Introduction
Sarcoma is a complex group of malignant diseases
consisting of >100 disease subtypes, most of which vary
according to the tissue and cell context, which include bone,
cartilage, kidney, adipose, colon, connective, subcutaneous and
other soft tissues. The prevalence of sarcoma varies across
different subgroups, but its overall prevalence among all
malignancies worldwide is <1% in adults and 6% in teenagers.
Among all subtypes, soft tissue sarcomas (STSs) are likely to have
a higher incidence than other subtypes. STSs can affect any part of
the human body, with the most frequent locations being the upper
and lower limbs (1). The distant
metastases of STS generally occur in the lung, bone and liver,
while regional lymph node metastasis is less common (2).
The etiology of sarcoma is largely unknown, but
evidence suggests that genetic and epigenetic components contribute
significantly to disease phenotypes (3–5).
During the past 10 years, several genetic evaluations have revealed
that nucleotide variations, germline or somatic mutations, certain
indels or deletions, copy number variations and chromosomal
translocations (CNVs) are associated with sarcoma (6). For example, mutations in the tumor
protein 53, ATRX chromatin remodeler and RB transcriptional
corepressor 1 genes are commonly detected in all subtypes. These
three genes play roles in the regulation of the cell cycle and
chromosomal stability. Some CNVs in cyclin dependent kinase
inhibitor 2, CDK4 and MDM2 proto-oncogene also contribute to
the disease manifestations, according to a recent genetic
architecture report of sarcoma published by The Cancer Genome Atlas
(TCGA) (7).
Receptor interacting serine/threonine kinase 3
(RIPK3) encodes a receptor-interacting protein that acts a
regulator of necroptosis and apoptosis (8). RIPK3 has been implicated to
serve a critical role in several inflammatory disorders, including
bowel diseases, psoriasis, systemic inflammatory response syndrome
and severe cancers (9–11). Its capacity to activate necroptosis
through the inflammasome has been confirmed by several research
groups. However, its function in sarcoma remains largely unknown.
In the current study, by retrieving the TCGA dataset, a
comprehensive evaluation of the association of RIPK3
expression level with survival, transcriptome alteration,
signalling pathways, immune checkpoint therapy and immune cell
infiltration of sarcoma was performed. In addition, the regulatory
effect of RIPK3 on the immune checkpoint gene hepatitis A
virus cellular receptor 2 (HAVCR2) was investigated.
Materials and methods
Data retrieval
Data were retrieved from the Gene Expression Omnibus
dataset GSE49972 and TCGA dataset (accession no. phs000178.v11.p8).
GSE49972 includes data for 22 clear cell sarcoma of the kidney
tissues and 10 non-neoplastic kidney tissues, while phs000178
includes 260 samples of a wide range of sarcoma subtypes and a
large amount of comprehensive clinical data. In addition, RNA-seq
data were extracted from the Therapeutically Applicable Research To
Generate Effective Treatments (TARGET) program (https://target-data.nci.nih.gov/Public/OS/mRNA-seq/).
In GSE49972 the expression levels were evaluated using the
HumanHT-12 v4 Expression BeadChip (12). Transcriptome data in dataset
phs000178 were generated using a HiSeq 2000 sequencing system with
76 bp paired-end parallel sequencing. The RNA-seq data of 89
sarcoma samples in TARGET were assayed by parallel sequencing with
an Illumina Genome Analyzer IIx, Illumina HiSeq 2000 or Illumina
MiSeq sequencer.
Survival and prognosis
Transcriptome read levels of RIPK3, survival,
sex and age were extracted from TCGA (https://portal.gdc.cancer.gov/) or the National Center
for Biotechnology (NCBI) databank (https://www.ncbi.nlm.nih.gov). Individuals were
divided into high and low expression groups
(RIPK3higher and RIPK3lower)
according to the expression level of RIPK3. The Kaplan-Meier
algorithm was used to construct survival curves for the
RIPK3 high and low groups, and the nonparametric log-rank
test was used to calculate the statistical significance of
differences in survival. HR represents chance of death occurring in
the RIPK3higher group compared with the
RIPK3lowergroup. HR >1 indicates that
RIPK3 is a risk factor, while HR <1 indicates that
RIPK3 has a protective role. The median survival time was
set at the time of 50% survival for each group. Time-dependent
receiver operating characteristic (ROC) analysis was carried out to
predict the accuracy of the RIPK3 prediction. In this analysis, the
larger the area under the curve (AUC) the more robust the
prediction model.
Univariate and multivariate Cox proportional hazards
regression models were used to determine the independent prognostic
factors using the survminer_0.4.9 R package (https://github.com/kassambara/survminer). The
forestplot_2.0.1 package (https://cran.r-project.org/web/packages/forestplot/index.html)
was used to generate the P-value, HR and 95% CI of each variable. A
nomogram was created based on the results of the multivariate Cox
proportional hazards analysis to predict the 1-, 3- and 5-year
overall recurrence, which was indicated by the points associated
with each risk factor through the rms_6.3–0 R package (https://cran.r-project.org/web/packages/rms/index.html).
Identification of differentially
expressed genes
Genome-wide mRNA expression data were obtained from
the phs000178 and TARGET datasets. The expression levels between
the RIPK3higher and RIPK3lower
groups were compared using the limma_3.40.2 R package (http://www.bioconductor.org/packages/release/bioc/html/limma.html).
Significant differentially expressed genes were defined as having a
fold change (FC) of >2 or <-2 and false discovery rate (FDR)
adjusted P<0.05. A volcano plot was constructed with
log2(FC) on the x-axis against
-log10(FDR-adjusted P-value), in which each dot
represents a single individual. A heatmap was generated with the
top 50 up- and top 50 downregulated genes. For the differentially
expressed genes, Kyoto Encyclopedia of Genes and Genomes (KEGG) and
Gene Ontology (GO) analyses were performed using the
clusterProfiler_3.18.0 R package (https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html).
GO annotation was divided into three separate parts: Molecular
function, biological pathways and cellular components.
Cell culture and reverse transcription
(RT)-PCR
The KHOS and 143B human osteosarcoma cell lines were
gifts from the American Type Culture Collection cell bank. The cell
lines were authenticated by STR genotyping and comparison with the
relevant reference data in Cellosaurus (https://web.expasy.org/cellosaurus/). Cells were
cultured in a humidified incubator at 37°C with 5% CO2
in DMEM (Gibco; Thermo Fisher Scientific, Inc) supplied with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.), 0.015 mg/ml
5-bromo-2′-deoxyuridine, 100 U/ml penicillin and 100 µg/ml
streptomycin. Cells were harvested when they reached 70–80
confluence in 15-cm dishes. GSK872 at concentrations of 100 nM and
1, 2, 4, 8 and 10 µM was added to the cultured cells and incubated
for 2 h at 37°C. The HAVCR2 and GAPDH expression of
GSK872-treated cells were compared with untreated cells as a
control. The cells were then harvested and the expression levels of
HAVCR2 and GAPDH were assayed.
