Introduction
Breast cancer is the most commonly diagnosed type of
cancer, surpassing lung cancer, and therefore presents a risk to
patients worldwide (1). Breast
cancer accounts for 11.7% of all new cancer cases and 6.9% of all
cancer-related deaths in women worldwide (1). Although the overall survival and
prognosis of patients with breast cancer have significantly
improved in recent years (2), the
analysis of data from the U.S. National Center for Health
Statistics highlights that further investigations are required;
these data indicated that after 2010, the breast cancer mortality
rate continued to decline by 1.2-2.2% per year in women aged
between 40–79 years. However, the breast cancer mortality rate
stopped decreasing in women aged <40 years, whereas the
mortality rate for women aged between 20–29 years increased by 2.8%
per year (3). Therefore, improving
the health awareness of young women, identifying novel therapeutic
targets and developing effective biomarkers to prevent the
progression of breast cancer are of importance.
Cisplatin (DDP) is a non-specific, first-generation
platinum drug, which affects the cell cycle and can be used to
treat several types of cancer, including breast, testicular,
ovarian and lung cancer (4). The
major targets of DDP are nucleophilic DNA, proteins and RNA
(5). It has previously been
reported that patients commonly exhibit a good initial response to
DDP chemotherapy (6). However,
drug resistance is an intractable problem in breast cancer
treatment (7). Moreover, the
specific mechanism underlying DDP drug resistance remains largely
unknown. The clinical use of DDP is limited due to its significant
cytotoxicity to normal tissues and increased potential for drug
resistance in cancer cells (8).
Therefore, investigating the mechanism underlying the sensitivity
of breast cancer cells to DDP is of great importance.
Non-structural maintenance of chromosome (SMC)
condensin I complex subunit H (NCAPH) is a structural component of
chromosomes during mitosis (9).
NCAPH and other subunits (NCAPD2 and NCAPG) that form the condensin
I complex cooperate with SMC to regulate the structure of
chromosomes (10). Moreover, it
has been reported that NCAPH is involved in the progression of
several types of cancer. A recent study reported that NCAPH was
significantly upregulated in endometrial cancer (EC), thus acting
as an oncogene to promote the development of EC (11). Another study indicated that NCAPH
knockdown could inhibit cell proliferation, migration and invasion
and induce cell cycle arrest in non-small cell lung cancer (NSCLC)
(12). Furthermore, a
bioinformatic analysis study using the ONCOMINE, University of
Alabama at Birmingham Cancer Data Analysis Portal and Gene
Expression-Based Outcome for Breast Cancer Online databases,
determined that NCAPH is upregulated in breast cancer, which
therefore indicates that NCAPH may be a promising biomarker in
breast cancer (13).
The present study aimed to explore the role and
regulatory mechanism of NCAPH in the malignant phenotype and DDP
resistance of breast cancer cells. Overall, the results of the
present study have provided novel insights into the progression and
resistance of breast cancer cells to cancer therapeutics, which may
be used to prevent and treat breast cancer in the future.
Materials and methods
Bioinformatic analysis
The Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING; string-db.org; organism=Homo sapiens)
(14); HumanBase
(hb.flatironinstitute.org) (15)
using the default data types ‘Co-expression’ AND ‘Interaction’ AND
‘TF binding’ AND ‘GSEA microRNAs targets’ AND ‘GSEA perturbations’,
with the criteria value of mimimum interaction confidence as 0.86
and the maximum number of genes as 9; and GeneMANIA (genemania.org)
(16) databases using the default
networks ‘Physical Interactions’ AND ‘Co-expression’ AND
‘Predicted’ AND ‘Co-localization’ AND ‘Genetic Interactions’ AND
‘Pathway’ AND ‘Shared protein domains’, were used to evaluate the
interaction between NCAPH and aurora kinase (AURK)B. The Gene
Expression Profiling Interactive Analysis (GEPIA) database
(gepia.cancer-pku.cn) (17) was
used to analyze the positive or negative association between the
mRNA expression levels of NCAPH and AURKB. P≤0.05 indicates that
the result of the model is reliable; R>0 indicates a positive
correlation and the closer R2 is to 1, the more relevant
the correlation between NCAPH and AURKB is.
Cell culture
The human mammary epithelial MCF-10A cell line (cat.
no. MCF-10A), the breast cancer cell lines MDA-MB-231 (cat. no.
TCHu227), SUM190PT (cat. no. CVCL_3423), SK-BR-3 (cat. no. TCHu225)
and MCF-7 (cat. no. ACC 115) and the DDP-resistant cell line
MCF-7/DDP (cat. no. MCF-7/DDP) were purchased from Beijing Protein
Biotechnology Co., Ltd. MCF-10A, SUM190PT, MCF-7 and MCF-7/DDP
cells were cultured in DMEM (Gibco; Thermo Fisher Scientific,
Inc.), MDA-MB-231 cells in Leibovitz's L-15 medium (Gibco; Thermo
Fisher Scientific, Inc.) and SK-BR-3 cells in McCoy's 5A medium
(Gibco; Thermo Fisher Scientific, Inc.). The media were
supplemented with 10% FBS (Merck KGaA) and 1%
penicillin/streptomycin solution (Gibco; Thermo Fisher Scientific,
Inc.). All cell lines were cultured at 37°C with 5% CO2,
except MDA-MB-231 cells, which were cultured without
CO2.
Cell transfection
NCAPH knockdown was achieved by transfecting breast
cancer cells with 50 nM short hairpin RNAs (shRNAs/sh) targeting
NCAPH (sh-NCAPH-1/2). Cells transfected with scrambled shRNAs
served as the negative control (NC; sh-NC) group. For AURKB
overexpression (oe; oe-AURKB), MCF-7 cells were transfected with
the pcDNA3.1 plasmid (2 µg) encoding AURKB complementary DNA
(cDNA), whereas empty vectors served as the oe-NC group. All
plasmids were synthesized by Shanghai GenePharma Co., Ltd. and the
cells seeded in 6-well plates at a density of 1×106 were
transfected with the aforementioned plasmids/shRNAs using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 48 h at 37°C according to the manufacturer's
instructions. The cells were collected for subsequent experiments
48 h after transfection. The targeting sequences used were as
follows: sh-NCAPH-1, 5′-CCCAAGGATTAGACATCACAA-3′; sh-NCAPH-2,
5′-ACACGCAGATTACGGAACATT-3′; and sh-NC,
5′-GCACTACCAGAGCTAACTCAG-3′.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from untransfected or
transfected cells in 6-well plates at a density of 2×105
cells/well using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) and reverse transcribed into cDNA using
the PrimeScript RT Reagent Kit (Takara Biotechnology Co., Ltd.)
according to the manufacturer's protocol. qPCR was performed using
SYBR Premix Ex Taq™ II (Takara Bio, Inc.) with Thermal Cycler Dice™
Real Time System III (Takara Bio, Inc.). The PCR conditions were as
follows: 95°C for 10 min for initial denaturation, followed by 40
cycles of denaturation for 15 sec at 95°C, annealing for 30 sec at
60°C, elongation for 30 sec at 72°C and a final extension for 5 min
at 72°C. The relative mRNA expression levels were quantified using
the 2−∆∆Cq method (18)
following normalization with GAPDH. The primer sequences used were
as follows: NCAPH forward (F), 5′-AAACAACCTCAATGTCTCCGAAG-3′ and
reverse (R), 5′-ACAACCTAACTCTGGCAACTCG-3′; AURKB F,
5′-TCACCCCATCTGCACTTGTC-3′ and R, 5′-TGTGAAGTGCCGCGTTAAGA-3′; and
GAPDH F, 5′-GACTCATGACCACAGTCCATGC-3′ and R,
5′-AGAGGCAGGGATGATGTTCTG-3′.
