Introduction
Renal cell carcinoma (RCC) originates from the renal
tubular epithelium in the proximal convoluted tubule (1); it is the 14th most common cancer type
in females and the 9th most common in males (2). The global incidence of RCC is ~4% and
rises each year (3). The World
Health Organization proposed that the annual number of
RCC-associated deaths is as high as ~140,000 and it ranks 13th for
cancer-associated mortality (4).
The introduction of checkpoint inhibitor (CPI) therapy has led to a
paradigm change in advanced RCC; dual immune checkpoint inhibition
or the combination of CPI and tyrosine kinase inhibitors were
indicated to improve survival (5).
In addition, new data from trials of immune CPIs for advanced
kidney cancer confirm a survival benefit with the combination of
cabozantinib plus nivolumab, pembrolizumab plus axitinib and
ipilimumab plus nivolumab (6).
Although the diagnostic and therapeutic efficacies for treating RCC
have been markedly improved during the past few decades, it remains
one of the most lethal urinary system malignancies.
MicroRNAs (miRNAs/miRs) are a class of short
non-coding RNAs that have been recognized as vital tumor regulators
in recent years (7). Recent
studies have indicated that miRNAs are involved in the regulation
of multiple signaling pathways during the progression of RCC,
serving in both oncogenic and tumor-suppressor roles. Therefore,
miRNAs are thought to be potential therapeutic and prognostic
targets in RCC (8). Maher
(9) reviewed the changes in our
understanding of the genetics of RCC, such as Hereditary
BAP1-associated RCC (10) and
activating mutations in the MET proto-oncogene predispose to Type 1
hereditary papillary RCC (11),
while Huang et al (12)
reported that miR-33b-5p may function as a tumor-suppressive
regulator and prognostic biomarker in RCC progression and sunitinib
resistance, which may provide novel therapeutic targets for
sunitinib-resistant RCC. In recent years, the function of
miR-5590-3p in mediating gene transcription in cancers, including
breast cancer (13), prostate
cancer (14) and gastric cancer
(15), has been reported. Although
the involvement of miR-5590-3p in RCC has been previously indicated
(16), its regulatory mechanisms
require further exploration.
Rho-associated protein kinase (ROCK) is a kinase
belonging to the AGC family of serine-threonine kinases. The ROCK
family consists of two isotypes, namely ROCK1 and ROCK2 (17). The ROCK signaling pathway has been
well documented in various biological processes (18). Numerous studies have indicated that
ROCK is associated with the accelerated metastasis of multiple
types of tumors and reduced survival, which is expected from a
cancer treatment target (19). It
has been suggested that ROCK expression is negatively associated
with the survival rate of patients with RCC. ROCK2 mediates RCC
proliferation through the ROCK2/β-catenin pathway (20). Therefore, ROCK is of great
significance during the progression of RCC.
In the present study, it was predicted that
miR-5590-3p directly targeted ROCK2 using Targetscan 7.1 and that
this was abnormally expressed in clinical specimens and cell lines
of RCC. It was then investigated how miR-5590-3p and ROCK2
regulated RCC cell functions and their exact molecular
mechanism.
Materials and methods
Patients and samples
A total of six surgical RCC specimens were collected
from patients with RCC who received surgery between January 1,
2018, and July 1, 2018, at the Second Affiliated Hospital of
Nanchang University (Nanchang, China). Patients were not treated by
any other means. This study was approved by the Second Affiliated
Hospital of Nanchang University Medical Research Ethics Committee
(Nanchang, China; no. 2017-100). Written informed consent was
obtained from each participant prior to the study.
Cell culture
The RCC cell lines A498 (cat. no. BNCC350808) and
A704 (cat. no. BNCC342393), the kidney fibroblast cell line KFB
(cat. no. BNCC341253) and 293T human embryonic kidney cells (cat.
no. BNCC100409) were purchased from BeNa Bio. and cultivated in
DMEM containing 10% FBS (all from BeNa Bio.) and 1% penicillin and
streptomycin in a humidified atmosphere with 5% CO2 at
37°C.
Cell transfection
miR-5590-3p mimics and the negative control (NC)
miR-5590-3p mimic-NC were provided by RiboBio Co., Ltd. (mimics
sequence: 5′-AAUAAAGUUCAUGUAUGGCAA-3′; and mimic-NC sequence:
5′-UUCUCCGAACGUGUCACGUTT-3′). Cells were seeded in a 12-well plate
(1×105 cells/well) and cultivated to 80% confluence.
After 4-h cell starvation in serum-free medium, cells were treated
with a mixture containing 100 nM plasmid and
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) that was prepared at room temperature 20 min
previously. After 24 h of incubation, the culture supernatant was
replaced with fresh medium and the mimics expression efficacy was
examined by reverse transcription-quantitative (RT-q)PCR.
RT-qPCR
Total RNA was isolated from RCC cells using TRIzol
Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and cDNA was
obtained through RT using the Primescript™ RT reagent kit with gDNA
Eraser (Takara Bio, Inc.) according to the manufacturer's protocol.
SYBR Premix Ex Taq™II (Tli RNaseH Plus; Takara Bio, Inc.) was used
in a fluorescence quantitative PCR system (Bio-Rad, Inc.) to
perform qPCR, using 0.2 µl of cDNA as the template and the
following thermocycling program: 95°C for 30 sec, followed by 40
cycles at 95°C for 10 sec and 60°C for 30 sec, and finally
extension at 60°C for 10 min. The relative expression level was
calculated using the 2−ΔΔCq method (21). GAPDH or U6 served as the internal
reference. The primer sequences are listed in Table I.
 | Table I.PCR primers used in the present
study. |
Table I.
