be a new
marker for identifying patients who are most likely to benefit from
CIRT therapy (Fig. 3).
CircRNAs and the prognosis of patients with
lung cancer
The prognosis of patients with lung cancer is a
critical follow-up process for effective tumor treatment, as it
serves as a reference point for adjusting medication, changing
treatment regimens and evaluating patient survival time (82).
CircRNAs play an oncogenic role in the
development and progression of lung cancer
CircRNA expression levels have been indicated to
vary significantly between cancerous and paracancerous tissues. The
majority of circRNAs were overexpressed in cancerous tissues and
were associated with poor survival, late TNM Classification of
Malignant Tumors staging and positive lymph node metastasis. These
findings suggest that most circRNAs play an oncogenic role in lung
cancer, promoting tumor invasion and metastasis, which is
associated with patient survival. Dou et al (83) found that the expression of
circ_0001944 was significantly higher in NSCLC than in adjacent
healthy tissues in 47 paired NSCLC tissues. The Kaplan-Meier
survival curve revealed that patients with NSCLC who expressed high
circ_0001944 levels had a poor prognosis. In the 40th month, the
survival rate of the high expression group was only 50%, while the
low expression group had a higher rate of ~90%. Shi et al
(84) reported that patients with
lung squamous cell carcinoma (LUSC) who expressed high levels of
circ_PVT1 had a lower overall survival (OS) than patients with LUSC
who expressed low levels of circ_PVT1. Multivariate analysis
confirmed that circ_PVT1 expression is a potential independent
predictor of OS. Circ_PVT1 may therefore be used to predict the
prognosis of lung cancer. Wang et al (85) showed that circ_PRMT5 was highly
expressed in cancer tissues compared with para-cancer tissues
(P<0.001), and that this level of expression was negatively
associated with OS (P=0.019) and progression-free survival
(P=0.004). Kaplan-Meier survival curves revealed that other
circRNAs highly expressed in cancer tissues, including circ_0067934
(86), circ_0016760 (87), circ_001569 (88), circ_AASDH (89) and circ_MTHFD2 (32), indicated that patients with high
expression had a poor OS (P<0.05). Cox regression analysis
revealed that circRNA may be a significant independent prognostic
factor for patients with NSCLC.
CircRNAs have a role as tumor
suppressor genes in the development and progression of lung
cancer
The expression of a few circRNAs has been
demonstrated to be lower in cancerous tissues than in adjacent
tissues, and patients expressing high circRNA levels in cancerous
tissues had a longer survival duration than those expressing low
circRNA levels. Liu et al (90) used RT-qPCR on 53 cancerous and
adjacent tissues and found that circ_0001649 expression in NSCLC
tissues was significantly lower than in adjacent tissues.
Additionally, patients with low circ_0001649 expression had an
advanced clinical stage and lymph node metastasis. Kaplan-Meier
curve analysis revealed that patients with a high level of
circ_0001649 expression had a significantly longer OS than those
with low circ_0001649 expression (P=0.004). These findings
suggested that circ_0001649 may inhibit proliferation, migration
and invasion of NSCLC cells by acting as a tumor suppressor gene in
NSCLC. A tumor model of nude mice was then established to confirm
the tumor-suppressive effect. Tumors overexpressing circ_0001649
were smaller in volume and weight than those in the negative
control group. A lung metastatic tumor model was developed, and
circ_0001649 overexpression in mice resulted in a reduction in
pulmonary metastatic nodules (87). Therefore, circ_0001649 may be a
therapeutic target for lung cancer. Chen et al (91) found that circ_0000079 (CiR79)
expression level was significantly downregulated in the cancerous
tissues of patients with NSCLC compared with normal control lung
tissue, particularly those with cisplatin resistance (P<0.005).
In their study, the reduction of circ_0000079 level was related to
the decline of OS in patients with NSCLC. Cell experiments revealed
that high circ_0000079 expression inhibited cisplatin resistance in
A549/DDP and H460/DDP cells, implying that circ_0000079 could be
used as a prognostic indicator and therapeutic target for NSCLC.
Chi et al (92) showed that
overexpression of hsa_circ_103820 significantly inhibited the
proliferation, migration and invasion of lung cancer cells, while
knockdown of hsa_circ_103820 played a tumorigenic role. These
results suggested that hsa_circ_103820 has a potential novel
function as a tumor suppressor in lung cancer.
Prognostic value of circRNAs and TNM
staging of lung cancer
Notably, the associated prognosis for certain
circRNAs is related to their level of expression and is inversely
proportional to the TNM stage of patients. Through RT-qPCR analysis
of cancerous and paracancerous tissues of 284 patients, Zhang et
al (93) found that the
expression level of circ_0001946 in paracancerous tissues of
patients with NSCLC was more than three times higher than that in
cancerous tissues. High expression of circ_0001946 is related to a
reduction in lymph node metastasis, decreased TNM stage and an
improved prognosis in patients with NSCLC. Patients were divided
into quartile 1–25%, quartile 25–50%, quartile 50–75% and quartile
75–100% groups based on their expression level of circ_0001946,
which ranged from low to high. The survival analysis using the
Kaplan-Meier curve revealed that disease-free survival (DFS) and OS
were positively associated with circ_0001946 expression levels
(both P<0.001). High expression of circ_0001946 was associated
with improved DFS in stage I, II and III patients (all P<0.05),
however, only stage III patients had a prolonged OS (P=0.037).
These findings indicate that circ_0001946 may have a high
prognostic value in stage III patients. Tong et al (62) revealed lower circ_SMARCA5
expression in cancer tissues of patients with NSCLC than in
adjacent tissues, and circ_SMARCA5 expression was negatively
correlated with tumor size, lymph node metastasis and TNM stage,
but positively correlated with DFS and OS. The study also grouped
circRNA expression levels from low to high, defining the
circ_SMARCA5 high expression group as being within 50–100% of the
relative expression quantile and the circ_SMARCA5 low expression
group as being within 0–50% of the relative expression quantile.