Western blotting
Transfected or untransfected cells
(2×106) were lysed using RIPA lysis buffer (Beijing
Solarbio Science & Technology Co., Ltd.) and the protein
concentration was assessed using the BCA method (Beijing Solarbio
Science & Technology Co., Ltd.). Subsequently, 30 µg
protein/lane was separated via SDS-PAGE on a 10% gel and then
transferred onto PVDF membranes (Beijing Solarbio Science &
Technology Co., Ltd.). Subsequently the membranes were blocked with
5% skimmed milk for 2 h at room temperature followed by incubation
with specific primary antibodies at 4°C overnight. After being
washed in a TBST solution, the membranes were incubated with
HRP-conjugated secondary antibodies for 1 h at room temperature.
The protein bands were developed with MilliporeSigma™ Luminata™
Western HRP Chemiluminescence Substrates (MilliporeSigma) and all
data were analyzed using ImageJ software (version 1.52; National
Institutes of Health). The antibodies used in the present study are
presented in Table I.
 | Table I.Antibodies used for western
blotting. |
Table I.
Antibodies used for western
blotting.
| Antibody | Dilution | Catalog no. | Host | Company |
|---|
| NCAPH | 1:1,000 | PA5-80842 | Rabbit | Invitrogen; Thermo
Fisher Scientific, Inc. |
| Ki67 | 1:500 | Orb389335 | Rabbit | Biorbyt Ltd. |
| PCNA | 1:1,000 | Orb48485 | Rabbit | Biorbyt Ltd. |
| MMP2 | 1:2,000 | GTX59880 | Rabbit | GeneTex, Inc. |
| MMP9 | 1:1,000 | GTX100458 | Rabbit | GeneTex, Inc. |
| Bcl-2 | 1:1,000 | AB112 | Rabbit | Beyotime Institute
of Biotechnology |
| Bax | 1:2,000 | AF1270 | Rabbit | Beyotime Institute
of Biotechnology |
| Cleaved
caspase-3 | 1:500 | ab32042 | Rabbit | Abcam |
| Cleaved
caspase-9 | 1:1,000 | GTX132331 | Rabbit | GeneTex, Inc. |
| Caspase-3 | 1:5,000 | GTX110543 | Rabbit | GeneTex, Inc. |
| Caspase-9 | 1:1,000 | GTX112888 | Rabbit | GeneTex, Inc. |
| AURKB | 1:20,000 | ab45145 | Rabbit | Abcam |
| GAPDH | 1:50,000 | GTX100118 | Rabbit | GeneTex, Inc. |
| Anti-rabbit IgG
(HRP) | 1:1,000 | A0208 | Goat | Beyotime Institute
of Biotechnology |
Determination of cell
proliferation
The Cell Counting Kit-8 (CCK-8; Dojindo
Laboratories, Inc.) and colony formation assays were performed to
assess cell proliferation. For the CCK-8 assay, untreated or
transfected MCF-7 cells were seeded into a 96-well plate at a
density of 3×103 cells/well and incubated at 37°C for
24, 48 and 72 h following transfection. Following incubation for
the indicated time points, 10 µl CCK-8 solution was added into each
well and the cells were then incubated for an additional 3 h.
Finally, the absorbance at a wavelength of 450 nm was assessed in
each well using a microplate reader (Bio-Rad Laboratories, Inc.).
Cell viability was calculated as follows: (OD value-OD value at 0
h)/(OD value at 0 h) ×100. The results are presented with the
proliferation rate of the control group at 24 h set as 100%.
For the colony formation assay, the untreated and
transfected MCF-7 cells were seeded into culture dishes at a
density of 500 cells/dish. Subsequently, cells were cultured for
two weeks at 37°C and the medium was changed every three days.
Following incubation, cells were washed twice with PBS, fixed with
4% paraformaldehyde (Merck KGaA) at room temperature for 15 min and
stained with 0.5% crystal violet (Shanghai Yeasen Biotechnology
Co., Ltd.) at room temperature for 30 min. Colonies containing
>50 cells were imaged and counted manually using an inverted
light microscope (magnification, ×10; Olympus Corporation).
Determination of cell viability
The effect of different concentrations of DDP (0.1,
1, 5, 10, 25, 50 and 100 µM; Shanghai Yuanye Bio-Technology, Co.,
Ltd.) on MCF-7 and MCF-7/DDP cell viability was assessed using the
CCK-8 assay. Cells at a density of 5×103 cells/well were
seeded into 96-well plates and cultured with medium containing DDP
for 48 h at 37°C. The other experimental steps were performed as
aforementioned when describing the procedures of determination of
proliferation of untreated or transfected MCF-7 cells.
Wound healing assay
Transfected cells at a density of 5×105
cells/well were inoculated into a 6-well plate and incubated at
37°C for 24 h until a cell monolayer was created when cells were at
70–80% confluence. Subsequently, a 200-µl pipette tip was used to
introduce a straight scratch in the middle of the cell monolayer.
Following culturing with serum-free medium for 24 h at 37°C, images
of the migrated cells at 0 and 24 h were captured using an inverted
light microscope (magnification, ×100; Olympus Corporation) and
quantified using ImageJ software (version 1.52; National Institutes
of Health).
Transwell assay
The upper chamber of the Transwell insert (8-µm
pore; Corning Inc.) was first pre-coated with Matrigel
(MilliporeSigma) for 1 h at room temperature and was then
supplemented with 0.1 ml cell suspension (3×103 cells in
FBS-free DMEM). The lower chamber was filled with DMEM supplemented
with 20% FBS. Following incubation for 24 h at 37°C, the cells on
the lower surface of the membrane were fixed and stained using the
aforementioned methods from the colony formation assay. The
invasive cells were quantified using an inverted light microscope
(magnification, ×100; Olympus Corporation) and quantified using
ImageJ software (version 1.52; National Institutes of Health).
TUNEL assay
TUNEL staining was performed to assess cell
apoptosis using the One Step TUNEL Apoptosis Assay Kit (cat. no.
C1086; Beyotime Institute of Biotechnology). Briefly, transfected
or untransfected MCF-7(/DDP) cells at a density of 5×105
cells/well were fixed with 4% paraformaldehyde for 15 min at room
temperature, permeated in PBS supplemented with 0.3% Triton X-100
for 10 min at 4°C and blocked with 3% H2O2
for 15 min at room temperature. Before the cell nuclei were mounted
with Vectashield® mounting medium containing 1 mg/ml
DAPI for 10 min at room temperature, cells were incubated with 50
µl TUNEL detection solution for 1 h at 37°C in the dark. Dehydrated
transparent neutral gum was used to mount the sections.
TUNEL-positive cells in six randomly selected fields of view were
imaged using a fluorescence microscope (magnification, ×200;
Olympus Corporation).
Co-immunoprecipitation (Co-IP)
assay
The interaction between NCAPH and AURKB was assessed
using a Co-IP assay kit (cat. no. P2179; Beyotime Institute of
Biotechnology) according to the manufacturer's protocol. First,
MCF-7 cells were lysed in lysis buffer supplemented with protease
inhibitors. Subsequently, 250 µl of cell lysates containing 100 µg
Protein A + G magnetic beads were supplemented with antibodies
against NCAPH (1/20), AURKB (1/20) or IgG (cat. no. ab172730; 1/60;
Abcam) as the negative control, and a certain proportion of
supernatant without any antibody (Input) was used as the positive
control, followed by incubation for 2 h, with tumbling, at room
temperature. The magnetic beads were separated via magnetic force
followed by washing with ice-cold lysis buffer. Subsequently, the
magnetic beads were immersed in SDS-PAGE sample loading buffer and
boiled for 5 min. Following magnetic separation for 10 sec, the
supernatant was collected for western blotting, using the
aforementioned method.
Statistical analysis
All experiments were repeated independently three
times. The results are presented as the mean ± SD. All statistical
analysis was performed using GraphPad Prism 8.0 software (GraphPad
Software, Inc.). Unpaired Student's t-test and one-way ANOVA
followed by Tukey's post-hoc test were used to compare the
differences between two and three or more groups, respectively.