PCR primers used in the present
study.
| Gene | Primer (5′-3′) |
|---|
| miR-5590-3p-F |
ACACTCCAGCTGGGAATAAAGTTCATGTA |
| miR-5590-3P-R |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTTGCCATA |
| ROCK2-F |
TCAGAGGTCTACAGATGAAGGC |
| ROCK2-R |
CCAGGGGCTATTGGCAAAGG |
| β-catenin-F |
AGCTTCCAGACACGCTATCAT |
| β-catenin-R |
CGGTACAACGAGCTGTTTCTAC |
| AKT-F |
GAAGCTGAGCCCACCTTTCA |
| AKT-R |
CATCTTGATCAGGCGGTGTG |
| MMP2-F |
TGATCTTGACCAGAATACCATCGA |
| MMP2-R |
GGCTTGCGAGGGAAGAAGTT |
| GAPDH-F |
AATCCCATCACCATCTTCCAG |
| GAPDH-R |
GAGCCCCAGCCTTCTCCAT |
| U6-F |
GCTTCGGCAGCACATATACTAAAAT |
| U6-R |
CGCTTCACGAATTTGCGTGTCAT |
Western blot analysis
Total protein was isolated using RIPA buffer
(Beyotime Institute of Biotechnology, Inc.) and quantified using
the BCA method. The protein sample (50 µg per lane) was separated
on 10% gels using SDS-PAGE, transferred onto PVDF membranes
(MilliporeSigma) and immersed in Tris-buffered saline with Tween-20
containing 5% skimmed milk (Sangon Biotech, Inc.). After
immunoblotting with primary antibodies (1:1,000 dilution) at 4°C
overnight and secondary antibodies (1:1,000 dilution) at room
temperature for 1 h. ECL luminescence reagent (Sangon Biotech, Co.,
Ltd.) was added dropwise onto the PVDF membrane. Bands were
visualized using the Bio-Rad Universal Hood II Gel Doc Imaging
system (Bio-Rad Laboratories, Inc.) and grey values were analyzed
using Image J software version 1.8.0.112 [US National Institutes of
Health (NIH)]. All antibodies used were purchased from Abclonal as
follows: ROCK2 (cat. no. A2395); β-catenin (cat. no. A19657); AKT
(cat. no. A17909); MMP-2 (cat. no. A19080); GAPDH (cat. no.
A19056); and the secondary antibody (cat. no. AS014).
Lentivirus transfection
Target gene fragments were designed and synthesized
based on gene sequences retrieved from GenBank and a lentiviral
recombinant overexpression plasmid was synthesized. The
overexpression plasmid and the empty plasmid were GV367-ROCK2 and
GV367 (GeneChem, Inc.), respectively. The lentiviral plasmid,
packaging vector and envelope vector (GeneChem, Inc.) were mixed at
4:3:2, with a total DNA mass of 20 µg. The mixture was incubated
with 1 ml of Lenti-Easy Packaging Mix (Shanghai GeneChem Co., Ltd.)
for 15 min and Lipofectamine® 2000 for 20 min.
Subsequently, they were added to the culture medium of 293T cells
for 6 h at 37°C; the medium was replaced with fresh medium after 6
h of incubation to continue the culture. Three days later, the 293T
cells were filtered using a 0.45-µM mesh, centrifuged at 70,000 × g
at 4°C for 2 h and the supernatant was collected for detecting
viral titers. RCC cells with 80% of confluence were cultured with
diluted lentiviruses and green fluorescence protein-labeled cells
with a minimum lentivirus transfection rate of 80% at 72 h were
screened. The overexpression efficacy was verified by RT-qPCR.
Colony formation assay
Cells were seeded in a six-well plate
(5×102 cells/well) with 37°C. Cell culture was
terminated when the majority of visible colonies contained >50
cells and they were then fixed with 4% paraformaldehyde at room
temperature for 20 min and stained using 0.2% crystal violet at
room temperature for 10 min. After washing, images of each well
were captured for calculating the number of colonies in three
replicates per sample.
Wound-healing assay
Cells were seeded in a 12-well plate
(1×105 cells/well) and cultivated in serum-free medium
for 12 h. A sterile pipette tip was used to create an artificial
wound in the cell monolayer and cell migration at 24 h was assessed
by calculating the cell-free zone at 24 vs. 0 h using Image J
software version 1.8.0.112 (NIH).
Transwell assay
Cells (5×104 cells) were seeded in the
upper chambers of a Transwell insert (8 µm pore size; BD
Biosciences, Inc.) pre-coated with Matrigel® and 700 µl
of DMEM containing 10% FBS and 1% penicillin and streptomycin was
added to the bottom wells. After 24 h of cell culture, cells were
stained using crystal violet at 37°C for 5 min. After being washed
and air-dried, invaded cells were calculated in five randomly
selected fields per well.
Dual-luciferase reporter assay
Target prediction for ROCK2 3′-UTR and miR-5590-3p
was performed using TargetScan 7.2 (http://www.TargetScan.org/vert_72/). Complementary
sequences in the ROCK2 3′-UTR and miR-5590-3p promoter region were
cloned into the 3′-UTR of pGL3 (Genechem, Inc.) to construct the
wild-type plasmid, pGL3-ROCK2-wt, and the mutant-type plasmid,
pGL3-ROCK2-mut, was generated using the GeneTailer site-directed
mutagenesis kit (Invitrogen; Thermo Fisher Scientific, Inc.). They
were co-transfected into cells with miR-5590-3p mimics or
miR-5590-3p mimic-NC in a 96-well plate for 48 h. Luciferase
activities were measured using the Dual-Luciferase Reporter Gene
Assay Kit (Promega Corporation).
Immunohistochemical detection
The RCC sections were incubated in xylene (15 min,
three times), dehydrated in anhydrous ethanol (5 min, twice), 85%
ethanol (5 min, once) and 75% ethanol (5 min, once), and washed in
ddH2O. Antigen retrieval was performed by pouring
citrate buffer (pH 6.0; Sangon Biotech, Co., Ltd.; cat. no.
