Introduction
Hepatocellular carcinoma (HCC) is one of the most
lethal cancer types worldwide (1).
It is a highly vascularized tumour and angiogenesis plays a crucial
role in its development, invasion and metastasis. The induction of
angiogenesis has been recognized as a crucial step for HCC
progression and one of the hallmarks of HCC progression (2–5). The
process of angiogenesis comprises of an active proliferation and
new vessel formation of vascular endothelial cells, followed by the
release of several growth factors, including vascular endothelial
growth factor (VEGF), platelet-derived growth factor and
erythropoietin (6–8). One of the main associations between
angiogenesis and HCC growth is the transition of epithelial liver
cells to a mesenchymal phenotype, specified as
epithelial-mesenchymal transition (EMT) by angiogenic growth
factors, such as VEGF (9–12). EMT confers traits of mesenchymal
cells to hepatic tumour cells, which subsequently display a high
motility and aggressiveness. Thus, tumour cells acquire the ability
to easily enter the bloodstream by invading tumour tissues and
blood vessel walls, ultimately resulting in metastasis (8). Hepatic tumour growth, angiogenesis
and EMT have been mostly studied in two-dimensional (2D) cell line
models in vitro; however, they may not be the ideal system
for the elucidation of the underlying molecular mechanisms as they
do not mimic the complex in vivo tumour tissue architecture.
It has been reported in recent studies that three dimensional (3D)
models more closely resemble the in vivo tumour
microenvironment by exhibiting complex phenotypic heterogeneity. It
has been illustrated that cell-matrix interactions are better
recreated by complex aggregated cell populations in 3D culture
systems rather than simple 2D cell monolayers (13,14).
3D in vitro models of HCC recapitulate the phenotypic and
functional characteristics of the in vivo liver tissue and
hence may be best suited to comprehend the role of growth factors
in inducing EMT and tumour growth (15,16).
VEGF is one of the key cytokines that affects tumour survival,
spread and aggressiveness. The effect of VEGF has been evaluated on
several tumour cell lines using conventional two-dimensional (2D)
cell culture models; however, they have provided only limited
information. Provided that three-dimensional (3D) cell spheroid
cultures closely mimic the in vivo-like microenvironments,
the effects of exogenous treatment with VEGF on several tumorigenic
properties of hepatoma cell lines were investigated in the present
study.
Materials and methods
Cells and cell culture
Human liver hepatoma Huh7 (p53mut) and liver cancer
HepG2 (p53++) cell lines were obtained from the National
Centre for Cell Science (Pune, India). All cell lines were cultured
in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.) with 10% FBS (HyClone; Cytiva) and 100 µg/ml
streptomycin and 100 IU/ml penicillin (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C in incubator containing 5%
CO2. Human umbilical vein endothelial cells (HUVECs),
which are primary cell line (cat. no. CL019; Hi-media Laboratories,
LLC) were grown in endothelial medium (HiMedia Laboratories, LLC)
with growth factors and 1% antibiotics (100 µg/ml streptomycin and
100 IU/ml penicillin) on gelatin-coated plates.
Co-culture of cells
In order to examine the mechanisms through which
endothelial cells modulate the tumorigenic behaviour of
HBx-transfected hepatoma cells, direct and indirect co-cultures
were performed. For indirect co-cultures, Huh7 cells were treated
with conditioned medium (CM) from HUVECs. CMs were prepared after
serum starvation of these cells for 24 h and then collecting the
supernatants after centrifugation to remove cell debris. For
obtaining VEGF-induced cells, hepatoma cells were exposed to a 10
ng/ml VEGF concentration for 48 h (17).
Drug induction in cells
For the evaluation of the effects of the inhibition
of VEGF on hepatoma cells, the cells were exposed to a sorafenib
(BAY-BAYERS Corporation) at a concentration of 15 µm, dissolved in
0.1% dimethyl sulfoxide (DMSO, HiMedia Laboratories, LLC) for 24 h
(18).
Cell proliferation
Initially, Huh7 cells were seeded at a concentration
of 30,000 cells/well in a 6-well plate and incubated overnight at
37°C. On the following day, the cells were exposed to VEGF and
incubated for 24 h at 37°C. Following 24 h, the cells were
trypsinized, stained with trypan blue (HiMedia Laboratories, LLC)
at room temperature and immediately counted using a haemocytometer
chamber slide. Cell numbers were counted manually at 10X objective
using under an inverted light microscope (NIKON Eclipse Ti inverted
microscope, Nikon Corporation), and the average of three
independent experiments was used.
Transwell assays
Control and VEGF-induced hepatoma cells were
detached, harvested by centrifugation (75 g for 5 min at 25°C) and
resuspended in DMEM (without serum), and then 50,000 cells per
chamber placed in the upper chamber of a modified Boyden chamber
consisting of uncoated polycarbonate filter membranes of an 8-µm
pore size. For invasion, the Transwell insert was first coated with
Matrigel. The chamber was placed in a 24-well culture dish
containing DMEM with FBS in the lower chamber. After 24 h (for
chemotaxis) and 48 h (for invasion) incubation at 37°C, the lower
side of the filter was washed with PBS and fixed with 4%
paraformaldehyde (HiMedia Laboratories, LLC) for 2 min at room
temperature. The cells were then washed and permeabilized by 100%
methanol (HiMedia Laboratories, LLC) for 20 min at room
temperature. For quantification, cell nuclei were stained with 0.5%
crystal violet (HiMedia Laboratories, LLC) for 15 min at RT. The
upper side of the filter containing the non-migrating cells was
scraped with a cotton swab. Cells migrating towards the lower
chamber were counted manually at 4X objective in random fields
under an inverted light microscope (NIKON Eclipse Ti inverted
microscope, Nikon Corporation).
Wound healing assay
Control, VEGF-induced and sorafenib-treated hepatoma
cells were plated in 12-well plates (3×106 cells/well).
Following a 6-h exposure to serum-starved conditions, a scratch was
made on the cell layer using a 100 µl sterile micropipette tip, in
order to create a wound. The cells were photographed using a
phase-contrast microscope (NIKON Eclipse Ti inverted microscope,
Nikon Corporation), to determine the wound width at the 0 h time
point. Following a 24-h culture, the cells were photographed again.
Wound healing was visualized by comparing cell layers at 0 h with
those at 24 h, analysing the distance migrated by the leading edge
of the wound at each time point in all the study groups. The
relative wound width was measured as wound width at the 24-h
timepoint, divided by the wound width at the 0-h time point.
Measurements were made using Software NIS Elements (version 2.3) of
the NIKON Eclipse Ti inverted microscope (Nikon Corporation). The
width was measured in µm.
Tube formation assay
Huh7 cells were seeded at 50,000 cells/well with
HUVECs at a 1:1 ratio for direct co-culture on a growth
factor-reduced Matrigel-coated 24-well plate and were incubated
overnight at 37°C. For indirect co-cultures, hepatoma cells were
seeded at 50,000 cells/well with CM from HUVECs including VEGF (10
ng/µl) on Matrigel, as described above. The average number (from
4–5 fields) of tube networks and circles per field were counted
manually, under an inverted microscope (NIKON Eclipse Ti inverted
microscope, Nikon Corporation) and photographed at 10X
objective.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Control, VEGF-induced and sorafenib-treated hepatoma
cells were harvested using trypsin-EDTA solution (0.25%). Total RNA
was isolated by using Trizol® reagent (Thermo Fisher
Scientific, Inc.) and quantified at 260/280 nm using a Thermo
Scientific Nanodrop 2000 Spectrophotometer (Thermo Fisher
Scientific, Inc.). The absorption ratio A260/A280 nm between 1.90
and 2 was considered to indicate adequate RNA quality. First strand
cDNA was synthesized from 1 µg of total RNA using reverse
transcriptase (cat. no# AB1453B; Verso cDNA synthesis kit; Thermo
Fisher Scientific, Inc.) according to the manufacturer
instructions. qPCR was performed by using SYBR-Green PCR master mix
(Fermentas; Thermo Fisher Scientific, Inc.) on the ViiA7 instrument
PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The following cycling parameters were used: Start at 95°C for 5
min, denaturing at 95°C for 30 sec, annealing at 60°C for 30 sec,
elongation at 72°C for 30 sec, and a final 5 min extra extension at
the end of the reaction and repeated for 40 amplification cycles.
The primer pairs used in the present study are listed in Table SI.