P<0.05 was considered to indicate a statistically significant
difference.
Results
NCAPH knockdown attenuates breast
cancer cell proliferation, migration and invasion
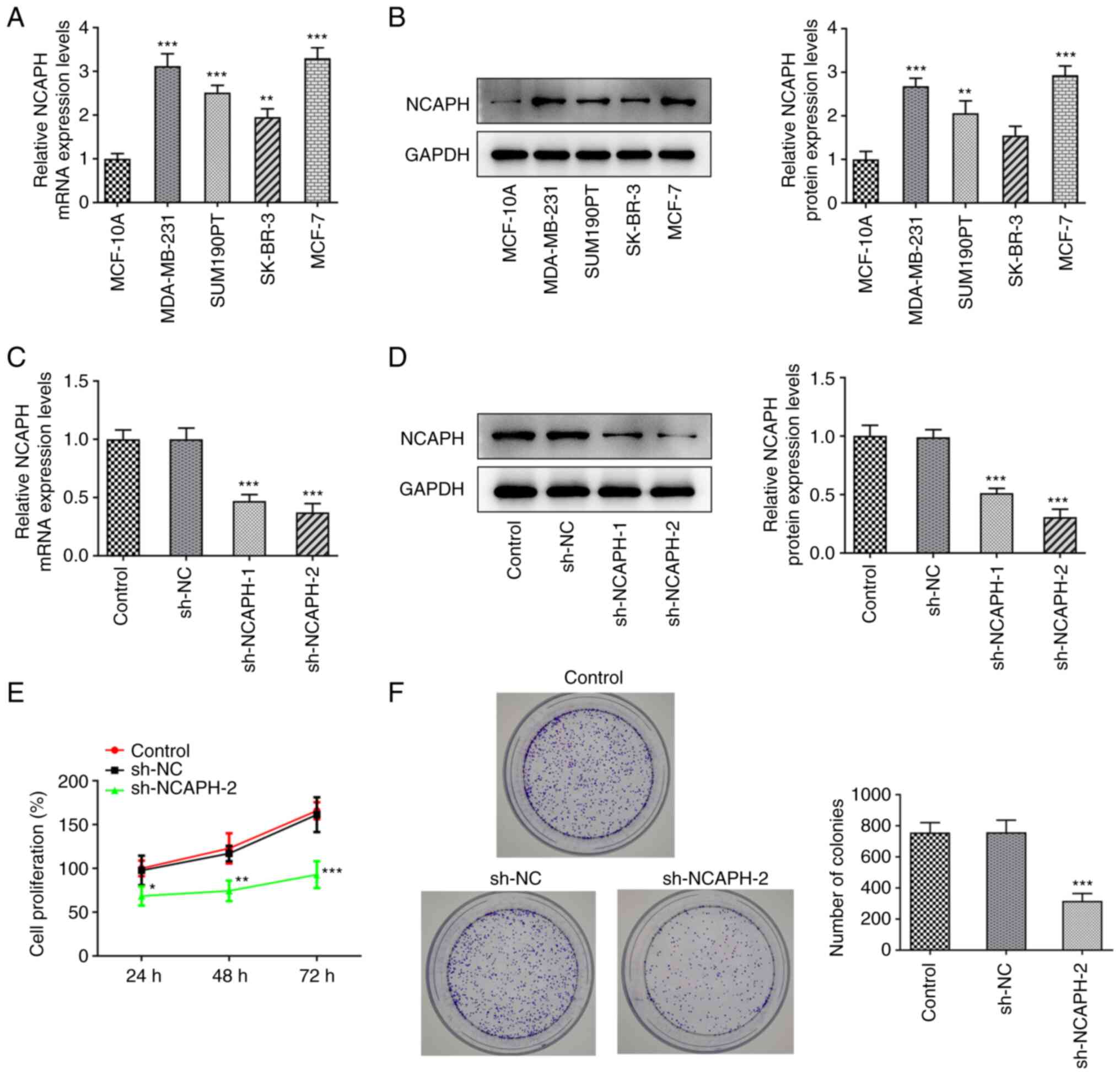
The mRNA and protein expression levels of NCAPH in
the MCF-10A, MDA-MB-231, SUM190PT, SK-BR-3 and MCF-7 cell lines
were determined using RT-qPCR and western blotting, respectively
(Fig. 1A and B). The results
demonstrated that the mRNA expression levels of NCAPH were
significantly increased in all of the breast cancer cell lines
compared with MCF-10A cells and the protein expression levels of
NCAPH were significantly increased in all of the breast cancer cell
lines except SK-BR-3 compared with MCF-10A cells. To highlight the
potential role of NCAPH in breast cancer, the widely used MCF-7
cell line was selected for use in the subsequent assays. These
cells demonstrated the highest NCAPH expression levels and were
therefore transfected with sh-NCAPH-1/2 and sh-NC as a control. The
transfection efficiency was assessed using RT-qPCR and western
blotting (Fig. 1C and D). The
results demonstrated that the mRNA and protein expression levels of
NCAPH were markedly reduced in the sh-NCAPH-2 group and sh-NCAPH-1
group compared with the sh-NC group. The interference efficiency of
sh-NCAPH-2 was more efficient than that of sh-NCAPH-1. Therefore,
the sh-NCAPH-2 construct was selected for use in the subsequent
assays. The effect of NCAPH knockdown on cell proliferation was
determined via CCK-8 and colony formation assays (Fig. 1E and F). These assays demonstrated
that NCAPH knockdown significantly reduced cell proliferation and
significantly reduced the colony formation ability of breast cancer
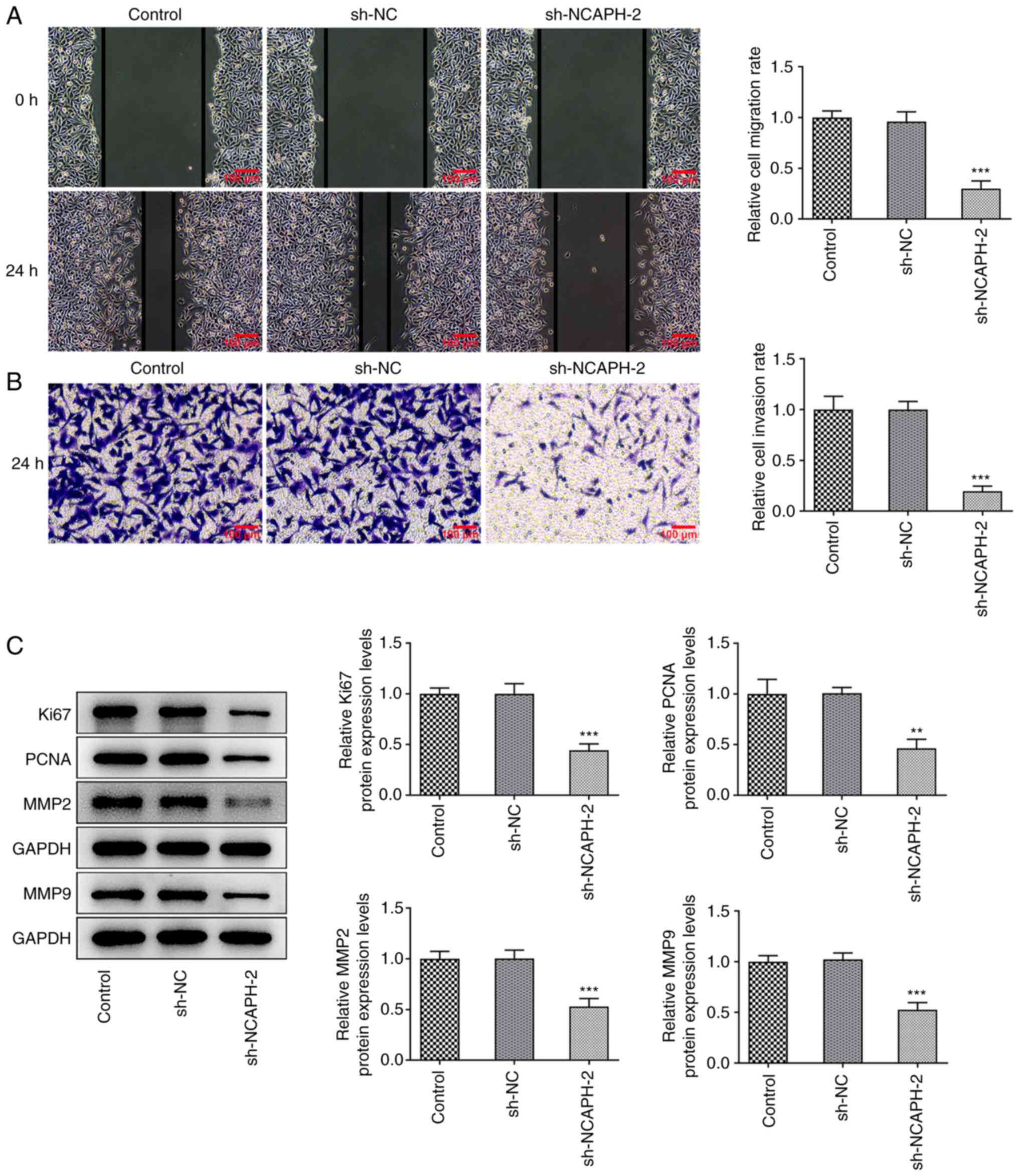
cells compared with sh-NC. Furthermore, the wound healing and
Transwell assays demonstrated that NCAPH knockdown significantly
reduced cell migration and invasion compared with the sh-NC group
(Fig. 2A and B). Moreover, western
blotting was performed to detect the protein expression levels of
proliferation- and migration-related proteins (Fig. 2C). The results demonstrated that
the protein expression levels of Ki67, proliferating cell nuclear
antigen (PCNA), MMP2 and MMP9 were all significantly reduced in the
sh-NCAPH-2 group compared with the sh-NC group. In summary, NCAPH
knockdown suppressed breast cancer cell proliferation, migration
and invasion.