Kaplan-Meier curve survival analysis revealed significantly longer
DFS and OS in the circ_SMARCA5 high expression group than in the
circ_SMARCA5 low expression group (P<0.001). High expression of
circ_SMARCA5 exacerbated the decline in NSCLC cell survival after
chemotherapy, implying that high expression of circ_SMARCA5
increased NSCLC cell sensitivity to chemotherapy. Previous studies
on the association of circRNA expression levels and survival in
patients with cancer have also divided patients into high and low
expression groups and found differences in the survival time of
patients. For example, Tong et al (62) and Zhang et al (93) further classified expression levels
and elucidated the relationship between expression levels and
prognosis. Additionally, Zhang et al further investigated
TNM staging and prognosis. The majority of previous studies did not
subdivide staging at the TNM level, which may have been attributed
to the small sample size (mostly 50-100 cases). This may have
resulted in poor statistical power, particularly for the subgroup
analysis of the association of circRNAs with the prognosis of stage
I, II and III patients.
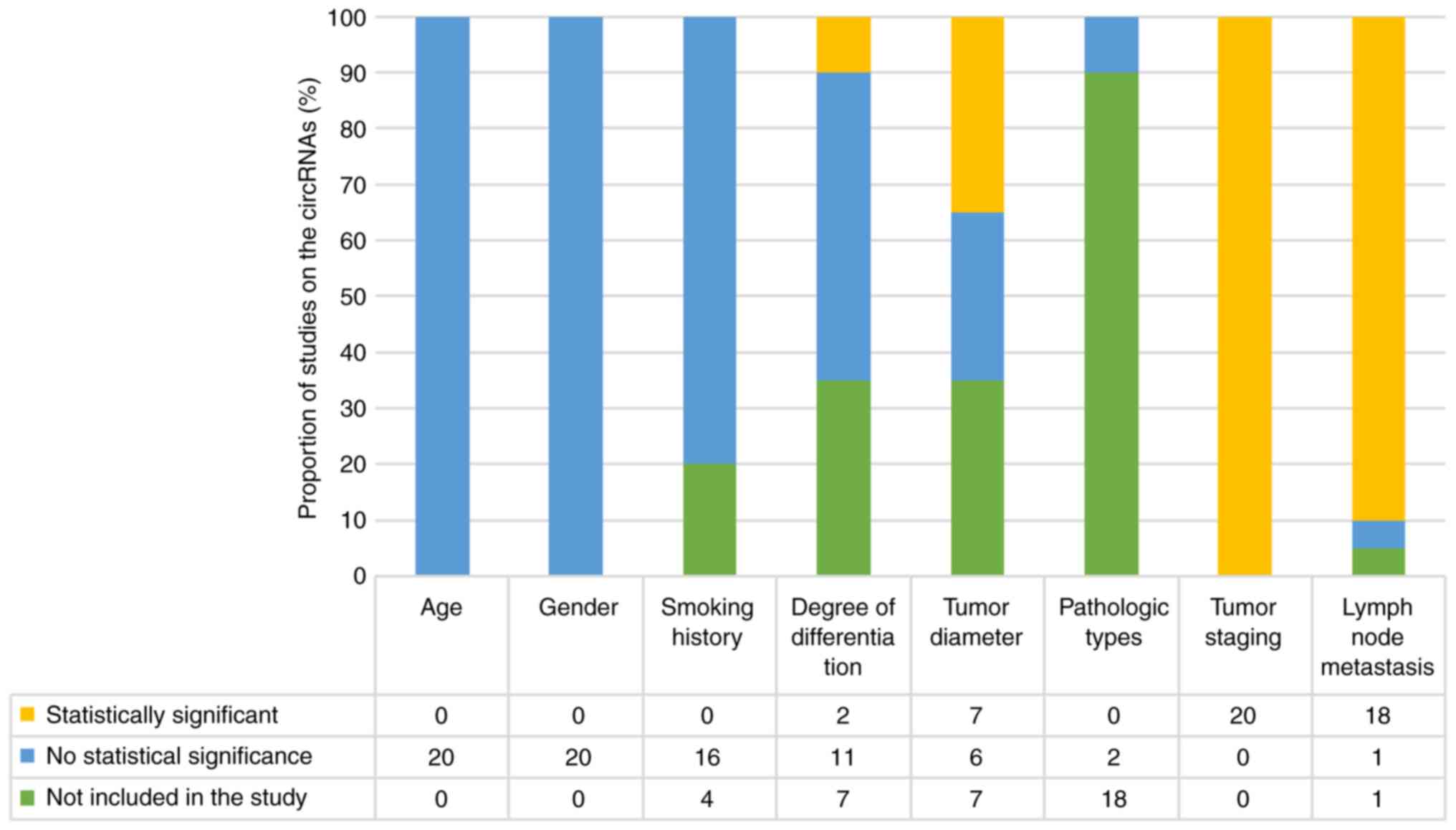
Prognostic value and clinical features
of circRNAs
The association between patients with lung cancer
and various circRNAs is shown in Table II, which also lists the number of
cases, as well as clinical features, namely age, sex, smoking
history, degree of tumor differentiation, tumor diameter, tumor
pathological type, tumor stage, lymph node metastasis and survival
duration of the high expression group. Most available literature
regarding research on circRNAs and lung cancer seems to be from
studies published in Asia. To avoid differences between races, the
present review uniformly selected studies with Asian patients
(23,39,62,83,85,87,93–106). Almost all circRNAs and tumor
prognoses were associated with TNM stage and lymph node metastasis
in the 20 groups of circRNAs listed in the table. Additionally,
high circRNA expression in cancerous tissues was associated with a
shortened OS, whereas low expression of circRNAs in cancerous
tissues exerted the opposite effect. Only one case was shown to be
associated with TNM stage and degree of differentiation, but not
with the other factors that were investigated. All 20 study groups
included age and sex as clinical features; however, there was no
statistically significant association between age, sex and circRNA
expression level. When smoking history was included as a clinical
characteristic, no statistical significance between smoking history
and circRNA expression was observed in any of the 16 groups. The
degree of tumor differentiation was included as a clinical
characteristic in 13 groups; however, only two groups showed a
statistically significant association between the degree of tumor
differentiation and the level of circRNA expression. Tumor diameter
was included as a clinical characteristic in 13 groups, and there
was a statistically significant difference between tumor diameter
and circRNA expression level in seven groups. Only two of the 20
groups included pathological type as a clinical characteristic, and
no statistically significant association between pathological type
and circRNA expression level was found. According to the findings
of the 20 studies, the level of circRNA expression level correlated
with TNM stage and lymph node metastasis, and the majority of the
highly expressed circRNAs in cancerous tissues were positively
correlated with late TNM stage and positive lymph node metastasis,
implying a poor prognosis for patients. Nearly all studies reported
that circRNA expression was unrelated to age, sex, smoking history
and pathological type (Fig. 4).