Total RNA was extracted from the cells with the
RNeasy Protect Mini Kit (Qiagen) according to the manufacturer's
protocols. Agarose gel electrophoresis was used to check the
integrity of the RNA. The RNA was then reverse transcribed with the
Third-Generation Reverse Transcription Master Mix according to the
manufacturer's instructions (MR05001S; Monad Biotech Co., Ltd.).
Quantitative expression primers for HAVCR2 were designed
using online tools from the Harvard website (https://pga.mgh.harvard.edu/primerbank/index.html).
The forward and reverse primer sequences were as follows. HAVCR2
forward CTGCTGCTACTACTTACAAGGTC and reverse,
GCAGGGCAGATAGGCATTCT. The forward primer was located at positions
40–62 and the reverse primer at positions 95–114 of the original
HAVCR2 mRNA sequence (accession no. NM_032782.5). The
primers for the reference gene were: GAPDH forward,
GGAGCGAGATCCCTCCAAAAT and reverse, GGCTGTTGTCATACTTCTCATGG. The
final content of the qPCR product was 1X master mix, 0.2 µM forward
and reverse primers, 1X low ROX dye, 50 ng cDNA and nuclease-free
water to a total volume of 20 µl. The PCR steps were set as
follows: Initial denaturation, 95°C for 10 min; denaturation, 95°C
for 10 sec; and annealing and extension, 60°C for 30 sec. There
were 40 cycles of denaturation, annealing and extension. RT-PCR
analysis was performed using an Applied Biosystems 7900HT Fast
Real-Time PCR System.
RIPK3 and immunity
Immune checkpoint genes were selected from
high-impact publication (13–15).
The xCell algorithm combined bulk RNA-seq data as the sum of the
expression in several single cell types in these samples
(https://xcell.ucsf.edu). Then, reference gene
expression profiles were established for each major
tumor-infiltrating immune cell type, i.e., CD4 T+, CD8
T+, B, natural killer (NK), neutrophils and macrophages
(16). Spearman's correlation
analysis was used to determine the correlation between quantitative
variables and RIPK3 expression. Tumor Immune Dysfunction and
Exclusion (TIDE) scores were used to evaluate immune checkpoint
blockade (ICB) therapy response. The TIDE scores were based on 189
human cancer studies and 33,197 samples (http://tide.dfci.harvard.edu/download/).
Statistical analysis
All statistical analyses were performed with R
project version 4.0.3 (https://www.r-project.org). The ggrisk_1.3 (https://cran.r-project.org/web/packages/ggrisk/index.html),
survival_3.3-1 (https://cran.r-project.org/web/packages/survival/index.html),
survminer, timeROC_0.4 (https://cran.r-project.org/web/packages/timeROC/index.html)
and rms packages were used for survival and prognosis analysis.
Differences among multiple groups were analyzed using one-way ANOVA
followed by Dunnett's post hoc test. Differences between two groups
were analyzed using Wilcoxon's rank-sum test or two-sample
(unpaired) t-tests.
Results
Higher RIPK3 is associated with
improved survival in sarcoma
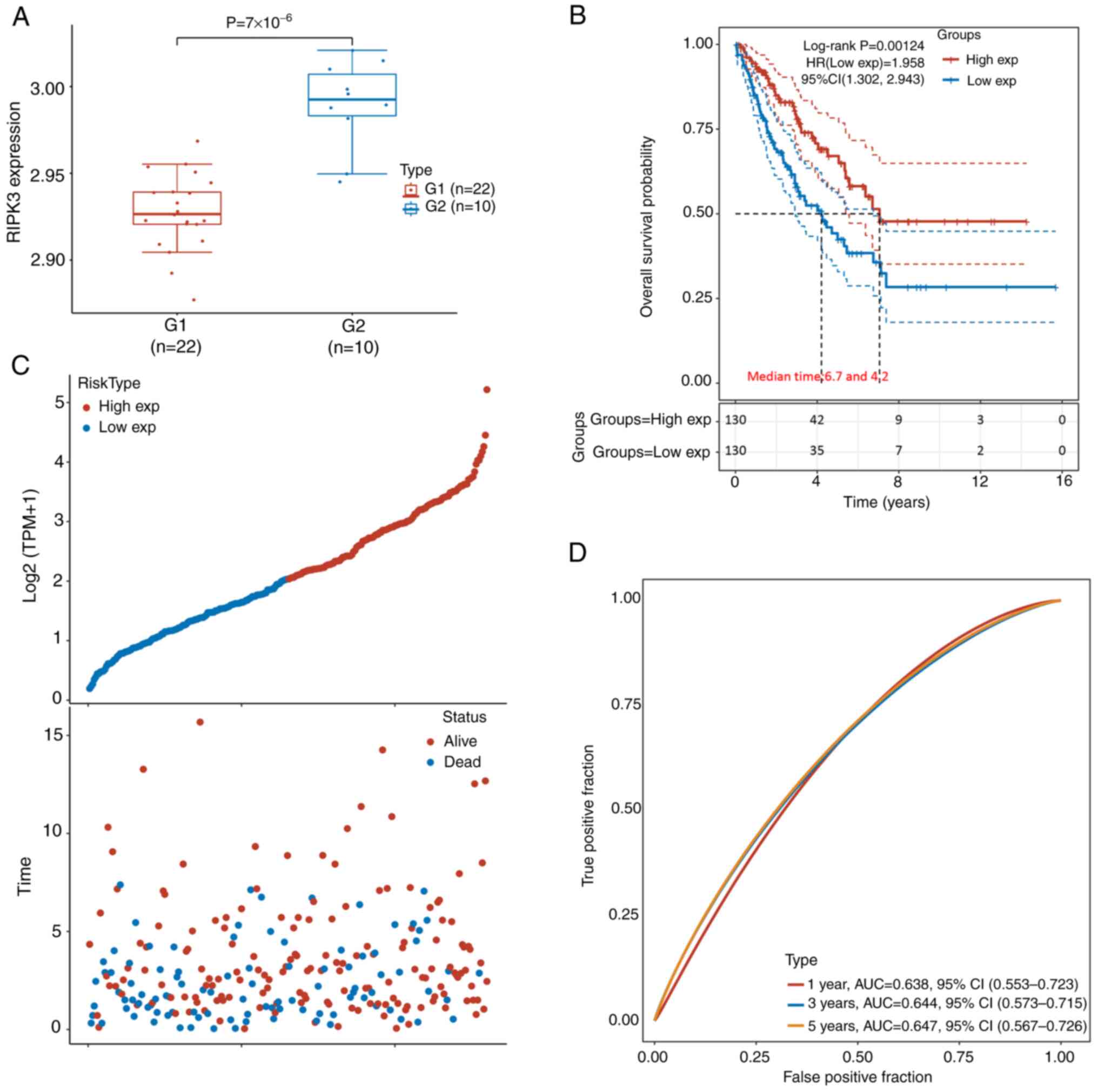
Expression data were retrieved from GSE49972,
including data for 22 kidney sarcomas, 6 adult normal nonneoplastic
kidneys and 4 fetal samples. The expression of RIPK3 was
significantly lower in the sarcoma patient group than in the normal
group (P=7×10−6; Fig.