NCAPH knockdown attenuates the
resistance of breast cancer cells to DDP
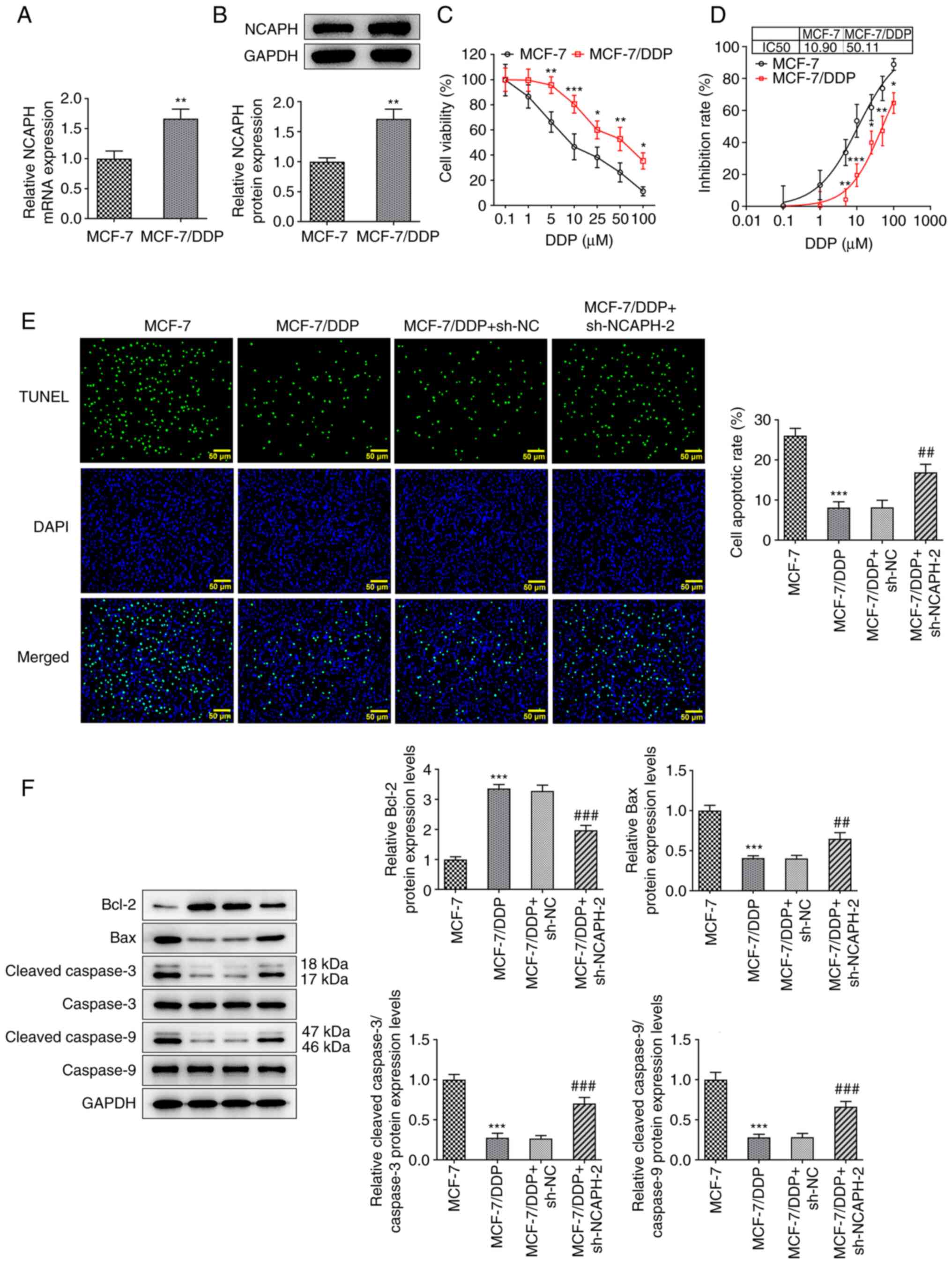
The mRNA and protein expression levels of NCAPH in
the MCF-7 and MCF-7/DDP cells were determined using RT-qPCR and
western blotting, respectively. NCAPH mRNA and protein expression
levels in the MCF-7/DDP cells were significantly higher compared
with the MCF-7 cells (Fig. 3A and
B). The effect of DDP (0.1, 1, 5, 10, 25, 50 and 100 µM) on
MCF-7 and MCF-7/DDP cell viability was assessed using the CCK-8
assay (Fig. 3C). Treatment of
MCF-7 and MCF-7/DDP cells with different concentrations of DDP
markedly decreased cell viability in a dose-dependent manner. The
decrease in cell viability was significantly less in the MCF-7/DDP
cell line compared with the MCF-7 cell line. The IC50
values for DDP were 10.90 and 50.11 µM in MCF-7 and MCF-7/DDP
cells, respectively (Fig. 3D).
Furthermore, MCF-7, MCF-7/DDP and transfected MCF-7/DDP cells were
treated with 10 µM DDP and cell apoptosis was detected using the
TUNEL assay (Fig. 3E) and western
blotting (Fig. 3F). The apoptotic
rate of cells in the MCF-7/DDP group was significantly lower
compared with the MCF-7 group. Furthermore, NCAPH knockdown
significantly increased the apoptotic rate of MCF-7/DDP cells
compared with the MCF-7/DDP + sh-NC group. The protein expression
levels of Bax, cleaved caspase-3 and cleaved caspase-9 were
significantly decreased, whereas those of Bcl-2 were significantly
increased in MCF-7/DDP cells compared with MCF-7 cells.
Furthermore, NCAPH knockdown partially restored their levels
compared with MCF-7/DDP + sh-NC cells. Overall, NCAPH knockdown
reduced the resistance of breast cancer cells to DDP.