E673000) on the sections for 15 min. After incubation in 3%
H2O2 in the dark for 25 min and blocking with
3% BSA (Sangon Biotech, Co., Ltd.; cat. no. E661003) at 37°C for 30
min, the sections were incubated with primary antibodies against
ROCK2 diluted at 1:50 (cat. no. A2395; Abclonal, Inc.) at 4°C
overnight. Next, the sections were washed in PBS (pH 7.4, 5 min,
three times) and incubated with HRP-labeled secondary antibody
diluted at 1:500 (cat. no. AS002; Abclonal, Inc.) at room
temperature for 2 h. The sections were then counterstained with DAB
and hematoxylin for 3 min. After dehydration in ddH2O,
75% ethanol (5 min, once), 85% ethanol (5 min, once), anhydrous
ethanol (5 min, twice) and n-butanol (5 min, once)
sequentially, and permeabilization in xylene (5 min, once), neutral
gum was used for mounting. The positive staining of cells was
observed under a microscope (XSP-8CA; Shanghai Optical Instrument
Co., Ltd.).
Statistical analysis
GraphPad Prism 8.0 (GraphPad Software, Inc.) was
used for statistical analyses. Values are expressed as the mean ±
standard deviation. All data conformed to a normal distribution. In
the cell experiment, comparisons between two groups were performed
using the unpaired t-test, while for comparisons among multiple
groups, one-way analysis of variance followed by Tukey's or
Bonferroni's post-hoc test were used. For the data comparison of
tumor tissues, a paired t-test was used. The correlation analysis
was performed by Spearman's correlation test. P<0.05 was
considered to indicate statistical significance. Each experiment
was performed in triplicate.
Results
Aberrant expression of miR-5590-3p and
ROCK2 in RCC samples and cell lines
RCC samples and paracancerous tissues (~4 cm from
the edge of the cancer and confirmed as non-cancerous by
histological examination) were collected from 6 cases of RCC (4
male and 2 female patients; age range, 47–55 years old) for
isolating total RNA and protein (Table II). The analyses indicated that
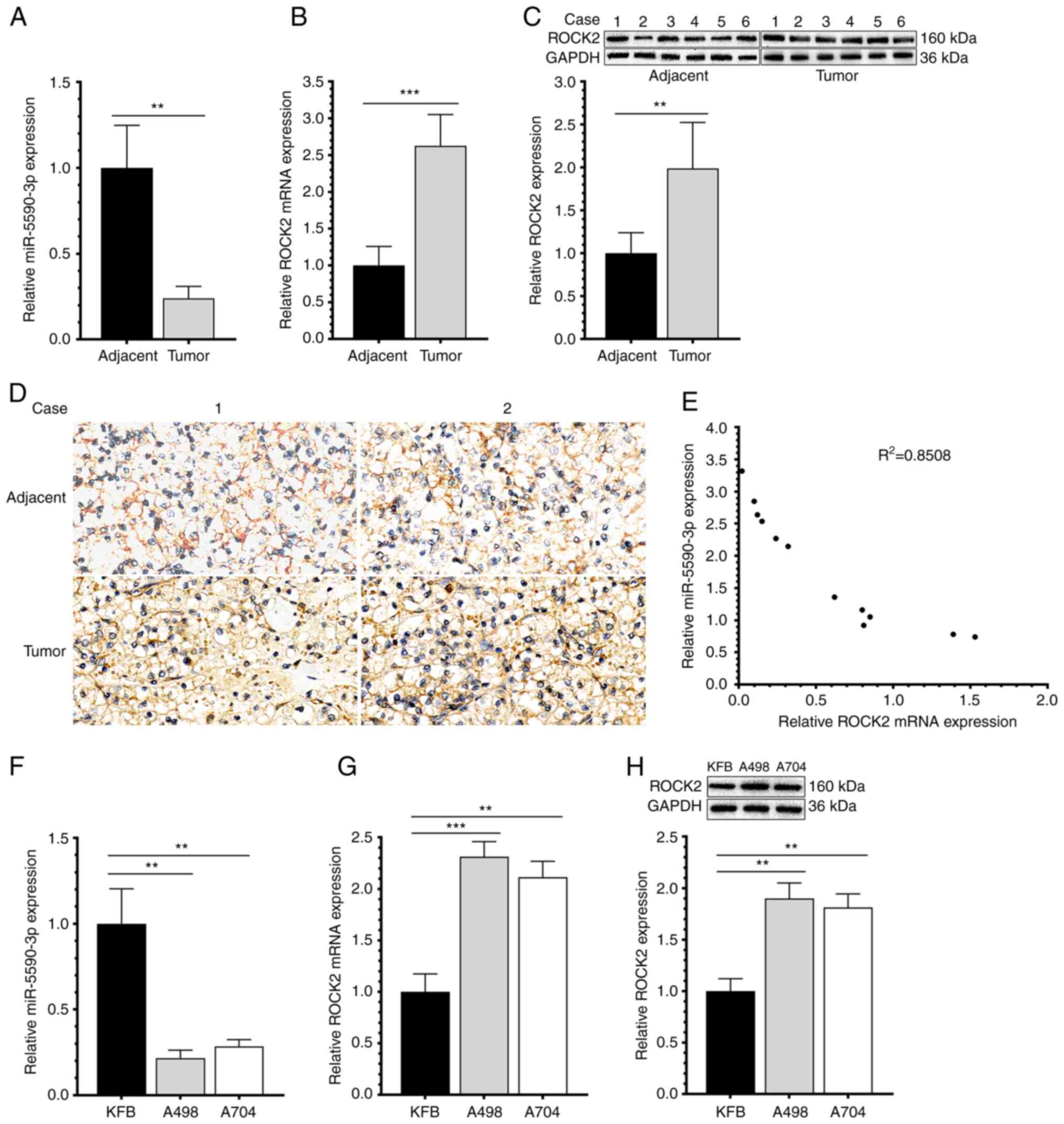
miR-5590-3p was significantly downregulated (Fig. 1A). By contrast, ROCK2 was
upregulated in RCC samples compared to adjacent controls both at
the mRNA and protein levels (Fig. 1B
and C). Immunohistochemical detection of ROCK2 expression in
tumor and adjacent tissues provided similar results (Fig. 1D). Furthermore, the expression of
miR-5590-3p and ROCK2 exhibited a significant negative correlation
(Fig. 1E). Downregulated
miR-5590-3p and upregulated ROCK2 were also detected in RCC cell
lines (Fig. 1F-H). Those results
suggested that miR-5590-3p and ROCK2 are abnormally expressed in
RCC.
 | Table II.Patient information. |
Table II.