The relationship between circRNA expression levels, tumor diameter
and degree of tumor differentiation is still debatable. The tumor
diameter limit in these studies may differ compared with that
established in previous studies (3 vs. 5 cm as the dividing point).
However, only 20 circRNAs are listed in the table, which may relate
to a low statistical significance. Therefore, these findings
suggest that circRNAs may be useful as prognostic markers for lung
cancer, but additional research is needed to establish their
validity as independent prognostic factors.
 | Table II.Association of circRNAs with
clinicopathological features of patients with lung cancer. |
Table II.
Association of circRNAs with
clinicopathological features of patients with lung cancer.
| CircRNA | Age | Sex | Smoking
history | Degree of
differentiation | Tumor diameter | Tumor pathological
type | TNM staging | Lymph node
metastasis | Survival duration
of patients with high circRNA expression | Number of cases
included in the study | (Refs.) |
|---|
|
hsa_circ_0016760 | P=0.504 | P=0.420 | P=0.412 | P=0.513 |
P=0.039a | - |
P=0.023a |
P=0.037a | Short OS | 80 | (87) |
|
hsa_circ_0001421 | P=0.107 | P=0.116 | - | - |
P=0.015a | - |
P=0.018a | P=0.153 | Short OS | 48 | (94) |
| circ_0003645 | P=0.435 | P=0.784 | P=0.293 | P=0.607 | - | - |
P=0.019a |
P=0.010a | Short OS | 59 | (95) |
| circ_000984 | P>0.05 | P>0.05 | - | - | P>0.05 | - |
P=0.004a |
P=0.005a | Short DFS/OS | 155 | (96) |
| circ_0001649 | P=0.572 | P=0.544 | P=0.406 | P=0.557 | - | - |
P=0.010a |
P=0.029a | Long OS | 53 | (97) |
| circ_0020123 | P=0.270 | P=0.788 | P=0.781 | P=0.391 | - | - |
P=0.037a |
P=0.029a | Short OS | 55 | (98) |
| circ_0074027 | P=0.579 | P=0.393 | P=0.404 |
P=0.025a | - | - |
P=0.011a | P=0.095 | Short OS | 52 | (99) |
| circ_CRIM1 | P=0.209 | P=0.676 | P=0.400 | - | P=0.204 | - |
P=0.021a |
P=0.022a | Short OS | 92 | (100) |
| circ_PRMT5 | P=0.205 | P=0.822 | P=0.607 | P=0.274 |
P=0.002a | - |
P=0.028a |
P=0.018a | Short DFS/OS | 90 | (85) |
|
hsa_circ_0007534 | P=0.837 | P=0.828 | P=0.529 | P=0.298 | - | - |
P=0.039a |
P=0.024a | Short OS | 98 | (101) |
| circ_0016760 | P=0.825 | P=0.820 | P=0.503 | P=0.128 | - | - |
P=0.044a |
P=0.003a | Short OS | 83 | (102) |
| circ_MEMO1 | P>0.05 | P>0.05 | - | - | P>0.05 | - |
P<0.05a |
P<0.05a | Short OS | 52 | (39) |
| circ_SMARCA5 | P=0.209 | P=0.178 | P=0.190 | P=0.260 |
P=0.001a | - |
P=0.001a |
P=0.004a | Long DFS/OS | 460 | (62) |
| circ_0001944 | P=0.191 | P=0.181 | P=0.058 | - | P=0.110 | - |
P=0.011a |
P=0.002a | Short DFS | 47 | (83) |
| circ_PVT1 | P=0.3216 | P=0.1936 | P=0.1733 | P=0.1151 |
P=0.0468a | - |
P=0.0001a |
P=0.0014a | Short OS | 104 | (103) |
| circ_0001946 | P=0.446 | P=0.187 | P=0.670 | P=0.185 | P=0.097 | - |
P=0.001a |
P<0.001a | Long DFS/OS | 284 | (93) |
| circ_TADA2A | P=0.791 | P=0.284 | P=0.787 | - |
P=0.020a | P=0.301 |
P=0.038a |
P=0.001a | Short OS | 60 | (104) |
| circ_0000517 | P=0.592 | P=0.793 | P=0.438 | P=0.071 | - | - |
P=0.024a |
P=0.020a | Short OS | 60 | (23) |
| circ_0005909 | P=0.988 | P=0.241 | - | - | P=0.217 | - |
P=0.023a | - | Short OS | 102 | (105) |
|
hsa_circ_0003222 | P>0.05 | P>0.05 | P>0.05 |
P<0.05a |
P<0.05a | P>0.05 |
P<0.05a |
P<0.05a | Short OS | 30 | (106) |
Limitations of circRNAs as diagnostic
markers for lung cancer
CircRNAs are non-coding RNAs. Several studies have
confirmed that circRNAs regulate the expression of cancer-related
genes through circRNA-miRNA-mRNA pathways, thus affecting the
proliferation, invasion and migration of cells in lung cancer.
Their unique biological characteristics cause circRNAs to be
regarded as potential biomarkers in the diagnosis of lung cancer.
However, the exact mechanism of circRNAs in the development of lung
cancer has not been fully elucidated. There are several studies on
the carcinogenesis and tumor suppression of circRNAs in the
development of lung cancer, but most of the studies first verified
the differential expression of circRNAs in cancerous tissues and
adjacent tissues, and then verified their expression in cells and
animal models. On the other hand, differentially expressed circRNAs
were reported in different studies. Specific circRNAs with higher
diagnostic values in lung cancer have not been established, and no
expert consensus has been formed on the diagnostic value of certain
circRNAs in lung cancer (107).
Certain circRNAs, such as CDR1as, are differentially expressed in
NSCLC, liver cancer (C22.001), stomach cancer (C16.902), esophageal
cancer (C15.901), bile duct cancer (C22.101) and other types of
cancer (108), which could be
used as biomarkers for lung cancer screening. However, the
diagnostic specificity of these circRNAs is not high.