NCAPH knockdown downregulates AURKB
expression levels
The mRNA and protein expression levels of AURKB in
MCF-10A and MCF-7 cells were detected using RT-qPCR and western
blotting, respectively (Fig. 4A and
B). mRNA and protein expression levels of AURKB were
demonstrated to be significantly upregulated in MCF-7 cells
compared with MCF-10A cells. Furthermore, the STRING, HumanBase and
GeneMANIA databases were used to assess the association between
NCAPH and AURKB (Fig. 4C-E).
Bioinformatic analysis using the GEPIA database predicted that
NCAPH was positively associated with AURKB mRNA expression
(Fig. 4F). The co-IP assay was
used to verify the association between NCAPH and AURKB, which
demonstrated that NCAPH and AURKB could interact as they
coprecipitated (Fig. 4G). Finally,
RT-qPCR and western blotting demonstrated that the mRNA and protein
expression levels of AURKB in MCF-7 cells transfected with
sh-NCAPH-2 were significantly reduced compared with those in the
sh-NC group (Fig. 4H and I). In
conclusion, NCAPH bound to AUKRB and NCAPH silencing downregulated
AURKB expression levels.
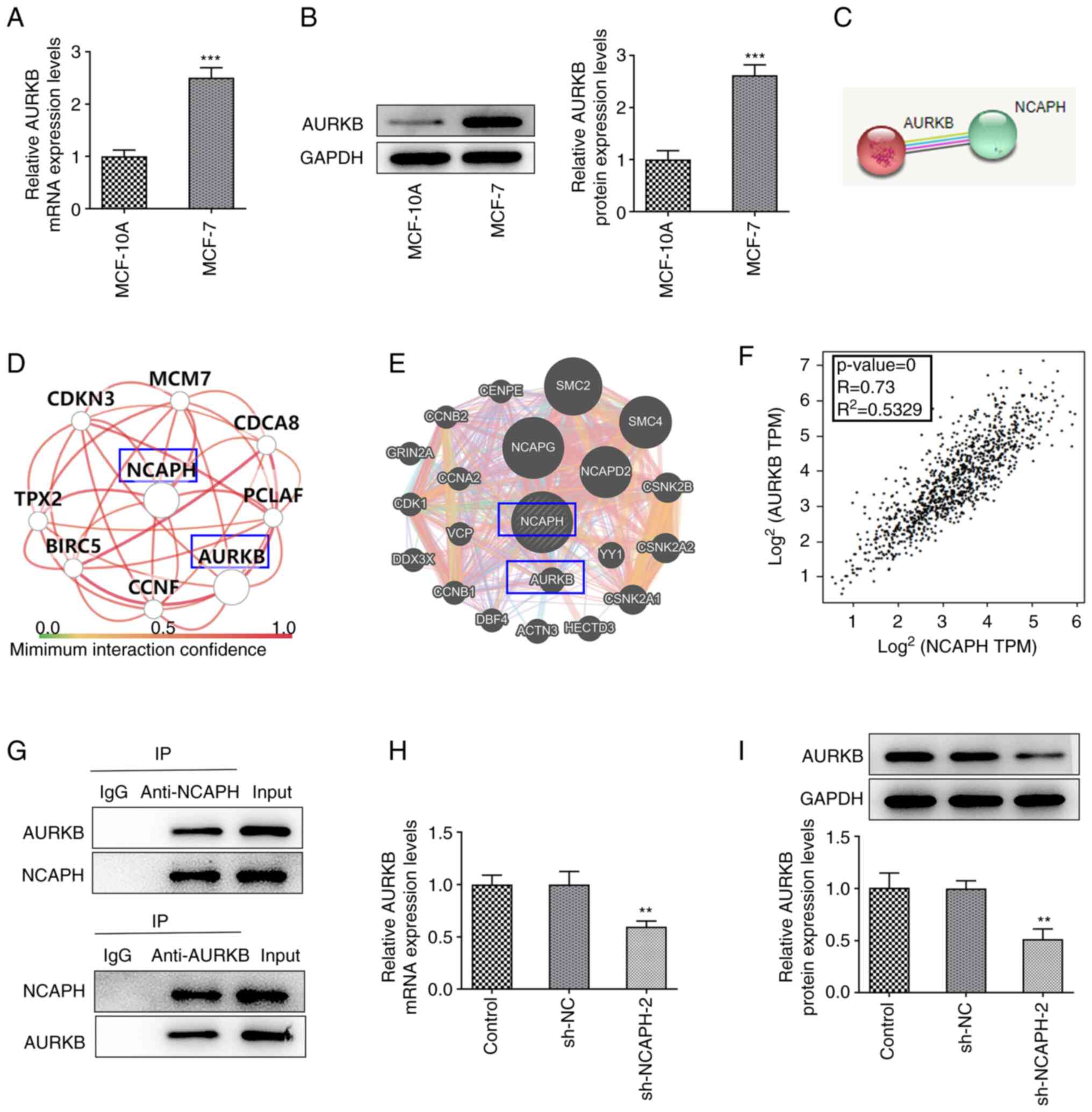 | Figure 4.NCAPH knockdown downregulates AURKB.
mRNA and protein expression levels of AURKB in MCF-10A and MCF-7
cells were detected via (A) RT-qPCR and (B) western blotting,
respectively. (C) Search Tool for the Retrieval of Interacting
Genes/Proteins, (D) HumanBase and (E) GeneMANIA databases were used
to predict the association between NCAPH and AURKB. (F)
Bioinformatics analysis in the Gene Expression Profiling
Interactive Analysis database predicted that NCAPH was positively
associated with AURKB. P≤0.05 indicates that the result of the
model is reliable; R>0 indicates a positive correlation and the
closer R2 is to 1, the more relevant the correlation
between NCAPH and AURKB is. (G) Co-immunoprecipitation assay was
performed to verify the association between NCAPH and AURKB. mRNA
and protein expression levels of AURKB in transfected MCF-7 cells
were determined via (H) RT-qPCR and (I) western blotting
respectively. **P<0.01 and ***P<0.001 vs. MCF-10A or sh-NC.
NCAPH, non-SMC condensin I complex subunit H; AURKB, aurora kinase
B; RT-qPCR, reverse transcription quantitative PCR; sh, short
hairpin RNA; NC, negative control; TPM, transcript per million;
CDKN3, cyclin dependent kinase inhibitor 3; MCM7, mini-chromosome
maintenance complex component 7; CDCA8, cell division
cycle-associated 8; TPX2, targeting protein for Xenopus plus
end-directed kinesin-like protein 2; BIRC5, Baculoviral inhibitor
of apoptosis domain repeat containing 5; PCLAF, proliferating cell
nuclear antigen clamp associated factor; CCN, cyclin; SMC,
structural maintenance of chromosome; CENPE, centromere-associated
protein E precursor; GRIN2A, glutamate ionotropic receptor
N-methyl-D-aspartate type subunit 2A; NCAPG, non-SMC condensin I
complex; CSNK, casein kinase; VCP, valosin-containing protein;
DDX3X, DEAD-box helicase 3 X-linked; YY1, YY1 transcription factor;
DBF4, dumbbell former 4 protein ACTN3, α-actinin-3; HECTD2, HECT
domain E3 ubiquitin protein ligase 2; IP, immunoprecipitation. |
AURKB overexpression partially
reverses the effects of NCAPH knockdown on cell proliferation,
migration and invasion
To assess the role of AURKB in the regulation of
NCAPH, MCF-7 cells were transfected with oe-AURKB. The transfection
efficiency was assessed via RT-qPCR and western blotting, and the
results revealed that AURKB expression was significantly elevated
in the oe-AURKB compared with the oe-NC group (Fig. 5A and B). Furthermore, MCF-7 cells
were co-transfected with sh-NCAPH-2 and oe-AURKB and the AURKB mRNA
and protein expression levels were assessed. The mRNA and protein
expression levels of AURKB were significantly elevated in the
sh-NACPH-2 + oe-AURKB group compared with the sh-NACPH-2 + oe-NC
group (Fig. 5C and D). The CCK-8
and colony formation assays demonstrated that AURKB overexpression
significantly increased cell proliferation compared with the
sh-NACPH-2 + oe-NC group, which suggested that AURKB potentially
reduced the effect of NCAPH knockdown on cell proliferation
(Fig. 5E and F). Furthermore, the
wound healing and Transwell assays demonstrated that AURKB
overexpression significantly increased cell migration and invasion
compared with the sh-NCAPH-2 + oe-NC group (Fig. 5G-J). Moreover, the western blotting
results demonstrated that AURKB overexpression significantly
increased the protein expression levels of Ki67, PCNA, MMP2 and
MMP9 compared with the sh-NACPH-2 + oe-NC group (Fig. 5K). Collectively, AURKB
overexpression partially reversed the effects of NCAPH knockdown on
breast cancer cell proliferation, migration and invasion.