Patient information.
| Case no. | Age, years | Sex | Stagea |
|---|
| 1 | 47 | Male | T2N0M0, II |
| 2 | 48 | Male | T2N2cM0, IVb |
| 3 | 52 | Male | T3N2bM0, IVb |
| 4 | 55 | Male | T3N2bM0, IVb |
| 5 | 51 | Female | T2N2cM0, IVb |
| 6 | 54 | Female | T3N2bM0, IVb |
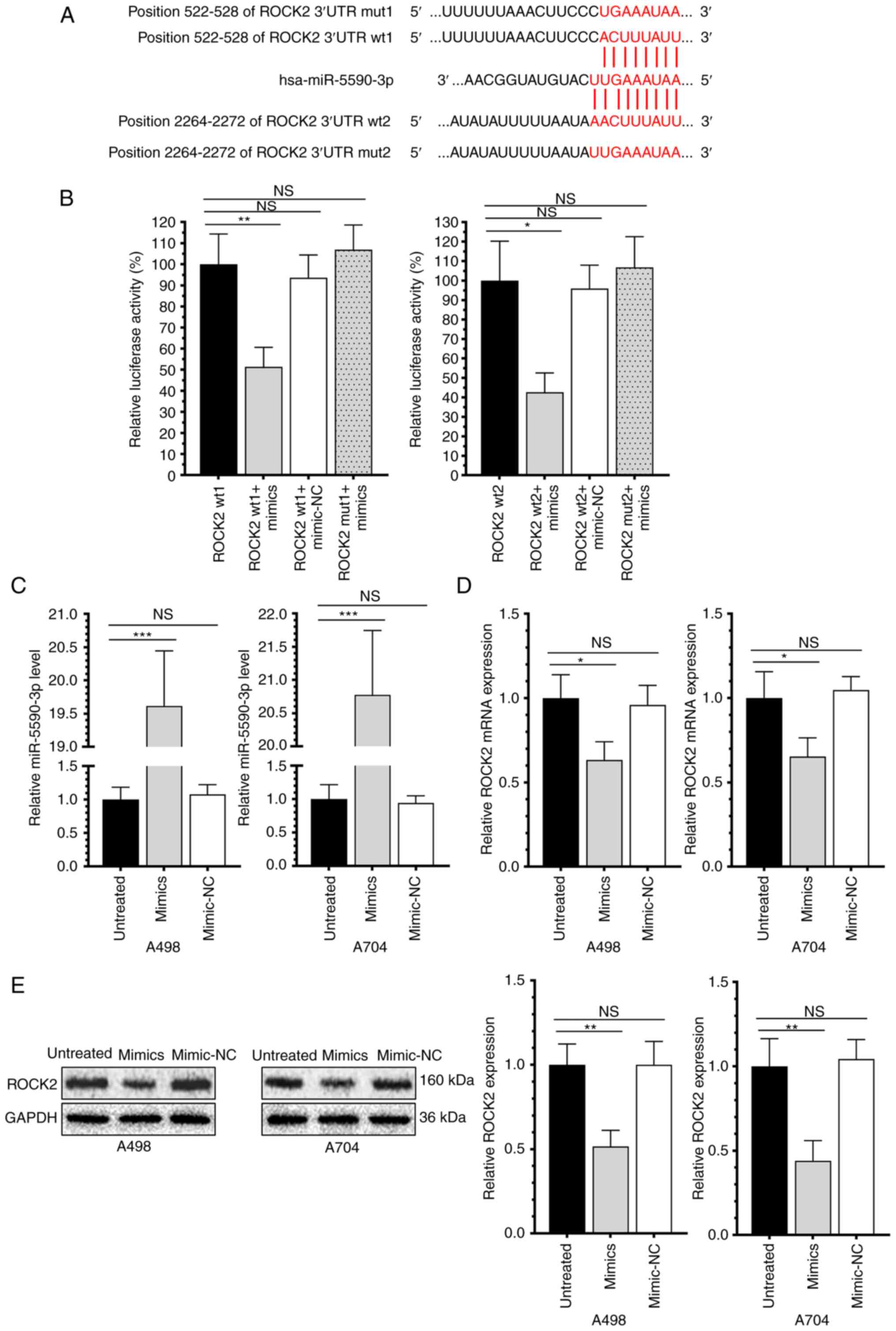
miR-5590-3p targets and downregulates
ROCK2
As predicted by Targetscan 7.1, complementary
binding sites were present in the miR-5590-3p promoter region and
ROCK2 3′-UTR (Fig. 2A).
Subsequently, luciferase activity in cells co-transfected with
pcDNA3.1-ROCK2-wt and miR-5590-3p mimics was only 51.33 and 42.68%
of that in those co-transfected with pcDNA3.1-ROCK2-wt and
miR-5590-3p mimics-NC. Overexpression of miR-5590-3p, however, did
not significantly change the luciferase reporter activity in cells
transfected with pcDNA3.1-ROCK2-mut (Fig. 2B). In RCC cells overexpressing
miR-5590-3p, the mRNA and protein levels of ROCK2 were both
downregulated (Fig. 2C-E). This
suggests that miR-5590-3p directly targets and downregulates ROCK2
in RCC cell lines.