1A). Due to the relatively limited sample size in this dataset,
other datasets with larger sample sizes were also searched for.
Under TCGA accession number phs000178, a transcriptome dataset of
260 patients with sarcoma was found. As the expression data of only
two normal tissues were available in this dataset, it was not
possible to compare the differences between case and normal groups.
The transcripts per million expression datasets were retrieved and
plotted against survival time and survival status for all patients
(Fig. 1C). The samples were then
classified into two groups: RIPK3higher (above
the median expression level) and RIPK3lower
(below the median expression level). The Kaplan-Meier survival
curve indicated that patients in the RIPK3higher
survived for a longer time than those in the
RIPK3lowe group, with median survival times of
6.7 and 4.2 years, respectively [log-rank P=0.0012, HR(low
exp)=1.958, 95% CI (1.302, 2.943); Fig. 1B)]. The AUC values were 0.64, 0.64
and 0.65 for 1 year, 3 years and 5 years, respectively (Fig. 1D). A similar trend was also
identified using data from the TARGET database, which showed that
patients in the RIPK3higher group were likely to
survive for a longer time than those in the
RIPK3lower group (median 9.2 vs. 7.2 years,
respectively; Fig. S1), although
the difference in survival between the groups was not found to be
statistically significant. Since other RIPKs could
potentially be involved in the necroptosis and apoptosis system,
whether RIPK1, RIPK2 and RIPK4 were associated with
the overall survival probability was also examined. However, there
was no evidence to suggest that these other RIPKs were
associated with improved or worse survival (Fig. S2). These data indicate that
RIPK3 may be associated with longer survival in sarcoma.
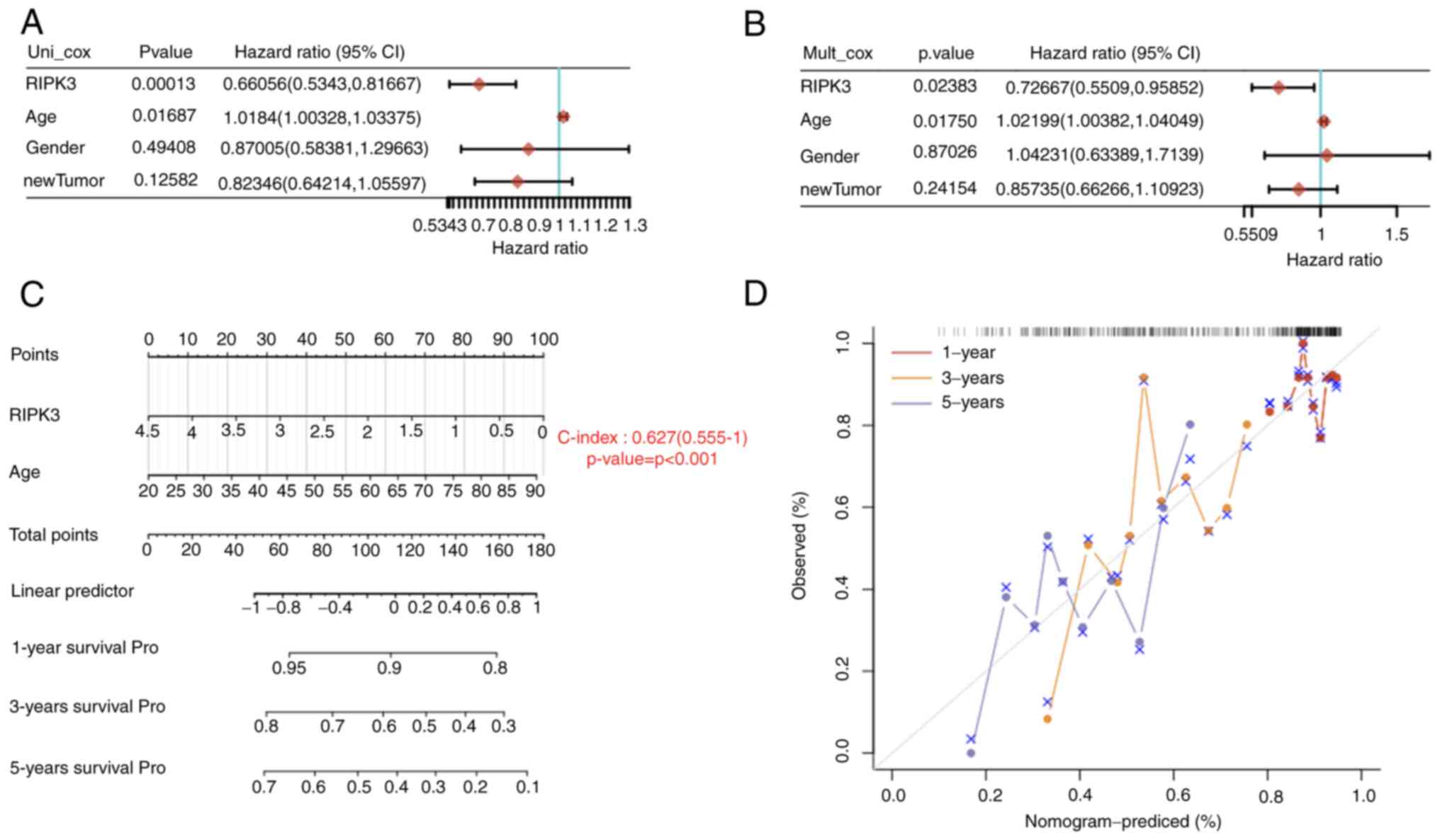
To further confirm the aforementioned findings,
prognostic analysis was performed for the RIPK3 gene, using
age, sex and new tumor type as covariates. The results suggested
that RIPK3 may independently protect patients, following
analysis using univariate and multivariate Cox regression models
(Puni-cox=1.3×10−4, HR=0.66;
Pmulti-cox=0.024, HR=0.73; Fig. 2A and B). Based on the findings of
the multivariate Cox proportional hazards analysis, a nomogram was
built to predict the 1-, 3- and 5-year overall recurrence of each
individual based on the assignment of points associated with each
risk factor and the concordance index (C-index) was used to
evaluate the predictive model. The C-index of 0.63 indicated a good
predictive outcome (P<0.001; Fig.
2C and D). These data suggest that higher RIPK3 may be a
protective factor that is associated with an improved
prognosis.