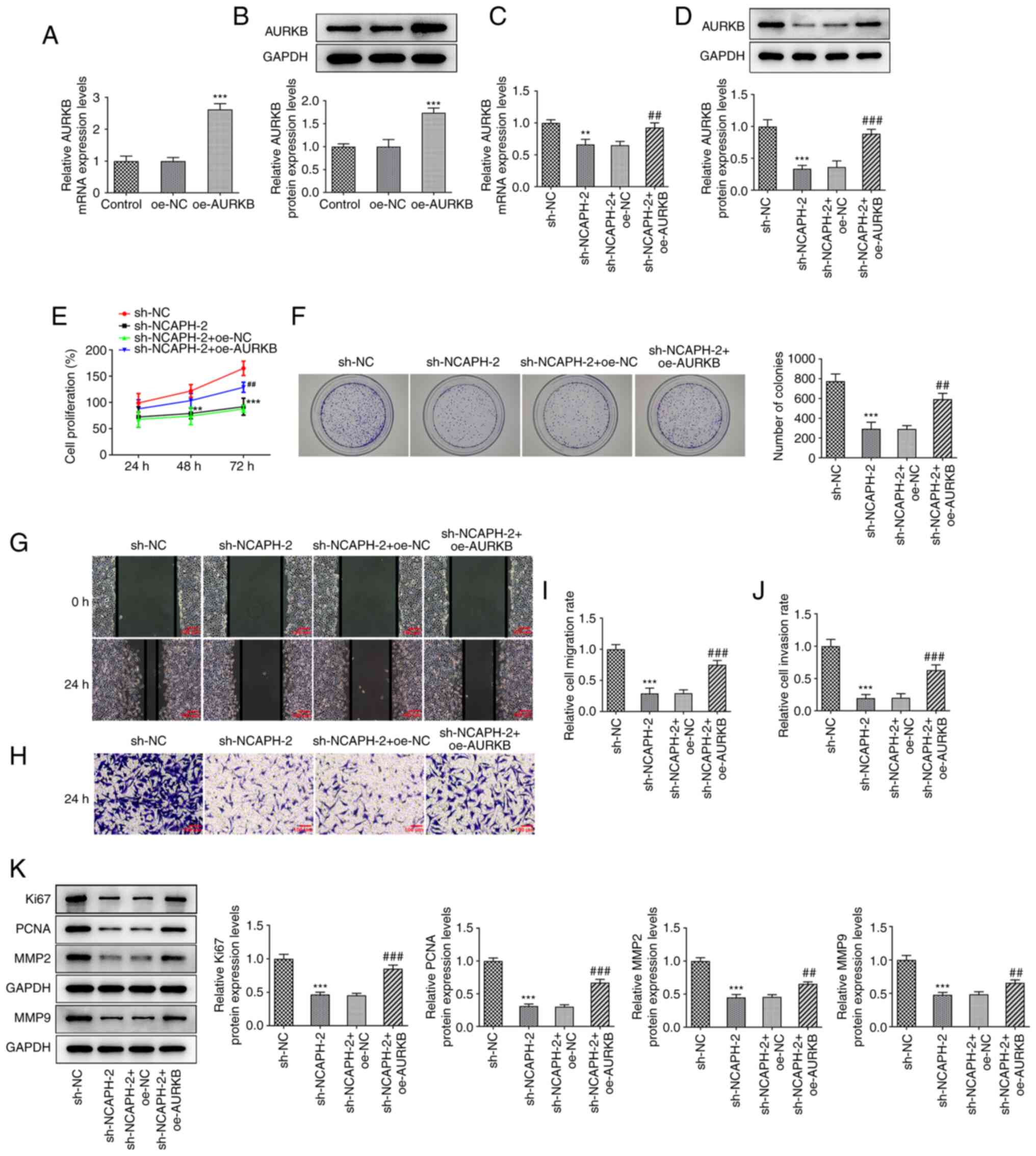 | Figure 5.AURKB overexpression partially
reverses the effects of NCAPH silencing on cell proliferation,
migration and invasion. Transfection efficiency of MCF-7 cells
transfected with oe-AURKB was assessed via (A) RT-qPCR and (B)
western blotting; respectively. MCF-7 cells were co-transfected
with sh-NCAPH and oe-AURKB and the AURKB mRNA and protein
expression levels were assessed via (C) RT-qPCR and (D) western
blotting respectively. Cell proliferation was assessed using the
(E) Cell Counting Kit-8 and (F) colony formation assays.
Magnification, ×10. (G) Cell migration was determined via wound
healing assays. Magnification, ×100. (H) Cell invasion was assessed
via Transwell assay. Magnification, ×100. Quantitative histograms
of (I) wound healing and (J) Transwell assays. (K) Western blotting
was performed to determine the expression levels of proliferation-
and migration-related proteins. **P<0.01 and ***P<0.001 vs.
oe-NC or sh-NC; ##P<0.01 and ###P<0.001
vs. sh-NCAPH-2 + oe-NC. AURKB, aurora kinase B; NCAPH,
non-structural maintenance of chromosome condensin I complex
subunit H; oe, overexpression; NC, negative control; RT-qPCR,
reverse transcription-quantitative PCR; sh, short hairpin RNA;
PCNA, proliferating cell nuclear antigen. |
AURKB overexpression partially
reverses the effects of NCAPH knockdown on DDP resistance and the
Akt/mTOR signaling pathway
Subsequently, the effect of AURKB on the resistance
of breast cancer cells to DDP was further assessed. MCF-7/DDP cells
were co-transfected with sh-NCAPH and oe-AURKB and were then
treated with 10 µM DDP. The results demonstrated that the apoptotic
rate in the MCF-7/DDP + sh-NCAPH-2 + oe-AURKB group was
significantly reduced compared with the MCF-7/DDP + sh-NCAPH-2 +
oe-NC group (Fig. 6A). Furthermore
the reduced apoptotic rate in the MCF-7/DDP + sh-NCAPH-2 + oe-AURKB
group was further supported by the significantly decreased protein
expression levels of Bax, cleaved caspase-3 and cleaved caspase-9
levels and the significantly increased protein expression levels of
Bcl-2, compared with the MCF-7/DDP + sh-NCAPH-2 + oe-NC group
(Fig. 6B). The protein expression
levels of Akt/mTOR signaling pathway-related proteins were detected
using western blotting. NCAPH knockdown significantly downregulated
the protein expression levels of both phosphorylated (p)-Akt and
p-mTOR compared with the sh-NC group, which were significantly
restored following AURKB overexpression compared with the
sh-NACPH-2 + oe-NC group (Fig.
6C). Taken together, AURKB overexpression partially reversed
the effects of NCAPH knockdown on breast cancer cell DDP resistance
and the Akt/mTOR signaling pathway.