The diagnostic value of circRNAs in lung cancer is
currently limited to the laboratory stage. The subsequent clinical
application also faces problems, such as diagnostic accuracy and
economic burden. Currently, biomarkers developed for lung cancer
screening in clinical practice include the carcinoembryonic
antigen, squamous cell carcinoma antigen, neuron-specific enolase,
pro-gastrin-releasing peptide, carcinoma antigen 125, cytokeratin
fragment 19, human epididymis protein 4 and others (109). The diagnostic value of circRNAs
in lung cancer has not been compared with such traditional
biomarkers. In conclusion, further studies are needed to confirm
the clinical application of circRNAs, but circRNAs are promising as
pot0ential biomarkers for lung cancer.
Conclusion
Lung cancer is one of the most common malignant
tumors in China, with the highest mortality rates among patients
with cancer (110,111). Patients with advanced lung cancer
have a very poor prognosis, necessitating the development of early
diagnostics and therapies for effective management of the disease
(112). CircRNAs are found in
abundance in cells, urine, saliva, plasma and other tissues, and
are characterized by their stability, universality, conservatism
and specificity. Because of their abundance in human tissues,
sampling is associated with minimal trauma, speed, minimal risk,
low cost, wide usage, and has high operability, implying their
potential use as a marker for the early diagnosis of patients with
lung cancer. Although circRNAs are abnormally expressed in a
variety of tumors, their biological function remains largely
unknown. Currently, the majority of studies on circRNAs have mainly
focused on their abnormal expression in lung cancer and their
regulatory role in gene expression via the miRNA ‘sponge’
mechanism. However, findings from related studies indicate that
circRNAs have a variety of functions in other diseases, including
protein binding and regulating transcription and translation. It
would be useful to further investigate these findings in relation
to other diseases, such as lung cancer. While the majority of
studies have demonstrated the diagnostic value of circRNAs in lung
cancer, their clinical application is still constrained by their
low abundance in blood and high detection cost. More in-depth and
comprehensive clinical trials are required to verify the value of
circRNAs as diagnostic markers. At present, the use of circRNAs for
lung cancer detection is primarily based on their differential
expression between cancerous and adjacent normal tissues, with few
studies incorporating tumor staging and pathological typing as
grouping criteria. Therefore, additional research is required to
identify highly specific and sensitive circRNAs during the early
developmental stages of lung cancer. Additionally, elucidating the
underlying biological mechanism and identifying molecular targets
would pave the way for effective lung cancer diagnosis, treatment
and prognosis.
Acknowledgements
Not applicable.
Funding
This work was supported by funds from the National Natural
Science Foundation of China (grant nos. 81860337 and
81960326,82060384), Natural Science Foundation of Jiangxi Province
(grant no. 20202ACBL206014), Natural Science Foundation of Jiangxi
Province (grant no. 20192BAB205009), National Health Commission
Science & Technology Development Research Center (grant no.
2019ZH-07E-003), Science and Technology Plan of Jiangxi Provincial
Health Commission (grant no. 20201080).
Availability of data and materials
Not applicable.
Authors' contributions
CS was responsible for the conception and design of
the study, the collection and assembly of data, as well as the data
analysis and interpretation. ZT and DR were responsible for the
provision of study materials. ZT was also responsible for
administrative support. CS, ZT, DR, CW, YX and MS contributed to
writing the manuscript. All authors read and approved the final
version of the manuscript. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Suster D and Mino-Kenudson M: Molecular
pathology of primary non-small cell lung cancer. Arch Med Res.
51:784–798. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Woodard G, Jones K and Jablons D: Lung
cancer staging and prognosis. Cancer Treat Res. 170:47–75. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao S, Li S, Liu W, Wang Y, Li X, Zhu S,
Lei X and Xu S: Circular RNA signature in lung adenocarcinoma: A
MiOncoCirc database-based study and literature review. Front Oncol.
10:5233422020. View Article : Google Scholar
|
|
5
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar
|
|
8
|
Wilusz JE: A 360° view of circular RNAs:
From biogenesis to functions. Wiley Interdiscip Rev RNA.
9:e14782018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peng Z, Hu Q, Fang S, Zhang X, Hong X, Tao
L, Pan J, Jiang M, Bai H, Wu Y, et al: Circulating circTOLLIP
serves as a diagnostic biomarker for liquid biopsy in non-small
cell lung cancer. Clin Chim Acta. 523:415–422. 2021. View Article : Google Scholar
|
|
10
|
Passiglia F, Bertaglia V, Reale ML,
Delcuratolo MD, Tabbò F, Olmetto E, Capelletto E, Bironzo P and
Novello S: Major breakthroughs in lung cancer adjuvant treatment:
Looking beyond the horizon. Cancer Treat Rev. 101:1023082021.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wan J, Ling XA, Wang J, Ding GG and Wang
X: Inhibitory effect of Ubenimex combined with fluorouracil on
multiple drug resistance and P-glycoprotein expression level in
non-small lung cancer. J Cell Mol Med. 24:12840–12847. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao Y, Wang J and Zhao F: CIRI: An
efficient and unbiased algorithm for de novo circular RNA
identification. Genome Biol. 16:42015. View Article : Google Scholar
|
|
13
|
Liang ZZ, Guo C, Zou MM, Meng P and Zhang
TT: circRNA-miRNA-mRNA regulatory network in human lung cancer: An
update. Cancer Cell Int. 20:1732020. View Article : Google Scholar
|
|
14
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015.
|
|
15
|
Petkovic S and Müller S: RNA
circularization strategies in vivo and in vitro. Nucleic Acids Res.
43:2454–2465. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016.
View Article : Google Scholar
|
|
17
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen B and Huang S: Circular RNA: An
emerging non-coding RNA as a regulator and biomarker in cancer.
Cancer Lett. 418:41–50. 2018. View Article : Google Scholar
|
|
19
|
Westholm JO, Miura P, Olson S, Shenker S,
Joseph B, Sanfilippo P, Celniker SE, Graveley BR and Lai EC:
Genome-wide analysis of drosophila circular RNAs reveals their
structural and sequence properties and age-dependent neural
accumulation. Cell Rep. 9:1966–1980. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Han B, Chao J and Yao H: Circular RNA and
its mechanisms in disease: From the bench to the clinic. Pharmacol
Ther. 187:31–44. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar
|
|
22
|
Bachmayr-Heyda A, Reiner AT, Auer K,
Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW,
Zeillinger R and Pils D: Correlation of circular RNA abundance with
proliferation-exemplified with colorectal and ovarian cancer,
idiopathic lung fibrosis, and normal human tissues. Sci Rep.