Expression profile of the
RIPK3higher and RIPK3lower groups
Exploration of the expression landscape of the
RIPK3higher and RIPK3lower
groups will improve understanding of the functional role of
RIPK3 in sarcoma development. Therefore, the differential
expression profiles were compared between the
RIPK3higher and RIPK3lower
groups from the phs000178 dataset. Applying stringent statistical
criteria of a FC of >2 or <-2 and FDR adjusted P<0.05, a
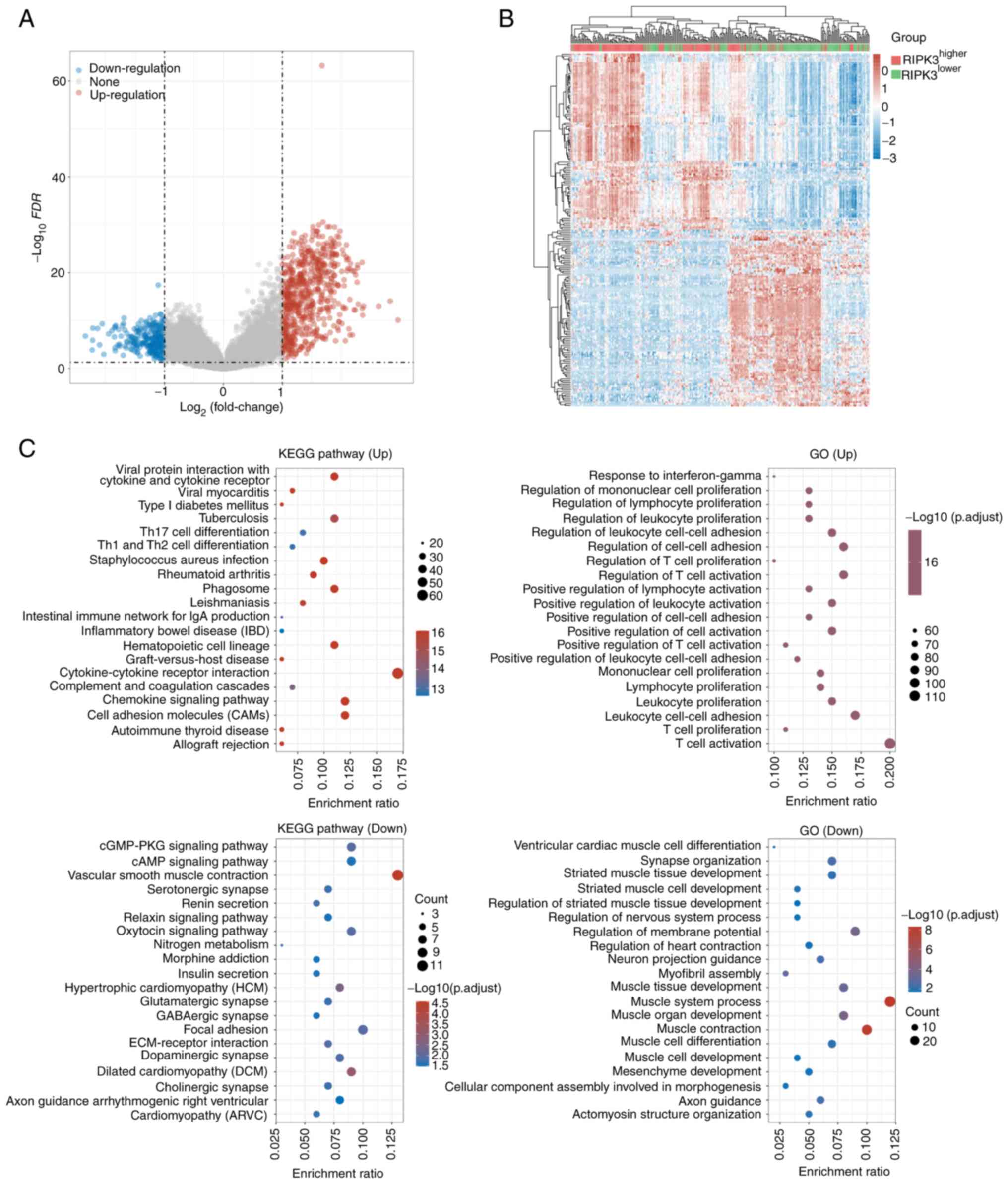
total of 863 differentially expressed genes were identified,
including 603 up- and 260 downregulated genes (Fig. 3A, Table SI). Notably, the number of genes
with upregulated expression was 2.3-fold that of genes with
downregulated expression. For the 50 fifty up- and downregulated
differentially expressed genes, 74.1% (192/259) of the sarcoma
samples were successfully classified into
RIPK3higher and RIPK3lower
groups (Fig. 3B). The top up- and
downregulated genes were transmembrane channel like 8 (TMC8;
FDR-adjusted P=2.62×10−31, log(FC)=1.7) and
transmembrane protein 97 (TMEM97; FDR-adjusted
P=3.91×10−18, log(FC)=−1.1), respectively. To confirm
these findings, the expression of these genes in the TARGET dataset
was determined, and the results revealed that the expression of
TMC8 was significantly increased while the expression of
TMEM97 was decreased in the RIPK3higher
group compared with the RIPK3lower group
(Fig. S3). TMC8 and
TMEM97 have both been implicated in cancer pathogenesis
(17,18); however, they have not previously
been identified as having an association with sarcoma. Therefore,
these are suggested new targets that deserve further investigation
in the future.
KEGG and GO functional annotation of the 603 up- and
260 downregulated genes revealed several signaling pathways that
may be involved in RIPK3-associated disease pathogenesis.
For the upregulated genes, 68 signaling pathways showed strong
enrichment against background with the pathway hsa04640
‘hematopoietic cell lineage’ having the lowest P-value
(GeneRatio=0.11, FDR-corrected P=3.49×10−24). This
pathway contains several cell surface markers (CD2, CD4, CD5,
CD7, CD14, CD33, CD37, CD55 and CD1C) and major
histocompatibility-related haplotypes [human leukocyte antigen
(HLA)-DPB1, HLA-DRB1, HLA-DRA/HLA-DPA1, HLA-DOA,
HLA-DQA1, HLA-DRB5, HLA-DQB1, HLA-DOB and HLA-DQA2].
Other top pathways included hsa04060 ‘cytokine-cytokine receptor
interaction’ and hsa04514 ‘cell adhesion molecules’ (Table SII). The 260 downregulated genes
were enriched in 25 pathways, with the top one being hsa04270
‘vascular smooth muscle contraction’ (GeneRatio=0.13, FDR-corrected
P=2.97×10−5; Fig. 3C).
To further validate these findings, the expression profiles of the
RIPK3higher and RIPK3lower
groups in the TARGET dataset were checked. A similar expression
pattern was observed, which verified several of the enriched
pathways or GO terms, particularly for the top pathway hsa04060
‘cytokine-cytokine receptor interaction’ and the GO terms ‘response
to interferon-gamma’ and ‘T cell activation’ (Table SIII; Fig. S4). These data indicate that
RIPK3 may modulate gene expression and immune signaling
pathways in sarcoma, reflecting some key features of sarcoma
etiology.