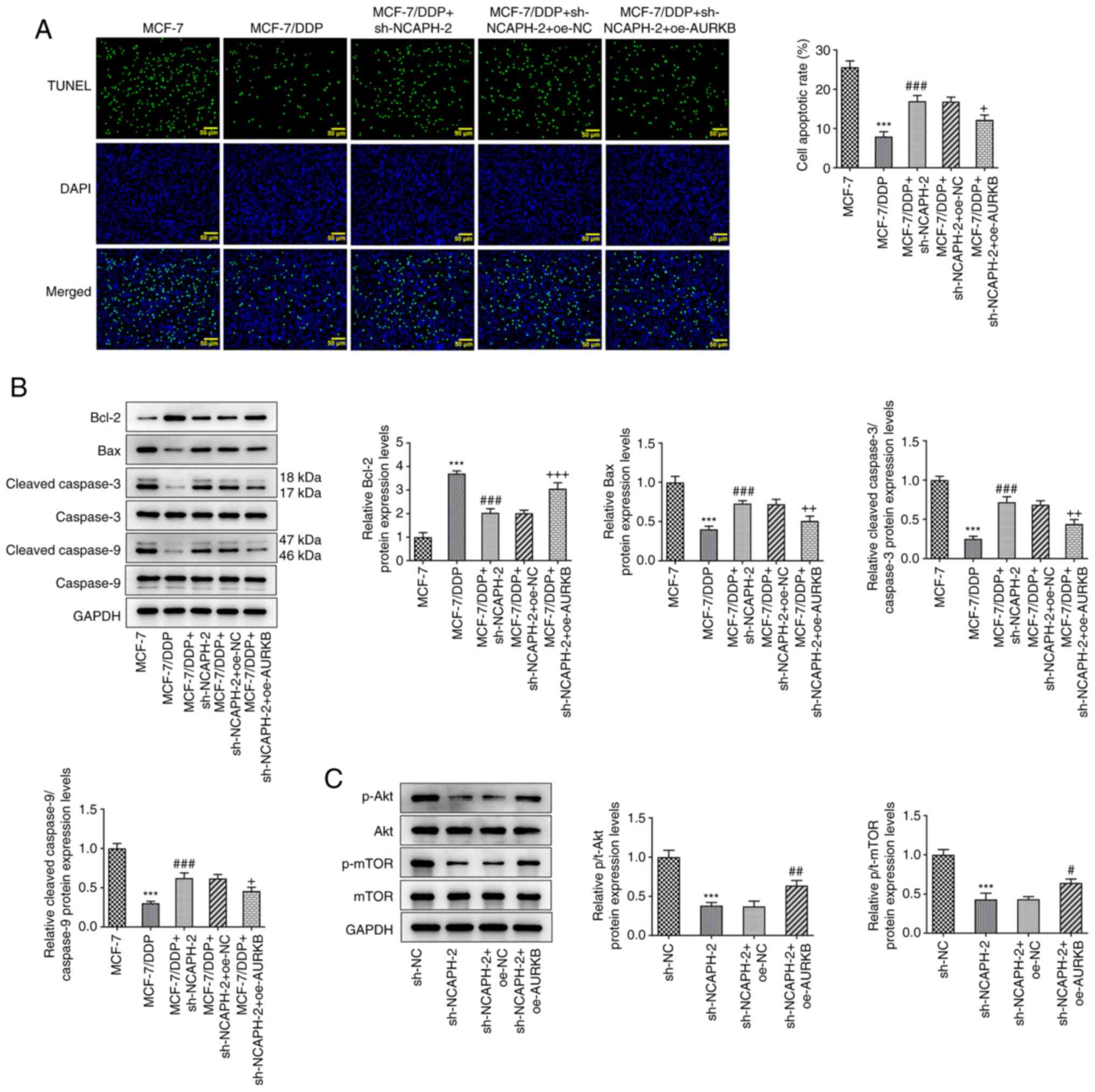 | Figure 6.AURKB overexpression partially
reverses the effects of NCAPH knockdown on DDP resistance and the
Akt/mTOR signaling pathway. (A) Cell apoptotic rate in each group
was assessed using the TUNEL assay. Magnification, ×200. (B)
Protein expression levels of apoptosis-related proteins were
detected using western blotting. (C) Protein expression levels of
the Akt/mTOR signaling pathway-related proteins were detected via
western blotting. ***P<0.001 vs. MCF-7 or sh-NC;
#P<0.05, ##P<0.01 and
###P<0.001 vs. MCF-7/DDP or sh-NCAPH-2 + oe-NC;
+P<0.05, ++P<0.01 and
+++P<0.001 vs. MCF-7/DDP + sh-NCAPH-2 + oe-NC. AURKB,
aurora kinase B; NCAPH, non-structural maintenance of chromosome
condensin I complex subunit H; sh, short hairpin RNA; DDP,
cisplatin; oe, overexpression; NC, negative control; p,
phosphorylated; t, total. |
Discussion
Breast cancer is the leading cause of cancer-related
deaths in women and is principally classified into three subtypes
based on the presence of estrogen (ER) and progesterone (PR)
receptors and human epidermal growth factor receptor 2 (HER2), in
the breast cancer tissue (19). In
total, ~90% of all breast cancer cases are not metastatic at the
time of diagnosis (20).
Therefore, the goal for non-metastatic breast cancer treatment is
to eradicate the tumor and prevent recurrence (21). Endocrine therapy, chemotherapy and
combined chemotherapy accompanied by tumor resection, can be
performed depending on the breast cancer subtype (22). Triple-negative breast cancer is
most likely to recur and is characterized by a 5-year survival rate
of 85% for stage I cancer compared with >94% for PR-positive and
HER2-positive breast cancer (21).
Instead of preventing recurrence, the aim of metastatic breast
cancer treatment is to prolong life and relieve symptoms.
Therefore, chemotherapy and radiotherapy are most commonly adopted
(23). According to a previously
published study, ER-positive/PR-positive/HER2-negative cancer
accounts for the majority of all breast cancer cases (24) and is clinically known as luminal A
breast cancer. It has previously been reported that MCF-7 cells,
mimic luminal breast cancer cells (25). In the present study, the results
demonstrated that NCAPH mRNA and protein expression levels were
significantly upregulated in breast cancer cells. Furthermore,
NCAPH knockdown significantly decreased the proliferation,
migration and invasion abilities of MCF-7 cells and reduced DDP
resistance in MCF-7/DDP cells.
Bioinformatic analysis indicated that AURKB was a
downstream regulatory target of NCAPH. AURKB belongs to a group of
highly conserved AURK isoforms that together with AURKA and AURKC
regulate chromosome arrangement and segregation during mitosis and
meiosis in mammals (26). Previous
studies have reported that during the chromosome segregation stage,
abnormal AURKB expression can promote the formation of abnormal
binucleate daughter cells via cytoplasmic bridges, which results in
tumorigenesis (27,28). The results of the present study
demonstrated that the mRNA and protein expression levels of AURKB
were significantly upregulated in MCF-7 cells. Furthermore, AURKB
overexpression could potentially reverse the inhibitory effect of
NCAPH knockdown on the progression of breast cancer and on the
resistance of breast cancer cells to DDP. The aforementioned
results suggested that AURKB could potentially trigger the
tumor-promoting and regulatory effects of NCAPH. It can therefore
be hypothesized that this process may be caused via AURKB
regulation of the phosphorylation of NCAPH-containing condensin and
histone ligation, suggesting a role for AURKB in histone
phosphorylation (29). Abnormal
NCAPH expression may lead to the unregulated expression of AURKB,
which may cause the dysregulation of mitosis. However, the specific
regulatory mechanism of this merits further research. Furthermore,
a previous study reported that AURKB is upregulated in gastric
cancer, whereas AURKB silencing can inhibit the invasion and
migration of gastric cancer cells (30). Moreover, AURKB is associated with
the resistance of NSCLC cells to DDP, which could be associated
with a poor prognosis in patients with breast cancer (31).
Mechanistically, mTOR serves an integral role in
signal transduction pathways involved in the regulation of cell
proliferation, protein synthesis and survival. mTOR is also
involved in several cellular processes that may lead to the
uncontrolled proliferation of cancer cells (32,33).