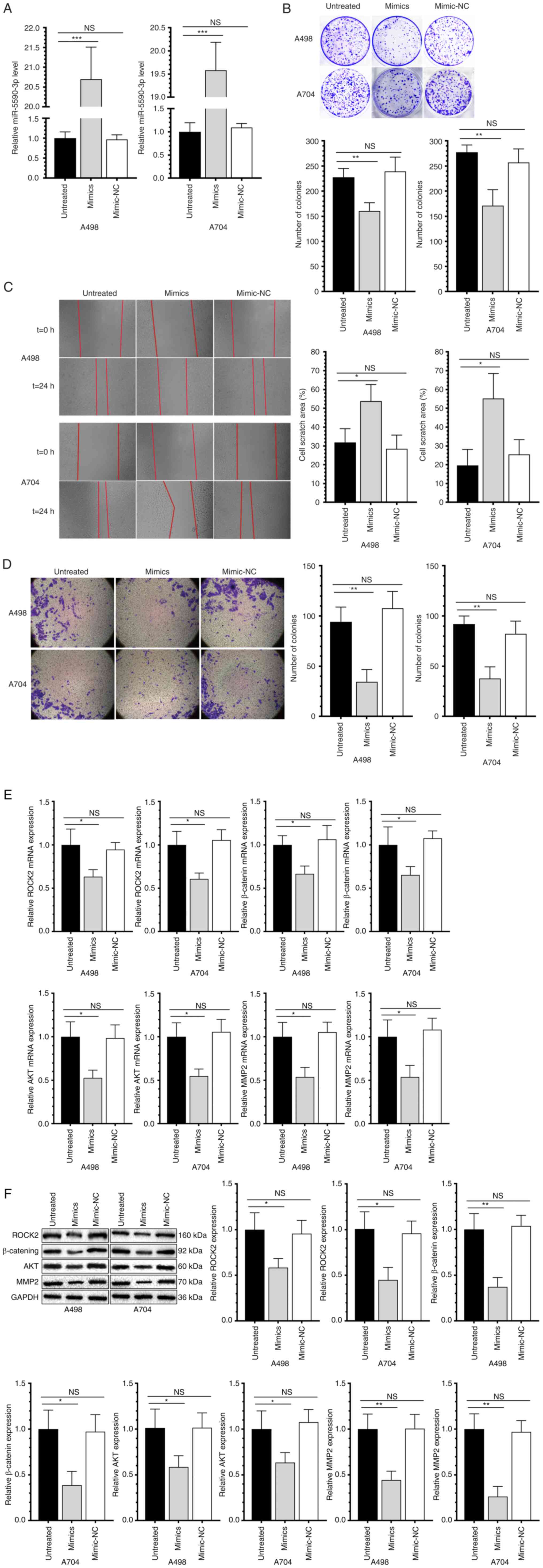
miR-5590-3p suppresses the
proliferative, migratory and invasive capacities of RCC cells
After transfection of miR-5590-3p mimics for 48 h,
cells were collected to examine the overexpression efficacy by
RT-qPCR (Fig. 3A). Compared with
those transfected with NC, RCC cells transfected with miR-5590-3p
mimics had reduced proliferative (Fig.
3B), migratory (Fig. 3C) and
invasive capacities (Fig. 3D). In
addition, the mRNA and protein levels of ROCK2 and β-catenin were
significantly downregulated in RCC cell lines overexpressing
miR-5590-3p. Genes associated with cell proliferation, migration
and invasion, were also examined and it was indicated that AKT and
MMP-2 were also significantly downregulated (Fig. 3E and F). Collectively,
overexpression of miR-5590-3p significantly downregulated
β-catenin, AKT and MMP-2, and suppressed the proliferation and
metastasis of RCC cells.
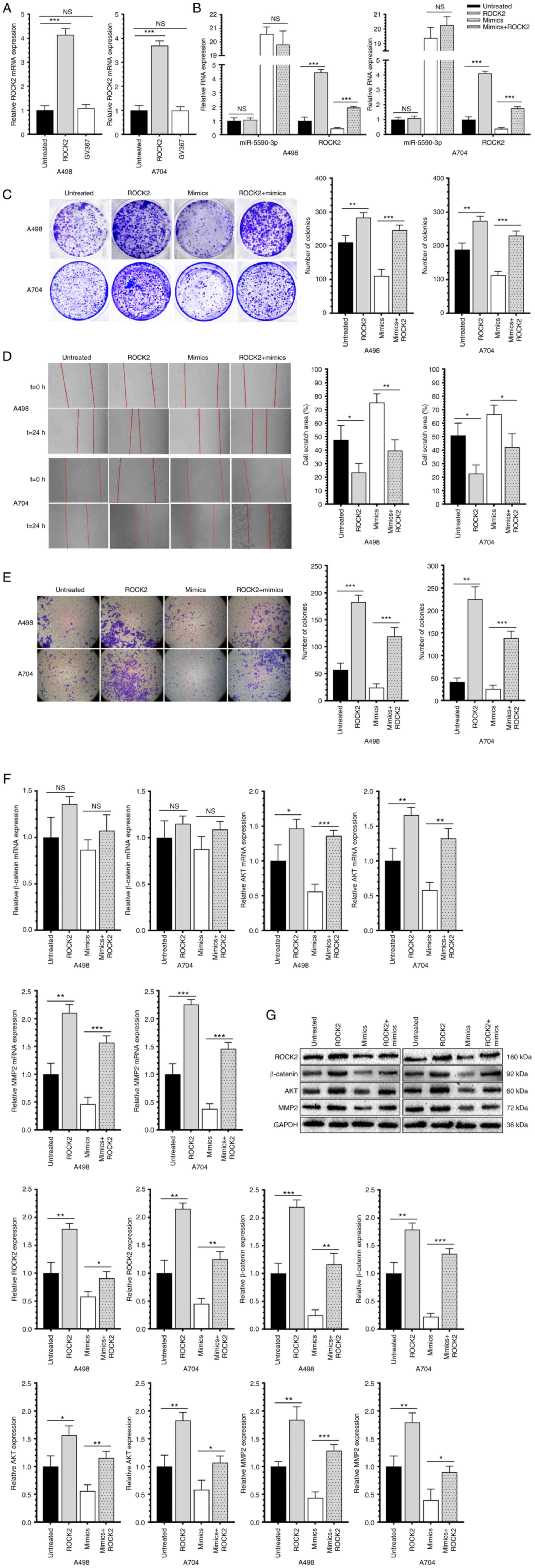
Overexpression of ROCK2 promotes the
proliferative, migratory and invasive capacities of RCC cells and
reverses the regulatory effects of miR-5590-3p
The overexpression efficacy of the lentivirus
GV367-ROCK2 was examined first (Fig.