3D spheroid cultures
Tumour spheroids were generated by seeding 4,000
cells/well in six-well ultra-low attachment culture plates (Corning
Inc.), culturing them in DMEM/F12 (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 5% FBS (HyClone; Cytiva) then incubated at
37°C in a 5% CO2 incubator for 2–3 days; after 3 days
the medium was replaced with serum-free medium (with or without
VEGF; 10 ng/µl). For HUVEC direct co-culture spheroids, Huh7 and
HUVECs were grown at a 1:1 ratio with DMEM plus endothelial medium
at a ratio of 1:1 and for indirect co-cultures, the medium was
replaced with CM of HUVECs. Spheroids were further grown on
ultra-low attachment plates for 2 weeks in order to establish 3D
anchorage-independent models. The 3D anchorage-independent models
were imaged using a Nikon Eclipse Ti inverted microscope (Nikon
Corporation) and the average size of the spheroids was measured
using ImageJ Software (version 2.3, National Institutes of
Health).
Hanging drop cultures
The hanging drop method was utilized to evaluate the
tumorigenic potential of HCC tumour spheroids. Untreated and
treated cells (with VEGF, 10 ng/µl and/or sorafenib, 15 µM) were
seeded in a hanging-drop plate at a concentration of 1,000 cells/µl
in growth medium. Images of drops were obtained at 24 and 72 h
post-treatment. Tumour spheroid adhesion occurrence was scored on a
0–2 scale, with ‘0’ score depicting no spheroid formation, ‘1’ the
formation of a loose spheroid, and ‘2’ signifying compact spheroid
formation. Migration was visualized by comparing the 0-h with the
24- and 72-h images and analyzing the distance migrated by the
cells to aggregate at each time point in all the study groups.
Relative migration percentage was measured as the migrated cell
area at 24 and 72 h, divided by the total area covered by seeded
cells at 0-h time point. All measurements were performed using
Software NIS Elements (version 2.3) from NIKON Eclipse Ti (Nikon
Corporation). In order to obtain morphometric data, two to three
individual plates of each condition were imaged, and four to six
images were obtained from each plate. Calibrated images were
exported using ImageJ software, (version, 1.53e, National
Institutes of Health) for morphometric analysis. The polygon tool
was used to outline the spheroids and projected area was measured
as [4π × (area)]/(perimeter)2. The measurement unit was
µm (19–21).
Spheroid invasion assay
Spheroids (1-week-old; both VEGF-induced and
control) were seeded in 24-well cell culture plates on which the
Matrigel (5 mg/ml) was coated (Life technologies; Thermo Fisher
Scientific, Inc.). Images were obtained at various time points
(days 2, 5, 7 and 10) at 20× magnification using an inverted
microscope (Nikon Ti Eclipse; Nikon Corporation). Area invaded by
the cells was evaluated by assessing the sprouting areas per
spheroid which were quantified using ImageJ software (version
1.53e) (13,19,22).
Immunofluorescence and histological
analysis
Spheroids were fixed in 4% paraformaldehyde (HiMedia
Laboratories, LLC) over night at 4°C. Subsequently, the spheroids
were rinsed with PBS (HiMedia Laboratories, LLC) and stained with
0.1% eosin (HiMedia Laboratories, LLC) for 15 min at room
temperature. For cryosectioning, the spheroids were embedded in
cryostat freezing mix (Sakura Finetek, USA) at −20°C. Sections of 4
µm thickness were mounted on pre-warmed slides. Standard
haematoxylin- and eosin spheroid-stained slides (staining at room
temperature for 8 and 5 min, respectively) were first evaluated to
confirm spheroid quality. For immunofluorescence staining,
4-µm-thick cryosections were cut and hydrated with 0.5% BSA-PBS
(HiMedia) for 15 min. The sections were exposed to blocking buffer
(0.1% donkey serum-PBS; Sigma-Aldrich; Merck KGaA) for 30 min. The
spheroids were then incubated overnight using primary mouse
Vimentin (VIM) antibody (1:50, Cat no# sc-6260; Santa Cruz
Biotechnology, Inc.; 1:50) at 4°C. After washing, cells were
incubated with anti-mouse Alexa Fluor 594 (Dilution 1:500, Cat
no#sc-516608, Santa Cruz Biotechnology, Inc.) secondary antibody
for 45 min at RT. Finally, the sections were mounted with DAPI
(HiMedia) for 5 min at RT.-Stained images were acquired using a
fluorescent microscope (Nikon Ti Eclipse; Nikon Corporation). For
2D culture immunofluorescence, Huh7 cells were first permeabilized
with Triton-X (0.1%, HiMedia) and followed by 4% paraformaldehyde.
Following this, cells were blocked with serum (1% FBS (Hyclone) in
PBS (HiMedia) for 1 h. All remaining staining protocol steps were
performed as previously described. Number of fluorescent cells were
counted manually (n=3) using ImageJ software (version 1.53e,
National Institutes of Health) by applying a ‘sharp edges cell
filter’ and the percentage was calculated according to the ratio of
average number of VIM-positive cells to the average number of
DAPI-stained cells per field (in 2D culture).
Cell adhesion assay
Briefly, 48-well plates pre-coated with fibronectin
(10 ng/ul; FN, Himedia) and bovine serum albumin (1% BSA; both
HiMedia) were washed with PBS twice and blocked for 1 h at 37°C
with DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing 0.1%
BSA (HiMedia) before plating the cells. A cell suspension
containing 2×105 cells/ml (control and VEGF-induced Huh7
cells) was prepared in serum-free medium. The cell suspension (150
µl) was added to each well (BSA-coated wells acting as a negative
control). Cells were allowed to adhere for 1 h at 37°C.
Subsequently, the unadhered cells were removed by gentle washing
three times with PBS. Adherent cells were stained with 0.5% crystal
violet for 10 min at room temperature and the optical density
values were measured at 405 nm wavelength using microplate reader
(Synergy/H1 Hybrid Multimode Plate Reader; Agilent Technologies,
Inc.).
MTT assay
For the assay, 10,000 cells/well were plated
[control, VEGF-induced and sorafenib (15 µM) treated Huh7 cells] in
a 96-well plate and incubated at 24 and 48 h. Cells were washed
with PBS and 20 µl MTT (0.5 mg/ml; HiMedia) were added to each well
of the plate. The plates were incubated at 37°C for 4 h.
Subsequently, 200 µl DMSO was added for formazan solubilization and
mixed thoroughly and left in the dark for 10 min. Intensity of the
colour developed was recorded at 570 nm using a fluorescence
microplate reader (Synergy/H1 Hybrid Multimode Plate Reader;
Agilent Technologies, Inc.).
Statistical analysis
All quantitative data are expressed as the mean ±
standard deviation.
An unpaired Student's t-test was used for analyses
and comparisons between two groups. For multiple group analyses and
comparisons, one-way ANOVA was used, with the subsequent
application of Tukey's post hoc test. All experiments were repeated
at least three times. P<0.05 was considered to indicate a
statistically significant difference.