RIPK3 is positively associated with
HAVCR2
Since RIPK3 is indicated to be involved in
cytokine-cytokine receptor interactions and to have a potential
role in regulation of the immune response, the correlations between
RIPK3 and several immune checkpoint genes were evaluated.
First, the expression levels of seven immune checkpoint genes
[sialic acid binding Ig like lectin 15 (SIGLEC15), CD274,
HAVCR2, T cell immunoreceptor with Ig and ITIM domains
(TIGIT), cytotoxic T-lymphocyte associated protein 4
(CTLA4), lymphocyte-activation 3 and programmed cell death 1
ligand 2) were extracted. Spearman's correlation analysis showed
that RIPK3 was most strongly correlated with HAVCR2,
TIGIT and CTLA4 in the TCGA dataset (correlation
coefficient=0.63, 0.59 and 0.47, P=2.00×10−14,
6.71×10−16 and 3.21×10−6, respectively,
Fig. 4A and B). Evaluation of the
correlation between the above three genes and RIPK3 in the
TARGET dataset validated the positive correlation of HAVCR2
with RIPK3 (correlation coefficient=0.41,
P=3.50×10−5; Fig. 4C and
D).
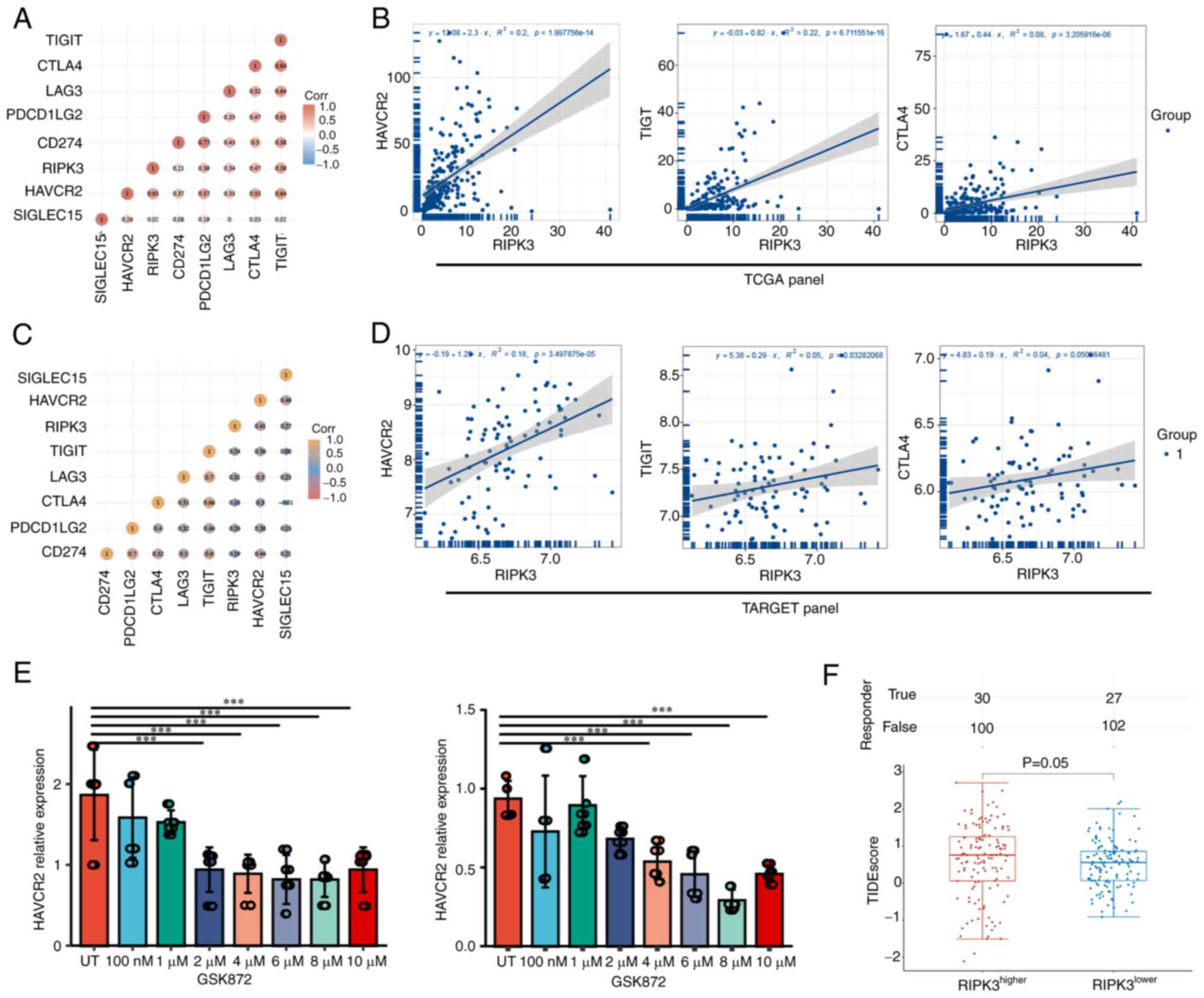 | Figure 4.RIPK3 may modulate the immune
checkpoint through HAVCR2. (A) Correlation matrix and (B)
Spearman's correlation analysis for RIPK3 and immune
checkpoint genes in The Cancer Genome Atlas. (C) Correlation matrix
and (D) Spearman's correlation analysis for RIPK3 and immune
checkpoint genes in the Therapeutically Applicable Research To
Generate Effective Treatments database. (E) HAVCR2
expression in cells treated with the RIPK3 inhibitor GSK872.
Data were analyzed using one-way ANOVA followed by Dunnett's post
hoc test. (F) Immune checkpoint blockade response, represented by
TIDE scores, in the RIPK3higher and
RIPK3lower groups. Data were analyzed using
Wilcoxon's rank-sum test. ***P<0.001. RIPK3, receptor
interacting serine/threonine kinase 3; TIGIT, T cell
immunoreceptor with Ig and ITIM domains; CTLA4, cytotoxic
T-lymphocyte associated protein 4; LAG3,
lymphocyte-activation 3; PDCD1LG2, lymphocyte-activation 3
and programmed cell death 1 ligand 2; HAVCR2, hepatitis A
virus cellular receptor 2; SIGLEC15, sialic acid binding Ig
like lectin 15; TIDE, Tumor Immune Dysfunction and Exclusion; UT,
untreated. |
To determine whether RIPK3 induces the
expression of HAVCR2, two osteosarcoma cell lines, KHOS and
143B, were treated with the RIPK3 inhibitor GSK872 and
HAVCR2 expression was detected using RT-qPCR. Information
was sought regarding the GSK872 concentration that would
sufficiently inhibit RIPK3 activity. A literature search on
NCBI indicated that 1–10 µM GSK872 fully inhibited RIPK3 activity
(19–21). The HAVCR2 expression level
was empirically tested following treatment of the two cell lines
with 100 nM −10 µM GSK872. The results indicate that HAVCR2
expression was decreased in the KHOS and 143B lines when the cells
were treated with ≥2 µM GSK872, suggesting that RIPK3 might
positively regulate HAVCR2 expression (Fig. 4E).