4A), followed by examination of ROCK2 and miR-5590-3p levels in
RCC cells co-transfected with GV367-ROCK2 and miR-5590-3p mimics
(Fig. 4B). Of note, overexpression
of ROCK2 significantly stimulated the proliferative, migratory and
invasive capacities of RCC cells, and it reversed the inhibitory
effects of miR-5590-3p on the above-mentioned cell behaviors
(Fig. 4C-E). Furthermore,
β-catenin, AKT and MMP-2 were upregulated in RCC cells
overexpressing ROCK2 (Fig. 4F and
G). Therefore, it was indicated that miR-5590-3p downregulated
β-catenin, AKT and MMP-2, and inhibited the proliferation and
metastasis of RCC cells through targeting ROCK2.
Discussion
RCC is the most common malignant tumor type of the
urinary system. RCC may be classified by pathological subtype,
among which clear cell renal cell carcinoma (ccRCC) is the most
prevalent, accounting for 75–80% of RCC cases (22). Approximately 30% of patients with
ccRCC develop metastatic RCC after curative nephrectomy (23) and are the high-risk population for
RCC-associated death (24). The
morbidity and mortality of advanced RCC are high and the five-year
survival is only 18% (25).
Although surgical resection, chemotherapy and radiotherapy are
preferred treatments for patients with RCC, their therapeutic
efficacies are far away from satisfying. Thus, it is of great
significance to seek novel biomarkers in order to facilitate early
diagnosis and improve the prognosis of RCC.
miRNAs are highly conserved endogenous RNAs.
Following a series of complex biological processes, miRNAs form the
RNA-induced silencing complex and bind to the target gene's 3′-UTR
to regulate their post-transcriptional levels (26). miRNAs are highly associated with
tumor cell functions, including proliferation, apoptosis and
migration. The role of miRNAs in the progression of RCC has been
previously reported. Çaykara et al (27) indicated that abnormally
downregulated miR-124 was associated with the tumor stage, tumor
size and neutrophil count in patients with RCC, and restoration of
miR-124 expression may be effective in the treatment of RCC. He
et al (28) reported that
miR-125b was significantly upregulated in RCC tissues, which was
negatively correlated with vitamin D receptor (VDR) levels. Through
mediating VDR, miR-125 is capable of enhancing proliferative and
migratory rates of RCC cells, suggesting miR-125 may be a potential
therapeutic target of RCC. Chen et al (29) determined that miR-26a-5p is lowly
expressed in RCC samples. Overexpression of miR-26a-5p inhibited
the migration, invasion and metastasis of RCC, and induced
apoptosis of RCC cells by targeting E2F7. miR-5590-3p is a recently
discovered miRNA exerting important functions in tumor signaling.
Zhang et al (30) reported
that miR-5590-3p exerts a sponge effect on SOX9-antisense 1 (AS1)
and thus upregulates SOX9, and the activated downstream
Wnt/β-catenin signaling pathway drives the growth and metastasis of
hepatocellular carcinoma through triggering epithelial-mesenchymal
transition (EMT). Wu et al (15) compared miR-5590-3p levels in
gastric cancer tissues with adjacent normal tissues and determined
that it is significantly downregulated in the former. They
additionally validated that miR-5590-3p inhibits gastric cancer
growth by directly targeting DEAD-box helicase 5 in an in
vivo xenograft nude mouse model, as well as in gastric cancer
cell line models in vitro. Yang et al (31) demonstrated that lncRNA FYVE, RhoGEF
and PH domain containing 5-AS1 competitively interacts with
miR-5590-3p, thereby mediating ccRCC cell proliferation and
metastasis by activating ERK/AKT signaling. In the present study,
miR-5590-3p was significantly downregulated in RCC samples compared
with that in normal tissues, and as expected, it was lowly
expressed in RCC cell lines. Transfection of miR-5590-3p mimics
effectively inhibited the proliferative, migratory and invasive
capacities of RCC cells, and downregulated the mRNA and protein
levels of β-catenin, AKT and MMP-2. It was concluded that
miR-5590-3p was important in the progression of RCC and its
overexpression effectively inhibited the proliferation and
metastasis of RCC cells.
ROCK2 is the downstream effector of the Rho family
of GTPases, which is responsible for mediating gene expression by
regulating their activities or phosphorylation (32). ROCK2 is closely linked with
tumorigenesis and tumor development by influencing cancer cell
functions (33), and as a result,
it has become a well-studied target for designing novel anti-cancer
drugs. Multiple studies have detected high expression of ROCK2 in
cancer tissues. Deng et al (34) reported that ROCK2 was upregulated
in osteosarcoma tissues compared to those of adjacent ones, and it
stimulated the malignant growth of osteosarcoma by upregulating
HKII through phosphorylating the PI3K/AKT signaling pathway. Luo
et al (35) discovered that
ROCK2 was upregulated in the bladder cancer cell line T24 under
hypoxic conditions. It inhibited apoptosis and induced
proliferation, migration, invasion and EMT by activating the Wnt
signaling pathway. Compared with non-cancerous tissues, Qiu et
al (36) detected a higher
level of ROCK2 in colorectal cancer (CRC) tissues; the high level
of ROCK2 was significantly associated with metastasis and poor
prognosis of CRC. ROCK2 stabilizes β-catenin by preventing its
ubiquitination, thereby stimulating metastasis of RCC cells. Xu
et al (20) revealed that
ROCK2 is significantly upregulated in clinical samples of RCC.