Results
VEGF promotes tumour growth in 2D and
3D cultures
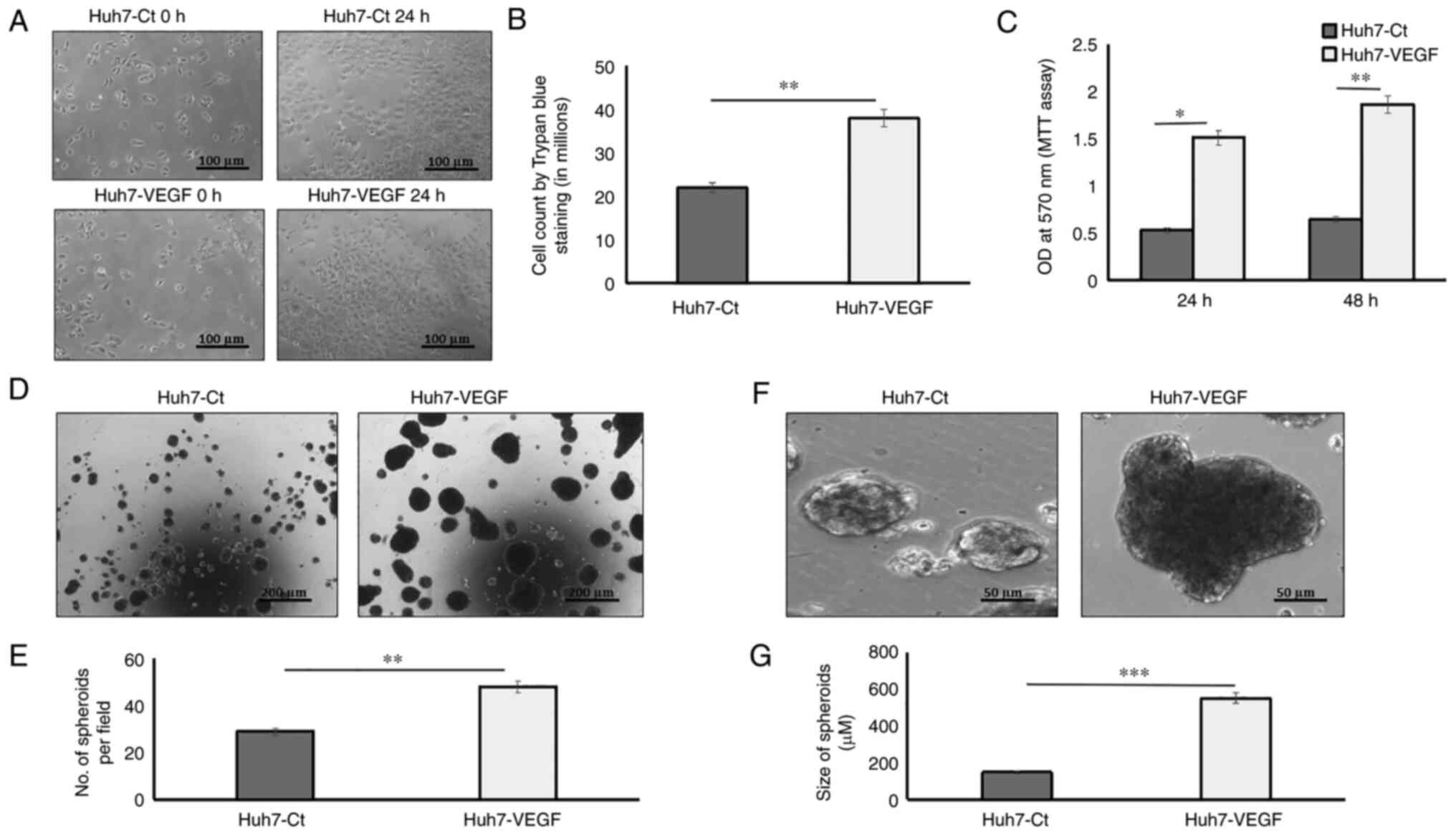
To examine the effects of VEGF on tumour growth, the
hepatoma cells were incubated with VEGF in 2D and 3D tumour cell
models. In 2D models, cell proliferation was assessed by counting
the actual number of cells using trypan blue exclusion experiments
and also using MTT cell viability assay. Following a 24-h
incubation with VEGF, the cell numbers were found to be
significantly higher in the VEGF-induced Huh7 cells as compared to
the control Huh7 cells (Fig. 1A and
B; P<0.05). MTT assay also revealed that VEGF induction
enhanced the proliferation of Huh7 cells both after 24 h
(P<0.05) and 48 h (P<0.01) as compared to the control Huh7
cells (Fig. 1C). In 3D models,
tumour growth was assessed using spheroid growth in ultra-low
attachment plates as suspension cultures, for 2 weeks,
continuously, to avoid sub-culturing. Huh7 cells (4,000 cells/well)
without or with VEGF induction formed distinct spheroids within 14
days of suspension culture (Fig. 1D
and F). The number of spheroids and their sizes revealed that
the VEGF-induced Huh7 spheroids were significantly increased in
number (Fig. 1E; P<0.01) and
also larger in size (551 µm; 3.7-fold), as compared to the
spheroids formed by hepatoma cells without VEGF induction (146 µm;
Fig. 1G; P<0.001).
Tumorigenic properties in the presence
of VEGF in 2D vs. 3D cultures
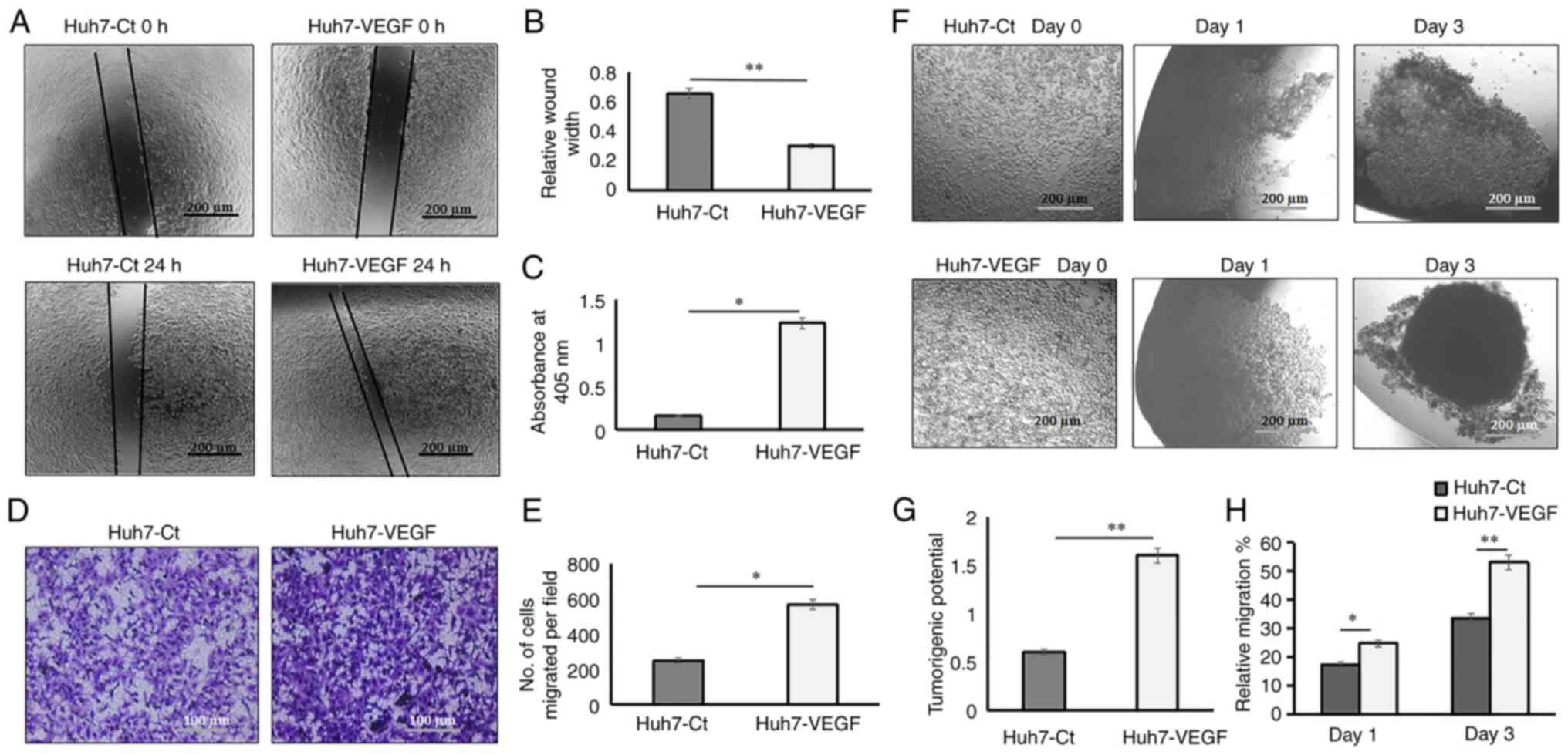
In 2D hepatoma models, the wound healing, adhesion
and migration potential of the hepatoma cells were examined, in
order to investigate the tumorigenic properties. The results
depicted that VEGF-induced Huh7 cells exhibited a relatively
smaller wound width or increased migration (Fig. 2A and B; P<0.01), increased
adhesion (Fig. 2C) and higher
chemotaxis (Fig. 2D and E;
P<0.05), as compared to the control Huh7 cells. In 3D models,
non-adherent hanging drop cultures for spheroid formation were used
to evaluate the tumorigenic property, on which cellular aggregation
is promoted based on gravity and there is complete absence of a
substratum. The adhesion property by the formation of 3D spheroids
was evaluated within a stringent time point i.e, on days 0 to 3,
and the adhesion of cells to form spheroids and the spheroid
compactness was considered as a measure of tumorigenic property. On
day 3, cell-to-cell adhesion and spheroid compaction were
significantly increased in spheroids which were treated with VEGF,
as compared to spheroids derived from control cells in the absence
of VEGF (Fig. 2F and G;
P<0.01). Migration was assessed by the distance covered by cells
within a confined space to form a spheroid from days 0 to days 1
and 3. The results revealed that on day 3, the percentage of
migrating cells in the hanging drop able to form a compact spheroid
was 53.10% in the VEGF-induced cells as compared to 33.64% in the
control cells (Fig. 2H;
P<0.01). The average spheroid sizes are presented in Table SII.