To confirm the important roles of RIPK3 in
sarcoma, the TIDE algorithm was used to predict potential ICBs. The
TIDE algorithm is a statistical method for the modelling of immune
evasion from two relevant perspectives: i) the induction of T-cell
dysfunction with high infiltration of cytotoxic T lymphocytes
(CTLs) and ii) the prevention of T-cell infiltration with low CTL
levels. The higher the TIDE score, the worse the response to ICB
therapy (22). The TIDE scores in
the RIPK3higher group were slightly higher than those in
the RIPK3lower group, indicating that
RIPK3high patients would benefit more from ICB
therapy (P=0.05; Fig. 4F). In
summary, these data suggest that RIPK3 may regulate
HAVCR2 and could serve as an ICB response marker.
RIPK3 is associated with macrophage
and monocyte infiltration
Next, whether immune cell subtypes contribute to
sarcoma development was investigated. Using an xCell algorithm to
calculate immune cell infiltration, the abundance of immune cell
types was successfully derived using a reference set with 46 immune
cell subtypes for the RIPK3higher and
RIPK3lower groups (including endothelial cells,
macrophages, B cells, NK cells, CD4+ T cells and
CD8+ T cells; Table
SIV). xCell is based on a spillover compensation technique in
which reference gene signatures of each immune cell type are built
from RNA-Seq samples of various human innate and adaptive
circulating immune cells (23).
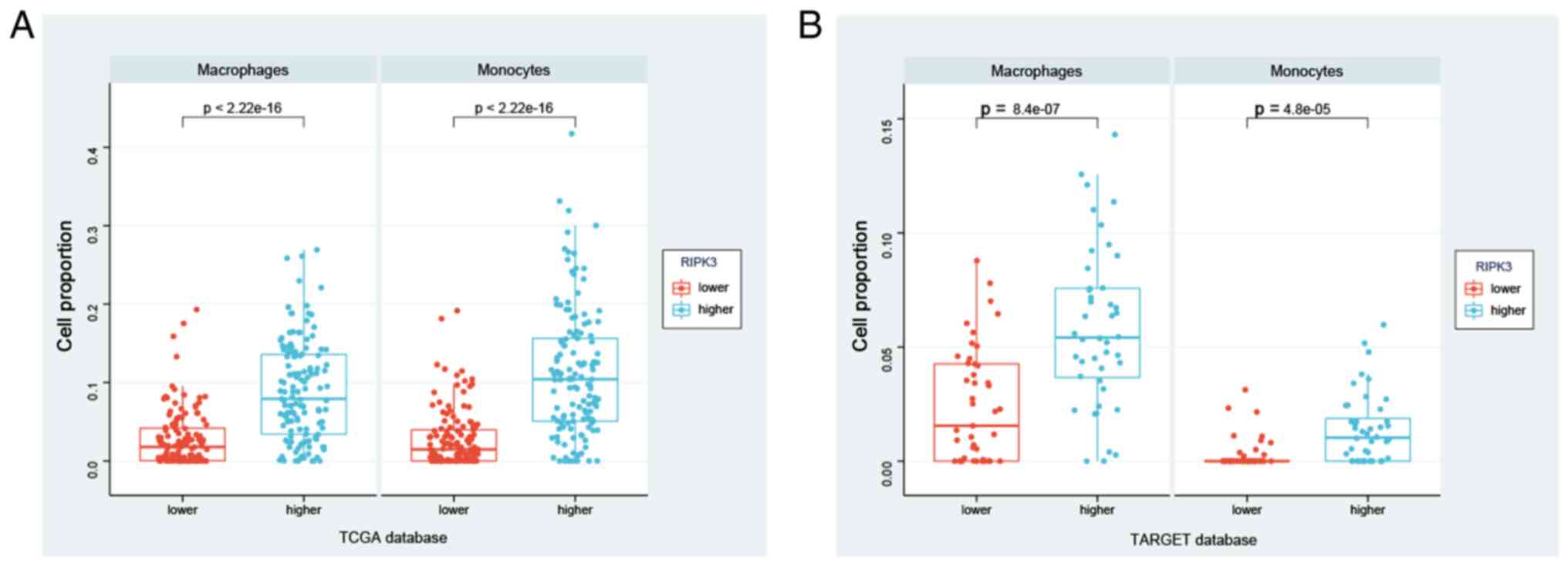
Using this strategy, significant differences in macrophage and
monocyte infiltration were observed between the
RIPK3higher and RIPK3lower
groups in the TCGA database (P<2.2×10−16 for both
monocytes and macrophages; Fig.
5A). These findings were verified in the TARGET database, with
significantly higher macrophage and monocyte infiltration in the
RIPK3higher group (Fig.
5B, Table SV). In summary,
these data suggest that RIPK3 may modulate disease through
effects on macrophages and monocytes, suggesting a potential target
for the regulation of sarcoma development.
Discussion
In the current study, evidence that RIPK3
might modulate sarcoma development through the immune checkpoint
HAVCR2 was presented. Notably, RIPK3 expression was
associated with immune scores in macrophages and monocytes,
suggesting its involvement in the regulation of immune checkpoints
in these cell types.
Sarcoma is an aggressive malignant cancer with
strong genetic and epigenetic components (24,25).
Several publications have reported that mutations or genetic
variations contribute to sarcoma; however, none of these variants
are located within or near RIPK3. Furthermore, a search for
mutations in TCGA datasets identified <10 RIPK3 mutations
in sarcoma cases, with no significant difference compared with the
control group (data not shown). This finding indicates that
RIPK3 is not a trigger factor for sarcoma but is indirectly
involved in disease regulation. This point was also supported by
the epigenetic findings of sarcoma. In a previous study, the DNA
methylation level of RIPK3 was found to be associated with a
poor prognosis in osteosarcoma (26). In line with the aforementioned
findings, the TCGA data analysis in the present study revealed
improved survival and prognosis in the osteosarcoma groups with
higher RIPK3 expression. DNA methylation would be expected
to suppress the expression of RIPK3, and the methylation of
RIPK3 could potentially serve as a therapeutic or survival
predictor. Interestingly, another study observed that upregulation
of RIPK3 in U2OS osteosarcoma cells led to cell death when
RIPK3-overexpressing U2OS cells were treated with
5-aminolevulic acid-mediated photodynamic therapy (27).
By comparing the gene expression profile of the
RIPK3higher and RIPK3lower
groups, 603 up- and 260 downregulated genes were identified. The
number of genes with increased expression was 2.3-fold that of
decreased genes, leading to the hypothesis that RIPK3 could
be universally involved in the modulation of gene expression, not
just the expression of one or two target genes such as
HAVCR2. Based on this hypothesis, it was considered that
RIPK3 may function as a transcription factor, although no
supporting data are available to support this. Most importantly,
the differentially expressed genes were involved in the pathways
‘Staphylococcus aureus infection’ and ‘cytokine-cytokine
receptor interaction’ (28).