ROCK2 promotes RCC proliferation by decreasing scavenger receptor
class A member 5 expression through β-catenin/transcription factor
4 signaling. The present study also consistently detected the
upregulation of ROCK2 at both the mRNA and protein levels in RCC
cells. It was indicated that ROCK2 was significant to the
progression of RCC and may be regulated by miR-5590-3p.
Next, the targeting relationship between miR-5590-3p
and ROCK2 was investigated. Through target sequence prediction
using Targetscan 7.1 and validation by a dual-luciferase reporter
assay, miR-5590-3p was confirmed to bind to the 3′-UTR of ROCK2.
Overexpression of miR-5590-3p significantly downregulated the mRNA
and protein levels of ROCK2, suggesting that miR-5590-3p targeted
and downregulated ROCK2, which was able to further downregulate the
expression of β-catenin, AKT and MMP-2. GV367-ROCK2 was
subsequently transfected into RCC cells, resulting in enhanced
proliferative and metastatic capacities. Furthermore,
overexpression of ROCK2 significantly promoted the proliferation,
migration and invasion of RCC cells, and, of note, reversed the
inhibitory effects of miR-5590-3p on the malignant behaviors of RCC
cells. It was concluded that miR-5590-3p directly targeted ROCK2
and this led to a decrease in the expression of β-catenin, AKT and
MMP-2, thereby inhibiting the proliferation and metastasis of
RCC.
In conclusion, miR-5590-3p inhibits the
proliferation, migration and invasion of RCC cells by targeting
ROCK2, which is a potential molecular biomarker and therapeutic
target of RCC.
Acknowledgements
Not applicable.
Funding
This project was funded by the National Natural Science
Foundation of China (grant no. 81760458).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QL, ZH and AZ contributed to the study conception
and design, and the acquisition of data. WG, ZH, YZ, XZ and FG
contributed to the analysis and interpretation of the data. QL and
ZH confirm the authenticity of all the raw data. All authors
contributed to the writing of the article and have read and
approved the manuscript.
Ethics approval and consent to
participate
This study was approved by the Second Affiliated
Hospital of Nanchang University Medical Research Ethics Committee
(Nanchang, China; approval no. 2017-100). Written informed consent
was obtained from each participant prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Koul H, Huh JS, Rove KO, Crompton L, Koul
S, Meacham RB and Kim FJ: Molecular aspects of renal cell
carcinoma: A review. Am J Cancer Res. 1:240–254. 2011.PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ahrens M, Scheich S, Hartmann A and
Bergmann L; IAG-N Interdisciplinary Working Group Kidney Cancer of
the German Cancer Society, : Non-clear cell renal cell
carcinoma-pathology and treatment options. Oncol Res Treat.
42:128–135. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Capitanio U, Bensalah K, Bex A, Boorjian
SA, Bray F, Coleman J, Gore JL, Sun M, Wood C and Russo P:
Epidemiology of Renal Cell Carcinoma. Eur Urol. 75:74–84. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bruchbacher A, Lemberger U, Hassler MR,
Fajkovic H and Schmidinger M: PD1/PD-L1 therapy in metastatic renal
cell carcinoma. Curr Opin Urol. 30:534–541. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bedke J, Albiges L, Capitanio U, Giles RH,
Hora M, Lam TB, Ljungberg B, Marconi L, Klatte T, Volpe A, et al:
Updated European Association of Urology guidelines on renal cell
carcinoma: Nivolumab plus cabozantinib joins immune checkpoint
inhibition combination Therapies for treatment-naive metastatic
clear-cell renal cell carcinoma. Eur Urol. 79:339–342. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang L, Zou X, Zou J and Zhang G: A review
of recent research on the role of MicroRNAs in renal cancer. Med
Sci Monit. 27:e9306392021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maher ER: Hereditary renal cell carcinoma
syndromes: Diagnosis, surveillance and management. World J Urol.
36:1891–1898. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Testa JR, Cheung M, Pei J, Below JE, Tan
Y, Sementino E, Cox NJ, Dogan AU, Pass HI, Trusa S, et al: Germline
BAP1 mutations predispose to malignant mesothelioma. Nat Genet.
43:1022–1025. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schmidt L, Duh FM, Chen F, Kishida T,
Glenn G, Choyke P, Scherer SW, Zhuang Z, Lubensky I, Dean M, et al:
Germline and somatic mutations in the tyrosine kinase domain of the
MET proto-oncogene in papillary renal carcinomas. Nat Genet.
16:68–73. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang G, Lai Y, Pan X, Zhou L, Quan J,
Zhao L, Li Z, Lin C, Wang J, Li H, et al: Tumor suppressor
miR-33b-5p regulates cellular function and acts a prognostic
biomarker in RCC. Am J Transl Res. 12:3346–3360. 2020.PubMed/NCBI
|
|
13
|
Chen FY, Zhou ZY, Zhang KJ, Pang J and
Wang SM: Long non-coding RNA MIR100HG promotes the migration,
invasion and proliferation of triple-negative breast cancer cells
by targeting the miR-5590-3p/OTX1 axis. Cancer Cell Int.