Invasive properties in the presence of
VEGF in 2D vs. 3D cultures
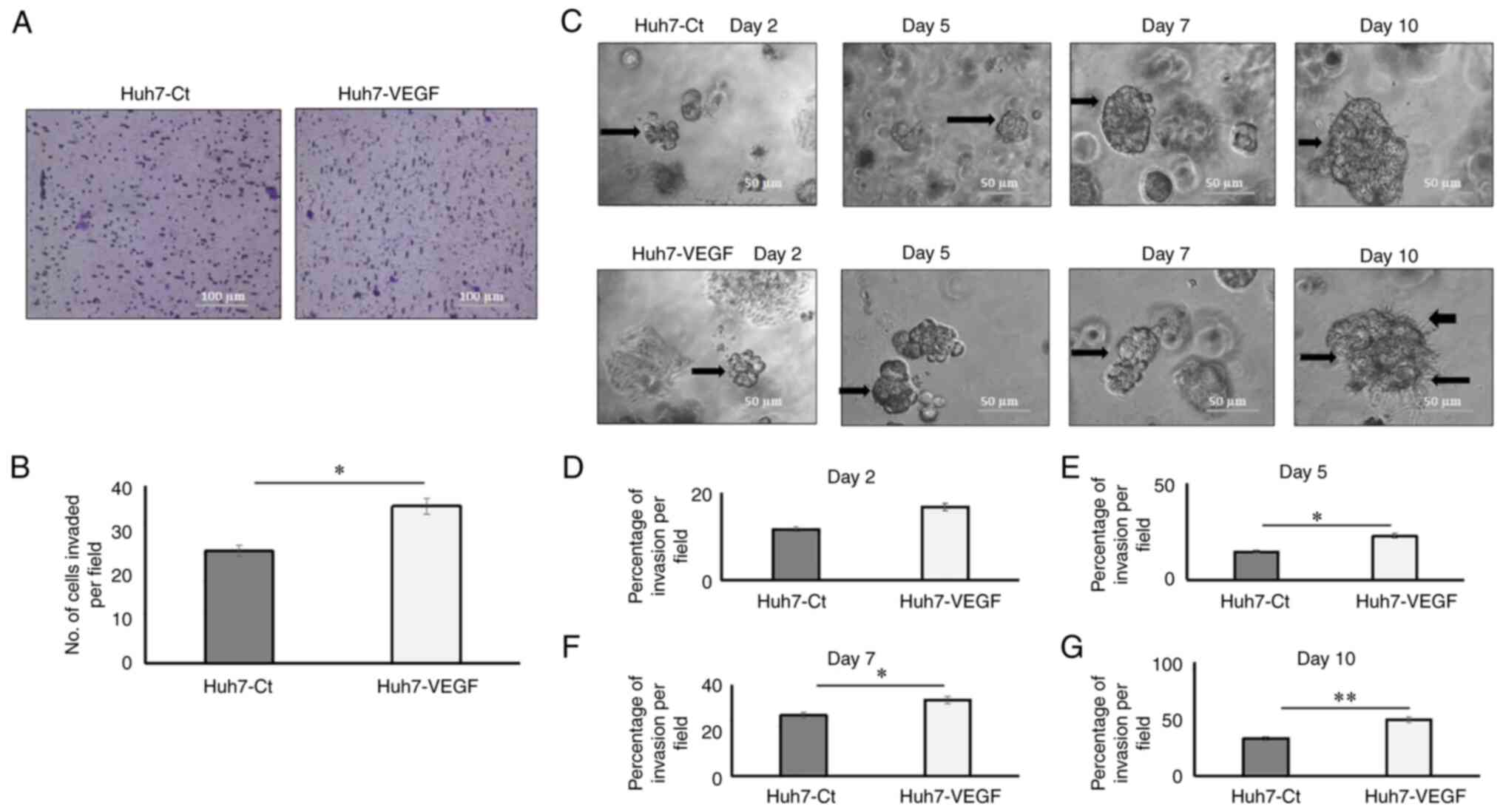
Tumour invasion in 2D cultures was measured as
number of cells invading through the extracellular matrix (ECM;
Matrigel-coated Transwell) per field, and the invasion in 3D models
was measured as the average percentage of area invaded by the
spheroids (n=4) through the ECM (Matrigel). In 2D models, the
results demonstrated that the numbers of Huh7 cells that invaded
through the Matrigel were increased when incubated with VEGF, as
compared to the control Huh7 cells after 48 h (Fig. 3A and B; P<0.05). In 3D models,
the VEGF-induced spheroids that invaded into the Matrigel (which
functioned as ECM) after day 2, demonstrated an enhanced invasive
potential at various time points, namely day 2 (Fig. 3D), day 5 (Fig. 3E; P<0.05), day 7 (Fig. 3F; P<0.05) and day 10 (Fig. 3G; P<0.01). The results also
revealed that after day 10, the VEGF-induced spheroids formed some
vessel-like structures (black arrows, Fig. 3C) inside the Matrigel. The average
spheroid sizes are presented in Table
SII.
VEGF enhances the stemness potential
and triggers EMT in the 2D monolayer and 3D spheroid model
In 2D models, gene expression data revealed an
upregulation of mesenchymal genes, including VIM (Fig. 4B; P<0.05), N-cadherin [or
Cadherin 2 (CDH2); Fig. S1A;
P<0.01], Thy-1 cell surface antigen (THY-1; Fig. S1B; P<0.05) and the cancer
stemness marker, CD133 (Fig. 4C;
P<0.05), while E-cadherin [or Cadherin 1 (CDH1); epithelial
marker; Fig. 4A] in VEGF-induced
Huh7 cells was downregulated in comparison to the control cells.
CDH1 gene expression data were also validated in the HepG2 liver
cancer cell line. The results revealed the upregulation of the
mesenchymal genes, VIM (Fig. S2B;
P<0.05), CDH2 (Fig. S2D),
THY-1 (Fig. S2E; P<0.01), and
the cancer stemness marker, CD133 (Fig. S2C; P<0.01), while CDH1
(Fig. S2A) in the VEGF-induced
HepG2 cells was downregulated in comparison to the control HepG2
cells in 2D cultures. In 3D models, gene expression analysis
demonstrated an upregulation in the expression of the mesenchymal
genes, VIM (Fig. 4F; P<0.05),
CDH2 (Fig. S3A, P<0.05), THY-1
(Fig S3B; P<0.01) and CD133
(Fig. 4G; P<0.01), while the
expression of CDH1 in the spheroids was only slightly reduced in
presence of VEGF (Fig. 4E). In 2D
culture models, the results of immunofluorescence staining revealed
the increased expression of the mesenchymal marker, VIM (1.49-fold
higher, Fig. S4A and B;
P<0.01) in the VEGF-induced Huh7 cells as compared to the
control cells (Fig. 4D). In 3D
spheroid cultures, haematoxylin and eosin staining revealed better
integrity and homogenous cell morphology in VEGF-induced spheroids,
as compared to the control spheroids (Fig. 4H). There was no marked difference
in the expression of the mesenchymal protein, VIM in the 3D
spheroids treated with and without VEGF (Fig. 4H).