5:80572015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bing ZX, Zhang JQ, Wang GG, Wang YQ, Wang
TG and Li DQ: Silencing of circ_0000517 suppresses proliferation,
glycolysis, and glutamine decomposition of non-small cell lung
cancer by modulating miR-330-5p/YY1 signal pathway. Kaohsiung J Med
Sci. 37:1027–1037. 2021. View Article : Google Scholar
|
|
24
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar
|
|
25
|
Yin H, Shen X, Zhao J, Cao X, He H, Han S,
Chen Y, Cui C, Wei Y, Wang Y, et al: Circular RNA CircFAM188B
encodes a protein that regulates proliferation and differentiation
of chicken skeletal muscle satellite cells. Front Cell Dev Biol.
8:5225882020. View Article : Google Scholar
|
|
26
|
Abdelmohsen K, Kuwano Y, Kim HH and
Gorospe M: Posttranscriptional gene regulation by RNA-binding
proteins during oxidative stress: Implications for cellular
senescence. Biol Chem. 389:243–255. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mi Z, Zhongqiang C, Caiyun J, Yanan L,
Jianhua W and Liang L: Circular RNA detection methods: A
minireview. Talanta. 238((Pt 2)): 1230662022. View Article : Google Scholar
|
|
28
|
Kalanjeri S, Holladay RC and Gildea TR:
State-of-the-Art modalities for peripheral lung nodule biopsy. Clin
Chest Med. 39:125–138. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang Q, Chen L, Yang L and Huang Y:
Diagnostic and prognostic values of circular RNAs for lung cancer:
A meta-analysis. Postgrad Med J. 97:286–293. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lin Y, Su W and Lan G: Value of circular
RNA 0007385 in disease monitoring and prognosis estimation in
non-small-cell lung cancer patients. J Clin Lab Anal.
34:e233382020.
|
|
31
|
Fu Y, Huang L, Tang H and Huang R:
hsa_circRNA_012515 is highly expressed in NSCLC patients and
affects its prognosis. Cancer Manag Res. 12:1877–1886. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Geng QQ, Wu QF, Zhang Y, Zhang GJ, Fu JK
and Chen NZ: Clinical significance of circ-MTHFD2 in diagnosis,
pathological staging and prognosis of NSCLC. Eur Rev Med Pharmacol
Sci. 24:9473–9479. 2020.
|
|
33
|
Li S, Sun X, Miao S, Lu T, Wang Y, Liu J
and Jiao W: hsa_circ_0000729, a potential prognostic biomarker in
lung adenocarcinoma. Thorac Cancer. 9:924–930. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan
X, Han S and Wu G: hsa_circ_0013958: A circular RNA and potential
novel biomarker for lung adenocarcinoma. FEBS J. 284:2170–2182.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li W, Liu JB, Hou LK, Yu F, Zhang J, Wu W,
Tang XM, Sun F, Lu HM, Deng J, et al: Liquid biopsy in lung cancer:
Significance in diagnostics, prediction, and treatment monitoring.
Mol Cancer. 21:252022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou R, Chen KK, Zhang J, Xiao B, Huang Z,
Ju C, Sun J, Zhang F, Lv XB and Huang G: The decade of exosomal
long RNA species: An emerging cancer antagonist. Mol Cancer.
17:752018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Y, Zhang H, Wang J, Li B and Wang X:
Circular RNA expression profile of lung squamous cell carcinoma:
Identification of potential biomarkers and therapeutic targets.
Biosci Rep. Apr 30–2020.(Epub ahead of print).
|
|
38
|
Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan
Y, Kong X, Bu J, Liu M and Xu S: circRNA-002178 act as a ceRNA to
promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis.
11:322020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ding C, Xi G, Wang G, Cui D, Zhang B, Wang
H, Jiang G, Song J, Xu G and Wang J: Exosomal Circ-MEMO1 promotes
the progression and aerobic glycolysis of non-small cell lung
cancer through targeting MiR-101-3p/KRAS axis. Front Genet.
11:9622020. View Article : Google Scholar
|
|
40
|
Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu
M, Dai X, Zhou H, Zhu J, Zhang H and Jiang Y: Circular RNA
circSATB2 promotes progression of non-small cell lung cancer cells.
Mol Cancer. 19:1012020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Song Y, Cao P and Li J: Plasma circular
RNA hsa_circ_0001821 acts as a novel diagnostic biomarker for
malignant tumors. J Clin Lab Anal. 35:e240092021.
|
|
42
|
Zhou X, Liu HY, Wang WY, Zhao H and Wang
T: Hsa_circ_0102533 serves as a blood-based biomarker for
non-small-cell lung cancer diagnosis and regulates apoptosis in
vitro. Int J Clin Exp Pathol. 1:4395–4404. 2018.
|
|
43
|
Liu Y, Li C, Liu H and Wang J:
Circ_0001821 knockdown suppresses growth, metastasis, and TAX
resistance of non-small-cell lung cancer cells by regulating the
miR-526b-5p/GRK5 axis. Pharmacol Res Perspect. 9:e008122021.
|
|
44
|
Guo C, Wang H, Jiang H, Qiao L and Wang X:
Circ_0011292 enhances paclitaxel resistance in non-small cell lung
cancer by regulating miR-379-5p/TRIM65 axis. Cancer Biother
Radiopharm. 37:84–95. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhao Y, Zheng R, Chen J and Ning D:
CircRNA CDR1as/miR-641/HOXA9 pathway regulated stemness contributes
to cisplatin resistance in non-small cell lung cancer (NSCLC).