20:5082020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luo ZF, Peng Y, Liu FH, Ma JS, Hu G, Lai
SL, Lin H, Chen JJ, Zou GM, Yan Q and Sui WG: Long noncoding RNA
SNHG14 promotes malignancy of prostate cancer by regulating with
miR-5590-3p/YY1 axis. Eur Rev Med Pharmacol Sci. 24:4697–4709.
2020.PubMed/NCBI
|
|
15
|
Wu N, Han Y, Liu H, Jiang M, Chu Y, Cao J,
Lin J, Liu Y, Xu B and Xie X: MiR-5590-3p inhibited tumor growth in
gastric cancer by targeting DDX5/AKT/m-TOR pathway. Biochem Biophys
Res Commun. 503:1491–1497. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang Y, Dong MH, Hu HM, Min QH and Xiao L:
LncRNA FGD5-AS1/miR-5590-3p axis facilitates the proliferation and
metastasis of renal cell carcinoma through ERK/AKT signalling. Eur
Rev Med Pharmacol Sci. 24:8756–8766. 2020.PubMed/NCBI
|
|
17
|
Sharma P and Roy K: ROCK-2-selective
targeting and its therapeutic outcomes. Drug Discov Today.
25:446–455. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei L, Surma M, Shi S, Lambert-Cheatham N
and Shi J: Novel insights into the roles of rho kinase in cancer.
Arch Immunol Ther Exp (Warsz). 64:259–278. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
de Sousa GR, Vieira GM, das Chagas PF,
Pezuk JA and Brassesco MS: Should we keep rocking? Portraits from
targeting Rho kinases in cancer. Pharmacol Res. 160:1050932020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu Z, Hong Z, Ma M, Liu X, Chen L, Zheng
C, Xi X and Shao J: Rock2 promotes RCC proliferation by decreasing
SCARA5 expression through β-catenin/TCF4 signaling. Biochem Biophys
Res Commun. 480:586–593. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Marchetti A, Rosellini M, Mollica V, Rizzo
A, Tassinari E, Nuvola G, Cimadamore A, Santoni M, Fiorentino M,
Montironi R and Massari F: The molecular characteristics of
non-clear cell renal cell carcinoma: What's the story morning
glory? Int J Mol Sci. 22:62372021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sharma R, Kadife E, Myers M, Kannourakis
G, Prithviraj P and Ahmed N: Determinants of resistance to VEGF-TKI
and immune checkpoint inhibitors in metastatic renal cell
carcinoma. J Exp Clin Cancer Res. 40:1862021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mlcochova H, Machackova T, Rabien A,
Radova L, Fabian P, Iliev R, Slaba K, Poprach A, Kilic E, Stanik M,
et al: Epithelial-mesenchymal transition-associated microRNA/mRNA
signature is linked to metastasis and prognosis in clear-cell renal
cell carcinoma. Sci Rep. 6:318522016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bhat S: Role of surgery in
advanced/metastatic renal cell carcinoma. Indian J Urol.
26:167–176. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Winter J, Jung S, Keller S, Gregory RI and
Diederichs S: Many roads to maturity: MicroRNA biogenesis pathways
and their regulation. Nat Cell Biol. 11:228–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Çaykara B, Ozturk G, Alsaadoni H,
Otunctemur A and Pence S: Evaluation of MicroRNA-124 expression in
renal cell carcinoma. Balkan J Med Genet. 23:73–78. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He X, Liao S, Lu D, Zhang F, Sun Y and Wu
Y: MiR-125b promotes migration and invasion by targeting the
vitamin D receptor in renal cell carcinoma. Int J Med Sci.
18:150–156. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cheng C, Guo L, Ma Y, Wang Z, Fan X and
Shan Z: Up-Regulation of miR-26a-5p Inhibits E2F7 to regulate the
progression of renal carcinoma cells. Cancer Manag Res.
12:11723–11733. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang W, Wu Y, Hou B, Wang Y, Deng D, Fu Z
and Xu Z: A SOX9-AS1/miR-5590-3p/SOX9 positive feedback loop drives
tumor growth and metastasis in hepatocellular carcinoma through the
Wnt/β-catenin pathway. Mol Oncol. 13:2194–2210. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang Y, Dong MH, Hu HM, Min QH and Xiao L:
LncRNA FGD5-AS1/miR-5590-3p axis facilitates the proliferation and
metastasis of renal cell carcinoma through ERK/AKT signalling. Eur
Rev Med Pharmacol Sci. 24:8756–8766. 2020.PubMed/NCBI
|
|
32
|
Du Y, Lu S, Ge J, Long D, Wen C, Tan S,
Chen L and Zhou W: ROCK2 disturbs MKP1 expression to promote
invasion and metastasis in hepatocellular carcinoma. Am J Cancer
Res. 10:884–896. 2020.PubMed/NCBI
|
|
33
|
Deng X, Yi X, Huang D, Liu P, Chen L, Du Y
and Hao L: ROCK2 mediates osteosarcoma progression and TRAIL
resistance by modulating O-GlcNAc transferase degradation. Am J
Cancer Res. 10:781–798. 2020.PubMed/NCBI
|
|
34
|
Deng B, Deng J, Yi X, Zou Y and Li C:
ROCK2 promotes osteosarcoma growth and glycolysis by up-regulating
HKII via Phospho-PI3K/AKT signalling. Cancer Manag Res. 13:449–462.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Luo J, Lou Z and Zheng J: Targeted
regulation by ROCK2 on bladder carcinoma via Wnt signaling under
hypoxia. Cancer Biomark. 24:109–116. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Qiu Y, Yuan R, Zhang S, Chen L, Huang D,
Hao H and Shao J: Rock2 stabilizes β-catenin to promote tumor
invasion and metastasis in colorectal cancer. Biochem Biophys Res
Commun. 467:629–637. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Amin MB, Edge S, Greene F, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, et al: AJCC Cancer Staging Manual. 8th edition.
Springer; Berlin, Germany: pp. 79–81. 2017, PubMed/NCBI
|