 | Figure 4.Epithelial-mesenchymal transition and
cancer stemness gene expression and mesenchymal protein expression
in 2D and 3D co-cultures. Relative mRNA expression of (A) CDH1 (B)
VIM (C) CD133 in Huh7 cells in presence of control medium and VEGF
in 2D models (D) Staining of mesenchymal protein, VIM and DAPI in
control and VEGF-induced Huh7 cells (magnification, ×10). Relative
mRNA expression of (E) CDH1 (F) VIM (G) CD133 in Huh7 cells in
presence of control medium and VEGF in 3D models. (H) H&E
images of control and VEGF-induced spheroid (magnification, ×20)
and staining of mesenchymal protein, VIM and DAPI in control and
VEGF-induced spheroid (magnification, ×20). Data are presented as
tbe mean ± SD (n=3 each). *P<0.05 and **P<0.01. VIM,
vimentin; Ct, control; VEGF, vascular endothelial growth factor;
CDH1, cadherin 1 (E-cadherin); H&E, haematoxylin and eosin. |
Tumor growth in 2D and 3D cultures of
hepatoma cells co-cultured with endothelial cells
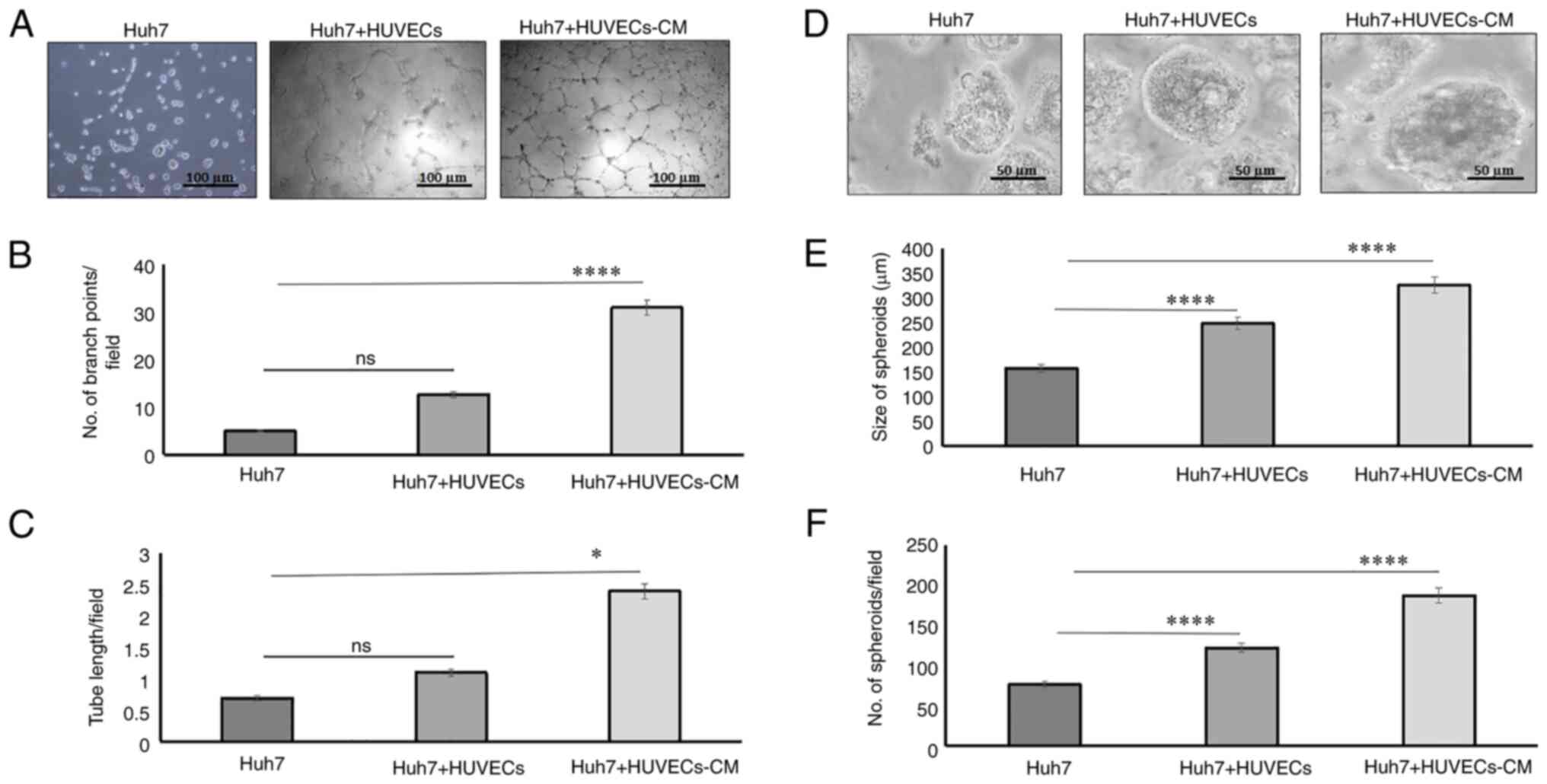
In order to determine the mechanisms through which
heterogenous cell cultures functioned in 2D and 3D hepatoma models,
HUVECs and Huh7 cells were concurrently cultured in direct and
indirect cultures on Matrigel-coated wells. In 2D cultures, the
results depicted that branch points (Fig. 5A and B; P<0.0001) and tube
lengths (Fig. 5A and C; P<0.05)
were significantly enhanced when Huh7 cells were co-cultured with
HUVEC-CM, as compared to single-Huh7 cultures (Fig. 5A-C). In direct 2D cultures, where
Huh7 cells were cultured with HUVECs at a 1:1 ratio, the tubes were
not well-formed on the Matrigel (Fig.
5A-C). Hence, we next developed anchorage-independent
heterologous 3D models of Huh7 cells and HUVECs. Size and number of
the spheroids was significantly increased in Huh7 cells cultured
with HUVECs (n=122; P<0.0001, 248.184 µm; 1.5-fold; P<0.0001)
or when Huh7 cells were co-cultured with HUVEC-CM (n=187;
P<0.0001, 325.631 µm; 2-fold; P<0.0001), as compared to
single Huh7 cultures (n=76; P<0.0001, 157.846 µm; Fig. 5D-F).
Sorafenib, a multikinase inhibitor
reduces migratory properties in VEGF-induced 2D and 3D
cultures
The cytotoxicity of sorafenib on Huh7 cells in the
absence or presence of VEGF was determined using MTT assay. It was
demonstrated that the viability of VEGF-untreated and VEGF-treated
Huh7 cells was reduced by 19.8 and 29% respectively, following
treatment with sorafenib for 24 h (Fig. S5; P<0.01). The effect of
sorafenib on the 2D hepatoma cell culture migratory ability was
evaluated by wound healing assay. In 2D and 3D cultures, in the
presence of VEGF, sorafenib treatment resulted in a considerable
reduction in migration in 2D cultures; i.e, a decreased wound
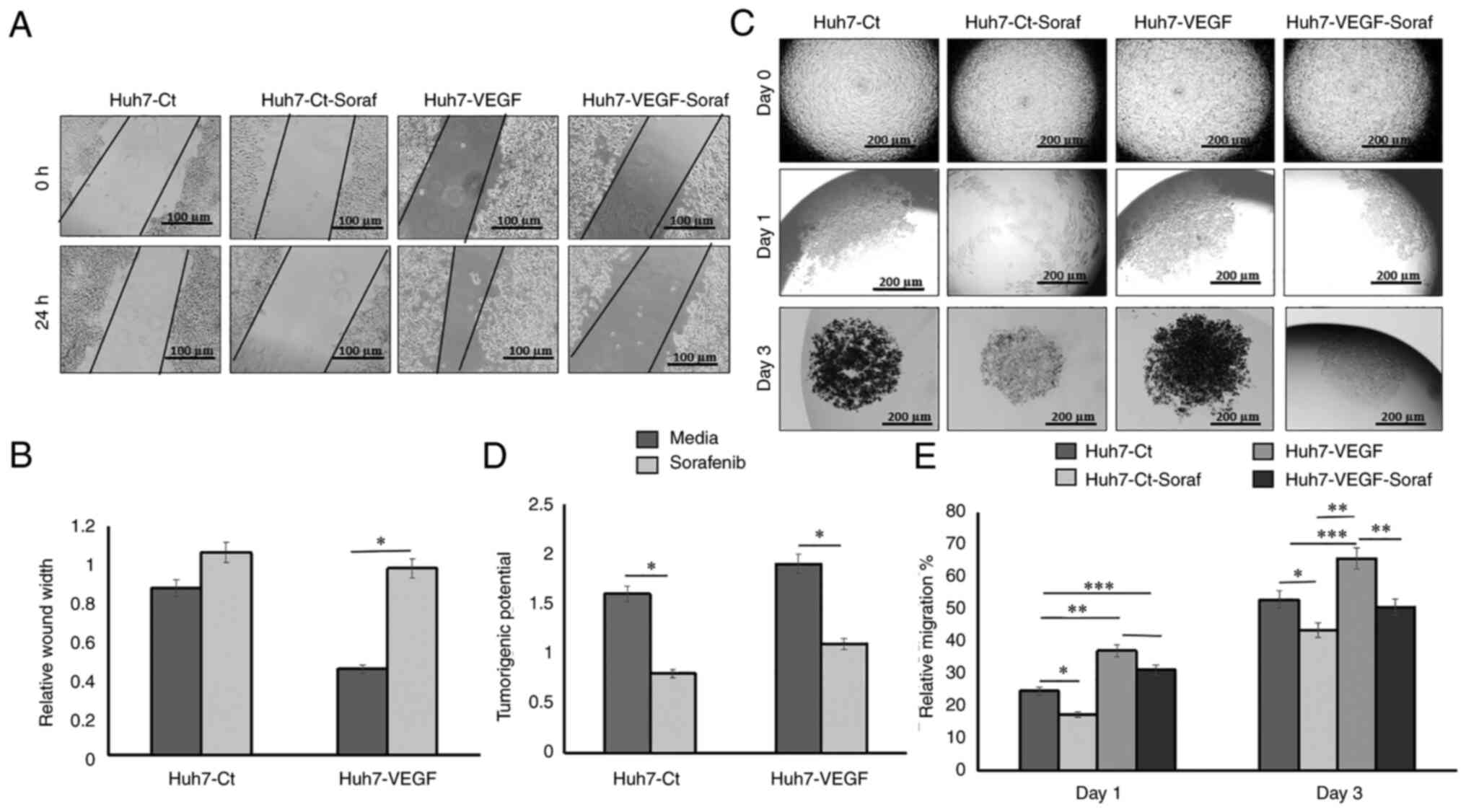
healing ability as compared with untreated cells (Fig. 6A and B; P<0.05). In 3D cultures,
on day 3, cell to cell adhesion and spheroid compaction were
significantly decreased in the sorafenib-treated spheroids as
compared to the spheroids derived from control cells in presence of
VEGF (Fig. 6C and D, P<0.05).