Furthermore, the top upregulated gene TMC8 has been
demonstrated to be a risk factor for head and neck squamous cancer
(16) and renal cell carcinoma
(29), while the top downregulated
gene TMEM97 is a transmembrane protein with largely unknown
functions. The role of TMEM97 in cancer appears to be
diverse. It may be a putative tumor suppressor for pancreatic or
prostate cancer (30), but it has
also been shown to be associated with progression and poor survival
in squamous cell carcinoma of the lung (18,31)
and breast cancer (32). These
genes and pathways have been confirmed as critical factors in
sarcoma and other cancer types, indicating that RIPK3 is a
modulator of these biological processes.
An inflammatory microenvironment increases the risk
of promoting tumors by mediating immune checkpoints in various
types of cancer (33,34). Coupled with RIPK1, RIPK3
induces cell death by assembling necrosomes, which is dependent on
the presence of caspase-8 and occurs in a cell-type-specific manner
(8). Apoptosis proteins (IAPs) are
E3 ubiquitin ligases that ubiquitinate RIPK1 or
RIPK2, inducing an inflammatory response by the NF-κB or
MAPK signalling pathways, suppressing the formation of the
RIPK3 necroptosis-inducing complex, or inhibiting the
inflammatory pathways activated by the RIPK1-RIPK3 complex
(35). IAP antagonists, also known
as Smac mimetics, have been shown to effectively sensitize cells
from osteosarcomas and kill these cells following the
administration of low levels of TNF-α. However, this function was
not found to be dependent on the expression level of RIPK3
in the cells, indicating that RIPK3 might not act through
classical inflammatory pathways (36). RIPK3 has also shown
inflammation-modulating effects in certain types of immune cells,
such as monocytes and macrophages (37); however neither of these cell types
were assayed in sarcomas conducted by Shekhar et al
(36).
Another critical point is the immune checkpoint
HAVCR2, which was first identified as an immune suppressor
(38), is widely expressed in
various cancer cells and can be induced by macrophage
colony-stimulating factor. As the present study lacks functional
data, it is not possible to evaluate whether RIPK3 directly
regulates the inflammatory response through HAVCR2. However,
we hypothesize that this is likely due to the critical role of
RIPK3 in the activation of inflammation, the prediction of
improved survival in cases with higher HAVCR2 and the
implied role of HAVCR2 in tumor suppression as a target of
cancer immunotherapy.
In conclusion, the present study analyzed TCGA data
and identified that RIPK3 was associated with improved
survival of sarcoma, mostly likely through the immune checkpoint
HAVCR2. The study findings suggest that RIPK3, as a
critical molecular marker of necroptosis and apoptosis, serves as
an immune regulator in sarcoma, which expands our understanding of
disease etiology. This previously unreported finding will help us
to better understand the etiology and treatment of sarcoma.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Professor Fusheng
Zhou (Anhui Medical University, Hefei, China) for their statistical
assistance.
Funding
This project was supported by the Affiliated Hospital of
Yangzhou University.
Availability of data and materials
All data generated or analyzed during this study
are included in this published article.
Authors' contributions
JD conceived the project and designed the study. CQ
wrote the manuscript and performed most of the statistical
analysis. DW helped to perform the statistical analysis, write the
draft, and revise and approve the final version of the manuscript.
All authors read and approved the final version of the manuscript.
CQ and JD confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Popovich JR, Kashyap S and Cassaro S:
Sarcoma. In: StatPearls. Treasure Island (FL). 2022.
|
|
2
|
Bui NQ, Wang DS and Hiniker SM:
Contemporary management of metastatic soft tissue sarcoma. Curr
Probl Cancer. 43:289–299. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Howitt BE, Sholl LM, Dal Cin P, Jia Y,
Yuan L, MacConaill L, Lindeman N, Kuo F, Garcia E, Nucci MR and
Quade BJ: Targeted genomic analysis of mullerian adenosarcoma. J
Pathol. 235:37–49. 2015. View Article : Google Scholar
|
|
4
|
Mirabello L, Zhu B, Koster R, Karlins E,
Dean M, Yeager M, Gianferante M, Spector LG, Morton LM, Karyadi D,
et al: Frequency of pathogenic germline variants in
cancer-susceptibility genes in patients with osteosarcoma. JAMA
Oncol. 6:724–734. 2020. View Article : Google Scholar
|
|
5
|
Koelsche C, Schrimpf D, Stichel D, Sill M,
Sahm F, Reuss DE, Blattner M, Worst B, Heilig CE, Beck K, et al:
Sarcoma classification by DNA methylation profiling. Nat Commun.
12:4982021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Steele CD and Pillay N: The genomics of
undifferentiated sarcoma of soft tissue: Progress, challenges and
opportunities. Semin Cancer Biol. 61:42–55. 2020. View Article : Google Scholar
|
|
7
|
Cancer Genome Atlas Research Network, .
Electronic address. simpleelizabeth.demicco@sinaihealthsystem.caCancer
Genome Atlas Research Network: Comprehensive and integrated genomic
characterization of adult soft tissue sarcomas. Cell.
171:950–965.e28. 2017. View Article : Google Scholar
|
|
8
|
Jensen S, Seidelin JB, LaCasse EC and
Nielsen OH: SMAC mimetics and RIPK inhibitors as therapeutics for
chronic inflammatory diseases. Sci Signal. 13:eaax82952020.
View Article : Google Scholar
|
|
9
|
Duan X, Liu X, Liu N, Huang Y, Jin Z,
Zhang S, Ming Z and Chen H: Inhibition of keratinocyte necroptosis
mediated by RIPK1/RIPK3/MLKL provides a protective effect against
psoriatic inflammation. Cell Death Dis. 11:1342020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Otsubo K, Maeyashiki C, Nibe Y, Tamura A,
Aonuma E, Matsuda H, Kobayashi M, Onizawa M, Nemoto Y, Nagaishi T,
et al: Receptor-Interacting Protein Kinase 3 (RIPK3) inhibits
autophagic flux during necroptosis in intestinal epithelial cells.
FEBS Lett. 594:1586–1595. 2020. View Article : Google Scholar
|
|
11
|
Wu S, Xu C, Xia K, Lin Y, Tian S, Ma H, Ji
Y, Zhu F, He S and Zhang X: Ring closure strategy leads to potent
RIPK3 inhibitors. Eur J Med Chem. 217:1133272021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Karlsson J, Holmquist Mengelbier L,
Ciornei CD, Naranjo A, O'Sullivan MJ and Gisselsson D: Clear cell
sarcoma of the kidney demonstrates an embryonic signature
indicative of a primitive nephrogenic origin. Genes Chromosomes
Cancer. 53:381–391. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ravi R, Noonan KA, Pham V, Bedi R,
Zhavoronkov A, Ozerov IV, Makarev E, Artemov A, Wysocki PT, Mehra
R, et al: Bifunctional immune checkpoint-targeted antibody-ligand
traps that simultaneously disable TGFβ enhance the efficacy of
cancer immunotherapy. Nat Commun. 9:7412018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zeng D, Li M, Zhou R, Zhang J, Sun H, Shi
M, Bin J, Liao Y, Rao J and Liao W: Tumor microenvironment
characterization in gastric cancer identifies prognostic and
immunotherapeutically relevant gene signatures. Cancer Immunol Res.