Cancer Cell Int. 20:2892020. View Article : Google Scholar
|
|
46
|
Zheng F and Xu R: CircPVT1 contributes to
chemotherapy resistance of lung adenocarcinoma through
miR-145-5p/ABCC1 axis. Biomed Pharmacother. 124:1098282020.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dong Y, Xu T, Zhong S, Wang B, Zhang H,
Wang X, Wang P, Li G and Yang S: Circ_0076305 regulates cisplatin
resistance of non-small cell lung cancer via positively modulating
STAT3 by sponging miR-296-5p. Life Sci. 239:1169842019. View Article : Google Scholar
|
|
48
|
Ma J, Qi G and Li L: A novel serum
exosomes-based biomarker hsa_circ_0002130 facilitates
osimertinib-resistance in non-small cell lung cancer by sponging
miR-498. Onco Targets Ther. 13:5293–5307. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Shao N, Song L and Sun X: Exosomal
circ_PIP5K1A regulates the progression of non-small cell lung
cancer and cisplatin sensitivity by miR-101/ABCC1 axis. Mol Cell
Biochem. 476:2253–2267. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhou Y, Zheng X, Xu B, Chen L, Wang Q,
Deng H and Jiang J: Circular RNA hsa_circ_0004015 regulates the
proliferation, invasion, and TKI drug resistance of non-small cell
lung cancer by miR-1183/PDPK1 signaling pathway. Biochem Biophys
Res Commun. 508:527–535. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huang Y, Dai Y, Wen C, He S, Shi J, Zhao
D, Wu L and Zhou H: circSETD3 contributes to acquired resistance to
gefitinib in non-small-cell lung cancer by targeting the
miR-520h/ABCG2 pathway. Mol Ther Nucleic Acids. 21:885–899. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Joseph NA, Chiou SH, Lung Z, Yang CL, Lin
TY, Chang HW, Sun HS, Gupta SK, Yen L, Wang SD and Chow KC: The
role of HGF-MET pathway and CCDC66 cirRNA expression in EGFR
resistance and epithelial-to-mesenchymal transition of lung
adenocarcinoma cells. J Hematol Oncol. 11:742018. View Article : Google Scholar
|
|
53
|
Liu Y, Zhai R, Hu S and Liu J: Circular
RNA circ-RNF121 contributes to cisplatin (DDP) resistance of
non-small-cell lung cancer cells by regulating the miR-646/SOX4
axis. Anticancer drugs. 33:e186–e197. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li J, Fan R and Xiao H: Circ_ZFR
contributes to the paclitaxel resistance and progression of
non-small cell lung cancer by upregulating KPNA4 through sponging
miR-195-5p. Cancer Cell Int. 21:152021. View Article : Google Scholar
|
|
55
|
Zhu X, Han J, Lan H, Lin Q, Wang Y and Sun
X: A novel circular RNA hsa_circRNA_103809/miR-377-3p/GOT1 pathway
regulates cisplatin-resistance in non-small cell lung cancer
(NSCLC). BMC Cancer. 20:11902020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ye Y, Zhao L, Li Q, Xi C, Li Y and Li Z:
circ_0007385 served as competing endogenous RNA for miR-519d-3p to
suppress malignant behaviors and cisplatin resistance of non-small
cell lung cancer cells. Thorac Cancer. 11:2196–2208. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen SW, Zhu SQ, Pei X, Qiu BQ, Xiong D,
Long X, Lin K, Lu F, Xu JJ and Wu YB: Cancer cell-derived exosomal
circUSP7 induces CD8+ T cell dysfunction and anti-PD1
resistance by regulating the miR-934/SHP2 axis in NSCLC. Mol
Cancer. 20:1442021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ge W, Chi H, Tang H, Xu J, Wang J, Cai W
and Ma H: Circular RNA CELF1 drives immunosuppression and anti-PD1
therapy resistance in non-small cell lung cancer via the
miR-491-5p/EGFR axis. Aging (Albany NY). 13:24560–24579. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liu Z, Wang T, She Y, Wu K, Gu S, Li L,
Dong C, Chen C and Zhou Y: N6-methyladenosine-modified
circIGF2BP3 inhibits CD8+ T-cell responses to facilitate
tumor immune evasion by promoting the deubiquitination of PD-L1 in
non-small cell lung cancer. Mol Cancer. 20:1052021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yang J, Jia Y, Wang B, Yang S, Du K, Luo
Y, Li Y and Zhu B: Circular RNA CHST15 Sponges miR-155-5p and
miR-194-5p to promote the immune escape of lung cancer cells
mediated by PD-L1. Front Oncol. 11:5956092021. View Article : Google Scholar
|
|
61
|
Li X, Yang B, Ren H, Xiao T, Zhang L, Li
L, Li M, Wang X, Zhou Ha and Zhang W: Hsa_circ_0002483 inhibited
the progression and enhanced the Taxol sensitivity of non-small
cell lung cancer by targeting miR-182-5p. Cell Death Dis.
10:9532019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Tong S: Circular RNA SMARCA5 may serve as
a tumor suppressor in non-small cell lung cancer. J Clin Lab Anal.
34:e231952020.
|
|
63
|
Huang W, Yang Y, Wu J, Niu Y, Yao Y, Zhang
J, Huang X, Liang S, Chen R, Chen S and Guo L: Circular RNA cESRP1
sensitises small cell lung cancer cells to chemotherapy by sponging
miR-93-5p to inhibit TGF-β signalling. Cell Death Differ.
27:1709–1727. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhang W, Song C and Ren X: Circ_0003998
regulates the progression and docetaxel sensitivity of
DTX-resistant non-small cell lung cancer cells by the
miR-136-5p/CORO1C axis. Technol Cancer Res Treat. Jan 29–2021.(Epub
ahead of print).
|
|
65
|
Wang Y, Li L, Zhang W and Zhang G:
Circular RNA circLDB2 functions as a competing endogenous RNA to
suppress development and promote cisplatin sensitivity in
non-squamous non-small cell lung cancer. Thorac Cancer.
12:1959–1972. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Li S, Liu Y, Qiu G, Luo Y, Li X, Meng F,
Li N, Xu T, Wang Y, Qin B and Xia S: Emerging roles of circular
RNAs in non-small cell lung cancer (Review). Oncol Rep. 45:172021.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yu HA, Arcila ME, Rekhtman N, Sima CS,
Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M and Riely GJ:
Analysis of tumor specimens at the time of acquired resistance to
EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers.