In addition, on day 3, the percentage of migration and compact
spheroid formation was reduced in the hanging drops by
sorafenib-treated cells as compared to that observed in the control
cells (50.77 vs. 65.82%; P<0.01, Fig. 6E). The average size of spheroids is
presented in Table SII.
Furthermore, the effect of the angiogenesis inhibitor, sorafenib,
on the expression of EMT-related genes in 2D and 3D VEGF-induced
hepatoma models was analysed. In 2D models, the gene expression
data revealed the upregulated expression levels of mesenchymal VIM
(Fig. S6B) and CDH2 (Fig. S6C, P<0.05) and cancer stemness
marker, CD133 (Fig. S6D), whereas
there was no significant difference in the expression of CDH1
(epithelial marker; Fig. S6A) in
the sorafenib-treated cells, in comparison with the control cells.
In the 3D models, sorafenib treatment resulted in downregulation in
the expression of the mesenchymal genes, VIM (Fig. S6B; P<0.01), CDH2 (Fig. S6C; P<0.01) and CD133 (Fig. S6D; P<0.01), while the
differences in CDH1 expression in spheroids were not statistically
significant between groups in the absence or presence of sorafenib
(Fig. S6A).
In the context of migration, there was a 0.46-fold
inhibition in 2D models while in 3D models, the inhibition in cell
migration was 1.29-fold (Fig.
6A-E). Similarly, in terms of mesenchymal gene expression, in
the 3D models, VIM expression was reduced by 2.27-fold in
sorafenib-treated cells as compared to that observed in untreated
cells, while in 2D models, VIM expression was decreased only by
1.23-fold in sorafenib-treated cells as compared to that in
untreated cells (Fig. S6B).
Expression of the cancer stemness gene, CD133 was also reduced by
2.25-fold in the 3D models, while in 2D models, it decreased only
by 1.03-fold (Fig. S6D, in
comparison with untreated cells in respective conditions.
Discussion
In the present study, the role of VEGF in inducing
EMT and cancer stemness in both 2D and 3D models of HCC was
investigated. In 2D models, the only known measure for tumour
growth and invasion is the increase in cell numbers, whereas in
anchorage-independent 3D spheroid models, the estimation of
spheroid sizes may also reflect in vivo aspects of tumour
growth (23). Hepatoma cells
cultured in anchorage-independent 3D models have been demonstrated
to exhibit increased stemness and gene expression in previous
studies. Jung et al (24)
previously reported Huh7 hanging drop models and Khawar et
al (25) used Huh7 liquid
overlay cultures for 3D tumour-related studies, in which tumour
cells aggregate spontaneously, without an exogenous supply of ECM.
These spheroids are heterogeneous cell populations (e.g., hypoxic
vs. normoxic, quiescent vs. replicating cells), have a well-defined
geometry and undergo optimal physiological cell-to-cell
interactions (26–28). In 3D models, hepatoma cells formed
well-defined spheroids (551 µm in diameter) in the presence of VEGF
in a time period of 2 weeks. In hanging drop cultures, cells
migrated towards each other, ultimately aggregating in the drop,
and then formed a compact spheroid which was a result of
cell-to-cell adhesion (20).
Hence, the migration and adhesion of hepatoma cells in these
spheroids, considered as a measure of tumorigenic property, were
considerably enhanced in cell cultures treated with VEGF, similar
to that reported in a previous study utilizing 2D cell culture
models (29). 3D model is an
improved model for the study of tumorigenic properties, since both
migration and adhesion can be studied in a single experimental
model. By contrast, in the 2D monolayer model, two sets of
experiments are required to study both characteristics. The
invasive properties of HCC models were studied in presence of
Matrigel, in 2D and 3D formats. The presence of Matrigel, which
represents the ECM, adds another level of complexity to the 2D vs.
3D differences. Matrigel invasion assays demonstrated that hepatoma
cells were capable of invading efficiently in the presence of VEGF
in comparison to cells in the control media. In 3D models, it was
feasible to efficiently monitor spheroid invasion in different
days, whereas invasion in 2D models could be recorded only after 48
h. Similar to 3D models, it was attempted to study tumour invasion
on different days for 2D models; however, the results were not
consistent, possibly as only the number of cells invading through
the ECM matrix (Matrigel) were counted, as a parameter for the
evaluation of invasion. In 3D models, sprouting in the spheroids
treated with VEGF after the 10th day was also observed, indicating
that 3D spheroids may be a promising model to simultaneously study
invasion and angiogenesis. However, angiogenesis was not
extensively studied on Matrigel in the 3D models, since a
significant formation of tubes and capillaries in 3D conditions,
requires media provision to the spheroids by using a
perfusion/fluidic system, which was not applied in the present
study. HCC is a fairly unique type of carcinoma, in which fibrosis
and chronic inflammation precedes the development of HCC in >90%
of cases (30). An excessive
deposition of the ECM has been reported as a key hallmark of HCC
(31). Hence, studying the
tumorigenic potential and invasiveness of hepatoma cells in
presence of ECM substrates, such as Matrigel is relevant (25).
The present study then investigated whether the 3D
tumour spheroids also exhibit a gene expression profile,
characterized by EMT and stemness. In our previous study, we
demonstrated that TGF-β imparted mesenchymal traits to the hepatoma
cells (32). In another study,
Takai et al (33) cultured
different hepatoma cells on varied matrices, demonstrating that
cells in porous alginate scaffolds may be able to generate
organoid-like spheroids that mimic numerous in vivo
features. It was stated in their study that EpCAM+
hepatoma cells cultured as spheroids may be more sensitive to
TGFβ-induced EMT and possess increased tumorigenic and metastatic
potential, as compared to conventional 2D cultures. The variability
of TGFβ-mediated tumorigenic effects on 2D and 3D cultures have
also been reported in ovarian cell lines (34). In the present study, the effects of
VEGF on the tumour properties of hepatoma cells were evaluated in a
2D, as well as in a 3D microenvironment. The results revealed that
hepatoma cells formed 3D spheroids, which differed in size and
density in absence and presence of the same VEGF concentration. In
all spheroids, tumour invasion and angiogenesis were more
aggressive in 3D cultures in comparison to 2D conditions, following
treatment with VEGF. In terms of gene expression, the VEGF-mediated
increase in the levels of the EMT markers, VIM, CDH2 and THY-1 was
observed in both 2D and 3D cultures; however, the increase was more
notable in the 2D conditions as compared to that observed in 3D
conditions, which may be attributed to the fact that tumour cells
in 3D conditions may be differentially exposed to VEGF. CDH1, a
cell-to-cell adhesion receptor is an important determinant of
tumour progression and it has been reported to be downregulated in
numerous HCC tumours (35). In the
presence of VEGF, no change in CDH1 gene expression was observed in
3D models, whereas in 2D models, a decreased expression of CDH1 was
observed, which was in line with a previously published study
(36). Of note, a proportionate
increase in the remaining gene expression levels was not observed
in the 3D models, in comparison to the 2D models. These findings
suggested that 3D cultures potentially avoid the overestimation of
mechanistic observations in 2D cultures, which may be ascribed to
many reasons. Firstly, cells in 2D are all at the same cell cycle
stage, whereas 3D cells are often in different cell cycle stages
resembling to cells in vivo, due to oxygen and nutrient
gradients. 3D models have limited media permeability, which may
have an impact on cell viability and gene expression. Secondly,
spheroid cells at the leading edge are metabolically active and
show the greatest invasive and proliferative capacities. By
contrast, the cells that are located away from this leading edge,
towards the tumour centre have been reported to be more quiescent
and less proliferative (37).