7:737–750. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang J, Sun J, Liu LN, Flies DB, Nie X,
Toki M, Zhang J, Song C, Zarr M, Zhou X, et al: Siglec-15 as an
immune suppressor and potential target for normalization cancer
immunotherapy. Nat Med. 25:656–666. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tirosh I, Izar B, Prakadan SM, Wadsworth
MH II, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G,
et al: Dissecting the multicellular ecosystem of metastatic
melanoma by single-cell RNA-seq. Science. 352:189–196. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin B, Wang S, Yao Y, Shen Y and Yang H:
Comprehensive co-expression analysis reveals TMC8 as a prognostic
immune-associated gene in head and neck squamous cancer. Oncol
Lett. 22:4982021. View Article : Google Scholar
|
|
18
|
Ding H, Gui XH, Lin XB, Chen RH, Cai HR,
Fen Y and Sheng YL: Prognostic value of MAC30 expression in human
pure squamous cell carcinomas of the lung. Asian Pac J Cancer Prev.
17:2705–2710. 2016.PubMed/NCBI
|
|
19
|
Mandal P, Berger SB, Pillay S, Moriwaki K,
Huang C, Guo H, Lich JD, Finger J, Kasparcova V, Votta B, et al:
RIP3 induces apoptosis independent of pronecrotic kinase activity.
Mol Cell. 56:481–495. 2014. View Article : Google Scholar
|
|
20
|
André S, Picard M, Cezar R, Roux-Dalvai F,
Alleaume-Butaux A, Soundaramourty C, Cruz AS, Mendes-Frias A, Gotti
C, Leclercq M, et al: T cell apoptosis characterizes severe
Covid-19 disease. Cell Death Differ. 29:1486–1499. 2022. View Article : Google Scholar
|
|
21
|
Chen X, Zhu R, Zhong J, Ying Y, Wang W,
Cao Y, Cai H, Li X, Shuai J and Han J: Mosaic composition of
RIP1-RIP3 signalling hub and its role in regulating cell death. Nat
Cell Biol. 24:471–482. 2022. View Article : Google Scholar
|
|
22
|
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X,
Li Z, Traugh N, Bu X, Li B, et al: Signatures of T cell dysfunction
and exclusion predict cancer immunotherapy response. Nat Med.
24:1550–1558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Racle J, de Jonge K, Baumgaertner P,
Speiser DE and Gfeller D: Simultaneous enumeration of cancer and
immune cell types from bulk tumor gene expression data. Elife.
6:e264762017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bieging KT, Mello SS and Attardi LD:
Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev
Cancer. 14:359–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Y, Sánchez-Tilló E, Lu X, Clem B,
Telang S, Jenson AB, Cuatrecasas M, Chesney J, Postigo A and Dean
DC: Rb1 family mutation is sufficient for sarcoma initiation. Nat
Commun. 4:26502013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tian W, Li Y, Zhang J, Li J and Gao J:
Combined analysis of DNA methylation and gene expression profiles
of osteosarcoma identified several prognosis signatures. Gene.
650:7–14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Coupienne I, Fettweis G and Piette J: RIP3
expression induces a death profile change in U2OS osteosarcoma
cells after 5-ALA-PDT. Lasers Surg Med. 43:557–564. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tuohy JL, Somarelli JA, Borst LB, Eward
WC, Lascelles BD and Fogle JE: Immune dysregulation and
osteosarcoma: Staphylococcus aureus downregulates TGF-β and
heightens the inflammatory signature in human and canine
macrophages suppressed by osteosarcoma. Vet Comp Oncol. 18:64–75.
2020. View Article : Google Scholar
|
|
29
|
Castro FA, Ivansson EL, Schmitt M,
Juko-Pecirep I, Kjellberg L, Hildesheim A, Gyllensten UB and
Pawlita M: Contribution of TMC6 and TMC8 (EVER1 and EVER2) variants
to cervical cancer susceptibility. Int J Cancer. 130:349–355. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ramalho-Carvalho J, Gonçalves CS, Graça I,
Bidarra D, Pereira-Silva E, Salta S, Godinho MI, Gomez A, Esteller
M, Costa BM, et al: A multiplatform approach identifies miR-152-3p
as a common epigenetically regulated onco-suppressor in prostate
cancer targeting TMEM97. Clin Epigenetics. 10:402018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ding H, Gui X, Lin X, Chen R, Ma T, Sheng
Y, Cai H and Fen Y: The prognostic effect of MAC30 expression on
patients with non-small cell lung cancer receiving adjuvant
chemotherapy. Technol Cancer Res Treat. 16:645–653. 2017.
View Article : Google Scholar
|
|
32
|
Xiao M, Li H, Yang S, Huang Y, Jia S, Wang
H, Wang J and Li Z: Expression of MAC30 protein is related to
survival and clinicopathological variables in breast cancer. J Surg
Oncol. 107:456–462. 2013. View Article : Google Scholar
|
|
33
|
Workenhe ST, Nguyen A, Bakhshinyan D, Wei
J, Hare DN, MacNeill KL, Wan Y, Oberst A, Bramson JL, Nasir JA, et
al: De novo necroptosis creates an inflammatory environment
mediating tumor susceptibility to immune checkpoint inhibitors.
Commun Biol. 3:6452020. View Article : Google Scholar
|
|
34
|
Bagchi S, Yuan R and Engleman EG: Immune
checkpoint inhibitors for the treatment of cancer: Clinical impact
and mechanisms of response and resistance. Annu Rev Pathol.
16:223–249. 2021. View Article : Google Scholar
|
|
35
|
Gyrd-Hansen M and Meier P: IAPs: From
caspase inhibitors to modulators of NF-kappaB, inflammation and
cancer. Nat Rev Cancer. 10:561–574. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shekhar TM, Miles MA, Gupte A, Taylor S,
Tascone B, Walkley CR and Hawkins CJ: IAP antagonists sensitize
murine osteosarcoma cells to killing by TNFα. Oncotarget.
7:33866–33886. 2016. View Article : Google Scholar
|
|
37
|
Wu L, Chung JY, Cao T, Jin G, Edmiston WJ
III, Hickman S, Levy ES, Whalen JA, Abrams ES, Degterev A, et al:
Genetic inhibition of RIPK3 ameliorates functional outcome in
controlled cortical impact independent of necroptosis. Cell Death
Dis. 12:10642021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kandel S, Adhikary P, Li G and Cheng K:
The TIM3/Gal9 signaling pathway: An emerging target for cancer
immunotherapy. Cancer Lett. 510:67–78. 2021. View Article : Google Scholar
|