Clin Cancer Res. 19:2240–2247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wang Y, Wang Y, Qin Z, Cai S, Yu L, Hu H
and Zeng S: The role of non-coding RNAs in ABC transporters
regulation and their clinical implications of multidrug resistance
in cancer. Expert Opin Drug Metab Toxicol. 17:291–306. 2021.
View Article : Google Scholar
|
|
69
|
Wen C, Xu G, He S, Huang Y, Shi J, Wu L
and Zhou H: Screening circular RNAs related to acquired gefitinib
resistance in non-small cell lung cancer cell lines. J Cancer.
11:3816–3826. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kalluri R and LeBleu VS: The biology,
function, and biomedical applications of exosomes. Science.
367:eaau69772020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yang B, Teng F, Chang L, Wang J, Liu DL,
Cui YS and Li GH: Tumor-derived exosomal circRNA_102481 contributes
to EGFR-TKIs resistance via the miR-30a-5p/ROR1 axis in non-small
cell lung cancer. Aging (Albany NY). 13:13264–13286. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Dongre A and Weinberg RA: New insights
into the mechanisms of epithelial-mesenchymal transition and
implications for cancer. Nat Rev Mol Cell Biol. 20:69–84. 2019.
View Article : Google Scholar
|
|
73
|
Luo YH, Yang YP, Chien CS, Yarmishyn AA,
Ishola AA, Chien Y, Chen YM, Huang TW, Lee KY, Huang WC, et al:
Plasma level of circular RNA hsa_circ_0000190 correlates with tumor
progression and poor treatment response in advanced lung cancers.
Cancers (Basel). 12:17402020. View Article : Google Scholar
|
|
74
|
Reck M, Rodríguez-Abreu D, Robinson AG,
Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe
S, et al: Pembrolizumab versus Chemotherapy for PD-L1-positive
non-small-cell lung cancer. N Engl J Med. 375:1823–1833. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhang R, Guo Y, Yan Y, Liu Y, Zhu Y, Kang
J, Li F, Sun X, Xing L and Xu Y: A propensity-matched analysis of
survival of clinically diagnosed early-stage lung cancer and
biopsy-proven early-stage non-small cell lung cancer following
stereotactic ablative radiotherapy. Front Oncol. 11:7208472021.
View Article : Google Scholar
|
|
76
|
Huang Q: Predictive relevance of ncRNAs in
non-small-cell lung cancer patients with radiotherapy: A review of
the published data. Biomark Med. 12:1149–1159. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zhang H, Si J, Yue J and Ma S: The
mechanisms and reversal strategies of tumor radioresistance in
esophageal squamous cell carcinoma. J Cancer Res Clin Oncol.
147:1275–1286. 2021. View Article : Google Scholar
|
|
78
|
Jin X, Yuan L, Liu B, Kuang Y, Li H, Li L,
Zhao X, Li F, Bing Z, Chen W, et al: Integrated analysis of
circRNA-miRNA-mRNA network reveals potential prognostic biomarkers
for radiotherapies with X-rays and carbon ions in non-small cell
lung cancer. Ann Transl Med. 8:13732020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Huang M, Li T, Wang Q, et al: Silencing
circPVT1 enhances radiosensitivity in non-small cell lung cancer by
sponging microRNA-1208. Cancer Biomark. 31:263–279. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zhang C, Li Y, Feng X and Li D: Circular
RNA circ_0001287 inhibits the proliferation, metastasis, and
radiosensitivity of non-small cell lung cancer cells by sponging
microRNA miR-21 and up-regulating phosphatase and tensin homolog
expression. Bioengineered. 12:414–425. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Jin X, Yuan L, Liu B, et al: Integrated
analysis of circRNA-miRNA-mRNA network reveals potential prognostic
biomarkers for radiotherapies with X-rays and carbon ions in
non-small cell lung cancer. Ann Transl Med. 8:13732020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Giaj-Levra N, Ricchetti F and Alongi F:
What is changing in radiotherapy for the treatment of locally
advanced nonsmall cell lung cancer patients? A review. Cancer
Invest. 34:80–93. 2016. View Article : Google Scholar
|
|
83
|
Dou Y, Tian W, Wang H and Lv S:
Circ_0001944 contributes to glycolysis and tumor growth by
upregulating NFAT5 through acting as a decoy for miR-142-5p in
non-small cell lung cancer. Cancer Manag Res. 13:3775–3787. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Shi J, Lv X, Zeng L, et al: CircPVT1
promotes proliferation of lung squamous cell carcinoma by binding
to miR-30d/e. J Exp Clin Cancer Res: CR. 40:1932021. View Article : Google Scholar
|
|
85
|
Wang Y, Li Y, He H and Wang F: Circular
RNA circ-PRMT5 facilitates non-small cell lung cancer proliferation
through upregulating EZH2 via sponging miR-377/382/498. Gene.
720:1440992019. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zou Q, Wang T, Li B, Li G, Zhang L, Wang B
and Sun S: Overexpression of circ-0067934 is associated with
increased cellular proliferation and the prognosis of non-small
cell lung cancer. Oncol Lett. 16:5551–5556. 2018.
|
|
87
|
Zhu Z, Wu Q, Zhang M, Tong J, Zhong B and
Yuan K: Hsa_circ_0016760 exacerbates the malignant development of
non-small cell lung cancer by sponging miR-145-5p/FGF5. Oncol Rep.
45:501–512. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Ding L, Yao W, Lu J, Gong J and Zhang X:
Upregulation of circ_001569 predicts poor prognosis and promotes
cellproliferation in non-small cell lung cancer by regulating the
Wnt/β-catenin pathway. Oncol Lett. 16:453–458. 2018.
|
|
89
|
Wang Y, Wo Y, Lu T, Sun X, Liu A, Dong Y,
Du W, Su W, Huang Z and Jiao W: Circ-AASDH functions as the
progression of early stage lung adenocarcinoma by targeting
miR-140-3p to activate E2F7 expression. Transl Lung Cancer Res.
10:57–70. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Liu T, Song Z and Gai Y: Circular RNA
circ_0001649 acts as a prognostic biomarker and inhibits NSCLC
progression via sponging miR-331-3p and miR-338-5p. Biochemical and
biophysical research communications. 503:1503–1509. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Chen C, Min Z, Yan Z, et al: Circ_0000079
decoys the RNA-binding protein FXR1 to interrupt formation of the
FXR1/PRCKI complex and decline their mediated cell invasion and
drug resistance in NSCLC. Cell transplantation.