Furthermore, spheroids often present in uneven sizes, with several
areas including great numbers of growing tumour cells, and other
areas including reduced tumour cell numbers. Thus, it may be
difficult to acquire a 3D culture with all spheroids having
identical features, particularly when they are grown without a
substratum. Hence, at a given time, the number and type of cells
exposed to an exogenous factor may vary in different wells, which
may have resulted in heterogenous gene expression values. By
contrast, under 2D adherent cell culture conditions, cell lines
derived from all three stages appeared phenotypically alike and
presented with similar motility characteristics. This may possibly
be the main reason for gene expression heterogeneity in 3D
cultures, as compared to 2D cultures. However, since this similar
scenario also exists in vivo, it is pertinent that the
results obtained in 2D models should be cautiously extrapolated to
in vivo models in future studies.
To further validate the role of VEGF in inducing the
tumorigenic properties of hepatoma cells, 2D and 3D culture studies
were also performed using the VEGFR-inhibitor, sorafenib (at a
recommended concentration of 15 µM), in VEGF-treated Huh7 cells for
24 h. Sorafenib has been suggested to inhibit hepatoma growth in
the presence of hypoxia and hypoxia-induced angiogenesis (38). A parallelism in the efficacy of
sorafenib in well-differentiated cells cultured in both 3D and 2D
models was described in another study (39). Cheng et al (40) reported that combined treatment with
sorafenib and dasatinib (another kinase inhibitor) effectively
inhibited HCC cell-induced angiogenesis. In particular, they
demonstrated that the administration of a regular 10 µM-dose of
sorafenib in both 3D and 2D cultures may be effective at reducing
cell proliferation and a reduction in the spheroid area in both
Huh7 and HepG2 cell lines (40).
In accordance with these findings, in the present study, sorafenib
treatment culminated in a marked reduction in cell migration in
both 2D and 3D cultures and of spheroid size in 3D cultures.
However, a higher reduction of cell migratory properties and a
decrease in the expression of mesenchymal genes was observed in 3D
cultures, as compared to the 2D cultures, following sorafenib
treatment, indicating that 3D cultures may serve as a more
promising and reliable model for screening the effectiveness of
tumour-inhibiting therapeutics. The use of 2D and 3D models was
also analysed to study the interactions between HUVECs and hepatoma
cells in both direct (with HUVECs) and indirect cultures (with
HUVEC-CM). HUVECs have been previously reported to promote EMT
(41). VEGF and angiogenin
secreted by the HUVECs have also been reported to induce
proliferation and invasion of hepatoma cells (24). Hence, both direct and indirect
cultures (both 2D and 3D) of HUVECs with hepatoma cells were used
to examine the mechanisms through which HUVECs may affect tumour
invasion. In 2D monolayer conditions, although indirect cultures of
hepatoma cells in the presence of HUVEC-CM revealed an increase in
tube-like structures, direct cultures of hepatoma cells with HUVECs
did not demonstrate significant changes in branch point numbers, as
well as the tube length of cells, in comparison to that observed in
single hepatoma cell culture. In 3D spheroid culture models, the
number of spheroids was significantly increased in both direct and
indirect cultures, as compared to HUVECs alone, possibly suggesting
that 2D monolayer models are not suitable to study
organotypic/heterogenous cell tumour cell models, with 3D models
being more reliable and predictive. In the present study, Huh7 cell
lines were used to examine the effects of VEGF after VEGF
induction, in 2D and 3D cultures. HepG2 cell lines have not been
previously used, to the best of our knowledge, in 3D cultures, but
only in several 2D culture-related studies. Fukuyama et al
(42) stated in their study that
three liver cancer cell lines (Huh7, HepG2 and Hep3B) fell in the
same hierarchical cluster and shared a common origin. These cell
lines and primary hepatocytes appeared next to each other in
cluster and demonstrated similar gene expression patterns. However,
due to various gene mutations, several genetic and growth
heterogeneities in different hepatic cell lines have been reported
(43). Hence, the use of
additional liver cancer cell lines, including HepG2 and Hep3B, is
required to determine whether the effects of VEGF on 3D conditions
are cell-specific.
The observations of the present study suggested that
the effects of growth factors as predicted in 2D cultures may
markedly differ than those in 3D cultures and hence, in the in
vivo setting. Taken together, the present study indicated
subtle differences between the 2D and 3D culture systems, in terms
of invasive phenotype and EMT-associated gene expression profile of
hepatoma cells in the presence of VEGF.
In conclusion, the present study demonstrated that
the tumorigenic properties of invasion and migration in response to
growth factors and/or therapeutics can be most effectively studied
in 3D setting as compared to the 2D setting. Additionally, gene
expression changes obtained in 2D cultures in response to different
treatment regimens require careful interpretations. Numerous
anti-tumour therapeutics appearing to be promising in a 2D cell
culture setup, fail heavily during clinical studies. A better
understanding of HCC with patient-specific cells in 3D cultures
in vitro models would certainly provide an improved
comprehension of HCC biology in vivo, the prediction of the
response of hepatoma cells to targeted therapy, as well as the
development of novel therapeutic concepts.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the DBT-RGYI grant
(BT/PR6356/GBD/27/407/2012).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PR performed cell culture experiments, migration,
chemotaxis, invasion, immunophenotyping, RT-qPCR and 3D culture
assays, collected and analysed data and drafted the manuscript. DMT
designed the experiments. VN analysed data. SK designed the study
and performed data analysis. All authors have read and approved the
final manuscript. PR and SK confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar
|
|
2
|
Zhang Y, Gao X, Zhu Y, Kadel D, Sun H,
Chen J, Luo Q, Sun H, Yang L, Yang J, et al: The dual blockade of
MET and VEGFR2 signaling demonstrates pronounced inhibition on
tumor growth and metastasis of hepatocellular carcinoma. J Exp Clin
Cancer Res. 37:932018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ma S, Pradeep S, Hu W, Zhang D, Coleman R
and Sood A: The role of tumor microenvironment in resistance to
anti-angiogenic therapy. F1000Res. 7:3262018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jiang Z, Zhai X, Shi B, Luo D and Jin B:
KIAA1199 overexpression is associated with abnormal expression of
EMT markers and is a novel independent prognostic biomarker for
hepatocellular carcinoma. Onco Targets Ther. 11:8341–8348. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li L and Li W: Epithelial-mesenchymal
transition in human cancer: Comprehensive reprogramming of
metabolism, epigenetics, and differentiation. Pharmacol Ther.
150:33–46. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Silvestre JS, Lévy BI and Tedgui A:
Mechanisms of angiogenesis and remodelling of the microvasculature.
Cardiovasc Res. 78:201–202. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhao W, Zhao T, Huang V, Chen Y, Ahokas RA
and Sun Y: Platelet-derived growth factor involvement in myocardial
remodeling following infarction. J Mol Cell Cardiol. 51:830–838.
2011. View Article : Google Scholar
|
|
8
|
Crivellato E, Nico B, Vacca A, Djonov V,
Presta M and Ribatti D: Recombinant human erythropoietin induces
intussusceptive microvascular growth in vivo. Leukemia. 18:331–336.
2004. View Article : Google Scholar
|
|
9
|
Zhan X, Wang F, Bi Y and Ji B: Animal
models of gastrointestinal and liver diseases. Animal models of
acute and chronic pancreatitis. Am J Physiol Gastrointest Liver
Physiol. 311:G343–G355. 2016. View Article : Google Scholar
|
|
10
|
Liu S, Sun Y, Jiang M, Li Y, Tian Y, Xue
W, Ding N, Sun Y, Cheng C, Li J, et al: Glyceraldehyde-3-phosphate
dehydrogenase promotes liver tumorigenesis by modulating
phosphoglycerate dehydrogenase. Hepatology. 66:631–645. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Karunagaran D, Rashmi R and Kumar TR:
Induction of apoptosis by curcumin and its implications for cancer
therapy. Curr Cancer Drug Targets. 5:117–129. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fantozzi A, Gruber DC, Pisarsky L, Heck C,
Kunita A, Yilmaz M, Meyer-Schaller N, Cornille K, Hopfer U,
Bentires-Alj M and Christofori G: VEGF-mediated angiogenesis links
EMT-induced cancer stemness to tumor initiation. Cancer Res.
74:1566–1575. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takai A, Dang H, Oishi N, Khatib S, Martin
SP, Dominguez DA, Luo J, Bagni R, Wu X, Powell K, et al:
Genome-wide RNAi screen identifies PMPCB as a therapeutic
vulnerability in EpCAM+ hepatocellular carcinoma. Cancer
Res. 79:2379–2391. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kozyra M, Johansson I, Nordling Å, Ullah
S, Lauschke VM and Ingelman-Sundberg M: Human hepatic 3D spheroids
as a model for steatosis and insulin resistance. Sci Rep.