In the present study, the protein expression levels of mTOR and
those of its upstream target Akt were detected and the results
demonstrated that NCAPH knockdown significantly inhibited the
activation of the Akt/mTOR signaling pathway. However, AURKB
overexpression significantly activated Akt/mTOR signaling. These
findings suggested that AURKB could potentially mediate the
regulation of the Akt/mTOR signaling pathway via NCAPH. A previous
study reported that AURKB cooperates with histone deacetylases to
regulate the Akt signaling pathway (34); however, the specific underlying
regulatory mechanism requires future exploration. To the best of
our knowledge the present study was the first to present the
mechanism of NCAPH in breast cancer cells, as well as its role in
DDP resistance. However, the present study was limited to in
vitro experiments and therefore further in vivo
experiments are required to confirm the aforementioned
findings.
In conclusion, the present study demonstrated that
NCAPH knockdown significantly downregulated AURKB and significantly
inhibited breast cancer cell proliferation, migration, invasion,
DDP resistance and the Akt/mTOR signaling pathway. These findings
have provided novel insights into the identification of novel
therapeutic targets in breast cancer.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LL and HC designed the study and performed the
experiments. SL performed the experiments and data analysis, and
made considerable contributions to the drafting of the manuscript.
All authors read and approved the final manuscript. LL and SL
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pruitt SL, Zhu H, Heitjan DF, Rahimi A,
Maddineni B, Tavakkoli A, Halm EA, Gerber DE, Xiong D and Murphy
CC: Survival of women diagnosed with breast cancer and who have
survived a previous cancer. Breast Cancer Res Treat. 187:853–865.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hendrick RE, Helvie MA and Monticciolo DL:
Breast cancer mortality rates have stopped declining in U.S. women
younger than 40 years. Radiology. 299:143–149. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tchounwou PB, Dasari S, Noubissi FK, Ray P
and Kumar S: Advances in our understanding of the molecular
mechanisms of action of cisplatin in cancer therapy. J Exp
Pharmacol. 13:303–328. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu L and Bian K: Advance in studies on
molecular mechanisms of cisplatin resistance and intervention with
traditional Chinese medicines. Zhongguo Zhong Yao Za Zhi.
39:3216–3220. 2014.(In Chinese). PubMed/NCBI
|
|
6
|
Wang S, Li MY, Liu Y, Vlantis AC, Chan JY,
Xue L, Hu BG, Yang S, Chen MX, Zhou S, et al: The role of microRNA
in cisplatin resistance or sensitivity. Expert Opin Ther Targets.
24:885–897. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Safi A, Bastami M, Delghir S, Ilkhani K,
Seif F and Alivand MR: miRNAs modulate the dichotomy of cisplatin
resistance or sensitivity in breast cancer: An update of
therapeutic implications. Anticancer Agents Med Chem. 21:1069–1081.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hatzidaki E, Daikopoulou V, Apostolou P,
Ntanovasilis DA and Papasotiriou I: Increased breast cancer cell
sensitivity to cisplatin using a novel small molecule inhibitor. J
Cancer Res Ther. 16:1393–1401. 2020.PubMed/NCBI
|
|
9
|
Sun Y, Wang X, Wen H, Zhu B and Yu L:
Expression and clinical significance of the NCAPH, AGGF1, and FOXC2
proteins in serous ovarian cancer. Cancer Manag Res. 13:7253–7262.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim JH, Youn Y, Kim KT, Jang G and Hwang
JH: Non-SMC condensin I complex subunit H mediates mature
chromosome condensation and DNA damage in pancreatic cancer cells.
Sci Rep. 9:178892019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qiu X, Gao Z, Shao J and Li H: NCAPH is
upregulated in endometrial cancer and associated with poor
clinicopathologic characteristics. Ann Hum Genet. 84:437–446. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim B, Kim SW, Lim JY and Park SJ: NCAPH
is required for proliferation, migration and invasion of
non-small-cell lung cancer cells. Anticancer Res. 40:3239–3246.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu H, Shi C, Wang S, Wan X, Luo Y, Tian L
and Li L: Identification of NCAPH as a biomarker for prognosis of
breast cancer. Mol Biol Rep. 47:7831–7842. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szklarczyk D, Gable AL, Nastou KC, Lyon D,
Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al:
The STRING database in 2021: Customizable protein-protein networks,
and functional characterization of user-uploaded gene/measurement
sets. Nucleic Acids Res. 49((D1)): D605–D612. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Greene CS, Krishnan A, Wong AK, Ricciotti
E, Zelaya RA, Himmelstein DS, Zhang R, Hartmann BM, Zaslavsky E,
Sealfon SC, et al: Understanding multicellular function and disease
with human tissue-specific networks. Nat Genet. 47:569–576. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res 38(Web Server Issue). W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Loibl S, Poortmans P, Morrow M, Denkert C
and Curigliano G: Breast cancer. Lancet. 397:1750–1769. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Raigon-Ponferrada A, Recio MED,
Guerrero-Orriach JL, Malo-Manso A, Escalona-Belmonte JJ, Aliaga MR,
Fernández AR, García FJF, Conejo EA and Cruz-Mañas J: Breast cancer
and anesthesia. Curr Pharm Des. 25:2998–3004. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wöckel A, Albert US, Janni W, Scharl A,
Kreienberg R and Stüber T: The screening, diagnosis, treatment, and
follow-up of breast cancer. Dtsch Arztebl Int. 115:316–323.
2018.PubMed/NCBI
|
|
23
|
Harbeck N and Gnant M: Breast cancer.
Lancet. 389:1134–1150. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Abdel-Hafiz HA: Epigenetic mechanisms of
tamoxifen resistance in luminal breast cancer. Diseases. 5:162017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu S, Kim T, Yoo KH and Kang K: The T47D
cell line is an ideal experimental model to elucidate the
progesterone-specific effects of a luminal A subtype of breast
cancer. Biochem Biophys Res Commun. 486:752–758. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Marima R, Hull R, Penny C and Dlamini Z:
Mitotic syndicates Aurora Kinase B (AURKB) and mitotic arrest
deficient 2 like 2 (MAD2L2) in cohorts of DNA damage response (DDR)
and tumorigenesis. Mutat Res Rev Mutat Res. 787:1083762021.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiao J and Zhang Y: AURKB as a promising
prognostic biomarker in hepatocellular carcinoma. Evol Bioinform
Online. 17:117693432110575892021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nie M, Wang Y, Yu Z, Li X, Deng Y, Wang Y,
Yang D, Li Q, Zeng X, Ju J, et al: AURKB promotes gastric cancer
progression via activation of CCND1 expression. Aging (Albany NY).
12:1304–1321. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tada K, Susumu H, Sakuno T and Watanabe Y:
Condensin association with histone H2A shapes mitotic chromosomes.
Nature. 474:477–483. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang Z, Yu Z, Wang GH, Zhou YM, Deng JP,
Feng Y, Chen JQ and Tian L: AURKB promotes the metastasis of
gastric cancer, possibly by inducing EMT. Cancer Manag Res.
12:6947–6958. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu J, Zhou J, Xu F, Bai W and Zhang W:
High expression of Aurora-B is correlated with poor prognosis and
drug resistance in non-small cell lung cancer. Int J Biol Markers.
33:215–221. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mossmann D, Park S and Hall MN: mTOR
signalling and cellular metabolism are mutual determinants in
cancer. Nat Rev Cancer. 18:744–757. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Alzahrani AS: PI3K/Akt/mTOR inhibitors in
cancer: At the bench and bedside. Semin Cancer Biol. 59:125–132.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang C, Chen J, Cao W, Sun L, Sun H and
Liu Y: Aurora-B and HDAC synergistically regulate survival and
proliferation of lymphoma cell via AKT, mTOR and Notch pathways.
Eur J Pharmacol. 779:1–7. 2016. View Article : Google Scholar : PubMed/NCBI
|