29:9636897209610702020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Chi Y, Zheng W, Bao G, et al: Circular RNA
circ_103820 suppresses lung cancer tumorigenesis by sponging
miR-200b-3p to release LATS2 and SOCS6. Cell Death Dis. 12:1852021.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zhang M, Wen F and Zhao K: Circular
RNA_0001946 is insufficiently expressed in tumor tissues, while its
higher expression correlates with less lymph node metastasis, lower
TNM stage, and improved prognosis in NSCLC patients. J Clin Lab
Anal. 35:e236252021. View Article : Google Scholar
|
|
94
|
Zhang K, Hu H, Xu J, Qiu L, Chen H, Jiang
X and Jiang Y: Circ_0001421 facilitates glycolysis and lung cancer
development by regulating miR-4677-3p/CDCA3. Diagn Pathol.
15:1332020. View Article : Google Scholar
|
|
95
|
An J, Shi H, Zhang N and Song S: Elevation
of circular RNA circ_0003645 forecasts unfavorable prognosis and
facilitates cell progression via miR-1179/TMEM14A pathway in
non-small cell lung cancer. Biochem Biophys Res Commun.
511:921–925. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Li XY, Liu YR, Zhou JH, Li W, Guo HH and
Ma HP: Enhanced expression of circular RNA hsa_circ_000984 promotes
cells proliferation and metastasis in non-small cell lung cancer by
modulating Wnt/β-catenin pathway. Eur Rev Med Pharmacol Sci.
23:3366–3374. 2019.
|
|
97
|
Liu T, Song Z and Gai Y: Circular RNA
circ_0001649 acts as a prognostic biomarker and inhibits NSCLC
progression via sponging miR-331-3p and miR-338-5p. Biochem Biophys
Res Commun. 503:1503–1509. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Wan J, Hao L, Zheng X and Li Z: Circular
RNA circ_0020123 promotes non-small cell lung cancer progression by
acting as a ceRNA for miR-488-3p to regulate ADAM9 expression.
Biochem Biophys Res Commun. 515:303–309. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Gao P, Wang Z, Hu Z, Jiao X and Yao Y:
Circular RNA circ_0074027 indicates a poor prognosis for NSCLC
patients and modulates cell proliferation, apoptosis, and invasion
via miR-185-3p mediated BRD4/MADD activation. J Cell Biochem.
121:2632–2642. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Zhang SJ, Ma J, Wu JC, Hao ZZ, Zhang YA
and Zhang YJ: Circular RNA circCRIM1 suppresses lung adenocarcinoma
cell migration, invasion, EMT, and glycolysis through regulating
miR-125b-5p/BTG2 axis. Eur Rev Med Pharmacol Sci. 25:33992021.
|
|
101
|
Qi Y, Zhang B, Wang J and Yao M:
Upregulation of circular RNA hsa_circ_0007534 predicts unfavorable
prognosis for NSCLC and exerts oncogenic properties in vitro and in
vivo. Gene. 676:79–85. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Li Y, Hu J, Li L, Cai S, Zhang H, Zhu X,
Guan G and Dong X: Upregulated circular RNA circ_0016760 indicates
unfavorable prognosis in NSCLC and promotes cell progression
through miR-1287/GAGE1 axis. Biochem Biophys Res Commun.
503:2089–2094. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Shi J, Lv X, Zeng L, Li W, Zhong Y, Yuan
J, Deng S, Liu B, Yuan B, Chen Y, et al: CircPVT1 promotes
proliferation of lung squamous cell carcinoma by binding to
miR-30d/e. J Exp Clin Cancer Res. 40:1932021. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zhang Y, Yao H, Li Y, Yang L, Zhang L,
Chen J, Wang Y and Li X: Circular RNA TADA2A promotes proliferation
and migration via modulating of miR-638/KIAA0101 signal in
non-small cell lung cancer. Oncol Rep. 46:2012021. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Song HM, Meng D, Wang JP and Zhang XY:
circRNA hsa_circ_0005909 predicts poor prognosis and promotes the
growth, metastasis, and drug resistance of non-small-cell lung
cancer via the miRNA-338-3p/SOX4 pathway. Dis Markers.
2021:83885122021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Li C, Zhang J, Yang X, Hu C, Chu T, Zhong
R, Shen Y, Hu F, Pan F, Xu J, et al: hsa_circ_0003222 accelerates
stemness and progression of non-small cell lung cancer by sponging
miR-527. Cell Death Dis. 12:8072021. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Liu Y, Ao Y, Yu W, Zhang Y and Wang J:
Biogenesis, functions, and clinical implications of circular RNAs
in non-small cell lung cancer. Mol Ther Nucleic Acid. 27:50–72.
2022. View Article : Google Scholar
|
|
108
|
Jian F, Yangyang R, Wei X, Jiadan X, Na L,
Peng Y, Maohong B, Guoping N and Zhaoji P: The prognostic and
predictive significance of circRNA CDR1as in tumor progression.
Front Oncol. 10:5499822021. View Article : Google Scholar
|
|
109
|
Wang J, Chu Y, Li J and Zeng F, Wu M, Wang
T, Sun L, Chen Q, Wang P, Zhang X and Zeng F: Development of a
prediction model with serum tumor markers to assess tumor
metastasis in lung cancer. Cancer Med. 9:5436–5445. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Wei J, Yan Y, Chen X, Qian L, Zeng S, Li
Z, Dai S, Gong Z and Xu Z: The roles of plant-derived triptolide on
non-small cell lung cancer. Oncol Res. 27:849–858. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Yan Y, Xu Z, Hu X, Qian L, Li Z, Zhou Y,
Dai S, Zeng S and Gong Z: SNCA is a functionally low-expressed gene
in lung adenocarcinoma. Genes (Basel). 9:162018. View Article : Google Scholar
|
|
112
|
Wei J, Xu Z, Chen X, Wang X, Zeng S, Qian
L, Yang X, Ou C, Lin W, Gong Z and Yan Y: Overexpression of GSDMC
is a prognostic factor for predicting a poor outcome in lung
adenocarcinoma. Mol Med Rep. 21:360–370. 2020.PubMed/NCBI
|