8:142972018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Loessner D, Rizzi SC, Stok KS, Fuehrmann
T, Hollier B, Magdolen V, Hutmacher DW and Clements JA: A
bioengineered 3D ovarian cancer model for the assessment of
peptidase-mediated enhancement of spheroid growth and
intraperitoneal spread. Biomaterials. 34:7389–7400. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pingitore P and Romeo S: The role of
PNPLA3 in health and disease. Biochim Biophys Acta Mol Cell Biol
Lipids. 1864:900–906. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sarkanen JR, Kaila V, Mannerström B, Räty
S, Kuokkanen H, Miettinen S and Ylikomi T: Human adipose tissue
extract induces angiogenesis and adipogenesis in vitro. Tissue Eng
Part A. 18:17–25. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chiang IT, Liu YC, Wang WH, Hsu FT, Chen
HW, Lin WJ, Chang WY and Hwang JJ: Sorafenib inhibits TPA-induced
MMP-9 and VEGF expression via suppression of ERK/NF-κB pathway in
hepatocellular carcinoma cells. In Vivo. 26:671–681.
2012.PubMed/NCBI
|
|
19
|
Bell CC, Hendriks DF, Moro SM, Ellis E,
Walsh J, Renblom A, Fredriksson Puigvert L, Dankers AC, Jacobs F,
Snoeys J, et al: Characterization of primary human hepatocyte
spheroids as a model system for drug-induced liver injury, liver
function and disease. Sci Rep. 6:251872016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Raghavan S, Mehta P, Horst EN, Ward MR,
Rowley KR and Mehta G: Comparative analysis of tumor spheroid
generation techniques for differential in vitro drug toxicity.
Oncotarget. 7:16948–16961. 2016. View Article : Google Scholar
|
|
21
|
Murad HY, Bortz EP, Yu H, Luo D,
Halliburton GM, Sholl AB and Khismatullin DB: Phenotypic
alterations in liver cancer cells induced by mechanochemical
disruption. Sci Rep. 9:195382019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
D'Angelo E, Natarajan D, Sensi F, Ajayi O,
Fassan M, Mammano E, Pilati P, Pavan P, Bresolin S, Preziosi M, et
al: Patient-derived scaffolds of colorectal cancer metastases as an
organotypic 3D model of the liver metastatic microenvironment.
Cancers (Basel). 12:3642020. View Article : Google Scholar
|
|
23
|
Kapałczyńska M, Kolenda T, Przybyła W,
Zajączkowska M, Teresiak A, Filas V, Ibbs M, Bliźniak R, Łuczewski
Ł and Lamperska K: 2D and 3D cell cultures-a comparison of
different types of cancer cell cultures. Arch Med Sci. 14:910–919.
2018.
|
|
24
|
Jung HR, Kang HM, Ryu JW, Kim DS, Noh KH,
Kim ES, Lee HJ, Chung KS, Cho HS, Kim NS, et al: Cell spheroids
with enhanced aggressiveness to mimic human liver cancer in vitro
and in vivo. Sci Rep. 7:104992017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Khawar IA, Park JK, Jung ES, Lee MA, Chang
S and Kuh HJ: Three dimensional mixed-cell spheroids mimic
stroma-mediated chemoresistance and invasive migration in
hepatocellular carcinoma. Neoplasia. 20:800–812. 2018. View Article : Google Scholar
|
|
26
|
Hirschhaeuser F, Menne H, Dittfeld C, West
J, Mueller-Klieser W and Kunz-Schughart LA: Multicellular tumor
spheroids: An underestimated tool is catching up again. J
Biotechnol. 148:3–15. 2010. View Article : Google Scholar
|
|
27
|
Kunz-Schughart LA, Freyer JP, Hofstaedter
F and Ebner R: The use of 3-D cultures for high-throughput
screening: The multicellular spheroid model. J Biomol Screen.
9:273–285. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin RZ and Chang HY: Recent advances in
three-dimensional multicellular spheroid culture for biomedical
research. Biotechnol J. 3:1172–1184. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bhattacharya R, Fan F, Wang R, Ye X, Xia
L, Boulbes D and Ellis LM: Intracrine VEGF signalling mediates
colorectal cancer cell migration and invasion. Br J Cancer.
117:848–855. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Refolo MG, Messa C, Guerra V, Carr BI and
D'Alessandro R: Inflammatory mechanisms of HCC development. Cancers
(Basel). 12:6412020. View Article : Google Scholar
|
|
31
|
Muenzner JK, Kunze P, Lindner P, Polaschek
S, Menke K, Eckstein M, Geppert CI, Chanvorachote P, Baeuerle T,
Hartmann A and Schneider-Stock R: Generation and characterization
of hepatocellular carcinoma cell lines with enhanced cancer stem
cell potential. J Cell Mol Med. 22:6238–6248. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rawal P, Siddiqui H, Hassan M, Choudhary
MC, Tripathi DM, Nain V, Trehanpati N and Kaur S: Endothelial
cell-derived TGF-β promotes epithelial-mesenchymal transition via
CD133 in HBx-infected hepatoma cells. Front Oncol. 9:3082019.
View Article : Google Scholar
|
|
33
|
Takai A, Fako V, Dang H, Forgues M, Yu Z,
Budhu A and Wang XW: Three-dimensional organotypic culture models
of human hepatocellular carcinoma. Sci Rep. 6:211742016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Al Ameri W, Ahmed I, Al-Dasim FM, Ali
Mohamoud Y, Al-Azwani IK, Malek JA and Karedath T: Cell
Type-specific TGF-β mediated EMT in 3D and 2D models and its
reversal by TGF-β receptor kinase inhibitor in ovarian cancer cell
lines. Int J Mol Sci. 20:35682019. View Article : Google Scholar
|
|
35
|
Schneider MR, Hiltwein F, Grill J, Blum H,
Krebs S, Klanner A, Bauersachs S, Bruns C, Longerich T, Horst D, et
al: Evidence for a role of E-cadherin in suppressing liver
carcinogenesis in mice and men. Carcinogenesis. 35:1855–1862. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Melissaridou S, Wiechec E, Magan M, Jain
MV, Chung MK, Farnebo L and Roberg K: The effect of 2D and 3D cell
cultures on treatment response, EMT profile and stem cell features
in head and neck cancer. Cancer Cell Int. 19:162019. View Article : Google Scholar
|
|
37
|
Smalley KS, Lioni M, Noma K, Haass NK and
Herlyn M: In vitro three-dimensional tumor microenvironment models
for anticancer drug discovery. Expert Opin Drug Discov. 3:1–10.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li F, Wang F and Wu J: Sorafenib inhibits
growth of hepatoma with hypoxia and hypoxia-driven angiogenesis in
nude mice. Neoplasma. 64:718–724. 2017. View Article : Google Scholar
|
|
39
|
Rodríguez-Hernández MA, Chapresto-Garzón
R, Cadenas M, Navarro-Villarán E, Negrete M, Gómez-Bravo MA, Victor
VM, Padillo FJ and Muntané J: Differential effectiveness of
tyrosine kinase inhibitors in 2D/3D culture according to cell
differentiation, p53 status and mitochondrial respiration in liver
cancer cells. Csell Death Dis. 11:3392020. View Article : Google Scholar
|
|
40
|
Cheng CC, Chao WT, Shih JH, Lai YS, Hsu YH
and Liu YH: Sorafenib combined with dasatinib therapy inhibits cell
viability, migration, and angiogenesis synergistically in
hepatocellular carcinoma. Cancer Chemother Pharmacol. 88:143–153.
2021. View Article : Google Scholar
|
|
41
|
Ou J, Guan D and Yang Y: Non-contact
co-culture with human vascular endothelial cells promotes
epithelial-to-mesenchymal transition of cervical cancer SiHa cells
by activating the NOTCH1/LOX/SNAIL pathway. Cell Mol Biol Lett.
24:392019. View Article : Google Scholar
|
|
42
|
Fukuyama K, Asagiri M, Sugimoto M,
Tsushima H, Seo S, Taura K, Uemoto S and Iwaisako K: Gene
expression profiles of liver cancer cell lines reveal two
hepatocyte-like and fibroblast-like clusters. PLoS One.
16:e02459392021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhao Y, Chen Y, Hu Y, Wang J, Xie X, He G,
Chen H, Shao Q, Zeng H and Zhang H: Genomic alterations across six
hepatocellular carcinoma cell lines by panel-based sequencing.
Transl Cancer Res. 7:231–239. 2018. View Article : Google Scholar
|